Abstract
The purpose of the study was to detect the AluI and DdeI polymorphisms within POU1F1 gene exon 6 and 3'UTR region in Turkish sheep breeds, and their association with milk productive traits. Jugular blood samples were collected from 50 Sakiz, 50 White Karaman, and 50 Awassi sheep using EDTA as an anticoagulant. PCR-RFLP and sequencing analysis were performed to investigate possible polymorphisms in the exon 6 and 3' flanking region of the sheep POU1F1 gene. The PCR products were digested with restriction endonuclease AluI and DdeI, and biallelic polymorphism was found with restriction endonuclease AluI and two genotypes (TT (296 bp and 173 bp) and CC (235 bp, 173 bp and 61 bp)) were detected. White Karaman and Awassi breeds did not show polymorphisms for AluI restriction sites. No polymorphism at the DdeI cleavage sites was detected in the three sheep breeds. Significant statistical results were found in milk yield (***P0.001), fat (***P0.001) and lactose (*P0.05) values with TT and CC genotypes, however no significant association of TT and CC genotypes with protein values was detected (P>0.05) and individuals with genotype TT had a superior milk yield in Sakiz sheep breeds. As sequence results, seven variation points were determined for exon 6 (g.185T>C) and 3'UTR (g.220G>A, g.229C>T, g.248C>T, g.250A>T, g.255T>C, g.258C>T) of the sheep POU1F1 gene. We have reported here for the first time single nucleotide polymorphisms of the POU1F1 gene for both exon 6 and 3'UTR and its effects on milk traits in Turkish sheep breeds were evaluated.
Key Words: Milk traits, POU1F1 gene, Polymorphism, Turkish sheep breeds
Introduction
POU class 1 homeobox 1 (POU1F1) is a member of the tissue-specific POU-containing transcription factor family. It contains a POU DNA-binding domain which consists of two regions, one is POU homoedomain required for DNA binding and the other is POU-specific domain, necessary for DNA-binding specifity and dimerization of POU1F1. This protein has an important role in the transcriptional regulation of growth hormone (GH) and prolactin (PRL) genes. Studies showed that the POU1F1 protein is expressed exclusively in soma-thotrophs, lactrotrophs and thyrotrophs and is required for the establishment and maintenance of these differentiated cell types, as well as for cell proliferation of somatotrophs and lactotrophs (Li et al., 1990 ▶; Tatsumi and Amino, 1999 ▶; Bastos et al., 2006 ▶). Sheep POU1F1 gene is located on chromosome 1q21-22 and consists of six exons which encode a protein of 33 kDa. Given the importance of POU1F1 gene in the control of gene expression of important hormones is directly associated with growth process and milk production, it is reasonable consider it as a good candidate gene for marker assisted selection (Bastos et al., 2006 ▶). Indeed, POU1F1 gene polymorphisms have been significantly associated with body weight, birth weight, litter size and milk production in cattle, goat and pig species (Yu et al., 1995 ▶; Renaville et al., 1997 ▶; Lan et al., 2007a ▶, b ▶; Daga et al., 2013 ▶).
Sakiz is a high milk yield local sheep breed in Turkey. Milk production varies from 120-250 kg of per lactation depending on management and husbandry conditions and lactation length is 175 days. White Karaman is another local sheep breed in Turkey and bred for both meat and milk production, milk production varies from 50-60 kg. per lactation and lactation length is 140 days. Awassi is principally a milk breed, and milk production varies from 120-160 kg, and lactation length is 165 days. This breed is reared in southeast Turkey (Akcapinar, 2000 ▶).
The purpose of the study was to detect the AluI and DdeI polymorphisms within POU1F1 gene exon 6 and 3'UTR region in Turkish sheep breeds, and their association with milk productive traits.
Materials and Methods
Animal resources and DNA isolation
Animals were chosen at random and were 4 years old, multiparous and lactating ewes and in their third lactation. During the day animals grazed on pasture and received as supplement 250 g/head/day concentrate commercial food (crude protein 20% and 2500 ME kcal/kg), animals were raised in semi-intensive conditions. Monthly individual milk yield has been recorded from day 30 of lactation until day 140, 165, 175 for White Karaman, Awassi and Sakiz, respectively (for a total 9, 10, 11 samples for each ewe for White Karaman, Awassi and Sakiz, respectively). Animals were milked once a day in the evening. Individual milk yield was recorded every fifteen days during lactation and each individual milk sample was analyzed for fat, protein and lactose using Lactoscan milk analyser.
Jugular blood samples (2 ml per ewe) were collected from 50 Sakiz, 50 White Karaman, 50 Awassi sheep using EDTA as an anticoagulant. Genomic DNA was extracted from the whole blood using the phenol chloroform method (Sambrook et al., 1989 ▶).
The study was approved by the Ethical Committee of Laboratory Animals, Firat University: 09/02, 04/10/ 2007.
PCR amplification
For this study according to the ovine (AJ549207) sequence of exon 4-6 of POU1F1 gene, the following PCR primers were designed: Forward: 5'- GTATTGCTG CTAAAGACGCC-3' and Reverse: 5'-GAGGGAAAGA TATAGTGAAAGGG-3'. For primer design, Primer 3 software (Rozen and Skaletsky, 2000 ▶) was used. PCR reaction was carried out in 50 μL of total volume, containing 10 X PCR buffer (50 mM/L KCl, 10 mM/L Tris-HCl (pH = 8.0), 0.1% Triton X-100), 1.5 mM MgCl2, 0.2 mM of each dNTP, 10 pM/L of each primer, 50 ng ovine genomic DNA and 1 U Taq DNA polymerase (Eppendorf AG, Hamburg, Germany). PCR conditions were as follows: denaturation at 94°C for 4 min, followed by 34 cycles of denaturation at 94°C for 1 min, annealing at 54°C for 1 min, extension at 72°C for 1 min and final extension at 72°C for 10 min, on Mastercycler® (Eppendorf AG, Hamburg, Germany). The PCR products were separated by electrophoresis on 2% agarose gels (Promega, Madison, WI, United States) in parallel with a 100 bp DNA marker.
Genotyping
20 μl of PCR product was digested separately with 10 U DdeI and 10 U AIuI (Fermentas GmbH, St. Leon-Rot, Germany) at 37°C overnight. PCR products of digestion were resolved by electrophoresis on a 4% agarose gel stained with ethidium bromide. Then, ten samples of each genotype were sequenced from both directions to confirm the results obtained with PCR-RFLP technique. Obtained sequence was aligned with the AJ549207 sequence of the GenBank.
Statistical analysis
Direct counting was used to estimate phenotype and allele frequencies of POU1F1 gene AluI genetic variants. ANOVA (one-way) was used to analyse association of POU1F1 variants with milk yield, fat and protein contents. Minitab statistical software was applied to evaluate the link between genotype and milk traits.
Results
In this study, the primers for the exon 6 and its flanking region of ovine POU1F1 gene were used for amplification of genomic DNA of three Turkish sheep breeds and uniform fragment was obtained after 2% agarose gel electrophoresis. The results showed that amplification fragment size (469 bp) was consistent with the target one and had good specificity, which could be directly analyzed by RFLP.
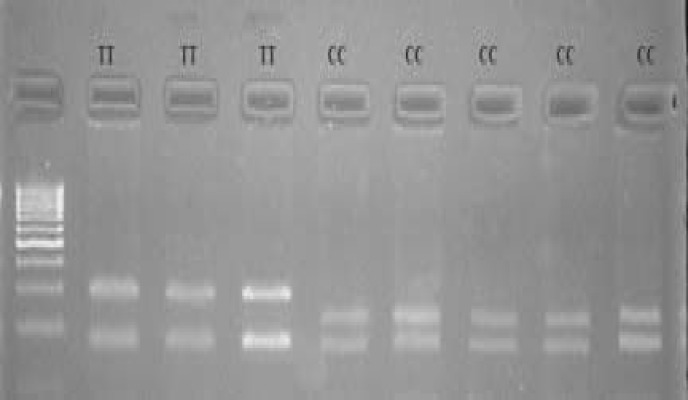
The results of this study indicated the presence of two AluI cleavage sites (235 and 296) within the POU1F1 gene exon 6 and its flanking region sequence. According to this, two genotypes were shown. Allele C restriction site resulted in 235 bp, 173 bp and 61 bp fragments, whereas T allele occured in the absence of the restriction site in the 236 bp, resulting in a three fragments 296 bp and 173 bp (Figs. 1 and 2). Therefore, genotype CC and TT demostrated three and two bands, respectively. While genotype TC showed four bands however, genotype TC was not found in Awassi, White Karaman and Sakiz sheep breeds. Allelic and genotypic frequencies of POU1F1 gene for AluI site in three Turkish sheep breeds were presented in Table 1. As a result, a biallelic polymorphism was found with restriction endonuclease AluI and two genotypes were detected, CC (235 bp, 173 bp and 61 bp) and TT (296 bp and 173 bp). Allelic frequency for Sakiz, White Karaman and Awassi breeds was 0.36, 1 and 1, respectively for allele C; 0.64, 0 and 0, respectively for allele T. The most frequently observed genotype was TT in Sakiz sheep breed. White Karaman and Awassi breeds did not show polymorphisms for AluI restriction sites.
Fig. 1.
Agarose gel electrophoresis band patterns after digestion with AluI endonuclease within the exon 6 and its flanking region sequence of the sheep POU1F1 gene. Lane M: 100 bp DNA marker. Lane 2-4: TT genotype (296 bp and 173 bp), and Lane 5-9: CC genotype (235 bp, 173 bp and 61 bp). As one small band (61 bp) was invisible on 4% agarose gel only two bands were visible for CC genotype
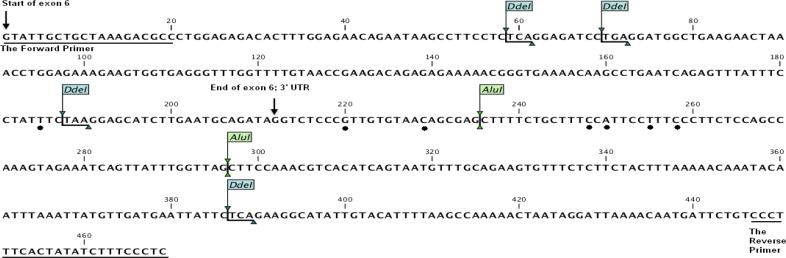
Fig. 2.
The structure of sheep POU1F1 gene and SNPs location for exon 6 and 3'UTR and AluI, DdeI enzyme restriction sites. Black dots (●) represent mutation points
Table 1.
Gene and genotype frequency of the exon 6 of POU1F1 gene for AluI site in three Turkish sheep breeds
| Sheep breeds | n | TT | TC | CC | T | C |
|---|---|---|---|---|---|---|
| Awassi | 50 | 0.00 | 0.00 | 1 | 0.00 | 1 |
| 0 | 0 | 50 | ||||
| W. Karaman | 50 | 0.00 | 0.00 | 1 | 0.00 | 1 |
| 0 | 0 | 50 | ||||
| Sakiz | 50 | 0.64 | 0.00 | 0.36 | 0.64 | 0.36 |
| 32 | 0 | 18 | ||||
| Total | 150 | 0.22 | 0.78 |
To determine the relationships between genotype TT and CC and milk traits in Sakiz sheep breed, statistical analyses were used. Significant statistical results were found in milk yield (***P 0.001), fat (***P 0.001) and lactose (*P 0.05) values with TT and CC genotypes, however, no significant association of TT and CC genotypes with protein values was detected (P>0.05). Individuals with genotype TT had a superior milk yield when compared to genotype CC, however, genotype CC had a superior fat value (Table 2).
Table 2.
Associations of AluI PCR-RFLP genotypes with milk traits in Sakiz sheep breeds
| Milk traits | Genotype |
P-value | ||
|---|---|---|---|---|
| TT (n=32) | CC (n=18) | |||
| Fat (%) | 5.78 ± 1.13*** | 8.60 ± 1.20*** | 0.000*** | |
| Proteins (%) | 6.446 ± 0.408 | 6.286 ± 0.737 | 0.325NS | |
| Lactose (%) | 5.482 ± 0.166* | 5.313 ± 0.398* | 0.040* | |
| Milk yield (kg) | 1.233 ± 0.202*** | 0.768 ± 0.160*** | 0.000** | |
Means of sheep milk traits with different letters are significantly different P0.05, P0.01, and P0.001, respectively
In spite of no polymorphisms being detected in White Karaman and Awassi sheep breeds, milk traits were analysed. From the milk analysis results, milk yield (**P 0.01), fat (**P 0.01), protein (**P 0.01) and lactose (**P 0.01) values were found as statistically important. Sakiz sheep breed showed the highest milk yeild, fat and protein values when compared to other sheep breeds, whereas White Karaman has shown superior lactose value (Table 3).
Table 3.
Means ± SD of milk traits in White Karaman, Awassi and Sakiz sheep breeds
| Milk traits | Sheep breeds |
P-value | ||
|---|---|---|---|---|
| Sakiz (n=50) | White Karaman (n=50) | Awassi (n=50) | ||
| Fat (%) | 6.792 ± 1.783c | 4.756 ± 1.289a | 5.597 ± 1.888b | 0.000*** |
| Proteins (%) | 6.3882 ± 0.5476a | 4.0468 ± 0.2096b | 3.9434 ± 0.1556b | 0.000*** |
| Lactose (%) | 5.4212 ± 0.2814a | 6.1256 ± 0.5164b | 5.8980 ± 0.2307b | 0.000*** |
| Milk yield (kg) | 1.0653 ± 0.2922a | 0.6505 ± 0.1422b | 0.9690 ± 0.2876a | 0.000*** |
Means of sheep milk traits with different letters were significantly different P0.001
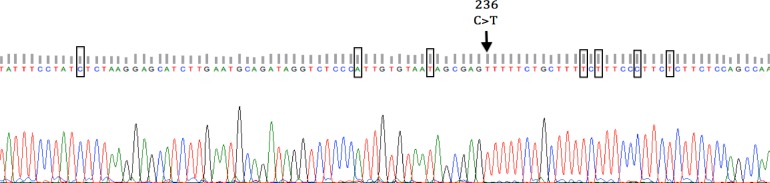
To confirm the PCR-RFLP results, ten PCR products were sequenced in both directions in an ABI 310 DNA sequencer. The sequence results showed the comparison between nucleotide sequence of AJ549207 (sheep POU1F1 gene), and seven variation points (g.185T>C, g.220G>A, g.229C>T, g.248C>T, g.250A>T, g.255T>C, g.258C>T) were detected in four samples, within these, one mutation g.185T>C was found in exon 6 and six other mutations were found in 3' UTR, these variation points cannot be determined to AluI restriction enzyme (Figs. 2 and 3).
Fig. 3.
DNA sequence of genotypes TT at the 236 locus and detected variation points in sheep POU1F1 gene exon 6 and 3'-UTR region
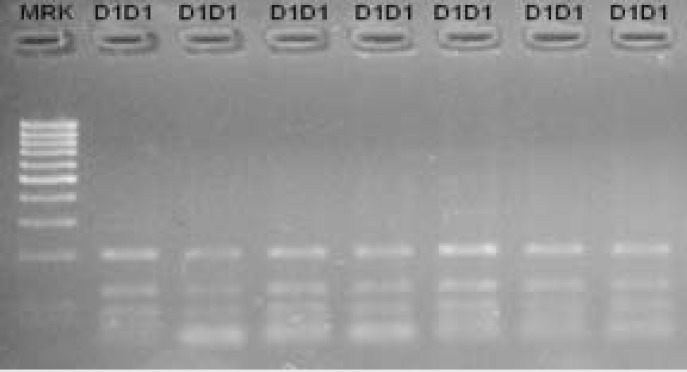
In this research, DdeI restriction enzyme showed four cleavage sites (58, 69, 187 and 386 bp) within the exon 6 and its flanking region sequence (Fig. 2), but none of these were shown to be polymorphic. Consequently, only one genotype was detected in investigated sheep breeds, which is D1D1 (199 bp, 118 bp, 83 bp, 58 bp and 11 bp) (Fig. 4).
Fig. 4.
Agarose gel electrophoresis band patterns after digestion with DdeI endonuclease within the exon 6 and its flanking region sequence of the sheep POU1F1 gene. Lane M: 100 bp DNA marker. Lane 2-7: D1D1 genotype (199 bp, 118 bp, 83 bp and 58 bp). As one small band (11 bp) was invisible on 4% agarose gel only four bands were visible for CC genotype
Discussion
The polymorphisms of sheep POU1F1 gene were first revealed by Bastos et al. (2006) ▶. Until now, only a few studies have been carried out on the sheep POU1F1 gene and their associations with production traits. In various Chinese goat breeds, polymorphisms in exon 6 of the POU1F1 gene have been shown. Lan et al. (2007a) ▶ found four mutation points in goat POU1F1 gene, within these two mutations g.60T>G and g.174T>C were found in exon 6 and two other mutations were found in 3'UTR. This g.174T>C mutation could be detected by AluI endonuclease. These authors found that individuals with genotype TC (*P 0.05) had a superior milk yield when compared to genotype TT and individuals with genotype TC showed significant associations with milk production. Lan et al. (2007a) ▶ suggested that genotype TC could be regarded as a molecular marker for superior milk yield.
Lan et al. (2007b) ▶ described a DdeI PCR-RFLP method for detecting a silent allele in goat POU1F1 gene, according to this method g.102T>C mutation point was detected. The g.102T>G mutation of the exon 6 region removes a DdeI endonuclease restriction site (CTNAG). Their results indicated that, the amplified DNA fragment with DdeI endonuclease digestion showed five fragments (200 bp, 118 bp, 102 bp, 20 bp and 11 bp) for POU1F1-D1 allele and four fragments (200 bp, 118 bp, 113 bp and 20 bp) for POU1F1-D2 allele. According to Lan et al’s. (2007b) ▶ results, the most observed genotype was D1D1 whereas D2D2 genotype was not detected and significant associations were found in milk yield between individuals with genotype D1D1 (*P 0.05). Genotype D1D1 has been recommended as a molecular marker for better performance in goat industry by Lan et al. (2007b) ▶.
Mura et al. (2012) ▶ performed PCR, SCCP and sequencing methods in order to examine polymorphism within POU1F1 gene in relation to milk production traits in Sarda sheep breed. Their results indicated that, only the 5'UTR of the fourth intron (g.121C>T) and 3'UTR of the six exon (g.249G>A) fragments displayed two novel SNPs, however, these polymorphisms were not found by Bastos et al. (2006) ▶. The statistical analysis showed no association between milk productive traits (milk yield, fat, proteins and lactose) and the found polymorphisms.
Bastos et al. (2006) ▶ found four substitutions in POU1F1 gene in Churra da Terra Quente sheep breed, one in the second exon in position g.150G>A, two substitutions in the third exon in position g.89G>A and g.105G>A, and one in the fourth intron in position g.93A>G.
In the present study, exon 6 and 3'UTR of the ovine POU1F1 gene polymorphisms were screened by PCR-RFLP and the sequencing technique. We have reported here for the first time single nucleotide polymorphisms of the POU1F1 gene for both exon 6 and 3'UTR in Turkish sheep breeds.
We observed that biallelic polymorphism was found with restriction endonuclease AluI, whereas no poly-morphism was found in DdeI. The AluI polymorphisms was only shown in Sakiz sheep breeds and TC genotype was not detected in genotype analysis in the three sheep breeds. We suggest that this difference is probably the consequence of different environmental conditions, selection and possibly even QTL or gene(s) regulating milk traits. The statistical results revealed significant associations between milk traits and genotypes in Sakiz sheep breeds. The individuals with genotype TT had better milk yield than individuals with genotype CC, on the contrary, individuals with genotype CC showed a higher fat value. Although no polymorphisms were detected in White Karaman and Awassi sheep breeds, milk traits were analysed. The milk analysis results show milk yield, fat, protein and lactose values were found to be statistically important between species. Sakiz sheep breed showed the highest milk yield, fat and protein values when compared to other sheep breeds whereas White Karaman has shown superior lactose value.
To confirm the PCR-RFLP results, ten PCR products were sequenced in both directions in an ABI 310 DNA sequencer. The sequence results show the comparison between nucleotide sequence of AJ549207 (sheep POU1F1 gene) revealed seven variation points detected in four samples; g.185T>C, g.220G>A, g.229C>T, g.248C>T, g.250A>T, g.255T>C, g.258C>T, within these, one mutation g.185T>C was found in exon 6 and six other mutations were found in 3'UTR. These detected polymorphisms are different from Bastos et al. (2006) ▶ and Mura et al. (2012) ▶. These sequences from Turkish sheep breeds have been deposited in the GenBank database (http://www.ncbi.nlm.nih.gov) under the accession No.: KC959950.
As a result, Sakiz sheep breed has a different genotype and has shown different mutation points in exon 6 and 3'UTR of the POU1F1 gene. The next step will be to analyse these SNPs and 3' flanking region of the POU1F1 gene in a larger sample of Sakiz sheep breeds and evaluate their association with productive traits. According to sequence results six mutation points were determined for 3'UTR, further analysis about 3' flanking region of the POU1F1 gene could clarify the role of this gene in regulating the productive traits.
Acknowledgment
The project was supported by Scientific Research Projects Council of Firat University (project code 2110).
References
- Akcapinar H. Sheep breeding. Ankara, Ismat Press; 2000. pp. 20–25. [Google Scholar]
- Bastos, E, Santos, I, Parentier, I, Castrillo, JL, Cravador, A, Guedes-Pinto, H, Renaville, R. Ovis aries POU1F1 gene: cloning, characterization and polymorphism analysis. Genetica. 2006;126:303–314. doi: 10.1007/s10709-005-0034-6. [DOI] [PubMed] [Google Scholar]
- Daga, C, Paludo, M, Luridiana, S, Mura, MC, Bodano, S, Pazzola, M, Dettori, ML, Vacca, GM, Carcangui, V. Identification of novel SNPs in the Sarda breed goats POU1F1 gene and their association with milk productive performance. Mol. Biol. Rep. 2013;40:2829–2835. doi: 10.1007/s11033-012-2298-0. [DOI] [PubMed] [Google Scholar]
- Lan, XY, Pan, CY, Chen, H, Lei, CZ, Hua, LS, Yang, XB, Qiu, GY, Zhang, RF, Lun, YZ. DdeI polymorphism in coding region of goat POU1F1 gene and its association with production traits. Asian Austral J. Anim. Sci. 2007b;20:1342–1348. [Google Scholar]
- Lan, XY, Pan, CY, Chen, H, Zhang, CL, Li, JY, Zhao, M, Lei, CZ, Zhang, AL, Zhang, L. An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits. Small Rum. Res. 2007a;73:8–12. [Google Scholar]
- Li, S, Crenshaw, EB, Rawson, EJ, Simmons, DM, Swanson, LW, Rosenfeld, MG. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene PIT-1. Nature. 1990;347:528–533. doi: 10.1038/347528a0. [DOI] [PubMed] [Google Scholar]
- Mura, MC, Daga, C, Paludo, M, Luridiana, S, Pazzola, M, Bodano, S, Dettori, ML, Vacca, GM, Carcangiu, V. Analysis of polymorphism within POU1F1 gene in relation to milk production traits in dairy Sarda sheep breed. Mol. Biol. Rep. 2012;39:6975–6979. doi: 10.1007/s11033-012-1525-z. [DOI] [PubMed] [Google Scholar]
- Renaville, R, Gengler, N, Vrech, E, Prandi, A, Massart, S, Corradini, C, Bertozzi, C, Mortiaux, F, Burny, A, Portetelle, D. PIT-1 gene polymorphism, milk yield, and conformation traits for Italian Holstein-Friesian bulls. J. Dairy Sci. 1997;80:3431–3438. doi: 10.3168/jds.S0022-0302(97)76319-7. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsk H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener, S, editors. Bioinformatics methods and protocols: methods in molecular biology. 1st Edn. Totowa: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sambrook, J, Fritsche, F, Maniatis, T. Molecular cloning: a laboratory manual. New York: Cold Spring HarbourLabPress, Cold Spring Harbour; 1989. pp. 1–99. [Google Scholar]
- Tatsumi, K, Amino, N. PIT1 abnormality. Growth Horm IGF Res. 1999;9:18–22. doi: 10.1016/s1096-6374(99)80076-8. [DOI] [PubMed] [Google Scholar]
- Yu, TP, Tuggle, CK, Schmitz, CB, Rothschild, MF. Associations of PIT1 polymorphisms with growth and carcass traits in pigs. J. Anim. Sci. 1995;73:1282–1288. doi: 10.2527/1995.7351282x. [DOI] [PubMed] [Google Scholar]