Abstract
Milk is considered a nutritious food because it contains several important nutrients including proteins and vitamins. Conversely, it can be a vehicle for several pathogenic bacteria such as Staphylococcus aureus. This study aimed to analyze the frequency of genes encoding the nine Staphylococcal enterotoxins (SEs) and enterotoxin gene profiles in S. aureus isolates derived from raw bovine milk. A total of 52 S. aureus isolates were obtained from 246 milk samples of 246 dairy cows from eight different farms in Qom, Iran. On the basis of cultural and biochemical properties as well as by amplification of the 23S rRNA specific to S. aureus, all isolates could be identified as S. aureus. Of the 52 isolates studied, 80.7% were positive for one or more genes encoding the enterotoxins, and 12 different genotypes were identified. The gene encoding for enterotoxin A (Sea) was the most frequent (16 isolates, 30.7%), followed by Seb (14 isolates, 26.9%) and Sed (8 isolates, 15.37%). Among the genes encoding the other enterotoxins, Seg and Seh were the most frequently observed (8 isolates each, 15.38%), followed by Sej (6 isolates, 11.5%) and Sei (1 isolates, 3.84%). With the recent identification of new SEs, the frequency of enterotoxigenic strains has increased, suggesting that the pathogenic potential of Staphylococci may be higher than previously thought. These results of enterotoxin genes positivity of milk-derived Staphylococci constitute a potential risk for consumers’ health.
Key Words: Staphylococcus aureus, Staphylococcal enterotoxins, Raw milk, PCR
Introduction
Staphylococcus aureus is a gram-positive bacterium, which produces many important virulence factors including Staphylococcal enterotoxins (SEs) which are responsible for Staphylococcal food poisoning (SFP), a major type of foodborne illness (Balaban and Rasooly, 2000 ▶). Classical SEs have been divided into five serological types (SEA through SEE) on the basis of their antigenicities. In recent years, the existence of new types of SEs (SEG-SEU, except SES and SET) have been reported (Ren et al., 1994 ▶; Su et al., 1995 ▶; Munson et al., 1998 ▶; Zhang et al., 1998 ▶; Jarraud et al., 2001 ▶; Orwin et al., 2001 ▶). Staphylococcal enterotoxins are resistant to inactivation by gastrointestinal proteases such as pepsin. In addition, they displayed strong thermo resistance, for example, SEA retains some biological activity after 28 min at 121°C (Anderson et al., 1996 ▶). Staphylococcal enterotoxins can be routinely detected by immunoassay, e.g. enzyme linked immunosorbent assay (ELISA), immunodiffusion, radioimmuno-assay and latex agglutination but the availability of these methods are usually limited to commercial tests for classical SEs (Omoe et al., 2002 ▶). Therefore, the DNA-based approach (PCR assays) is thought to be an essential tool for investigating SE genes (Omoe et al., 2002 ▶). The identification of S. aureus at the species level is based on amplification of target genes highly conserved within the species. In addition to the 16S rRNA gene, the well established standard target for the identification of bacterial species (Amann et al., 1995 ▶), the 23S rRNA genes has proven useful for identification of S. aureus at the species level (Akineden et al., 2001 ▶; Phuektes et al., 2003 ▶). The aim of this study was to identify S. aureus strains via amplification of the genes encoding the 23S rRNA and to analyze the genes encoding the Staphylococcal enterotoxins SEA, SEB, SEC, SED, SEE, SEG, SEH, SEI and SEJ in S. aureus strains isolated from raw bovine milk.
Materials and Methods
Sample collection and identification
A total of 52 S. aureus isolates were collected from milk sample of 246 cows from eight different farms in Qom, Iran. The identification of the isolates was performed by gram staining, catalase, hemolysis and tube coagulase test (Liofilchem, Italy) and was confirmed by PCR amplification of species specific parts of the S. aureus 23S rRNA gene.
Genotypic characterization
The DNA of isolates was prepared with the high pure PCR template preparation kit (Roche, Germany) as described by the manufacturer. The sequences of the oligonucleotide primers, the predicted PCR product sizes and the references are summarized in Table 1. For PCR amplification, the reaction mixture (25 µl) contained 15 ng DNA template, 1 µl of primer F (10 pmol), 1 µl of primer R (10 pmol), 2.5 µl 10 X PCR buffer (Fermentas, Lithuania), 0.5 µl dNTP (10 mM, Fermantas, Lithuania), 1.5 µl Mgcl2 (25 mM, Fermentas, Lithuania), 0.5 U Taq DNA polymerase (Fermentas, Lithuania) and double-distilled water to the final volume of 25 µl. DNA amplification was performed in a thermal cycler (Eppendorf, Germany) with initial denaturation at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 1 min, primer annealing 58°C for 1 min and extension at 72°C for 1 min, followed by a final extension at 72°C for 7 min. The amplified PCR products were electrophoresed in a 1% agarose gel (Fermentas, Lithuania) containing ethidium bromide (Fermentas, Lithuania) and visualized by trans illumination under UV. Molecular size marker (Vivantis, Malaysia) was included in each agarose gel. The S. aureus reference strains ATCC 19095, ATCC 23235, ATCC 14458, ATCC 700699, ATCC 27664, ATCC 25923 were used as positive controls. The reference strains Staphylococcus epidermidis ATCC 12228, Staphylococcus saprophyticus ATCC 14448 were used as negative controls.
Table 1.
Primers used for the detection of Staphylococcus aureus SE genes
| Gene | Primer | Sequence (5΄-3΄) | Size (bp) of PCR product | Reference |
|---|---|---|---|---|
| 23S rRNA | 23S rRNA F | ACG GAG TTA CAA AGG ACG AC | 1250 | Straub et al. 1999 |
| 23S rRNA R | AGC TCA GCC TTA ACG AGT AC | |||
| Sea | sea F | TAA GGA GGT GGT GCC TAT GG | 180 | Cremonesi et al. 2005 |
| sea R | CAT CGA AAC CAG CCA AAG TT | |||
| Seb | Seb F | TCG CAT CAA ACT GAC AAA CG | 478 | Johnson et al. 1991 |
| Seb R | GCA GGT ACT CTA TAA GTG CC | |||
| Sec | sec F | ACC AGA CCC TAT GCC AGA TG | 371 | Cremonesi et al. 2005 |
| sec R | TCC CAT TAT CAA AGT GGT TTC C | |||
| Sed | sed F | TCA ATT CAA AAG AAA TGG CTC A | 339 | Cremonesi et al. 2005 |
| sed R | TTT TTC CGC GCT GTA TTT TT | |||
| See | see F | AGG TTT TTT CAC AGG TCA TCC | 209 | Mehrotra et al. 2000 |
| See R | CTT TTT TTT CTT CGG TCA ATC | |||
| Seg | seg F | CCA CCT GTT GAA GGA AGA G | 432 | Cremonesi et al. 2005 |
| seg R | TGC AGA ACC ATC AAA CTC GT | |||
| Seh | seh F | TCA CAT CAT ATG CGA AAG CAG | 463 | Cremonesi et al. 2005 |
| seh R | TCG GAC AAT ATT TTT CTG ATC TTT | |||
| Sei | sei F | GGT GAT ATT GGT GTA GGT AAC | 454 | Omoe et al. 2002 |
| sei R | ATC CAT ATT CTT TGC CTT TAC CAG | |||
| Sej | sej F | GGT TTT CAA TGT TCT GGT GGT | 306 | Cremonesi et al. 2005 |
| sej R | AAC CAA CGG TTC TTT TGA GG |
Results
Staphylococcus aureus was observed in 52 (21.1%) samples of 246 raw milk samples. According to the results of PCR assay by amplification of the 23S rRNA gene specific to S. aureus, all 52 isolates contained 1250bp DNA fragments bands and showed positive PCR assay. Table 2 shows the results of molecular tests for the detection of gene encoding the enterotoxins SEA, SEB, SEC, SED, SEE, SEG, SEH, SEI and SEJ. Of the 52 isolates of S. aureus tested, 42 (80.7%) were positive for one or more SE genes, and 12 different genotypes were observed. Among the 52 S. aureus isolates, 28 isolates (53.8%) harbored only one enterotoxin gene, 8 isolates (15.38%) carried gene coding for two enterotoxin. Genotypes encoding three enterotoxins (Sea+Seb+Seh+, Sea+Seb+Sej+, Seb+Sed+Seg+) were detected in 6 isolates (11.53%). Genes encoding the enterotoxins SEC and SEE were not observed separately. Among the genes that code for classic enterotoxins (SEA-SEE), Sea was the most frequent, it was found in 16 isolates (30.7%) followed by Seb in 14 (26.9%) and Sed in 8 (15.37%) isolates. Regarding the other enterotoxins, Seg and Seh were the most frequently observed (8 isolates each, 15.38%), followed by Sej in 6 (11.5%) isolates and Sei in 1 (3.84%) isolate (Fig. 1).
Table 2.
Genotypic profile of Staphylococcus aureus strains isolated from raw milk according to SE genes
| Genotypic profile | Number of strains |
|---|---|
| Sea | 10 |
| Seb | 4 |
| Sed | 4 |
| Seg | 4 |
| Seh | 4 |
| Sei | 2 |
| Sea+Sed | 2 |
| Seb+Sej | 4 |
| Seg+Seh | 2 |
| Sea+Seb+Seh | 2 |
| Sea+Seb+Sej | 2 |
| Seb+Sed+Seg | 2 |
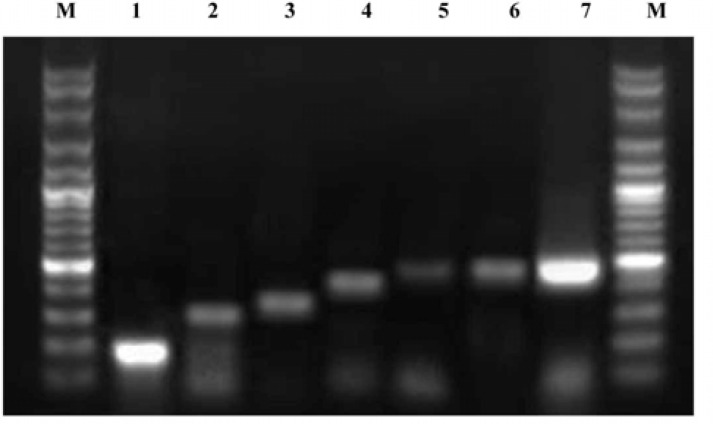
Fig. 1.
PCR amplification for the detection of staphylococcus aureus SE genes. Lane M: 100 bp marker. Lane 1: sea (180 bp), Lane 2: sej (306 bp), Lane 3: sed (339 bp), Lane 4: seg (432 bp), Lane 5: sei (454 bp), Lane 6: seh (463 bp), and Lane 7: seb (478 bp)
Discussion
Staphylococcus aureus produces a spectrum of extracellular protein toxins and virulence factors which are thought to contribute to the pathogenicity of the organism. The SEs are recognized agents of the Staphylococcal food poisoning syndrome (Straub et al., 1999 ▶). In the present work, nine major types of enterotoxin genes are investigated. Considering the genes encoding enterotoxins, 28 (53.8%) out of 52 isolates of S. aureus were positive for at least one enterotoxin gene. The most frequently observed gene was Sea, observed in 16 (30.7%) isolates. In spite of the great discrepancy in data concerning the prevalence of enterotoxigenic S. aureus isolates found in the literature, which is attributable to the different types of foods and strains involved (Mathieu et al., 1991 ▶), SEA is the most frequently observed enterotoxin in enterotoxigenic strains of S. aureus (Normanno et al., 2005 ▶). In the current study, it was revealed that genes of Sea and Seb, the newly described enterotoxin genes of Seg, Seh and Sej seemed to be the predominant enterotoxin genes of S. aureus isolated from milk of cows. These data were accordance with the finding of Akineden et al. (2001) ▶, too. Our study showed that 80.7% of 52 isolates were positive for the presence of genes coding for one or more enterotoxins. In Italy, Morandi et al. (2007) ▶ found that 67% of the S. aureus isolated from milk and dairy products were positive for the presence of toxin genes. In Japan, Omoe et al. (2002) ▶ observed that 77.4% of the S. aureus isolates were positive for the presence of genes that encode one or more enterotoxins, a frequency close to that observed in the present work (80.7%). With the discovery of the new enterotoxins, the percentage of enterotoxigenic or potentially enterotoxigenic S. aureus isolates increased. In this study, 28 (53.8%) isolates were positive for at least one Se gene, however, that number would decrease to 18 (34.6%) isolates if only the classic enterotoxins (Sea to See) were considered. Rosec and Gigaud (2002) ▶ also observed the increase in the number of enterotoxigenic isolates as a consequence of the discovery of the new SEs. In their study, 30% of the isolates had genes encoding the classic toxins in which frequency was found to be 57% when the new SEs were taken into account. In the present work, six isolates (11.53%) were positive for genes encoding three enterotoxins. Their individual genotypes were Sea+Seb+Seh+, Sea+Seb+Sej and Seb+Sed+Seg+. Nashev et al. (2002) ▶ have identified genetic profiles comprising multiple genes in S. aureus.
In conclusion, the genotypic results of the present study might help to understand the distribution of enterotoxigenic S. aureus clones among bovine isolates. This can aid in the investigation and control of S. aureus infections in dairy herds.
References
- Akineden, O, Annemuller, C, Hassan, AA, Lammler, C, Wolter, W, Zschock, M. Toxin genes and other characteristics of Staphylococcus aureus isolates from milk of cows with mastitis. Clin. Diagn. Lab. Immunol. 2001;8:959–964. doi: 10.1128/CDLI.8.5.959-964.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann, RL, Ludwig, W, Schleifer, KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, JE, Beelman, RR, Doores, S. Persistence of serological and biological activities Staphylococcal enterotoxin A in canned mushrooms. J. Food. Prot. 1996;59:1292–1299. doi: 10.4315/0362-028X-59.12.1292. [DOI] [PubMed] [Google Scholar]
- Balaban, N, Rasooly, A. Staphylococcal enterotoxins. Int. J. Food. Microbiol. 2000;61:1–10. doi: 10.1016/s0168-1605(00)00377-9. [DOI] [PubMed] [Google Scholar]
- Cremonesi, P, Luzzana, M, Brasca, M, Morandi, S, Lodi, R, Vimercati, C, Agnellini, D, Caramenti, G, Moroni, P, Castiglioni, B. Development of multiplex PCR assay for the identification of Staphylococcus aureus enterotoxigenic strains isolated from milk and dairy products. Mol. Cell. Probes. 2005;19:299–305. doi: 10.1016/j.mcp.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Jarraud, S, Peyrat, MA, Lim, A, Tristan, A, Bes, M, Mougel, C, Etienne, J, Vandenesch, F, Bonneville, M, Lina, G. A highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 2001;166:669–677. doi: 10.4049/jimmunol.166.1.669. [DOI] [PubMed] [Google Scholar]
- Johnson, WM, Tyler, SD, Ewan, FE, Ashton, FR, Pollard, DR, Rozee, KR. Detection of genes enterotoxins, exfoliative toxins and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J. Clin. Microbiol. 1991;29:426–430. doi: 10.1128/jcm.29.3.426-430.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, AM, Isigidi, BK, Devriese, LA, Godard, C, Vanhoof, R. Characterization of Staphylococcus aureus and Salmonella spp strains isolated from bovine meat in Zaire. Int. J. Food. Microbiol. 1991;14:119–126. doi: 10.1016/0168-1605(91)90098-a. [DOI] [PubMed] [Google Scholar]
- Mehrotra, M, Wang, G, Johnson, WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1 and methicillin resistance. J. Clin. Microbiol. 2000;38:1032–1035. doi: 10.1128/jcm.38.3.1032-1035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandi, S, Brasca, M, Lodi, R, Cremonesi, P, Castiglioni, B. Detection of classical enterotoxins and identification of enterotoxin genes in Staphylococcus aureus from milk and dairy products. Vet. Microbiol. 2007;124:66–72. doi: 10.1016/j.vetmic.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Munson, SH, Tremiane, MT, Beteley, MJ, Welch, RA. Identification and characterization of Staphylo-coccal enterotoxin type G and I from Staphylococcus aureus. Infect. Mmun. 1998;66:3337–3348. doi: 10.1128/iai.66.7.3337-3348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashev, D, Toshkova, K, Isrina, S, Salaisa, S, Hassan, AA, Lammler, C, Zschock, M. Distribution of virulence genes of Staphylococcus aureus isolated from stable nasal carriers. FEMS. Microbiol. Lett. 2004;233:45–52. doi: 10.1016/j.femsle.2004.01.032. [DOI] [PubMed] [Google Scholar]
- Normanno, G, Firinu, A, Virgilio, S, Mula, G, Dambrosio, A, Poggiu, A, Decastelli, L, Mioni, R, Sucuota, S, Bolzoni, G, Digiannatale, E, Salinetti, AP, Lasalandra, G, Bartoli, M, Zuccon, F, Pirino, T, Sias, S, Parisi, A, Quaglia, NC, Celano, GV. Coagulase positive Staphylococci and Staphylococcus aureus in foods products marketed in Italy. Food. Microbiol. 2005;98:73–79. doi: 10.1016/j.ijfoodmicro.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Omoe, K, Ishikawa, M, Shimoda, Y, Hu, DL, Ueda, S, Shinagawa, K. Detection of Seg, Seh and Sei genes in Staphylococcus aureus isolates and determination of enterotoxin productivities of Staphylococcus aureus isolates harboring Seg, Seh, or Sei genes. J. Clin. Microbiol. 2002;40:857–862. doi: 10.1128/JCM.40.3.857-862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orwin, P, Leung, D, Donahue, H, Novick, R, Schlievert, P. Biochemical and biological properties of Staphylococcal enterotoxin K. Infect. Immun. 2001;69:2916–2919. doi: 10.1128/IAI.69.1.360-366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuektes,P , Browning,F , Anderson, G, Mansell, P. Multiplex polymerase chain reaction as a mastitis screening test for Staphylococcus aureus, Streptococcus agalactiae, Streptococcus dysgalactiae and Streptococcus uberis in bulk milk samples. J. Dairy Res. 2003;70:149–155. doi: 10.1017/s0022029903006010. [DOI] [PubMed] [Google Scholar]
- Ren, K, Bannan, JD, Pancholi, V, Cheung, AL, Robbins, JC, Fischetti, VA, Zabriskie, JB. Characteri-zation and biological properties of a new Staphylococcal enterotoxin. J. Exp. Med. 1994;180:1675–1683. doi: 10.1084/jem.180.5.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosec, JP, Gigaud, O. Staphylococcal enterotoxin genes of classical and new types detected by PCR in France. Int. J. Food. Microbiol. 2002;77:61–70. doi: 10.1016/s0168-1605(02)00044-2. [DOI] [PubMed] [Google Scholar]
- Straub, JA, Hertel, C, Hammes, WP. A 23S rRNA targeted polymerase chain reaction based system for detection of Staphylococcus aureus in meat and dairy products. J. Food. Prot. 1999;62:1150–1156. doi: 10.4315/0362-028x-62.10.1150. [DOI] [PubMed] [Google Scholar]
- Su, YC, Wong, AC. Identification and purification of a new Staphylococcal enterotoxin H. Appl. Environ. Microbiol. 1995;61:1438–1443. doi: 10.1128/aem.61.4.1438-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S, Iandolo, JJ, Stewart, GC. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (Sej) FEMS. Microbiol. Lett. 1998;168:227–233. doi: 10.1111/j.1574-6968.1998.tb13278.x. [DOI] [PubMed] [Google Scholar]