Abstract
Cardiac progenitor cells (CPCs) are a crucial source of cells in cardiac development and regeneration. However, reported CPCs are heterogeneous, and no gene has been identified to transiently mark undifferentiated CPCs throughout heart development. Here we show that Spalt-like gene 1 (Sall1), a zing-finger transcription factor, is expressed in undifferentiated CPCs giving rise to both left and right ventricles. Sall1 was transiently expressed in precardiac mesoderm contributing to the first heart field (left ventricle precursors) but not in the field itself. Similarly, Sall1 expression was maintained in the second heart field (outflow tract/right ventricle precursors) but not in cardiac cells. In vitro, high levels of Sall1 at mesodermal stages enhanced cardiomyogenesis, whereas its continued expression suppressed cardiac differentiation. Our study demonstrates that Sall1 marks CPCs in an undifferentiated state and regulates cardiac differentiation. These findings provide fundamental insights into CPC maintenance, which can be instrumental for CPC-based regenerative medicine.
Keywords: cardiac progenitor, cardiac development, ES/iPS cells, cardiac transcription factors
1. Introduction
Cardiac progenitor cells (CPCs), identified from embryos or pluripotent stem cell (PSC) culture, hold tremendous regenerative potential with the unique capability to expand and differentiate into cardiac cells [1]. Cardiogenesis initiate as the basic-helix-loop-helix factor Mesp1 is expressed in precardiac mesoderm. Mesp1+ cells migrate and form the cardiac crescent (referred to as the first heart field (FHF)), which largely contribute to the left ventricle (LV). A subset of Mesp1+ cells remain undifferentiated in the second heart field (SHF), located dorsal to the FHF, and give rise to the outflow tract (OFT), right ventricle (RV), and part of atria [2]. Early CPCs can be identified by transient expression of the chromatin factor Smarcd3, prior to expression of known CPC markers [3, 4], including Islet1 (Isl1), fetal liver kinase 1 (Flk1), and Nkx2.5 [1]. While Isl1 is transiently expressed in the SHF, Smarcd3 and Nkx2-5 are continually expressed in cardiomyocytes [5, 6]. Although, CPCs can be identified by their stage-specific expression of various cell markers, no gene has been identified to mark undifferentiated pools of CPCs giving rise to four chambers throughout development. In the present study, we demonstrate that Sall1, a zinc-finger transcription factor, is transiently expressed and maintained in undifferentiated CPCs throughout early heart development in vivo. Sall1 has a biphasic role: early overexpression of Sall1 enhanced, but its late overexpression suppressed cardiomyogenesis in pluripotent stem cell (PSC) culture.
2. Materials and methods
2.1 Mice and lineage tracing
Sall1GFP, Sall1CreERT2, ROSAYFP mouse lines were generated as described [7, 8]. Experiments were conducted according to a protocol approved by the international Animal care and Use committee of IMCB, the University of Tokyo. To induce Cre activity of Sall1CreERT2/+, depending on embryonic stage, pregnant mice were fed 150ul/30g of Tamoxifen (10mg/ml: sigma) by injection into the abdominal cavity at desired time points.
2.2 PSC generation, maintenance, and differentiation
Sall1GFP/+ ESCs were derived from Sall1GFP/+ mice [9]. ESCs were maintained in Knockout DMEM supplemented with 15% KSR (Gibco), Penicillin-Streptomycin (Nacalai tesque), MEM-NEAA, GlutaMAX, Sodium Pylvate, 2-mercaptoethanol (Gibco), LIF (WAKO) on MEF. To differentiate Sall1-GFP ESCs, embryoid bodies (EB) were generated with DMEM (KOHJIN BIO) containing 20% FBS, 2.4mM L-Glutamine, MEM NEAA, 2-mercaptoethanol (Gibco), LIF (WAKO). After 2 days, EBs were cultured in DMEM (KOHJIN BIO) containing 20% FBS, 2.4mM L-Glutamine, 2-mercaptoethanol (Gibco). The medium was changed every two days. To generate time dependent Sall1 overexpressing cells, A DOX inducible SALL1 expressing piggybac vector and a PBEF1a-mSALL1-IRES-mcherry vector were co-electroporated with the piggybac transposase vector PBASE2 into 201B7 cells [10] with NEPA21 (NEPA GENE) [11]. DOX-SALL1 hiPSCs were maintained and differentiated as described [12].
2.3 Immunohistochemistry and flow cytometry
Antibodies used: mouse a-Sall1 (1:100, PPMX), rabbit a-Isl1 (1:200, Abcam), mouse a-Isl1 (1:100, hybridoma bank), goat a-Nkx2-5 (1:2000, Santa Cruz Biotechnology), rabbit a-GFP (1:400, MBL), chick a-GFP (1:400, Life technologies), rat a-CD31 (1:100, BD biosciences), mouse a-cTnT (1:10000, Thermo Fisherscientific), rabbit a-HCN4 (1:2000, alomone lab). Alexa Fluor secondary antibodies (Life technologies) were used for secondary detection and images were acquired with a KEYENCE BZ-9000 Fluorescence Microscope. For flow cytometry, ESCs/iPSCs were dissociated using 0.1% Trypsin or Accumax (Funakoshi). Cells were re-suspended in 0.1%FBS/D-PBS(-) without Ca2+ and Mg2+ and sorted using a FACSAriaIII (BD Biosciences). Cells were incubated with primary antibodies followed by secondary antibodies conjugated with Alexa Fluor 647 (Invitrogen).
2.4 Chromatin Immunoprecipitation (ChIP)
ChIP was done using antibodies against a-trimethylated H3K27 (Cell Signaling), and aacetylated H3K27 (Abcam) as described [13].
Immunoprecipitated DNA was amplified using the primer pairs:
Isl1 3.2F 5-CCAATCTAGTGAGCAGGCAAA-3,
Isl1 3.2R 5-TCAAGTTTCAGGAGGAACCAAG-3,
Isl1 3.1F 5-TCAGTGGGCACTGGCTCAA-3,
Isl1 3.1R 5-GCTAGCAGTGGATAAAGGGCATC-3,
Flk1 F 5-CAGGATAGGGAAGCCTTGGA-3
Flk1 F 5-CCACCATGCCCAGCTTACTT-3
3. Results
3.1. Sall1 is expressed in early CPCs during development
To examine Sall1 expression during heart development, we used Sall1GFP mice [9] and compared GFP expression with Mesp1-lineage cells by generating Sall1GFP; Mesp1cre; ROSARFP mice. Sall1 expression was initially observed at embryonic day 7 (E7.0) in the nascent mesoderm, prior to the emergence of embryonic Mesp1-lineage cells (Figure 1A). Unlike Mesp1-lineage cells, Sall1 was not expressed in the extraembryonic tissue (Figure 1A). At the crescent stage (E7.5), the Sall1 expression domain was adjacent to, but non-overlapping with, the domain of Nkx2-5 expression that represents the FHF (Figure 1B). Expression of Sall1 and Nkx2-5 continued in complementary patterns throughout heart development (Figure 1B, C). In contrast, Sall1 was expressed in the SHF where Isl1 is expressed (Figure 1D). Thus, Sall1 is expressed in the mesoderm at pre-heart field stages and in SHF cells from heart field stages, but is not expressed in FHF cells.
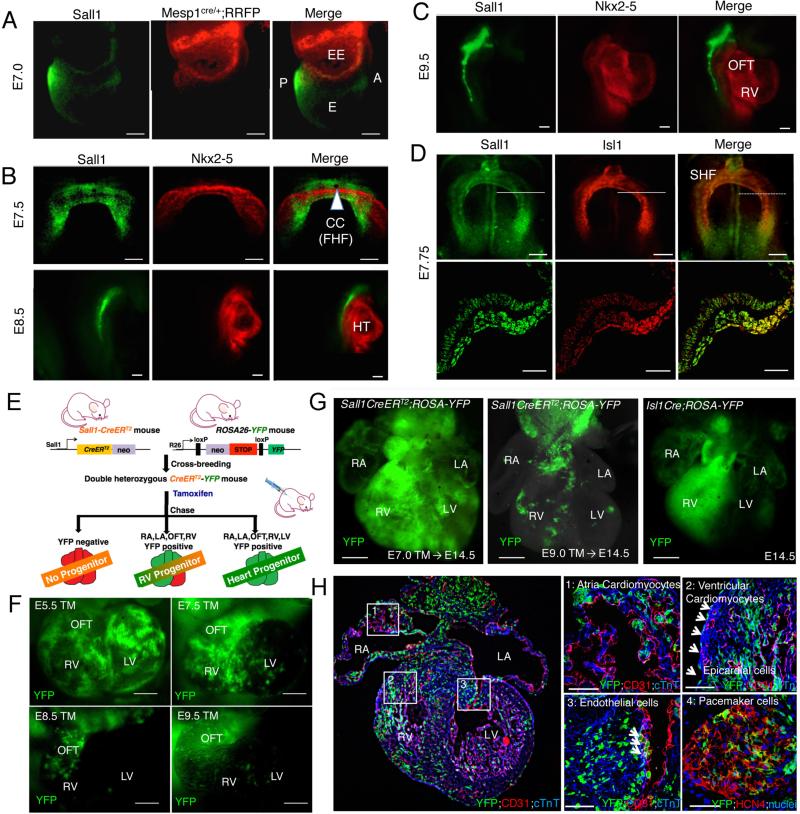
Figure 1. Sall1 is expressed in nascent mesoderm and SHF giving rise to four chamber cells.
(A) Sall1 expression was observed in nascent mesoderm at E7.0 when Mesp1 lineage cells appear in extraembryonic tissue. (B) Sall1 is not expressed in the FHF. Immunostaining shows that Sall1 expression (green) does not overlap with the FHF marker Nkx2-5 (red) at E7.5–8.5. (C, D) Sall1 overlaps with the SHF marker Isl1 (D, red) and remains in the SHF, but not expressed in the heart (C). (E) Schematic diagram of Sall1 lineage analysis. (F, G) Analysis of Sall1-lineage cells with YFP signal on E10.5 (F) or E14.5 (G) heart. Sall1+ cells from the early stages (E5.5/E7.0) contribute to the whole heart, whereas Sall1+ cells from the later stage (E7.5–9.5) contribute to the OFT and RV. Isl1 lineage was also traced for comparison (G). (H) Section of Sall1 lineage-traced heart showing its contribution to atria/ventricular cardiomyocytes (1), epicardial cells (2, arrows), endothelial cells merged with CD31 (3, red), and SAN cells with HCN4+ (4, red) analyzed at E14.5 heart. Scale bars: 20μm (B, C), 50μm (A, D, F), 100μm (H), 200μm (G). A, anterior; P, posterior; EE, extraembryonic; E, embryonic; CC, cardiac crescent; HT, heart tube; OFT, outflow tract; RV, right ventricle; LV, left ventricle, TM, tamoxifen.
3.2 Sall1+ cells give rise to distinct anatomical structures of the heart in vivo
To determine the fate of Sall1+ CPCs during heart development, we performed lineage-tracing experiments with Sall1creERT2; R26RYFP lineage reporter mice [7, 8] (Figure 1E). Cre activity was induced at different stages (E5.5–E9.5) by tamoxifen, and hearts were analyzed for YFP expression at E10.5 (Figure 1F). Cre activation prior to gastrulation (E5.5) resulted in widespread distribution of YFP positive cells in the heart. However, when Cre activity was induced at crescent stages (E7.5), YFP positive cells were mostly confined to the OFT and RV. Later stages of Cre activation (E8.5 and E9.5) resulted in further restriction of YFP positive cells to OFT/RV cells (Figure 1F). Next, we induced Cre activation at E7.0 and E9.0, collected hearts at E14.5, and analyzed YFP positive cells. Cre activation at the pre-crescent stage (E7.0) resulted in abundant appearance of YFP cells in the entire heart (Figure 1G). Histological analysis of the hearts revealed YFP positive progeny in atrial and ventricular cardiomyocytes (Figure 1H1 and 2), epicardial cells (Figure 1H2; arrowheads), endothelial cells (Figure 1H3), pacemaker cells in sinoatrial node (Figure 1H4). Cre induction at E9.0 resulted in a pronounced reduction of YFP cells within the left ventricle (Figure 1G), with YFP cells primarily restricted to the RV. These results suggest that Sall1 is expressed in precursors of FHF and SHF cells during gastrulation, and remains expressed in the SHF before giving rise to the OFT/RV.
3.3. Sall1 positively regulates CPC genes
To examine whether Sall1 affects cardiogenesis in vitro, we differentiated Sall1GFP ESCs into precardiac mesodermal cells and isolated GFP+/GFP− cells (Figure S1 A, B histogram). The GFP+ cells showed enrichment for Flk1 and Pdgfra along with Mesp1 compared to GFP− cells (Figure S1D), indicating that the Sall1+ cells contain early precardiac mesodermal cells. Isl1 was also enriched along with Foxa2/Foxc2 and Six2, which are involved in regulating progenitor renewal, suggesting the ESC-derived Sall1+ cells contain both early and SHF progenitors. The GFP+ cells were efficiently differentiated into cardiomyocytes (Figure S1B, C) with increased levels of contractile genes (Figure S1E). Isl1 levels were maintained even after 8 days of culture, but this is likely due to an ESC culture effect [14]. Through bioinformatics analyses, we identified putative Sall1-binding sites near loci of Isl1 and Flk1 (Figure S1F). Our ChIP assay showed that the sites were poorly associated with H3K27me3 or showed no change in GFP+ cells as compared to GFP− cells (Figure S1F). In contrast, the sites were highly associated with H3K27ac in GFP+ cells, suggesting they are transcriptionally active in Sall1+ CPCs (Figure S1F).
3.4 Mesodermal SALL1 expression promotes cardiac fate but its prolonged expression suppresses cardiac differentiation
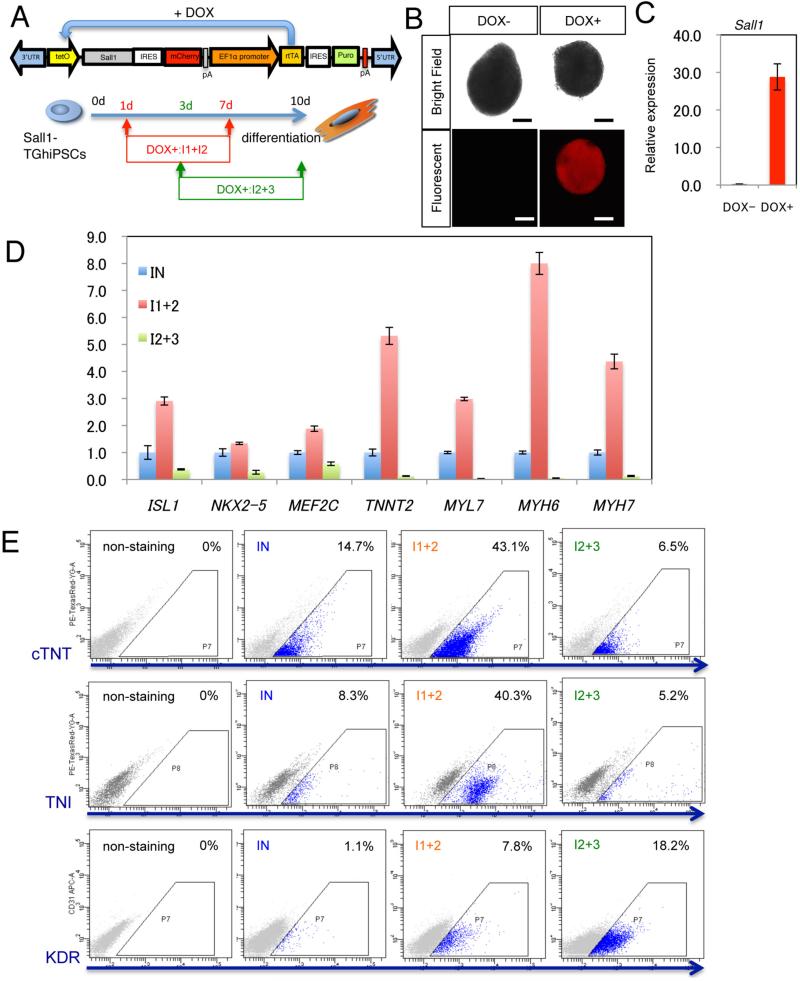
To temporally control Sall1 expression, we generated human PSCs that stably express a reverse tetracycline-controlled transactivator (rtTA) and a doxycycline (DOX)-inducible expression cassette containing SALL1 and mCherry (Sall1-TGhiPSCs) [11] (Figure 2A). Expression of exogenous SALL1 and mCherry was observed within 12 hours following DOX treatment (Figure 2B, C). Cardiac differentiation was performed with a step-wise developmental progression from a primitive streak-like population (days 1–3) to early precardiac mesoderm (days 3–4), CPC specification (days 4–7), and cardiomyocyte formation (after day 7) (Figure S2A). Molecular profiling showed that SALL1 is initially upregulated during mesodermal stages but downregulated near the end of CPC stages (Figure S2B). We found that DOX treatment during CPC stages markedly increased expression of cardiac genes (Figure 2D). However, continued DOX treatment at later stages strongly prevented cardiac gene expression (Figure 2D). Consistently, the number of cardiomyocyte was markedly increased with early DOX treatment and decreased with late DOX treatment (Figure 2E).
Figure 2. Mesodermal SALL1 expression promotes cardiac fate but its continued expression in CPCs suppresses cardiac differentiation.
(A) DOX-inducible SALL1 construct and strategy for temporal SALL1 overexpression in human PSCs (Sall1-TGhiPSc). (B, C) mCherry (B) or SALL1 (C) expression with/without DOX (after 12 hrs, 1d). This level of SALL1 expression is about six-fold higher than endogenous levels (Fig. S2B), which may be appropriate for the assay. Scale bars: 200μm. (D) Relative expression of cardiac transcription factors and myocardial genes after temporal DOX treatment shown in (A). (E) Flow cytometry analysis showing percentages of cTNT/TNI/KDR positive cells after temporal DOX treatment shown in (A).
4. Conclusion
The present study shows that Sall1 is a previously unrecognized CPC gene expressed specifically in undifferentiated cells during CPC development. The absence of Sall1 expression in the FHF suggests that Sall1 is a maker of undifferentiated CPCs as, unlike SHF cells, FHF cells express sarcomere genes [15]. Sall1 expression was also maintained in the SHF from mesodermal stages. These findings suggest that Sall1+ cells represent an undifferentiated population of CPCs. In agreement with this, a temporal increase in Sall1 levels at early stages enhanced the cardiogenic potential, whereas its prolonged expression suppressed cardiac differentiation. Genome-wide ChIP analysis may allow us to identify a set of CPC genes crucial for maintaining CPCs in an undifferentiated state.
Supplementary Material
Acknowledgement
This study was supported by JST PRESTO, Grants-in-Aid for Scientific Research (B), JSPS to J.K.T., AMED to J.K.T., A.K., Y.Y., a research fellowship for young scientist from JSPS (DC2) to Y.M. C.K. and P.A. were supported by NHLBI/NIH, MSCRF, and AHA grants.
Reference
- 1.Garry DJ, Olson EN. A common progenitor at the heart of development. Cell. 2006;127:1101–4. doi: 10.1016/j.cell.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 2.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–35. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–11. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Weerd JH, Koshiba-Takeuchi K, Kwon C, Takeuchi JK. Epigenetic factors and cardiac development. Cardiovasc Res. 2011;91:203–11. doi: 10.1093/cvr/cvr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devine WP, Wythe JD, George M, Koshiba-Takeuchi K, Bruneau BG. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. Elife. 2014:3. doi: 10.7554/eLife.03848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shenje LT, Andersen P, Uosaki H, Fernandez L, Rainer PP, Cho GS, et al. Precardiac deletion of Numb and Numblike reveals renewal of cardiac progenitors. Elife. 2014;3:e02164. doi: 10.7554/eLife.02164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue S, Inoue M, Fujimura S, Nishinakamura R. A mouse line expressing Sall1-driven inducible Cre recombinase in the kidney mesenchyme. Genesis. 2010;48:207–12. doi: 10.1002/dvg.20603. [DOI] [PubMed] [Google Scholar]
- 8.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takasato M, Osafune K, Matsumoto Y, Kataoka Y, Yoshida N, Meguro H, et al. Identification of kidney mesenchymal genes by a combination of microarray analysis and Sall1-GFP knockin mice. Mech Dev. 2004;121:547–57. doi: 10.1016/j.mod.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Kim SI, Oceguera-Yanez F, Sakurai C, Nakagawa M, Yamanaka S, Woltjen K. Inducible Transgene Expression in Human iPS Cells Using Versatile All-in-One piggyBac Transposons. Methods Mol Biol. 2015 doi: 10.1007/7651_2015_251. [DOI] [PubMed] [Google Scholar]
- 12.Miki K, Endo K, Takahashi S, Funakoshi S, Takei I, Katayama S, et al. Efficient Detection and Purification of Cell Populations Using Synthetic MicroRNA Switches. Cell Stem Cell. 2015;16:699–711. doi: 10.1016/j.stem.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguin P, Holloway AK, et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat Commun. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uosaki H, Cahan P, Lee DI, Wang S, Miyamoto M, Fernandez L, et al. Transcriptional Landscape of Cardiomyocyte Maturation. Cell Rep. 2015;13:1705–16. doi: 10.1016/j.celrep.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–89. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.