Abstract
Small peptides formed from non-ribosomal peptide synthetases (NRPS) are bioactive molecules produced by many fungi including the genus Aspergillus. A subset of NRPS utilizes tryptophan and its precursor, the non-proteinogenic amino acid anthranilate, in synthesis of various metabolites such as A. fumigatus fumiquinazolines (Fqs) produced by the fmq gene cluster. The A. fumigatus genome contains two putative anthranilate synthases - a key enzyme in conversion of anthranilic acid to tryptophan - one beside the fmq cluster and one in a region of co-linearity with other Aspergillus spp. Only the gene found in the co-linear region, trpE, was involved in tryptophan biosynthesis. We found that site-specific mutations of the TrpE feedback domain resulted in significantly increased production of anthranilate, tryptophan, p-aminobenzoate and fumiquinazolines FqF and FqC. Supplementation with tryptophan restored metabolism to near wild type levels in the feedback mutants and suggested that synthesis of the tryptophan degradation product kynurenine could negatively impact Fq synthesis. The second putative anthranilate synthase gene next to the fmq cluster was termed icsA for its considerable identity to isochorismate synthases in bacteria. Although icsA had no impact on A. fumigatus Fq production, deletion and over-expression of icsA increased and decreased respectively aromatic amino acid levels suggesting that IcsA can draw from the cellular chorismate pool.
Keywords: Aspergillus fumigatus, Tryptophan Metabolism, Secondary Metabolism, Metabolic Engineering, Metabolic Flux, Fumiquinazoline
Graphical Abstract
Introduction
The Kingdom Fungi constitutes an unparalleled genomic resource of natural product (also called secondary metabolite) pathways with many fungi containing up to 70 secondary metabolite gene clusters in their genome (Cacho et al., 2014). Although some natural products – both harmful and beneficial – are produced in copious quantities under laboratory growth conditions, most are produced either in small quantities or not at all. Numerous efforts have recently been aimed at inducing production of these lowly expressed metabolites, primarily through over-expression of the secondary metabolite cluster genes in endogenous and/or exogenous host systems (Anyaogu and Mortensen, 2015).
Not all over expression efforts have resulted in success. Many reasons could account for these failures, ranging from toxicity of the products to the host system, unknown post-transcriptional regulations, requirement of other genes not in the cluster, and lack of sufficient primary metabolite pools. All natural products originate from various primary metabolites. Polyketides are derived from acyl coenzyme A (CoA) and malonyl-CoA units, terpenes from isoprene units and non-ribosomal peptides from proteinogenic and non-proteinogenic amino acids (Keller et al., 2005).
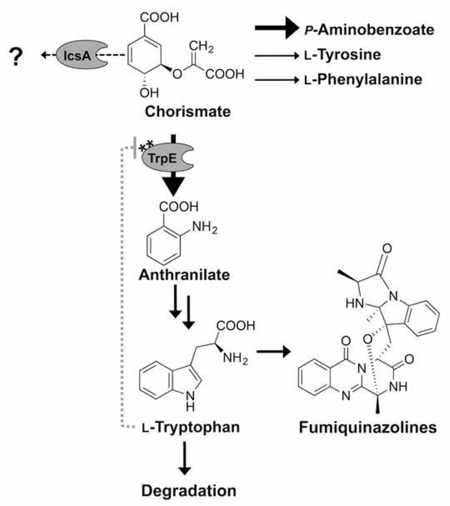
Non-ribosomal peptides are synthesized by non-ribosomal peptide synthetases (NRPS), which consist minimally of A (adenylation), PCP (peptide carrier protein), C (condensation) and releasing domains. The A domain determines which amino acid is incorporated into the growing chain. For example, Aspergillus fumigatus FmqA is a three A domain NRPS incorporating anthranilate, L-tryptophan and l-alanine into the precursor peptide fumiquinazoline F (FqF) (Ames et al., 2010). FqF is further processed by additional enzymes to the end metabolite FqC (Fig. 1). As a considerable subset of fungal NRPS utilize both anthranilate and l-tryptophan in production of valuable alkaloid peptides (Ames and Walsh, 2010), we became interested in the potential contribution of modulating tryptophan biosynthesis to increase production of alkaloid peptides using fumiquinazolines (Fqs) as the targeted metabolites.
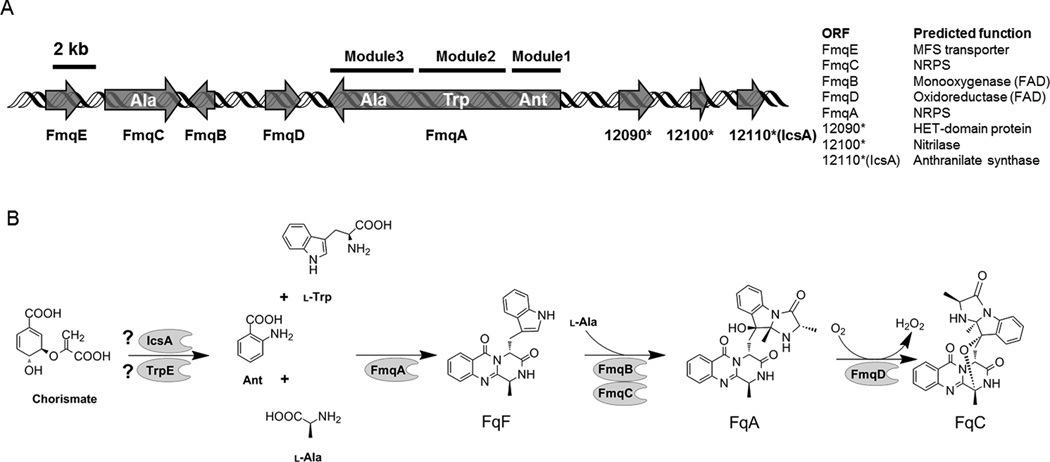
Figure 1. Fumiquinazoline biosynthesis in A. fumigatus.
(Modified from that of Ames et al. (2010) and Lim et al.(2014))
(A) Fumiquinazoline gene cluster in A. fumigatus with nearby putative anthranilate synthase gene Afu6g12110 (icsA).* Abbreviation: 12090, Afu6g12090; 12100, Afu6g12100; 12110, Afu6g12110.
(B) Biosynthetic route of fumiquinazolines in A. fumigatus. Abbreviations: Ant, anthranilate; l-Trp, l-tryptophan; l-Ala, l-alanine; FqF, fumiquinazoline F; FqA, fumiquinazoline A; FqC, fumiquinazoline C.
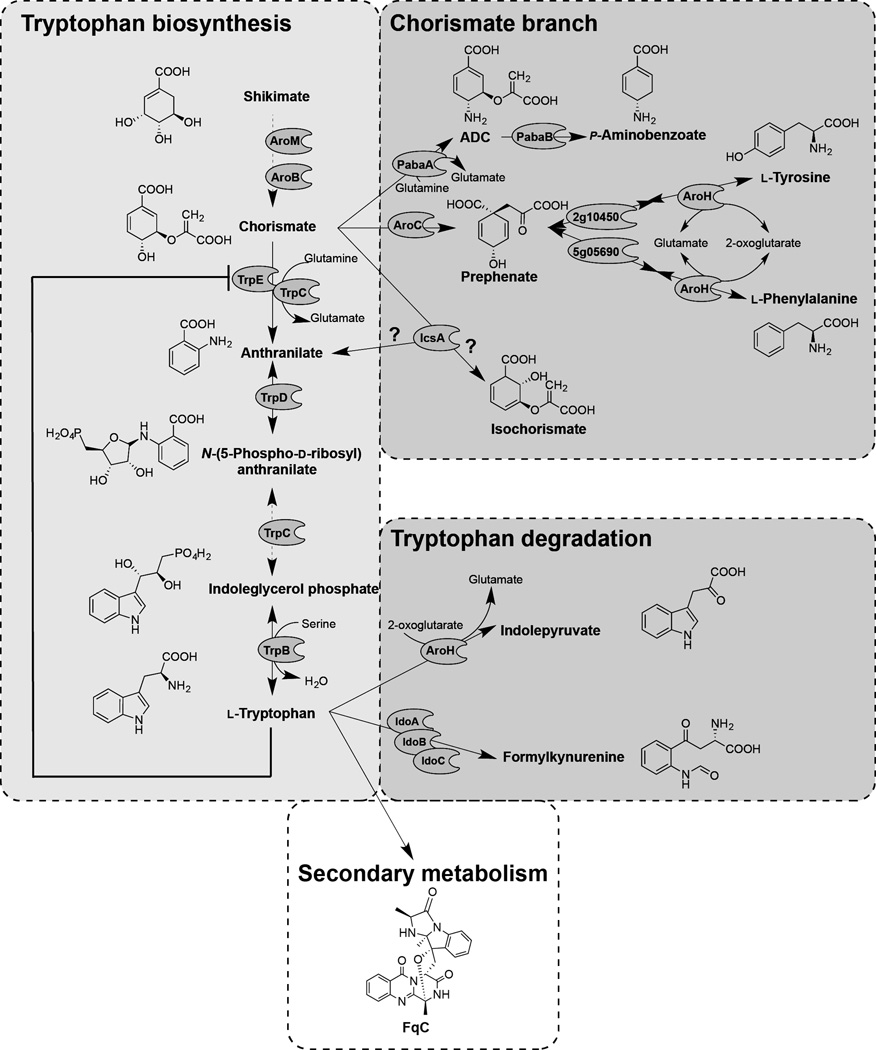
Figure 2 presents the known and predicted A. fumigatus enzymes involved in aromatic amino acid metabolism. Chorismate is a common intermediate for the aromatic amino acids anthranilate, l-tryptophan, l-phenylalanine and l-tyrosine in yeast (Braus, 1991). l-tryptophan is derived from chorismate by five enzymatic steps encoded by four genes in A. fumigatus (Fig. 2 and Table 1). Anthranilate synthase (AS) [EC 4.1.3.27] catalyzes the production of anthranilate from chorismate in the l-tryptophan biosynthesis branch. ASs have been characterized in bacteria (Bae et al., 1989; Ito et al., 1969; Palmer, G.C. et al., 2013), in plants (Morino et al., 2005; Saika et al., 2012; Tozawa et al., 2001) and in Saccharomyces cerevisiae (Braus, 1991; Graf et al., 1993). In yeast, AS consists of two subunits (AAS-I and AAS-II) that are necessary to yield anthranilate from chorismate by addition of an amino group from l-glutamine (Graf et al., 1993). AAS-I is responsible for chorismate binding, and AAS-II is a glutamine amidotransferase that catalyzes the transfer of the amino group (Graf et al., 1993). A tryptophan feedback loop specified by amino acids in the AAS-I subunit (Graf et al., 1993; Tozawa et al., 2001) makes AS the rate-limiting enzyme in l-tryptophan synthesis. Feedback inhibition resistant forms of this enzyme – allowing for increased tryptophan pools – have been associated with increases in several plant natural products including the alkaloid lochnericine in transgenic hairy root cultures of the periwinkle Catharanthus roseus (Hughes et al., 2004) and indole alkaloids in rice (Dubouzet et al., 2013). In Aspergillus spp., the AAS-II encoding gene, trpC, was first characterized in A. nidulans in 1977 (Käfer, 1977) but, until now, trpE encoding AAS-I has only been defined by predicted enzymatic activity. Here, we show that AAS-I feedback mutants in A. fumigatus enhance Fq production. The A. fumigatus genome contains two putative AAS-I proteins, one beside the fmq cluster and one in a region of co-linearity with other Aspergillus spp. Over-expression of the fmq-cluster-associated gene (icsA/Afu6g12110) could not complement the tryptophan auxotrophy of the trpE (Afu6g12580) deletion mutant, and had no significant difference on Fq production. However, over-expression of icsA resulted in significantly decreased anthranilate, l-tryptophan, l-phenylalanine, and l-tyrosine levels, suggesting that IcsA metabolizes chorismate into a yet unknown product in A. fumigatus. Phylogenetic analysis showed it to be closely related to bacterial isochorismate synthases. The second gene, trpE (Afu6g12580), encoded the canonical AAS-I required for l-tryptophan production. Creation of synthetic feedback resistant alleles of TrpE resulted in strains that produced approximately 1.5 fold more FqF and FqC, and displayed a 120+ fold increase in intracellular levels of anthranilate and up to 3+ fold more tryptophan than the control strain on minimal medium. Supplementation with l-tryptophan eliminated any gain in Fq production and in fact lowered production in the single site feedback mutant, potentially through increases in the l-tryptophan degradation product l-kynurenine.
Figure 2. Schematic outline of the l-tryptophan metabolism and regulation of enzyme in A. fumigatus.
Enzymes are indicated by their gene designations: AroM (Afu1g13740), shikimate kinase (EC:2.7.1.71), EPSP synthase (EC:2.5.1.19); AroB (Afu1g06940), chorismate synthase (EC:4.2.3.5); TrpC (Afu1g13090), TrpE (Afu6g12580), anthranilate synthase (EC:4.1.3.27); TrpD (Afu4g11980), anthranilate phosphoribosyltransferase (EC:2.4.2.18); TrpC (Afu1g13090), phosphoribosylanthranilate isomerase (EC:5.3.1.24), indole-3-glycerol-phosphate synthase (EC:4.1.1.48); TrpB (Afu2g13250), tryptophan synthase (EC:4.2.1.20); PabaA (Afu6g04820), ADC synthetase (EC:2.6.1.85); PabaB (Afu2g01650), ADC lyase (4.1.3.38); AroC (Afu5g13130), chorismate mutase (EC:5.4.99.5); 2g10450 (Afu2g10450), prephenate dehydrogenase (EC:1.3.1.13); 5g05690 (Afu5g05690), prephenate dehydratase (EC:4.2.1.51); AroH (Afu2g13630), aromatic aminotransferase (EC:2.6.1.27; 2.6.1.57; 2.6.1.5); IcsA (Afu6g12110), isochorismate synthetase (EC:5.4.4.2); IdoA (Afu3g14250), IdoB (Afu4g09830), IdoC (Afu7g02010), indoleamine 2,3-dioxygenase (EC:1.13.11.52). Abbreviations: EPSP, enolpyruvylshikimate-3-phosphate; ADC, 4-Amino-4-deoxychorismate.
Table 1.
Aspergillus fumigatus tryptophan metabolism genes and putative protein function
| Alias | Protein | Ortholog in S.cerevisiae |
GenBank accession |
EC number |
Protein functiona |
|---|---|---|---|---|---|
| Tryptophan biosynthesis | |||||
| Afu1g13740 | AroM | Aro1 | XM_747651 | 2.7.1.71 | Shikimate kinase |
| Afu1g13740 | AroM | Aro1 | 2.5.1.19 | EPSPb synthase | |
| Afu1g06940 | AroB | Aro2 | XM_745350 | 4.2.3.5 | Chorismate synthase |
| Afu6g12580 | TrpE | Trp2 | XM_746043 | 4.1.3.27 | Anthranilate synthase |
| Afu1g13090 | TrpC | Trp3 | XM_747586 | ||
| Afu4g11980 | TrpD | Trp4 | XM_746540 | 2.4.2.18 | Anthranilate phosphoribosyltransferase |
| Afu1g13090 | TrpC | Trp1 | XM_747586 | 5.3.1.24 | Phosphoribosylanthranilate isomerase |
| Afu1g13090 | TrpC | Trp3 | XM_747586 | 4.1.1.48 | Indoyl-glycerolphosphate synthase |
| Afu2g13250 | TrpB | Trp5 | XM_750564 | 4.2.1.20 | Tryptophan synthase |
| Chorismate branch | |||||
| Afu6g04820 | PabaA | Abz1 | XM_742503 | 2.6.1.85 | ADCb synthetase |
| Afu2g01650 | PabaB | Abz2 | XM_744207 | 4.1.3.38 | ADCb lyase |
| Afu6g12110 | IcsA | - | XM_745997 | 5.4.4.2 | Isochorismate synthase |
| Afu5g13130 | AroC | Aro7 | XM_748248 | 5.4.99.5 | Chorismate mutase |
| Tryptophan degradation | |||||
| Afu2g13630 | AroH | Aro8 | XM_750603 | 2.6.1.27 2.6.1.57 2.6.1.5 |
Aromatic amino acid transaminase |
| Afu3g14250 | IdoA | Bna2 | XM_749108 | ||
| Afu4g09830 | IdoB | Bna2 | XM_746750 | 1.13.11.52 | Indoleamine 2,3-dioxygenases |
| Afu7g02010 | IdoC | Bna2 | XM_741638 | ||
Prediction of protein function based on AspGD (http://www.aspgd.org/) and KEGG(http://www.genome.jp/kegg/)
Abbreviation: EPSP synthase, enolpyruvylshikimate-3-phosphate synthase; ADC, 4-Amino-4-deoxychorismate.
Material and Methods
Strains and medium
Strains used or created in this study are listed in Table 2. The genetic background of the primary strain used in this study is A. fumigatus AF293 (Osherov et al., 2001). All strains were maintained as glycerol stocks at −80 °C, and activated on solid glucose minimal media (GMM) at 37 °C (Shimizu and Keller, 2001). Growth media was supplemented with 1.26 g/L uridine and 0.56 g/L uracil for pyrG auxotrophs, 1 g/L l-arginine for argB auxotrophs, and 1 g/L l-tryptophan for trpE auxotrophs.
Table 2.
Fungal strains used in this study
| Name | ID | Parental strain |
Genotype | Reference |
|---|---|---|---|---|
| AF293 | Wild-type | (Osherov et al., 2001) | ||
| AF293.1 | AF293 | pyrG1 | (Osherov et al., 2001) | |
| AF293.1 comp | TJW55.2 | AF293.1 | A.p.pyrG | (Bok and Keller, 2004) |
| ΔicsA | TPMW1.13 | AF293.1 | ΔicsA::A.p.pyrG | This study |
| OE::icsA | TPMW1.70 | AF293.1 | gpdA(p)::icsA::A.p.pyrG | This study |
| AF293.6 | AF293.1 | pyrG1; argB1 | (Xue et al., 2004) | |
| ΔtrpE pyrG- | TPMW6.2 | AF293.6 | ΔtrpE::argB; pyrG1 | This study |
| ΔtrpE | TPMW12.4 | TPMW6.2 | ΔtrpE::argB; A.p.pyrG | This study |
| ΔtrpE ΔicsA | TPMW7.14 | TPMW6.2 | ΔtrpE::argB; ΔicsA::A.p.pyrG | This study |
| ΔtrpE OE::icsA | TPMW11.18 | TPMW6.2 | ΔtrpE::argB; gpdA(p)::icsA::A.p.pyrG | This study |
| trpEC | TPMW8.9 | TPMW6.2 | gpdA(p)::trpEC::A.p.pyrG; ΔtrpE::argB | This study |
| trpES77L | TPMW9.5 | TPMW6.2 | gpdA(p)::trpES77L::A.p.pyrG; ΔtrpE::argB | This study |
| trpES66R,S77L | TPMW10.9 | TPMW6.2 | gpdA(p)::trpES66R,S77L::A.p.pyrG; ΔtrpE::argB | This study |
Genetic manipulations
Fungal DNA extraction, gel electrophoresis, restriction enzyme digestion, Southern blotting, hybridization and probe preparation were performed according to standard methods (Sambrook and Russell, 2001). For DNA isolation, A. fumigatus strains were grown for 24 h at 37 °C in steady state liquid GMM, supplemented as needed for auxotrophs. DNA isolation was performed as described by Sambrook and Russell (2001). Gene deletion mutants in this study were constructed by targeted integration of the deletion cassette through transformation (Lim et al., 2012b; Szewczyk et al., 2006). The deletion cassettes were constructed using a double-joint fusion PCR (DJ-PCR) approach (Lim et al., 2012b; Szewczyk et al., 2006). A. fumigatus protoplast generation and transformation were carried out as previously described (Lim et al., 2012b; Szewczyk et al., 2006). The plasmids used in this work are listed in Table S1 and all primers are listed in Table S2.
An A. fumigatus trpE (Afu6g12580) disruption cassette consisted of the following: 1 kb DNA fragment upstream of the trpE start codon (primers DAf6g12580F1 and DAf6g12580R1), a 2.7 kb selectable auxotrophic marker, A. fumigatus argB, cloned from plasmid pJMP4 (Palmer, J.M. et al., 2013) (primers GF A. Fumi argB F and GF A. Fumi argB R), and 1 kb DNA fragment downstream of the trpE stop codon (primers DAf6g12580F2 and zDAf6g12580R2). The deletion construct was transformed into AF293.6 (pyrG1, argB1). Transformants were screened for arginine prototroph in media supplemented with l-tryptophan for trpE auxotrophs, uridine and uracil for pyrG auxotrophs. Transformants obtained were verified by PCR using primers PM-F6g12580F and PM-F6g12580R, and Southern analysis using a trpE probe SB1 (primers DAf6g12580F1 and DAf6g12580R2) (Fig. S1). TPMW6.2 (ΔtrpE, pyrG1-) was chosen for further experiments.
In order to complement the ΔtrpE mutant with a wild-type and feedback insensitive trpE copies, the plasmids pPMW1 (trpEC), pPMW2 (trpES77L) and pPMW3(trpES66R,S77L) were constructed using in vivo yeast recombination (Yin et al., 2013b) of 4 linear DNA fragments consisting of (1) the gpdA(p) promoter obtained by PCR from plasmid pJMP9.1 (Lim et al., 2012a) (with primers PM-yAup-gpdA F and PM-yAup-gpdA R), (2) a AscI-linearized 2µ-based yeast-Escherichia coli shuttle plasmid pYH-yA-riboA (Palmer, J.M. and Keller, unpublished), (3) the first half of wild-type or a Ser66Arg-mutated trpE gene (obtained by PCR from A. fumigatus wild-type genomic DNA with primers PM-F6g12580F and PM-S66-afu6g12580R for trpEC; and with primers PM-F6g12580F and PM-R66M–afu6g12580R for the S66R–mutation) and (4) the second half of wild-type or a Ser77Leu-mutated trpE gene (obtained by PCR from A. fumigatus wild-type genomic DNA with primers PM-S77 Afu6g12580F and PM-riboB-Afu6g12580R for trpEC;and with primers PM-L77M Afu6g12580F and PM-riboB-Afu6g12580R for the S77L–mutation), respectively. The above fragments contained 20–35 bp overlapping bases to the correct flanking fragments for in vivo yeast recombination to create the plasmids. All of the plasmids described above were verified by restriction digest profiles and sequencing, and then digested with AvrII and SacII to release the appropriate gpdA(p)-trpE fragment to be ligated into the AvrII/SacII sites of pJMP9.1 (Lim et al., 2012a). The resulting plasmids pPMW4(OE::trpEC), pPMW5 (OE::trpES77L), pPMW6(OE::trpES66R,S77L) and the parent plasmid pJMP9.1 were transformed into the auxotrophic ΔtrpE mutant TPMW6.2 (ΔtrpE, pyrG1) to yield mutants TPMW8.9 (trpEC), TPMW9.5(trpES77L), TPMW10.9 (trpES66R,S77L) and TPMW12.4 (ΔtrpE), respectively. These mutants were selected for tryptophan and pyrimidine prototrophy in media supplemented with l-tryptophan for trpE auxotrophs, in case site-mutation strain cannot produce l-tryptophan for growth. PCR screening (primers PM-F6g12580F and PM-F6g12580R) and Southern analysis were applied to obtain single-gene-copy replacement transformants, using probe SB2 (primer pair PM-F6g12580F/DAf6g12580R2) (Fig. S2A). Over-expression of trpE was verified via Northern analysis for trpEC, trpES77L, and trpES66R,S77L compared to the complemented parental strain TJW55.2 (Fig. S2B) icsA (Afu6g12110) was disrupted and over-expressed both in AF293.1 (pyrG1) and TPMW6.2 (ΔtrpE, pyrG1) using a pyrG marker cassette. To construct the icsA deletion cassette, two 1 kb fragments flanking the ORF were amplified from AF293 genomic DNA using the primer pair DAfu6gF1/ DAfu6gR1 and DAfu6gF2/ DAfu6gR2, respectively (Table S2). The selection marker, A. parasiticus pyrG, was PCR amplified from plasmid pJW24 (Calvo et al., 2004) using the primer pair ParapyrGF/ParapyrGR (Table S2). To construct an icsA over-expression strain at its locus, its native promoter was replaced with the constitutive promoter, gpdA(p), amplified from plasmid pJMP9.1 using the primer pair OEpyrGgpdF/OEpyrGgpdR. The two 1 kb fragments for the icsA over-expression cassette were amplified from AF293 genomic DNA using the primer pair DAfu6gF1/OEAfu6gR1 and OEAfu6gF2/DAfu6gR2, respectively (Table S2). Transformants were selected for pyrimidine prototrophy in media without any supplements and amended with l-tryptophan in case TPMW6.2 was used as the parental strain. Transformants were further screened by PCR (primer pair diAfu6gF1/diAfu6gR1 for ΔicsA mutants, OEdiAfu6gF/OEdiAfu6gR for OE::icsA mutants) and Southern analysis using probe SB3 (primer pair DAfu6gF1/DAfu6gR2) (Fig. S3). The obtained mutants were named TPMW1.13 (ΔicsA), TPMW1.70 (OE::icsA), TPMW7.14 (ΔtrpE ΔicsA), and TPMW11.18 (ΔtrpE OE::icsA). Over-expression of icsA was verified via Northern analysis for TPMW1.70 compared to the complemented parental strain TJW55.2, and the icsA deletion strain TPMW1.13 (Fig. S4).
Phylogenetic analysis
For phylogenetic analysis, reviewed and curated sequences from the Swiss-Prot database (www.uniprot.org) of proteins containing a chorismate binding domain (isochorismate synthase, anthranilate synthase and p-aminobenzoate synthase) were retrieved and aligned using ClustalW (MegAlign, DNAStar, Madison, WI, USA) together with the protein sequences of Afu6g12110/IcsA, Afu6g04820/PabaA, and Afu6g12580/TrpE (www.aspergillus.org) (Cerqueira et al., 2014). From the alignment, the chorismate binding domains were extracted based on the canonical chorismate binding domain from the Conserved Domain Database (CDD) (www.ncbi.nlm.nih.gov). Phylogenetic analysis of the chorismate binding domains was performed using MAFFT (http://mafft.cbrc.jp/alignment/software/) (Katoh et al., 2002) and (http://www.microbesonline.org/fasttree/) (Price et al., 2009). Results were visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/) collapsing branches with bootstrap values below 70% support value. For phylogenetic analysis of the indoleamine 2,3-dioxygenase (Ido) proteins, sequences from Aspergillus and Fusarium species were obtained from the National Center for Biotechnology Information (NCBI) by performing a protein blast search using the S. cerevisiae Bna2p sequence as query. Sequences were processed and visualized including Ido sequences previously analyzed by Yuasa and Ball (2011; 2012; 2013) as described for the chorismate binding domain proteins above.
Physiology experiments
Colony diameters of strains were measured after 3 days of growth at 37 °C on solidified GMM and GMM supplemented with 5 mM l-tryptophan, respectively. Strains were point-inoculated onto the media at 104 conidia total (in 5 µL). Conidial production studies were performed on solid GMM and tryptophan plates. For each plate, a 10 mL top layer of cool but molten agar that contained 107 spores of each strain, respectively, was added. Strains were cultured at 37 °C for 24 h. A core of 1.5 cm diameter was removed from the plates and homogenized in 2 mL 0.01% Tween 80 to release the spores. Spores were counted on a hemocytometer. Three replicates were used for each assay and statistical significance was calculated with by analysis of variance (ANOVA) using Prism 6 software (Graph Pad).
Primary metabolites extraction and analysis
A. fumigatus strains were inoculated into 50 mL of liquid GMM and GMM supplemented with 5mM l-tryptophan, respectively, at 107 conidia per mL and cultured at 25 °C and 250 rpm for 84 h in triplicates. Fungal tissue was collected by rapid filtration using miracloth and immediately frozen in liquid nitrogen. About 0.1 g of fungal tissue was transferred to a 15 mL conical vial containing 3 mL extraction solvent (2/2/1 (v/v/v) acetonitrile/methanol/water) cooled on dry ice. After homogenization and centrifugation, 2 mL of supernatant was filtered using a 0.45 µm PTFE Mini-UniPrep filter vial (Agilent). The supernatant of these hyphal metabolites were used for metabolite analysis. For metabolite measurement, samples were dried under N2 and resuspended in LC-MS grade water (Sigma-Aldrich). Samples were analyzed using an HPLC-MS system consisting of a Dionex UPHLC coupled by electrospray ionization (ESI; negative mode) to a hybrid quadrupole-high-resolution mass spectrometer (Q Exactive orbitrap, Thermo Scientific) operated in full scan mode. Liquid chromatography separation was achieved using an ACQUITY UPLC® BEH C18 (2.1 × 100 mm column, 1.7 µm particle size, Waters). Solvent A was 97:3 water:methanol with 10 mM tributylamine and 10 mM acetic acid, pH 8.2; solvent B was methanol. The gradient was: 0 min, 5% B; 1.5 min, 5% B, 11.5 min, 95% B; 12.5 min, 95% B; 13 min, 5% B; 14.5 min, 5% B. Autosampler and column temperatures were 4 °C and 25 °C, respectively. Metabolite peaks were identified by their exact mass and matching retention time to those of pure standards (Sigma-Aldrich).
RNA extraction and northern analysis
For northern expression analysis of trpE and icsA in A. fumigatus wild-type AF293 at different time point, wild-type strain AF293 was grown in liquid glucose minimal media (GMM) (replacing nitrate with 20mM glutamine as nitrogen source) at 37 °C and 250 rpm for 24 h. Then mycelia were collected, transferred into solid GMM and grown in duplicates for the indicated time at 29 °C. To verify over-expression of icsA via northern blot, TPMW1.13 (ΔicsA), TPMW1.70 (OE::icsA) and the complemented strain TJW55.2 were grown in liquid GMM at 37 °C and 250 rpm for 24 h. For northern analysis, strains TPMW8.9 (trpEC), TPMW9.5 (trpES77L), TPMW10.9 (trpES66R,S77L) and the complemented control strain TJW55.2 were inoculated into 50 mL of liquid GMM containing 20mM glutamine as nitrogen source as indicated in the text according to Schrettl et al. (2008). Strains were grown for 24 h at 37 °C, 250 rpm with an initial spore concentration of 106 conidia per mL. Mycelia were harvested by filtration through Miracloth (Calbiochem) and 2 g of mycelia were transferred into 50 mL of liquid GMM and GMM supplemented with 5mM l-tryptophan, respectively. Strains were grown in duplicates for 1 h at 29 °C and 250 rpm. Mycelia were harvested by filtration through Miracloth. Total RNA was extracted with Trizol reagent (Invitrogen) from freeze-dried mycelia, according to the manufacturer’s protocol. Northern analysis was performed as described by Sambrook and Russell (2001). Northern analysis for trpE used trpE probe SB1 (primers DAf6g12580F1 and DAf6g12580R2), for icsA used icsA probe SB3 (primer pair DAfu6gF1/DAfu6gR2). Other probes for northern analysis were constructed using primers listed in Table S2 and labeled with dCTP αP32.
Secondary metabolite extraction and profiling
Secondary metabolites of strains were extracted after 6 days of growth at 29 °C on solidified GMM and GMM supplemented with 5 mM l-tryptophan, respectively. Strains were point-inoculated onto the media at 104 conidia total (in 5 µL). Agar cores (1.5 cm in diameter) were prepared in triplicate for each strain cultured. Three cores from each plate were extracted with 2.5 mL of ethyl acetate. The solvent was evaporated and suspended in 500 µL 19.5/79.5/1 (v/v/v) acetonitrile/water/formic acid. Subsequently, the samples were filtered using a 0.45 µm PTFE Mini-UniPrep filter vial (Agilent) and 50 µL of the filtrate analyzed by high-performance liquid chromatography (HPLC) (Perkin Elmer) coupled to a photo diode array (PDA) as described by Wiemann et al. (2014). Fq quantification was performed as described by Ames and Walsh (2010).
Results
The A. fumigatus genome contains a canonical anthranilate synthase and a putative isochorismate synthase
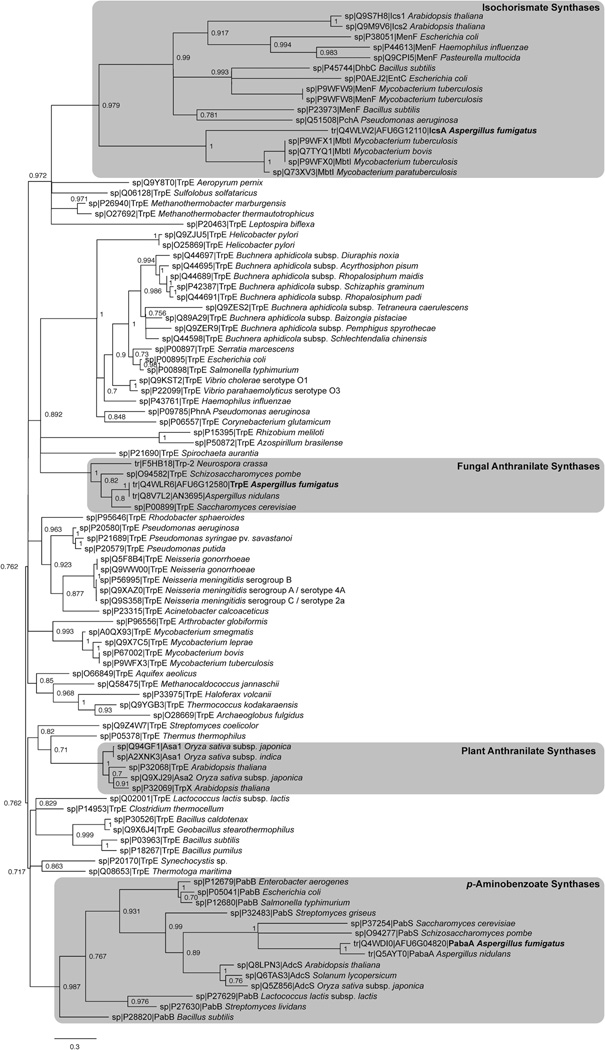
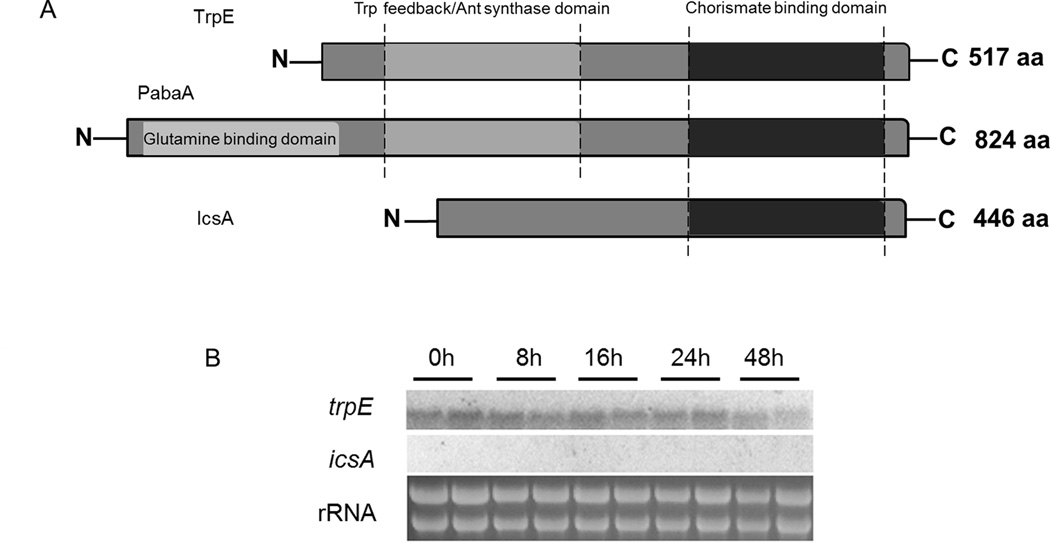
When characterizing the fmq cluster (Ames et al., 2010; Lim et al., 2014), a putative anthranilate synthase subunit I (AAS-I), Afu6g12110, was found 8.3 kb downstream of the fmq gene cluster (Fig. 1A). Considering the requirement for both l-tryptophan and anthranilate for Fq synthesis (Fig. 1B), it seemed reasonable that Afu6g12110 could be involved in synthesis of both amino acids. Further analysis of the genome, however, revealed a canonical AAS-I, Afu6g12580, which was present in region of co-linearity in all sequenced Aspergillus species. In contrast genes displaying significant homology to Afu6g12110 were only detected in a subset of Aspergillus species located at dispersed genomic locations (http://www.aspergillusgenome.org). A comparison of the two proteins indicated they shared 30% identity (E value=6×10−23), primarily along the conserved chorismate binding domain. As a first step toward determining the function of the two potential anthranilate synthases in A. fumigatus, a phylogenetic tree was created. Our results show that Afu6g12580, is closely related to characterized AAS-I of fungi including A. nidulans, Neurospora crassa, and S. cerevisiae (Fig. 3) and thus was named TrpE following terminology used in A. nidulans. By contrast, the protein sequence of Afu6g12110 was most closely related to bacterial isochorismate synthases (Fig. 3) and named IcsA for isochorismate synthase. Notably IcsA lacked the feedback inhibition domain found in TrpE and all known AAS-I (Fig. 4A) but contained a chorismate binding domain also found in PabaA (Afu6g04820). Under culture conditions with nitrate as sole nitrogen source, trpE but not icsA was strongly expressed (Fig. 4B).
Figure 3. Maximum likelihood phylogenetic analysis of chorismate-binding domains extracted from 99 characterized protein sequences.
Monophyletic leaves of relevant proteins are boxed in grey. A. fumigatus proteins are indicated in bold.
Figure 4. Domain architecture and expression analysis of chorismate-binding domain proteins in A. fumigatus.
(A) Domain comparison of A. fumigatus TrpE, PabaA and IcsA. Abbreviations: Ant, Anthranilate; Trp, Tryptophan.
(B) Northern expression analysis of trpE and icsA in A. fumigatus wild-type AF293. Ribosomal RNA was visualized by ethidium bromide staining as loading control. A. fumigatus strain AF293 was grown in liquid GMM containing 20mM glutamine as nitrogen source at 37 °C and 250 rpm for 24 h. Then mycelia were collected, transferred into solid GMM and grown in duplicates for the indicated time at 29 °C.
To determine if these proteins were required for tryptophan biosynthesis, we deleted and over-expressed both genes in the A. fumigatus AF293 genetic background. Southern analysis and Northern analysis of transformants for each gene identified correct gene replacement and over-expression constructs for both genes (Fig. S1–Fig. 4). One representative deletion mutant for each gene, TPMW12.4 (ΔtrpE) and TPMW1.13 (ΔicsA), one representative over-expression (OE) strain, TPMW8.9 (trpEC) and TPMW1.70 (OE::icsA), as well as the double mutants TPMW7.14 (ΔtrpE ΔicsA) and TPMW11.18 (ΔtrpE OE::icsA) were chosen for further studies (Table 2). The ΔtrpE mutant could not grow unless supplemented with l-tryptophan. Complementation of ΔtrpE with a trpE allele restored wild-type-like growth (Fig. 5A). In contrast neither the ΔicsA nor the OE::icsA strains exhibited a phenotypical difference to the respective control strains on any media examined in this study (Fig. 5A and Fig. S5). Furthermore, the OE::icsA allele could not complement the tryptophan auxotrophy of ΔtrpE (Fig. 5A) thus suggesting that icsA played no major role in tryptophan or anthranilate biosynthesis.
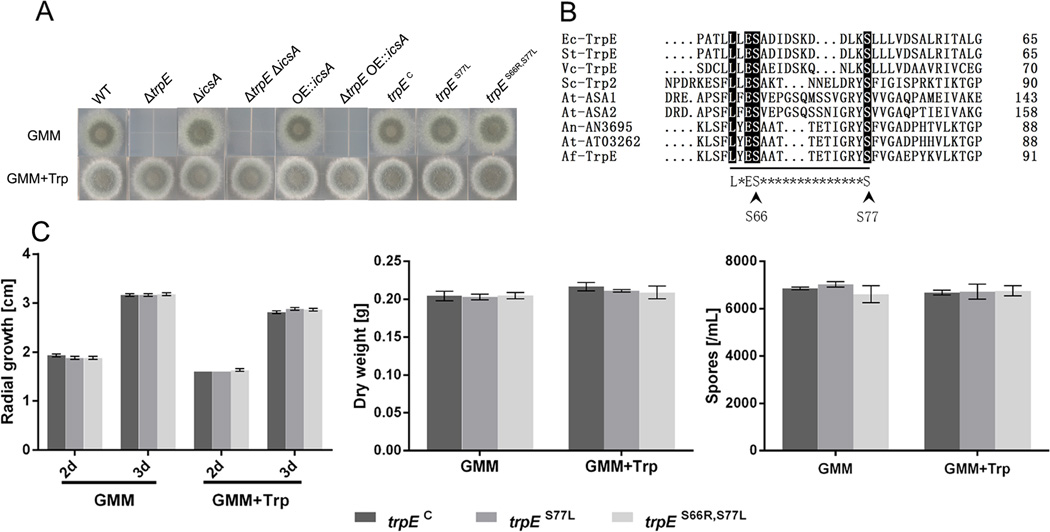
Figure 5. Physiological analysis of A. fumigatus mutants used in this study and the alignment of anthranilate synthases.
(A) Radial growth of the A. fumigatus wild-type strain (WT), ΔtrpE, ΔicsA, double deletion mutant ΔicsAΔtrpE, icsA over-expression strain OE::icsA, double mutant ΔtrpE OE::icsA, ΔtrpE complemented strain trpEC, and site mutants trpES77L and trpES66R,S77L and on solid GMM and GMM amended with 5mM l-tryptophan (Trp) medium at 37 °C.
(B) Alignment of the L*ES*nS regions of anthranilate synthases from various organisms. The conserved motif is indicated on the bottom line. The sequences shown are: Ec-TrpE, Escherichia coli TrpE (CAA23666); St-TrpE, Salmonella enterica TrpE(WP_001194371); Vc-TrpE, Vibrio cholerae TrpE (WP_001030227); Sc-Trp2, Saccharomyces cerevisiae Trp2 (NP_011014); At-ASA1, Arabidopsis thaliana ASA1 (NP_001190231); At-ASA2, Arabidopsis thaliana ASA2 (NP_180530); An-AN3695, Aspergillus nidulans AN3695 (CBF75591); At-AT03262, Aspergillus terreus AT03262 (XP_001212440); Af-TrpE, Aspergillus fumigatus TrpE (XP_751136). Identical residues among the various proteins are indicated by dark shading, and respective amino acid residue numbers are shown at the C-termini. Arrows mark amino acids of Af-TrpE mutated in this study. (C) Quantification of radial growth on solid media GMM and GMM+5mM l-tryptophan (Trp) at 37 °C, dry weight of mycelia from culture in liquid medium GMM and GMM+5mM l-tryptophan (Trp) at 37 °C, and spore production on solid GMM and GMM+5mM l-tryptophan (Trp) at 37 °C. No phenotype was observed for tryptophan-feedback mutants trpES77L and trpES66R,S77L compared with trpEC.
TrpE feedback mutants exhibit increased production of anthranilate and fumiquinazolines FqF and FqC and IcsA mutants alter amino acid pools
To create feedback insensitive mutants in A. fumigatus, we first aligned the TrpE sequence with known AAS-I proteins and readily located the amino acids involved in feedback inhibition (Fig. 5B). Two mutant alleles were created, one was designed to change serine residue 77 to leucine (S77L), and the other was designed to additionally change serine residue 66 to arginine (S66R). These residues were chosen as they have been shown essential for tryptophan feedback inhibition in yeast (Graf et al., 1993) and some plants (Kanno et al., 2005; Saika et al., 2012). Transformation of the ΔtrpE strain with a wild-type and these two mutant trpE alleles, respectively, yielded three strains TPMW8.9 (trpEC), TPMW9.5 (trpES77L) and TPMW10.9 (trpES66R,S77L) for comparison of aromatic acid metabolism and Fq synthesis (Fig. S2).
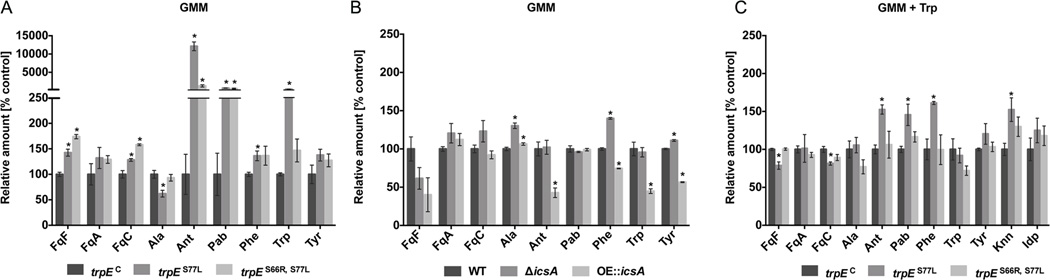
Although there were no significant differences in growth or sporulation among the three strains on either GMM or GMM supplemented with 5 mM l-tryptophan (Fig. 5C), we observed a large intracellular buildup of anthranilate (ca. 120 fold vs. control strain trpEC) in the feedback mutant trpES77L grown on GMM medium (Fig. 6A). The substantial increase in intracellular anthranilate was consistent with the loss of tryptophan feedback in this enzyme, which resulted in the increased biosynthesis of the related intermediates. In agreement with this, we also observed increased tryptophan levels (4 fold) in the trpES77L mutant. Surprisingly, we also found that p-aminobenzoate (7 fold) and to a small extent phenylalanine (1.4 fold) were increased in this mutant. The trpES66R,S77L feedback mutant displayed smaller but still significant increased levels of anthranilate (ca. 13 fold) and p-aminobenzoate (5.5 fold). Examination of Fq production in these three strains showed that FqC and FqF production was significantly increased by 1.3–1.8 fold in both feedback mutants grown on GMM medium (Fig. 6A).
Figure 6. Primary metabolites and fumiquinazoline production in A. fumigatus trpE and icsA mutants.
(A) Comparison of primary metabolites and fumiquinazoline production in the complemented control strain trpEC and the feedback site mutants trpES77L and trpES66R,S77L grown on GMM medium.
(B) Comparison of primary metabolites and fumiquinazoline production in wild-type control strain AF293.1 comp (TJW55.2) and icsA mutants grown on GMM medium.
(C) Comparison of primary metabolites and fumiquinazoline production in the complemented control strain trpEC and the feedback site mutants trpES77L and trpES66R,S77L grown on GMM supplemented with 5mM l-tryptophan (Trp) medium.
The amounts of metabolites were normalized to the relative amount of the complemented control strain trpEC (100%). Displayed are means ± SEM. Asterisk indicates p < 0.05 using an ANOVA test for statistical significance with Prism 6 software, comparing mutants to the control strain TJW55.2 or trpEC.
Abbreviations: FqF, Fumiquinazoline F; FqC, Fumiquinazoline C; FqA, Fumiquinazoline A; Ala, l-alanine; Ant, Anthranilate; Pab, p-aminobenzoate; Phe, l-phenylalanine; Trp, l-tryptophan; Tyr, l-tyrosine; Knn, l-kynurenine; Idp, Indolepyruvate.
We then assessed the metabolome of the icsA mutants. Although there was no significant impact of either mutant on Fq production, deletion of icsA resulted in a significant increase in phenylalanine and tyrosine levels (as well as alanine) whereas OE::icsA exhibited significantly decreased anthranilate, l-tryptophan, l-phenylalanine, and l-tyrosine levels (Fig. 6B). Together this data suggested that IcsA metabolizes chorismate into a yet unknown metabolite thereby making the common substrate less available for TrpE and AroC, respectively (Fig. 2).
To assess if the TrpE feedback mutants could be pushed to further increase Fq production, media was supplemented with l-tryptophan. We found this treatment primarily restored metabolite levels of the feedback mutants to those of wild type. There were no differences between wild type and the trpES66R,S77L mutant (Fig. 6C). The trpES77L mutant still accumulated significantly more anthranilate, p-aminobenzoate and phenylalanine than wild type but at much less fold than in GMM medium. Furthermore, in contrast to results on GMM, this mutant produced less FqC and FqF than wild type (Fig. 6C). Finally, two tryptophan degradation products, l-kynurenine and indolepyruvate were detected in all three strains grown on tryptophan medium. The single site mutant produced significantly more l-kynurenine than either the wild type or the double site mutant, possibly suggesting that increases in l-kynurenine production could be related to decreases in Fq production (Fig. 6C).
Transcriptional profiling of tryptophan metabolism genes
To help interpret the changes in primary and secondary metabolite synthesis on GMM and GMM supplemented with l-tryptophan, we next examined a transcriptional profile of l-tryptophan metabolic genes in both GMM and GMM medium supplemented with l-tryptophan for the two feedback mutants (trpES77L and trpES66R,S77L) and complemented control trpEC (Fig. 7). Genes were identified through blast analysis using characterized genes from tryptophan metabolic enzymes in S. cerevisiae and KEGG l-tryptophan metabolism (http://www.genome.jp/kegg/pathway.html) to identify putative genes and encoded proteins illustrated in Fig. 2 and Table 1.
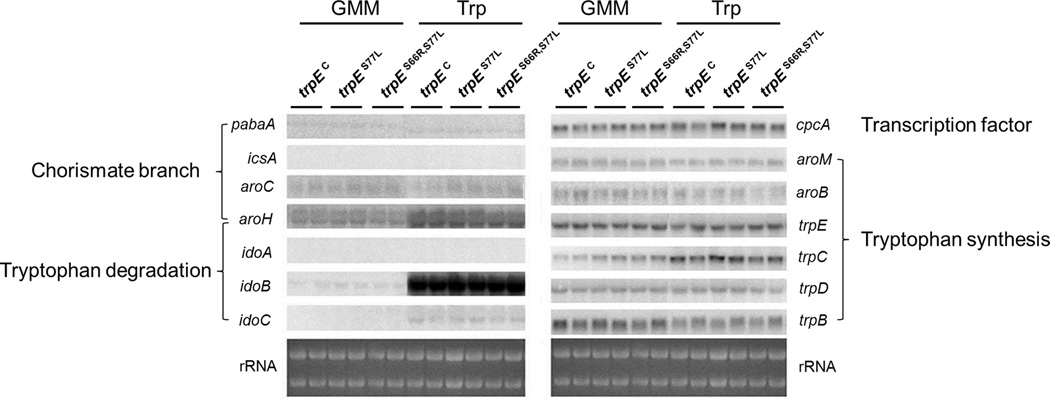
Figure 7. Northern expression analysis of genes related to tryptophan metabolism.
Northern expression analysis of indicated genes comparing the A. fumigatus ΔtrpE complemented strain trpEC, the trpE tryptophan-feedback mutant trpES77L, and trpES66R,S77L. Ribosomal RNA was visualized by ethidium bromide staining as loading control. The indicated strains were grown in liquid glucose minimal media (GMM) (replacing nitrate with 20 mM glutamine as sole nitrogen source) at 37 °C and 250 rpm for 24 h. After the initial incubation period, 2 g of mycelia were transferred into liquid GMM and GMM supplemented with 5mM l-tryptophan (Trp), respectively. The indicated strains were grown in duplicates for 1 h at 29 °C and 250 rpm.
Chorismate is a common precursor for metabolic pathways leading to the formation of l-tryptophan, p-aminobenzoate, salicylate and l-phenylalanine/l-tyrosine in microbes and plants (Braus, 1991; Maeda and Dudareva, 2012; Tzin and Galili, 2010). Four putative enzymes, encoded by pabaA, aroC, icsA, and trpE/trpC (Fig. 2, Table 1), theoretically could compete for and utilize chorismate as a substrate to catalyze the committed step of the respective pathways to produce p-aminobenzoate, l-phenylalanine/l-tyrosine, isochorismate, and l-tryptophan in A. fumigatus (Fig. 2). Although we did not detect isochorimate in our analysis, over-expression of icsA decreased anthranilate, l-tryptophan, l-phenylalanine, and l-tyrosine levels suggesting that IcsA can reduce the cellular chorismate pool (Fig. 6B). Furthermore, our metabolic data (Fig. 6) supported the activities of the other enzymes and suggested that pabaA and aroC may be up-regulated in the feedback-domain site mutants grown on GMM medium. However, this was not borne out at the transcriptional level as there were no differences in pabaA or aroC expression between the wild type and feedback mutants in either GMM or l-tryptophan supplemented medium (Fig. 7). In fact, there were no differences in gene expression between these three strains for any of the genes assessed. Tryptophan supplementation, however, did impact gene expression. Two putative l-tryptophan degradation genes, aroH and idoB were highly up-regulated in all three strains (Fig. 7). aroH (Afu2g13630) encodes a putative aromatic aminotransferase involved in transamination of l-tryptophan to generate indolepyruvate and idoB (Afu4g09830) encodes a putative indoleamine 2,3-dioxygenase which could be involved in dioxygenation of l-tryptophan as the first step to produce l-kynurenine. This matched well with production of these two products by A. fumigatus grown on l-tryptophan supplemented medium (Fig. 6B). Two other genes, Afu3g14250 and Afu7g02010, also encode putative indoleamine 2,3-dioxygenases with Afu7g02010 weakly expressed under these conditions. Based on the phylogenetic analysis of indoleamine 2,3-dioxygenases (Fig. S6), we name these genes idoA (Afu3g14250), idoB (Afu4g09830), and idoC (Afu7g02010). l-tryptophan supplementation also affected trpC expression (increased) and trpB (decreased) but had no impact on expression of cpcA, encoding the ortholog of Gcn4p in yeast, the transcription factor globally modulating Aspergillus spp. amino acid biosynthesis (Krappmann et al., 2004).
Discussion
Aspergillus fumigatus is an opportunistic human pathogen renown for its secondary metabolites that are thought to contribute to disease development (Bok et al., 2005). The fumiquinazolines are signature metabolites of A. fumigatus and comprise a family of cytotoxic peptidyl alkaloids, which have received considerable interest due to their complex biochemistry, antitumor properties and cellular localization (Ames et al., 2010; Han et al., 2007; Lim et al., 2014) but until now, nothing is known of the effect of primary metabolism on their biosynthesis. Here our efforts focused on the effect of manipulating tryptophan biosynthesis on Fq production.
Anthranilate is a non-proteinogenic aryl β-amino acid generated from enzymatic amination of chorismate by anthranilate synthases (ASs), and serves as the framework for the indole ring of the amino acid l-tryptophan (Ames and Walsh, 2010). Along with l-tryptophan, anthranilate is incorporated into several fungal peptidyl alkaloids including Fq, the benzodiazepine dione, ardeemin and asperlicin (Ames and Walsh, 2010; Walsh et al., 2013). AS consists of two subunits, anthranilate synthase subunit I (AAS-I) termed TrpE as described here, and AAS-II termed TrpC (Käfer, 1977), respectively in Aspergillus species. We found trpE was present in a region of co-linearity in all sequenced Aspergillus spp. while a second protein, IcsA, encoded by a gene near the fmq cluster and with considerable homology to TrpE was more closely related to bacterial isochorismate synthases and lacked the tryptophan feedback domain found in TrpE and all known AAS-I (Figs. 3 and 4A). The ΔtrpE mutant required exogenous l-tryptophan for growth and could not be rescued by OE::icsA. However, over-expression of icsA decreased anthranilate, l-tryptophan, l-phenylalanine, and l-tyrosine levels suggesting that IcsA can draw from the cellular chorismate pool (Fig. 6B). A protein alignment of the A. fumigatus IcsA and TrpE with characterized bacterial ASS-I and isochorismate synthases revealed that important residues that changed enzymatic activity of the Salmonella typhimorium ASS-I from producing anthranilate to isochorismate (Plach et al., 2015) are identical to the A. fumigatus IcsA, suggesting that it likely is responsible for isochorismate formation although we did not identify this metabolite in our study. Identification of the putative IcsA metabolic product will be a future task.
The bacterium Pseudomonas aeruginosa possesses two ASs, TrpEG and PhnAB (Palmer, G.C. et al., 2013). Although only the former and not the latter is required for l-tryptophan biosynthesis in P. aeruginosa, over-expression of phnAB can rescue a trpEG deletion. A third protein harboring a chorismate binding domain is bifunctional PchA in P. aeruginosa (Fig. 3) and was shown to be involved in isochorismate and subsequently salicylic acid production, that is incorporated into the bacterium’s siderophores, mediating iron uptake (Serino et al., 1997). Based on the chorismate binding domains, IcsA from A. fumigatus is closely related to MbtI from Mycobacterium tuberculosis (Fig. 3), that is also responsible for salicylic acid formation (Harrison et al., 2006). Escherichia coli harbors two enzymes, EntC and MenF, that are responsible for isochorismate production (Buss et al., 2001), that also group with IcsA in our analysis (Fig. 3). Bioinformatic analysis shows some but not all Aspergilli contain putative IcsA homologs, all missing the feedback domain and often located near secondary metabolite clusters. Although fungal siderophores do not contain salicylic acid or isochorismate (Haas, 2014), A. fumigatus was shown to produce at least one other amino acid-derived small molecule with iron binding abilities (Wiemann et al., 2014; Yin et al., 2013a). What the role, if any, IcsA-like proteins play in fungal physiology has yet to be elucidated.
The importance of the feedback regulation of AS in the control of metabolic flow in l-tryptophan production and secondary metabolites has been demonstrated in many plants expressing feedback-resistant AS genes (Dubouzet et al., 2013; Hong et al., 2006; Hughes et al., 2004; Ishihara et al., 2006; Li and Last, 1996; Saika et al., 2012; Tozawa et al., 2001). Here, inactivation of the putative feedback domain in TrpE of A. fumigatus also had a significant effect on aromatic amino acid metabolism and Fq production with the single mutation strain trpES77L consistently having a greater impact on metabolism than the double site mutant. It was only this mutant which showed a significant increase in l-tryptophan pools, similar to reports of increased l-tryptophan pools in feedback mutants in plants (Hughes et al., 2004; Ishihara et al., 2006; Li and Last, 1996; Tozawa et al., 2001). This same strain also showed modest but significant increases in one of the two other aromatic amino acids, l-phenylalanine (Fig. 6A). As Fqs contain l-alanine, l-tryptophan, and anthranilate, the decreased l-alanine was consistent with the increased Fqs. However, both feedback mutants showed large increases in anthranilate and p-aminobenzoate that possibly diverted precursor pools from fumiquinazolines as reflected by the modest increases in FqF and FqC when grown on minimal medium (Fig. 6A). p-aminobenzoate is synthesized by PabaA and PabaB (Fig. 2 and Table 1) in Aspergillus spp. and strains with inactivation of either gene require p-aminobenzoate for growth (Brown et al., 2000; Mellado et al., 2015; Tang et al., 1994). Possibly less efficient versions of these enzymes could reduce p-aminobenzoate accumulation and enhance Fq synthesis. Studies in E. coli and Arabidopsis thaliana have shown that the p-aminobenzoate synthase complex is not inhibited by tryptophan, but by 2-fluorochorismate and methotrexate, respectively (Bulloch et al., 2004; Parsons et al., 2002; Sahr et al., 2006). Whether or not these or analogous inhibitors could direct metabolite flux towards Fq biosynthesis in A. fumigatus could be explored in future studies.
An interesting finding was that supplementation with tryptophan restored metabolite levels in the TrpE feedback mutants to those near the control strain. This suggests that there is an additional regulatory control mechanism(s) for tryptophan synthesis other than anthranilate synthase that is controlled differentially by externally added tryptophan and tryptophan produced by A. fumigatus itself. Intracellular amino acid pools are stored in vacuoles (Sekito et al., 2008) and their accessibility will differ from supplementation from external sources. Our data allude to different intra- and extracellular amino tryptophan sensing mechanisms in A. fumigatus, supporting a hypothesis of distinct nitrogen sensing mechanisms in other filamentous fungi (Wagner et al., 2013; Wiemann and Tudzynski, 2013). Our expression profiling data suggests that several tryptophan degradation pathways could be important in this regard (Fig. 7). The decrease of FqF and FqC in the TrpE single feedback mutant grown on l-tryptophan amended medium may be reflective of not only differential regulatory mechanisms and diversion of chorismate to p-aminobenzoate and l-phenylalanine but also to the increase in the degradation product l-kynurenine (Fig. 6C).
l-kynurenine is produced by indoleamine 2,3-dioxygenase (Ido) activity. Whereas S. cerevisiae has a single Ido gene (BNA2) (Panozzo et al., 2002), A. fumigatus, like many other filamentous Ascomycetes, possesses three putative Ido genes, idoA (Afu3g14250), idoB (Afu4g09830) and idoC (Afu7g02010) (Fig. 2 and Table 1) in its genome (Yuasa and Ball, 2013). One of them, idoB, was highly induced and a second, idoC, slightly induced by external l-tryptophan (Fig. 7). Enzymatic studies of A. oryzae Ido enzymes suggest two of the three enzymes, Idoa and Idopβ, may participate in tryptophan degradation (Yuasa and Ball, 2011). The Idoa showed higher activity and lower Km values (higher affinity for substrates), similar to the enzymatic properties of Bna2p in S. cerevisiae (Yuasa and Ball, 2011; 2012). However, idoA, the homolog of A. oryzae idoa, was not induced by tryptophan under our conditions (Fig. 7). Instead idoB, the homolog of A. oryzae idoβ was most highly induced by l-tryptophan. The third Ido in A. oryzae, Idoγ, had a closer relationship to bacterial Idos than fungal Idos (Fig. S6) and showed very high Km values (low affinity for substrates) and low catalytic efficiencies for l-tryptophan (Yuasa and Ball, 2012). idoC, the homolog of idoγ, was slightly induced by l-tryptophan. Our data suggests that A. fumigatus idoB is mostly likely to contribute to l-kynurenine production.
l-tryptophan can also be metabolized via the indolepyruvate pathway. AroH (termed Aro8 in S. cerevisiae (Iraqui et al., 1998) is a key enzyme in the transamination of l-tryptophan to generate indolepyruvate and aroH was highly induced by l-tryptophan feeding. Similar to S. cerevisiae, AroH could also participates in l-phenylalanine and l-tyrosine synthesis in A. fumigatus (Fig. 2) and therefore could direct metabolite pools to l-phenylalanine and l-tyrosine as well as towards l-tryptophan degradation. Thus it is likely that both AroH and IdoB activities could divert pathways from fumiquinazoline synthesis, a hypothesis we will explore in future studies.
In summary, we have genetically characterized two putative A. fumigatus chorismate binding proteins, TrpE and IcsA. TrpE was required for l-tryptophan production and its deletion results in tryptophan auxotroph, and over-expression of the fmq-cluster-associated gene (icsA/Afu6g12110) could not complement the tryptophan auxotroph of deletion trpE mutant or significantly change the Fq production. However, deletion and over-expression of icsA resulted in significantly changed anthranilate, l-tryptophan, l-phenylalanine, and l-tyrosine levels, suggesting that IcsA metabolizes chorismate into a yet unknown product in A. fumigatus. Synthetic feedback resistant alleles of TrpE had a significant effect in aromatic amino acid metabolism, resulting in anthranilate and l-tryptophan accumulation and modest increases in Fq synthesis on minimal growth medium. Inhibition of tryptophan-feedback effect in TrpE eliminated a roadblock during anthranilate biosynthesis, as a conduit to increase the anthranilate and tryptophan related secondary metabolism in A. fumigatus.
Supplementary Material
Highlights.
-
-
Inactivation of the tryptophan feedback domain in TrpE increases anthranilate levels
-
-
Elevated intracellular anthranilate and tryptophan levels are positively correlated with fumiquinazoline production
-
-
Re-routing metabolic flux of amino acids has the potential to boost NRPS-derived secondary metabolite production
Acknowledgements
P.M.W. was supported by grant from the National Natural Science Foundation of China NSFC No. 41406141 and NIH (R01 AI065728) to N.P.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ames BD, Liu X, Walsh CT. Enzymatic processing of fumiquinazoline F: a tandem oxidative-acylation strategy for the generation of multicyclic scaffolds in fungal indole alkaloid biosynthesis. Biochemistry. 2010;49:8564–8576. doi: 10.1021/bi1012029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BD, Walsh CT. Anthranilate-activating modules from fungal nonribosomal peptide assembly lines. Biochemistry. 2010;49:3351–3365. doi: 10.1021/bi100198y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anyaogu DC, Mortensen UH. Heterologous production of fungal secondary metabolites in Aspergilli . Front Microbiol. 2015;6:77. doi: 10.3389/fmicb.2015.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae YM, Holmgren E, Crawford IP. Rhizobium meliloti anthranilate synthase gene: cloning, sequence, and expression in Escherichia coli . J. Bacteriol. 1989;171:3471–3478. doi: 10.1128/jb.171.6.3471-3478.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Balajee SA, Marr KA, Andes D, Nielsen KF, Frisvad JC, Keller NP. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot Cell. 2005;4:1574–1582. doi: 10.1128/EC.4.9.1574-1582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braus GH. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol. Rev. 1991;55:349–370. doi: 10.1128/mr.55.3.349-370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JS, Aufauvre-Brown A, Brown J, Jennings JM, Arst H, Holden DW. Signature -tagged and directed mutagenesis identify PABA synthetase as essential for Aspergillus fumigatus pathogenicity. Mol. Microbiol. 2000;36:1371–1380. doi: 10.1046/j.1365-2958.2000.01953.x. [DOI] [PubMed] [Google Scholar]
- Bulloch EM, Jones MA, Parker EJ, Osborne AP, Stephens E, Davies GM, Coggins JR, Abell C. Identification of 4-amino-4-deoxychorismate synthase as the molecular target for the antimicrobial action of (6 S)-6-fluoroshikimate. J. Am. Chem. Soc. 2004;126:9912–9913. doi: 10.1021/ja048312f. [DOI] [PubMed] [Google Scholar]
- Buss K, Müller R, Dahm C, Gaitatzis N, Skrzypczak-Pietraszek E, Lohmann S, Gassen M, Leistner E. Clustering of isochorismate synthase genes menF and entC and channeling of isochorismate in Escherichia coli . Biochim. Biophys. Acta, Gene Struct. Expr. 2001;1522:151–157. doi: 10.1016/s0167-4781(01)00325-6. [DOI] [PubMed] [Google Scholar]
- Cacho RA, Tang Y, Chooi YH. Next-generation sequencing approach for connecting secondary metabolites to biosynthetic gene clusters in fungi. Front Microbiol. 2014;5:774. doi: 10.3389/fmicb.2014.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo AM, Bok J, Brooks W, Keller NP. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 2004;70:4733–4739. doi: 10.1128/AEM.70.8.4733-4739.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira GC, Arnaud MB, Inglis DO, Skrzypek MS, Binkley G, Simison M, Miyasato SR, Binkley J, Orvis J, Shah P, Wymore F, Sherlock G, Wortman JR. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014;42:705–710. doi: 10.1093/nar/gkt1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Matsuda F, Ishihara A, Miyagawa H, Wakasa K. Production of indole alkaloids by metabolic engineering of the tryptophan pathway in rice. Plant Biotechnol. J. 2013;11:1103–1111. doi: 10.1111/pbi.12105. [DOI] [PubMed] [Google Scholar]
- Graf R, Mehmann B, Braus GH. Analysis of feedback-resistant anthranilate synthases from Saccharomyces cerevisiae . J. Bacteriol. 1993;175:1061–1068. doi: 10.1128/jb.175.4.1061-1068.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas H. Fungal siderophore metabolism with a focus on Aspergillus fumigatus . Nat. Prod. Rep. 2014;31:1266–1276. doi: 10.1039/c4np00071d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Xu X, Cui C, Gu Q. Alkaloidal compounds produced by a marine-derived fungus, Aspergillus fumigatus H1-04, and their antitumor activities. Chinese J. Medicinal Chemistry. 2007;17:232–237. [Google Scholar]
- Harrison AJ, Yu M, Gårdenborg T, Middleditch M, Ramsay RJ, Baker EN, Lott JS. The structure of MbtI from Mycobacterium tuberculosis, the first enzyme in the biosynthesis of the siderophore mycobactin, reveals it to be a salicylate synthase. J. Bacteriol. 2006;188:6081–6091. doi: 10.1128/JB.00338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-B, Peebles CA, Shanks JV, San K-Y, Gibson SI. Expression of the Arabidopsis feedback-insensitive anthranilate synthase holoenzyme and tryptophan decarboxylase genes in Catharanthus roseus hairy roots. J. Biotechnol. 2006;122:28–38. doi: 10.1016/j.jbiotec.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Hughes EH, Hong SB, Gibson SI, Shanks JV, San KY. Expression of a feedback-resistant anthranilate synthase in Catharanthus roseus hairy roots provides evidence for tight regulation of terpenoid indole alkaloid levels. Biotechnol. Bioeng. 2004;86:718–727. doi: 10.1002/bit.20081. [DOI] [PubMed] [Google Scholar]
- Iraqui I, Vissers S, Cartiaux M, Urrestarazu A. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 1998;257:238–248. doi: 10.1007/s004380050644. [DOI] [PubMed] [Google Scholar]
- Ishihara A, Asada Y, Takahashi Y, Yabe N, Komeda Y, Nishioka T, Miyagawa H, Wakasa K. Metabolic changes in Arabidopsis thaliana expressing the feedback-resistant anthranilate synthase α subunit gene OASA1D . Phytochemistry. 2006;67:2349–2362. doi: 10.1016/j.phytochem.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Ito J, Cox EC, Yanofsky C. Anthranilate synthetase, an enzyme specified by the tryptophan operon of Escherichia coli: purification and characterization of component I. J. Bacteriol. 1969;97:725–733. doi: 10.1128/jb.97.2.725-733.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käfer E. The anthranilate synthetase enzyme complex and the trifunctional trpC gene of Aspergillus . Can. J. Genet. Cytol. 1977;19:723–738. doi: 10.1139/g77-079. [DOI] [PubMed] [Google Scholar]
- Kanno T, Komatsu A, Kasai K, Dubouzet JG, Sakurai M, Ikejiri-Kanno Y, Wakasa K, Tozawa Y. Structure-based in vitro engineering of the anthranilate synthase, a metabolic key enzyme in the plant tryptophan pathway. Plant Physiol. 2005;138:2260–2268. doi: 10.1104/pp.105.062885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller NP, Turner G, Bennett JW. Fungal secondary metabolism - from biochemistry to genomics. Nat. Rev. Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Krappmann S, Bignell EM, Reichard U, Rogers T, Haynes K, Braus GH. The Aspergillus fumigatus transcriptional activator CpcA contributes significantly to the virulence of this fungal pathogen. Mol. Microbiol. 2004;52:785–799. doi: 10.1111/j.1365-2958.2004.04015.x. [DOI] [PubMed] [Google Scholar]
- Li J, Last RL. The Arabidopsis thaliana trp5 mutant has a feedback-resistant anthranilate synthase and elevated soluble tryptophan. Plant Physiol. 1996;110:51–59. doi: 10.1104/pp.110.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim FY, Hou Y, Chen Y, Oh JH, Lee I, Bugni TS, Keller NP. Genome-based cluster deletion reveals an endocrocin biosynthetic pathway in Aspergillus fumigatus . Appl. Environ. Microbiol. 2012a;78:4117–4125. doi: 10.1128/AEM.07710-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim FY, Sanchez JF, Wang CC, Keller NP. Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi. Meth. Enzymol. 2012b;517:303–324. doi: 10.1016/B978-0-12-404634-4.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim FY, Ames B, Walsh CT, Keller NP. Co-ordination between BrlA regulation and secretion of the oxidoreductase FmqD directs selective accumulation of fumiquinazoline C to conidial tissues in Aspergillus fumigatus . Cell Microbiol. 2014;16:1267–1283. doi: 10.1111/cmi.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Dudareva N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol. 2012;63:73–105. doi: 10.1146/annurev-arplant-042811-105439. [DOI] [PubMed] [Google Scholar]
- Mellado L, Calcagno-Pizarelli AM, Lockington RA, Cortese MS, Kelly JM, Arst HN, Jr, Espeso EA. A second component of the SltA-dependent cation tolerance pathway in Aspergillus nidulans . Fungal. Genet. Biol. 2015;82:116–128. doi: 10.1016/j.fgb.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino K, Matsuda F, Miyazawa H, Sukegawa A, Miyagawa H, Wakasa K. Metabolic profiling of tryptophan-overproducing rice calli that express a feedback-insensitive α subunit of anthranilate synthase. Plant Cell Physiol. 2005;46:514–521. doi: 10.1093/pcp/pci051. [DOI] [PubMed] [Google Scholar]
- Osherov N, Kontoyiannis DP, Romans A, May GS. Resistance to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14α-demethylase gene, pdmA. J. Antimicrob. Chemoth. 2001;48:75–81. doi: 10.1093/jac/48.1.75. [DOI] [PubMed] [Google Scholar]
- Palmer GC, Jorth PA, Whiteley M. The role of two Pseudomonas aeruginosa anthranilate synthases in tryptophan and quorum signal production. Microbiology. 2013;159:959–969. doi: 10.1099/mic.0.063065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JM, Bok JW, Lee S, Dagenais TR, Andes DR, Kontoyiannis DP, Keller NP. Loss of CclA, required for histone 3 lysine 4 methylation, decreases growth but increases secondary metabolite production in Aspergillus fumigatus . PeerJ. 2013;1:e4. doi: 10.7717/peerj.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panozzo C, Nawara M, Suski C, Kucharczyka R, Skoneczny M, Becam AM, Rytka J, Herbert CJ. Aerobic and anaerobic NAD+ metabolism in Saccharomyces cerevisiae . Febs. Lett. 2002;517:97–102. doi: 10.1016/s0014-5793(02)02585-1. [DOI] [PubMed] [Google Scholar]
- Parsons JF, Jensen PY, Pachikara AS, Howard AJ, Eisenstein E, Ladner JE. Structure of Escherichia coli aminodeoxychorismate synthase: architectural conservation and diversity in chorismate-utilizing enzymes. Biochemistry. 2002;41:2198–2208. doi: 10.1021/bi015791b. [DOI] [PubMed] [Google Scholar]
- Plach MG, Löffler P, Merkl R, Sterner R. Conversion of anthranilate synthase into isochorismate synthase: implications for the evolution of chorismate-utilizing enzymes. Angew. Chem. Int. Ed. Engl. 2015;54:11270–11274. doi: 10.1002/anie.201505063. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahr T, Ravanel S, Basset G, Nichols BP, Hanson AD, Rebeille F. Folate synthesis in plants: purification, kinetic properties, and inhibition of aminodeoxychorismate synthase. Biochem. J. 2006;396:157–162. doi: 10.1042/BJ20051851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saika H, Oikawa A, Nakabayashi R, Matsuda F, Saito K, Toki S. Changes in primary and secondary metabolite levels in response to gene targeting-mediated site-directed mutagenesis of the anthranilate synthase gene in rice. Metabolites. 2012;2:1123–1138. doi: 10.3390/metabo2041123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor; 2001. [Google Scholar]
- Schrettl M, Kim HS, Eisendle M, Kragl C, Nierman WC, Heinekamp T, Werner ER, Jacobsen I, Illmer P, Yi H. SreA-mediated iron regulation in Aspergillus fumigatus . Mol. Microbiol. 2008;70:27–43. doi: 10.1111/j.1365-2958.2008.06376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T, Fujiki Y, Ohsumi Y, Kakinuma Y. Novel families of vacuolar amino acid transporters. Iubmb Life. 2008;60:519–525. doi: 10.1002/iub.92. [DOI] [PubMed] [Google Scholar]
- Serino L, Reimmann C, Visca P, Beyeler M, Chiesa VD, Haas D. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa . J. Bacteriol. 1997;179:248–257. doi: 10.1128/jb.179.1.248-257.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Keller NP. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans . Genetics. 2001;157:591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Fusion PCR and gene targeting in Aspergillus nidulans . Nat. Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Tang CM, Smith JM, Arst HN, Jr, Espeso EA, Holden DW. Virulence studies of Aspergillus nidulans mutants requiring lysine or p-aminobenzoic acid in invasive pulmonary aspergillosis. Infect Immun. 1994;62:5255–5260. doi: 10.1128/iai.62.12.5255-5260.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozawa Y, Hasegawa H, Terakawa T, Wakasa K. Characterization of rice anthranilate synthase alpha-subunit genes OASA1 and OASA2.. Tryptophan accumulation in transgenic rice expressing a feedback-insensitive mutant of OASA1. Plant Physiol. 2001;126:1493–1506. doi: 10.1104/pp.126.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzin V, Galili G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol Plant. 2010;3:956–972. doi: 10.1093/mp/ssq048. [DOI] [PubMed] [Google Scholar]
- Wagner D, Wiemann P, Huss K, Brandt U, Fleissner A, Tudzynski B. A sensing role of the glutamine synthetase in the nitrogen regulation network in Fusarium fujikuroi . PLoS ONE. 2013;8:e80740. doi: 10.1371/journal.pone.0080740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CT, Haynes SW, Ames BD, Gao X, Tang Y. Short pathways to complexity generation: fungal peptidyl alkaloid multicyclic scaffolds from anthranilate building blocks. ACS Chem. Biol. 2013;8:1366–1382. doi: 10.1021/cb4001684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P, Tudzynski B. The nitrogen regulation network and its impact on secondary metabolism and pathogenicity. In: Proctor DWBa.R.H., editor. Fusarium: Genomics, Molecular and Cellular Biology. Norwich, UK: Caister Academic Press; 2013. pp. 111–142. [Google Scholar]
- Wiemann P, Lechner BE, Baccile JA, Velk TA, Yin WB, Bok JW, Pakala S, Losada L, Nierman WC, Schroeder FC, Haas H, Keller NP. Perturbations in small molecule synthesis uncovers an iron-responsive secondary metabolite network in Aspergillus fumigatus . Front Microbiol. 2014;5:530. doi: 10.3389/fmicb.2014.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T, Nguyen CK, Romans A, Kontoyiannis DP, May GS. Isogenic auxotrophic mutant strains in the Aspergillus fumigatus genome reference strain AF293. Arch. Microbiol. 2004;182:346–353. doi: 10.1007/s00203-004-0707-z. [DOI] [PubMed] [Google Scholar]
- Yin WB, Baccile JA, Bok JW, Chen Y, Keller NP, Schroeder FC. A nonribosomal peptide synthetase-derived iron(III) complex from the pathogenic fungus Aspergillus fumigatus . J. Am. Chem. Soc. 2013a;135:2064–2067. doi: 10.1021/ja311145n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin WB, Chooi YH, Smith AR, Cacho RA, Hu Y, White TC, Tang Y. Discovery of cryptic polyketide metabolites from dermatophytes using heterologous expression in Aspergillus nidulans . ACS Synth. Biol. 2013b;2:629–634. doi: 10.1021/sb400048b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa HJ, Ball HJ. Molecular evolution and characterization of fungal indoleamine 2,3-dioxygenases. J. Mol. Evol. 2011;72:160–168. doi: 10.1007/s00239-010-9412-5. [DOI] [PubMed] [Google Scholar]
- Yuasa HJ, Ball HJ. The evolution of three types of indoleamine 2,3 dioxygenases in fungi with distinct molecular and biochemical characteristics. Gene. 2012;504:64–74. doi: 10.1016/j.gene.2012.04.082. [DOI] [PubMed] [Google Scholar]
- Yuasa HJ, Ball HJ. Indoleamine 2, 3-dioxygenases with very low catalytic activity are well conserved across kingdoms: IDOs of Basidiomycota. Fungal. Genet. Biol. 2013;56:98–106. doi: 10.1016/j.fgb.2013.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.