Abstract
The aim of the present study was to screen the genotype profile of bovine kappa-casein gene among Frieswal (HF × Sahiwal) crossbred cattle developed in India. A total number of two hundred Frieswal cows were evaluated for HinfI RFLP based genotyping of kappa-casein gene. We observed that only two genotypes (AA and AB) exist among the studied population with the genotype frequency of 0.58 (n=117) and 0.42 (n=83), respectively. The calculated allele frequency for A and B was 0.79 and 0.21, respectively. Association of genotypes with certain milk production traits revealed that AB had significant (P<0.05) effect on total milk yield, peak yield, yield at 300 days and SNF% as compared to AA.
Key Words: Kappa-casein, Polymorphism, Frieswal, RFLP, Milk production traits
Introduction
Casein is a family of milk proteins that exists in several molecular forms and is the main protein present in the bovine milk (Alipanah et al., 2005 ▶). The bovine milk specific proteins include casein fractions: α s1 casein (CSN1S1), α s2 casein (CSN1S2), β casein (CSN2) and κ-caseins (CSN3) as insoluble fractions, α lactalbumin (LALBA) and β lactogloulin (LGB), which are classified as soluble fractions (Galila and Darwish, 2008 ▶). The κ-casein (CSN3) molecule is a single-chain polypeptide of 169 amino acids with a molecular weight of 19.2 kDa. CSN3 plays an important role in milk chemistry by providing colloidal to the casein micelle. In the micelle, κ-casein is mostly located at the periphery, with its hydrophilic C-terminal sequence protruding into the solvent (Rachagani and Gupta, 2008 ▶).
The κ-casein gene comprises a 13 kb sequence divided into 5 exons (Alexander et al., 1988 ▶). Point mutations in exon IV of the bovine kappa-casein (CSN3) gene determine two allelic variants, A and B (Alipanah et al., 2007 ▶). The A and B variants differ in the amino acids 136 and 148. At position 136, threonine is replaced by isoleucine, while at position 148, aspartic acid is replaced by alanine, for A and B, respectively (Alexander et al., 1988 ▶). This variation, associated with processing properties like cheese production technology (Alipanah et al., 2007 ▶) and physiological processes such as cytotoxic and antibacterial effects, enhances the immunity (Hamza et al., 2010 ▶). The B allele was found to be associated with thermal resistance, shorter coagulation time, better curdles and micelles of different sizes, which are preferable in cheese making (Azevedo et al., 2008 ▶). The cheese yield from cows with genotype BB is 10% higher when compared with AA cows (Azevedo et al., 2008 ▶). These variants were dis-tinguished by polymerase chain reaction (PCR) and restriction fragment length polymorphism (PCR-RFLP) analysis (Rachagani and Gupta, 2008 ▶). The aim of this study was to determine possible κ-casein gene polymorphism and their association with milk production traits among Frieswal cattle of Indian origin. Frieswal cows were developed in India using 62 percent exotic (HF) and 38 percent indigenous (Sahiwal) blood which exhibit high milk yielding and better fat percentage capability.
Materials and Methods
A total of 200 Frieswal heifers were included in the study. Genomic DNA was isolated from the venous blood using standard phenol chloroform extraction method (Sambrook et al., 1989 ▶). For detection of kappa-casein genotypes PCR amplification was done using primer CSN3-F: 5΄-TGTGCTGAGTAGGTATCCTAGT TATGG-3΄ and CSN3-R 5΄-GCGTTGTCTTCTTTGAT GTCTCCT-3΄ (Barroso et al., 1998). PCR was carried out from a starting template of approximately 50 ng of genomic DNA in a final reaction volume of 25 μl containing 1 X Taq DNA polymerase buffer (Sigma), 1.5 mM MgCl2 (Sigma), 200 μM dNTPs (Sigma), 0.5 μM of each primer and 1 U Taq polymerase (Sigma). The PCR reaction included pre-denaturation for 5 min at 95°C followed by 35 cycles 94°C for 1 min, 55°C for 1 min, 72°C for 1 min and a final extension of 10 min at 72°C. PCR products were visualized in 1.0% agarose gels. The amplicon was sequenced directly using automated DNA sequencer by Sanger’s dideoxy chain termination method.
For genotyping, PCR product was digested with HinfI restriction enzyme which was used for the determination of kappa-casein alleles. Gene fragments were subjected to digestion by restriction enzymes in a total volume of 20 μl (8 μl PCR product, 1 X enzyme buffers, 4 U enzymes and distilled water) and placed in the incubator at 37°C for 5 h. The restriction products were analyzed by electrophoresis on a 2% agarose gel. Gene (allele) and genotype frequencies were calculated as per Falconer and Mackay (1996) ▶. Data were analyzed by using SPSS statistical program (SPSS 10.0 for Windows; SPSS, Inc., Chicago, IL, USA). Significant differences were determined by one-way analysis of variance (ANOVA) using the SPSS program according to the following statistical model:
Yij = μ + Gi + Mj + eij
Where,
Yij: The analyzed trait of each cow
Μ: The overall mean
Gi: The fixed effect of the ith genotype
Mj: The fixed effect of jth season of calving
eij: The random error
Results
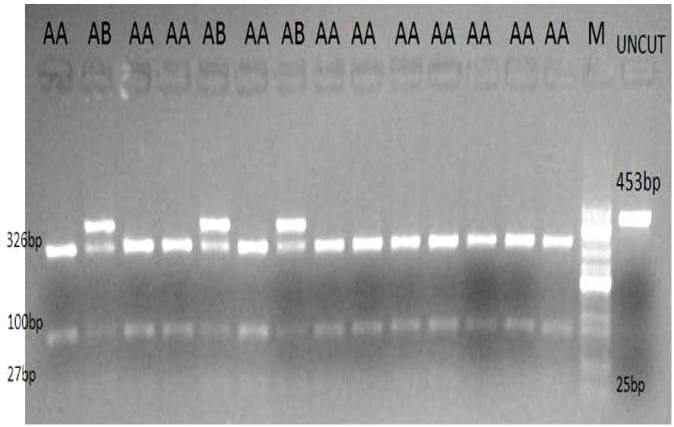
The PCR product of the kappa-casein gene using specific set of primers (CSN3-F and CSN3-R) was a fragment of 453 bp DNA. Digestion of 453 bp fragment of kappa-casein gene by HinfI restriction endonuclease generated four fragments, i.e. 453, 326, 100 and 27 bp (Fig. 1). Three fragments of 326, 100 and 27 bp represent homozygotes A allele wherein four fragments viz. 453, 326, 100 and 27 bp represent heterozygotes AB for kappa-casein gene. No BB genotypes were observed among the studied population. Estimated genotypic frequencies were 0.58 and 0.42 for AA and AB, respectively (Table 1).
Fig. 1.
PCR-RFLP product of kappa-casein gene. After the PCR product was digested by HinfI and visualized on 1% agarose gel, the results were the 453 bp fragment of uncut PCR product representing homozygotes B allele (not shown), three fragments of 326, 100 and 27 bp representing homozygotes A allele, and four fragments 453, 326, 100 and 27 bp representing heterozygotes (A/B) for kappa-casein gene
Table 1.
Determination of the relation between genotypes and its association with milk production traits among Frieswal cattle
| Genotype | First lactation milk yield (kg) | Peak yield (kg) | Yield 300 days (kg) | Fat% | Protein% | Lactose% | SNF% |
|---|---|---|---|---|---|---|---|
| AA (n=117) | 2492.75±146.01a | 11.30±0.64 | 2820.90±163.91e | 4.0319±0.03 | 3.005±0.01 | 4.570±0.01 | 8.56±0.05c |
| AB (n=83) | 2778.79±215.67b | 13.68± 0.65d | 3012.78±189.99f | 3.990±0.04 | 2.690±0.02 | 4.430±0.01 | 8.77±0.03d |
Number in parenthesis is the number of animals. Mean values with the different superscript lower case letters: (a, b, c, d, e and f) in the same column denote significant difference (P<0.05)
The least-squares means of total milk yield, peak yield, yield at 300 days, fat%, protein%, lactose% and SNF% for different genotypes are presented in Table 1. The animals with genotype AB had a significantly (P<0.05) higher total milk yield, peak yield and yield at 300 days than those with genotype AA. Table 1 also showed that AB genotypes have significantly (P<0.05) higher SNF percentage than AA genotypic animals. However, the statistical analysis revealed that, there was non-significant (P>0.05) difference between the geno-types with fat, protein and lactose percentage.
Discussion
The present study was aimed to screen the HinfI RFLP pattern of bovine kappa-casein among Frieswal cattle and, further, to identify its association with certain lactogenic traits. The present finding indicated that the A allele was more frequent than the B allele among the Frieswal population studied here. Interestingly no BB homozygous was observed among the studied population. Results from the present study was in agreement with the earlier observations reported by various authors. Kučerová et al. (2006) ▶ reported that A allele (0.60) are more frequently present than B (0.38) among Czech Simental cattle. Similar findings of the allele and genotype frequencies among Czech Fleckvieh cattle population was reported by Bartoňová et al. (2012). The observation made by Keating et al. (2007) showed that, A allele (0.80) was most frequently present among various dairy cattle population. Curi et al. (2005) ▶ identified a higher frequency of A allele among Simental and Aberdeen Angus cattle. Trakovická et al. (2012) ▶ also showed confirmed that existence of A allele among the crosses of Simmental and Holstein cattle breeds was more frequent than B allele. In their study, the predominant allele was A with the observed frequency of 0.76.
The present study showed that animals with AB genotype had a significantly (P<0.05) higher total milk yield, peak yield and yield at 300 days than those with genotype AA. However, Lin et al. (1986) ▶ found that the BB genotype had a higher average milk yield than AA and AB. In contast, Curi et al. (2005) ▶ reported that the κ-casein genotype AA was associated with higher milk production than BB, with the heterozygous AB being intermediate. In the present study, we could not achieve any significant difference between the two identified genotypes with respect to fat, protein and lactose percentage so far. However, Ng-Kwai-Hang et al. (1986) ▶ and Strzalkowska et al. (2002) ▶ reported that the κ-casein variant BB genotypes were associated with increased milk fat percentage.However, Bovenhuis et al. (1992) ▶, Strzalkowska et al. (2002) ▶ and Rachagani et al. (2008) ▶ found no significant difference in milk protein content between the two κ-casein variants among Ayrshire, Jersey, brown Swiss, Canadienne, Guernsey and Polish Black-and-White cattle. Interestingly, our study indicated that, AB genotypes had significantly (P<0.05) higher SNF percentage than AA among the Frieswal cows. Though, Rachagani et al. (2008) ▶ earlier reported that BB animals of Sahiwal and Tharparker cattle had higher monthly SNF percentage and yield than those with genotypes AA and AB.
In conclusion, AB genotypes may have greater influence on certain lactogenic traits among Frieswal crossbred cattle which require further study with a higher number of studied population.
Acknowledgements
The authors are thankful to the directorate for providing all the necessary facilities to conduct the present work. Authors are also thankful to the Incharge, Military Farm, Meerut, Uttar Pradesh, India for providing animals.
References
- Alexander, LJ, Stewart, AF, Mackinlay, AG, Kapelinskaya, TV. Isolation and characterization of the bovine kappa-casein gene. Eur. J. Biochem. 1988;178:395–401. doi: 10.1111/j.1432-1033.1988.tb14463.x. [DOI] [PubMed] [Google Scholar]
- Alipanah, M, Kalashnikova, L, Rodionov, G. Kappa-casein genotypic frequencies in Russian breeds Black and Red Pied cattle. Iran. J. Biotec. 2005;3:191–194. [Google Scholar]
- Azevedo, ALS, Nascimento, CS, Steinberg, RS, Carvalho,MRS , Peixoto, MGCD, Teodoro, RL, Verneque, RS, Guimarães, SEF, Machado, MA. Genetic polymorphism of the kappa-casein gene in Brazilian cattle. Gen. Mol. Res. 2008;7:623–630. doi: 10.4238/vol7-3gmr428. [DOI] [PubMed] [Google Scholar]
- Bartoňová, P, Vrtková, I, Kaplanová, K, Urban, T. Association between CSN3 and BCO2 gene polymorphisms and milk performance traits in the Czech Fleckvieh cattle breed. Gen. Mol. Res. 2012;11:1058–1063. doi: 10.4238/2012.April.27.4. [DOI] [PubMed] [Google Scholar]
- Bovenhuis, H, Van Arendonk, JAM, Korver, S. Associations between milk protein polymorphism and milk production traits. J. Dairy Sci. 1992;72:2549–2559. doi: 10.3168/jds.S0022-0302(92)78017-5. [DOI] [PubMed] [Google Scholar]
- Curi, RA, Oliveira, HND, Gimenes, MA, Silveira, AC, Lopes, CR. Effects of CSN3 and LGB gene polymorphisms on production traits in beef cattle. Gen. Mol. Biol. 2005;28:262–266. [Google Scholar]
- Falconer, DS, Mackay, TFC. Introduction to quantitative genetics. 4th Edn. Essex, UK: Longman Group Ltd; 1996. p. 464. [Google Scholar]
- Galila, ASE, Darwish, SF. A PCR-RFLP assay to detect genetic variants of kappa-casein in cattle and buffalo. Arab. J. Biotec. 2008;11:11–18. [Google Scholar]
- Hamza, AE, Wang, XL, Yang, ZP. Kappa casein gene polymorphism in Holstein Chinese cattle. Pak. Vet. J. 2010;30:203–206. [Google Scholar]
- Kučerová, J, Metějíčková, A, Jandurová, OM, Sørensen, P, Němcová, E, Stipková, M, Kott, T, Bouška, J, Frelich, J. Milk protein genes CSN1S1, CSN2, CSN3, LGB and their relation to genetic values of milk production parameters in Czech Fleckvieh. Czech J. Anim. Sci. 2006;51:241–247. [Google Scholar]
- Lin, CY, McAllister, AJ, Ng-Kwai-Hang, KF, Hayes, JF. Effects of milk protein loci on first lactation production in dairy cattle. J. Dairy Sci. 1986;69:704–712. doi: 10.3168/jds.S0022-0302(86)80459-3. [DOI] [PubMed] [Google Scholar]
- Ng-Kwai-Hang, KF, Hayes, JE, Moxley, JD, Monardes, HG. Association of genetic variants of casein and milk serum protein with milk fat and protein production in dairy cattle. J. Dairy Sci. 1984;67:835–840. doi: 10.3168/jds.S0022-0302(84)81374-0. [DOI] [PubMed] [Google Scholar]
- Rachagani, S, Gupta, ID. Bovine kappa-casein gene polymorphism and its association with milk pro-duction traits. Gen. Mol. Biol. 2008;31:893–897. [Google Scholar]
- Sambrook, J, Fritsch, EF, Maniatis, T. Molecular cloning: a laboratory manual. 2nd Edn. NY: Cold Spring Harbor Laboratory, Cold Spring Harbor Laboratory Press; 1989. p. 1659. [Google Scholar]
- Strzalkowska, N, Krzyzewski, J, Ryniewicz, Z. Effects of κ-casein and β-lactoglobulin loci polymorphism, cow’s age, stage of lactation and somatic cell count on daily milk yield and milk composition in Polish Black-and-White cattle. Anim. Sci. Paper Rep. 2002;20:21–35. [Google Scholar]
- Trakovická, A, Moravčíková, N, Navrátilová, A. Kappa-casein gene polymorphism (CSN3) and its effect on milk production traits. Acta fytotechnica et zootechnica. Nitra, Slovaca Universitas Agriculturae Nitriae; 2012. pp. 61–64. Suppl. 3/2012. [Google Scholar]