Abstract
Macroalgae (seaweeds) are a promising feedstock for the production of third generation bioethanol, since they have high carbohydrate contents, contain little or no lignin and are available in abundance. However, seaweeds typically contain a more diverse array of monomeric sugars than are commonly present in feedstocks derived from lignocellulosic material which are currently used for bioethanol production. Hence, identification of a suitable fermentative microorganism that can utilise the principal sugars released from the hydrolysis of macroalgae remains a major objective. The present study used a phenotypic microarray technique to screen 24 different yeast strains for their ability to metabolise individual monosaccharides commonly found in seaweeds, as well as hydrolysates following an acid pre-treatment of five native UK seaweed species (Laminaria digitata, Fucus serratus, Chondrus crispus, Palmaria palmata and Ulva lactuca). Five strains of yeast (three Saccharomyces spp, one Pichia sp and one Candida sp) were selected and subsequently evaluated for bioethanol production during fermentation of the hydrolysates. Four out of the five selected strains converted these monomeric sugars into bioethanol, with the highest ethanol yield (13 g L−1) resulting from a fermentation using C. crispus hydrolysate with Saccharomyces cerevisiae YPS128. This study demonstrated the novel application of a phenotypic microarray technique to screen for yeast capable of metabolising sugars present in seaweed hydrolysates; however, metabolic activity did not always imply fermentative production of ethanol.
Keywords: Macroalgae, Phenotypic microarray, Bioethanol, Fermentation, Yeast
Introduction
The European Commission has stated that European countries rely too heavily on imports of gas and fuel and energy sources need to be more diverse (www.europa.eu). The consumption of fossil fuels and effects of global warming are causing constraints on our planet, and the search to find alternative, sustainable and cleaner burning sources of energy has become an extensive area of research (Jeong et al. 2010).
Bioethanol derived from sugar-based biomass in a fermentation process is a potential way of generating energy-rich transportation fuels (Karakashev et al. 2007). First generation feedstock materials such as corn and sugarcane have already been widely exploited (Bothast and Schlicher 2005), and the current commercial production of bioethanol, mainly in the USA and Brazil, is based around the use of terrestrial plants such as maize or sugar cane. Controversy, however, has arisen over the utilisation of such substrates which are essential food sources, and attention has been diverted to lignocellulosic biomass (2nd generation biomass) which comprises mainly of agricultural waste products (Lu et al. 2009). However, the production of fuel from second generation crops has proven to be problematic with energy and chemical inputs required to breakdown the recalcitrant lignocellulosic complex highlighted as a significant technical challenge to the use of such biomass for ethanol production (Taherzadeh and Karimi 2008). The release of fermentable sugars from lignocellulosic materials usually therefore requires a physico-chemical pre-treatment to breakdown the native plant structures and enable cellulose degrading enzymes to have access to their substrates. Such pre-treatments have been found to generate a range of inhibitory compounds (e.g. furans, phenolic compounds or organic acids) which can impact on the viability of yeast in a subsequent fermentation processes (Singh et al. 2011; Mukherjee et al. 2014).
Macroalgae (seaweeds) have gained much attention in recent years as promising bioethanol feedstocks due to their fast growth rate and large biomass yields, seaweeds offer certain advantages over terrestrial crops which have previously been considered as favourable feedstocks for bioethanol production (Adams et al. 2009). Several species of seaweed are known to be rich in carbohydrates, and they contain little to no lignin (Yanagisawa et al. 2011), implying that less energy intensive processes might be established to produce bioethanol from seaweed.
Despite the many advantages seaweeds possess over terrestrial energy crops, there are potential bottlenecks which could hinder the development of bioethanol production from hydrolysates derived from seaweed. The aquaculture and adequate supply of desirable seaweed species for bioethanol production is certainly a factor. Additionally, the diverse array of carbohydrates typically found in seaweeds is also a further technical obstacle to their exploitation. This means that specialised enzyme blends will be required to liberate fermentable sugars from seaweeds and that specialised fermentative organisms are needed which can utilise the majority of the sugars present. Although seaweeds contain carbohydrates that are also found in terrestrial plants (e.g. some starch and cellulose), the majority of carbohydrate present is in the form of polysaccharides such as laminarin (glucose polymer with β1-3 linkages), mannitol (C6 sugar alcohol), agar (agarobiose polymer) and carrageenan (D-galactose and 3,6-anhydro-D-galactose monomers); hence, biomass processing which has been developed for terrestrial plants cannot be directly applied to seaweeds (Jung et al. 2013). Conversion methods to release monosaccharides from their various sources and structures and the choice of microorganisms for bioethanol production from seaweed are necessary pre-requisites for the development of a viable process utilising seaweed as a feedstock. Attempts have been made to identify natural strains or to generate genetically engineered microorganisms with the ability to ferment sugars inherent to seaweed into ethanol. Horn et al. (2000a) revealed the possibility of simultaneously fermenting laminarin and mannitol extracts from the brown species Laminaria hyperborea to ethanol using Pichia angophorea, where 0.43 g ethanol g−1 of substrate (both laminarin and mannitol) was recovered. Further research by Horn et al. (2000b) demonstrated that the bacterium Zymobacter palmae was able to ferment synthetic mannitol under different oxygen regimes, yielding 0.38 g ethanol g−1 mannitol. Additionally, Wargacki et al. (2012) engineered an Escherichia coli strain that was capable of co-fermenting glucose, mannitol and alginate from the brown algal species Saccharina japonica, yielding 0.41 g ethanol g−1 of carbohydrate sugars (alginate, mannitol and glucan). Similarly, Enquist-Newman et al. (2014) developed the first S. cerevisiae synthetic biology platform with the ability to produce ethanol from mannitol and the alginate monomer 4-deoxy-l-erythro-5-hexoseulose uronate (DEHU), achieving yields up to 83 % of the maximum theoretical yield from these sugars. These findings reveal that all major polysaccharides in seaweeds could be converted to ethanol and other value-added chemicals.
As the interest in bioethanol production from seaweeds is increasing, the search to find wild-type microorganisms with the ability to ferment sugars specific to the breakdown of seaweed has proven to be somewhat challenging. Natural strains may lack the relevant metabolic pathways for the conversion of certain sugars into ethanol (Roesijadi et al. 2010), and therefore, the maximum conversion of the mixed array of monosaccharides found in seaweed hydrolysates into bioethanol cannot be fully achieved. However, phenotypic screening of yeast strains may be employed and can be a useful tool with the potential to identify strains with desirable traits for bioethanol production (Greetham et al. 2014a, b). This was recently highlighted in the work of Wimalasena et al. (2014), where candidate strains for efficient second generation bioethanol production were identified by employing a phenotypic microarray (PM) analysis to measure tolerance to stresses on Saccharomyces spp yeast during fermentation. There are many advantages of utilising a PM approach, which include the high output of informative data in a short amount of time, and avoiding the need to use traditional yeast methodologies (Greetham et al. 2014a, b). The PM technique gives an insight into the yeast cell’s metabolic activity against various media (e.g. individual monosaccharide carbon sources). Although this is not directly indicative of fermentation activity, the approach can identify microorganisms which are able to metabolise different carbon sources. Even when an organism does not produce ethanol as a major metabolic product, it remains useful to identify organisms which can assimilate and metabolise the sugars which are liberated from macroalgae. Whilst not the focus of the current research, this could identify novel routes to the production of other platform chemicals.
This study reports the first application of PM analysis to the screening and selection of yeast strains able to metabolise a range of monosaccharides originating from the breakdown of seaweed polysaccharides. Strains which displayed promising metabolic activity when cultured on a wide cross-section of such carbon sources were further screened for ability to metabolise hydrolysates prepared from the acid hydrolysis of five native UK seaweeds. Following these initial screening processes, selected yeast strains were examined in laboratory-scale fermentations of hydrolysates derived from seaweed in which fermentation progression and ethanol formation was determined.
Materials and methods
Two brown algae (Laminaria digitata and Fucus serratus), two red algae (Chondrus crispus and Palmaria palmata) and green alga (Ulva lactuca) were collected from the coast around Plymouth, UK, during the spring of 2013. The seaweed samples were rinsed in distilled water to remove salt and debris, before being dried in a fan oven at 80 °C for a minimum of 48 h until perceived to be dry. The seaweed samples were then milled using a ball mill (Fritsch, Germany) to obtain a fine homogeneous powder and stored away from direct sunlight and moisture until further use.
Compositional analysis of the seaweed species
Moisture content was measured by drying 5 g of each seaweed species in a convection oven at 105 °C for 90 min (Santos et al. 2003). After heating, the samples were placed in a desiccator for 30 min to prevent rehydration before being weighed again.
Ash content was determined by heating in a muffle furnace at 580 °C for 24 h.
A Thermo Flash Nitrogen Analyser (ThermoFischer Scientific, USA) was used to determine the protein content of each species. Sample (50 mg) was sealed in a tin capsule and combusted at approximately 1800 °C. Combustion gases were passed into a reduction reactor (at 680 °C and containing reduced copper) where nitrogen oxides are converted to elemental nitrogen. Carbon dioxide, sulphur dioxide and water were removed via filters of soda lime, magnesium perchlorate and a molecular sieve. The effluent stream was passed through a nitrogen separation column (50 °C) and into a thermal conductivity detector. Quantitation was achieved with Eager 300 software using an L-aspartic acid standard. Protein was determined using the N × 6.25 conversion factor.
Lipid content was determined using an adaption of the Folch method (Folch et al. 1957). Powdered sample (400 mg) was added to a 50-mL glass centrifuge tube. To each sample, 12 mL of dichloromethane/methanol (2:1, v/v) was added and left for 2 h with occasional agitations. The glass tubes were then centrifuged at 1000 rpm for 5 min or until a pellet had formed at the bottom of the tube. Using a glass syringe, the upper organic phase was removed and transferred into a clean 50-mL centrifuge glass tube were 2.5 mL KCl (0.88 %, w/v) was added before being inverted, vortexed and centrifuged at 1000 rpm for 5 min. The lower organic phase was removed using a glass syringe and transferred into a pre-weighed glass tube. The lower organic phase was dried with nitrogen gas and left uncapped in a fume cupboard overnight until all the liquid had evaporated.
Carbohydrate content was measured following a modified version of the Dubois assay (Dubois et al. 1956) where 1 mL of 12 M H2SO4 was added to 30 mg of seaweed in a heat resistant screw cap glass tube and incubated at 37 °C for 1 h. Water (11 mL) was added to the sample to dilute the acid strength to 1 M, following which, samples were incubated at 100 °C for 2 h. Liberated monosaccharides (mannitol, fucose, arabinose, galactose, glucose and xylose) were analysed by HPLC as described in section 2.6. HPLC analysis was used instead of the colourimetric phenol-sulphuric acid method as for the purpose of this study as it was important to be able to identify individual monosaccharides in the different seaweed species and whether different yeast strains could utilise specific sugars.
Acid hydrolysis and preparation of seaweed hydrolysates
For the production of seaweed hydrolysates, acid hydrolysis was conducted where 200 g of seaweed powder was treated with 2 L of 5 % (w/v) H2SO4 in 2 L Duran bottles. The reaction took place in a benchtop autoclave at 121 °C for 15 min. After hydrolysis, the liquid fraction was separated from the residue by filtration using a Büchner funnel and flask connected to a pump. The seaweed hydrolysates were adjusted to pH 7 using NaOH and filter sterilised before being analysed for monosaccharides and concentrations of potential yeast-inhibitory compounds (see below). The remaining solid seaweed residues were washed repeatedly with excess water until a neutral pH was achieved, dried overnight at 60 °C and stored for future work. Free amino nitrogen (FAN) was determined by following the European Brewing Convention ninhydrin spectrophotometric method, which utilises glycine as a standard (Analytica—EBC, 2000).
Yeast strain screen using the Biolog-Omnilog
Yeast strains and growth conditions
All 24 yeast strains which were screened in this investigation (listed in Table 1) were taken from glycerol stocks stored at −80 °C and inoculated into a liquid medium containing 10 g L−1 yeast extract, 20 g L−1 peptone, and 20 g L−1 glucose (YPD) and placed in an orbital shaker (180 rpm; CERTOMAT BS-1 incubator, Germany) at 30 °C under aerobic conditions for 48 h. One hundred microlitres of these cells were then spread on YPD slopes and incubated at 30 °C under aerobic conditions for a further 48 h.
Table 1.
List of yeast strains investigated in this study
| Microorganism | Source | Equivalent strain designations/Ref | ID in this study |
|---|---|---|---|
| Kluyveromyces marxianus | TP | ATCC 22296, CBS 5671, NRRL Y8287, Phaff 71-15 | TP1 |
| Candida veronae | TP | – | TP2 |
| Pichia Mexicana | TP | ATCC 28874, CBS 5815, NRRL Y17672, Phaff 69-32 | TP3 |
| Candida utilitis var thermophila | TP | CBS 1517, NRRL Y-1082, Phaff 74-62, DBVPG 7306 | TP4 |
| Candida mogii | TP | – | TP5 |
| Candida sorboxylosa | TP | Phaff 91-491.3 | TP6 |
| Candida stellate | TP | Phaff 72-1034 | TP7 |
| Candida utilis | TP | ATCC 9950, CBS 5609, Phaff 74-61 | TP8 |
| Candida santjackobensis | TP | ATCC 58898, CBS 8183 | TP9 |
| Pichia stipitis | TP | ATCC 58785, CBS 6054, NRRL Y11545 | TP10 |
| Candida valida | TP | Phaff 94-150.3 | TP11 |
| Pichia anomala | TP | CBS 5759, NRRL Y-366, Phaff 76-71 T | TP12 |
| Saccharomyces cerevisiae | NCYC | Melbourne No. 1 strain, NCTC 4919 | NCYC 192 |
| Hanseniaspora valbyensis | NCYC | CBS 479, NRRL Y-1626, ATCC 10631, IFO 0670, NCTC 478 | NCYC 17 |
| Candida tenuis | NCYC | ATCC 58782, CBS 4285, CCRC 21772, NRRL Y-17106 | NCYC 2545 |
| Candida succiphila | NCYC | ATCC 46049, CBS 8003, CCRC 21410, IAM 12489, IFO 1911, JCM 9445, KL30, NRRL Y-11998 | NCYC 1403 |
| Saccharomyces cerevisiae | NCYC | CBS 1200, ATCC 4126, CCRC 21494, MUCL 39497, CLIB 409 | NCYC 2592 |
| Candida lyxosophila | NCYC | CBS 8194, CCY 29-173-1, JCM 7532 | NCYC 2379 |
| Pichia guilliermondii | NCYC | ATCC 46036, CCRC 21697, CCRC 22093, DBVPG 6571, IFO 10106, NRRL Y-2075 | NCYC 443 |
| Candida shehatae var lignosa | NCYC | CBS 4705, ATCC 58779, CCRC 21774, IGC 3590, JCM 9837, NRRL Y-12856, NRRL Y-17027 | NCYC 2389 |
| Candida arabinofermentans | NCYC | CBS 8468, NRRL YB-2248 | NCYC 2916 |
| Candida shehatae | BSYC | – | BSYC 1 |
| Saccharomyces cerevisiae | EL | Liti et al. (2009) | YPS128 |
| Saccharomyces cerevisiae | EL | Liti et al. (2009) | Y12 |
TP courtesy of Trevor Phister, NCYC National Collection of Yeast Cultures, BSYC Brewing Science Yeast Culture, EL courtesy of Ed. Louis
Phenotypic microarray analysis (Biolog-Omnilog) using synthetic minimal media and seaweed hydrolysates
The Biolog-Omnilog phenotypic microarray (PM) system (Biolog, Hayward, USA) was used according to the manufacturer’s instructions with minor modifications according to Greetham et al. (2014a) to screen for yeast strains that can metabolise different carbon sources, in particular monosaccharides liberated from the acid hydrolysis of seaweed species. Synthetic minimal media containing different individual carbon sources (arabinose, fucose, galactose, glucose, mannitol, rhamnose and xylose) at 6 % (w/v) were prepared and supplemented into 0.67 % (w/v) yeast nitrogen base (YNB), 2.6 μL of yeast nutrient supplement mixture (NSx48-24 mM Adenine-HCl, 4.8 mM L-histidine HCl monohydrate, 48 mM L-leucine, 24 mM L-lysine-HCl, 12 mM L-methionine, 12 mM L-tryptophan and 14.4 mM uracil) and 0.2 μL of dye D. Reverse osmosis (RO) sterile distilled water was added to make a final volume of 60 μL, which was then aliquoted into individual wells of the PM array plates. Fifty-six microlitres of seaweed hydrolysate which was generated in section 2.3 was supplemented with 0.4 μL of dye D due to the hydrolysates’ darker colour and then aliquoted into the wells of the PM plates. Sixty microlitres of hydrolysate without dye D was also aliquoted into specific wells in order to act as a control.
Yeast strains were prepared according to the method described by Greetham, et al. (2014a).
The conditions were made anaerobic by vacuum packing the PM plates in PM gas bags (Biolog) before being placed in the Omnilog reader at 30 °C for 95 h. The Omnilog reader was programmed to photograph the PM plates every 15 min for 95 h, converting the pixel density of each well into a signal data value that represents cell metabolic output and dye colour change. The reduction of the dye, producing a colour change from colourless to violet, results from the cells’ metabolic activity when in contact with the different media in each well; this has been defined here as redox signal intensity. The redox signal intensity data was assembled in the Biolog software and exported using Microsoft Excel. All assays were conducted in triplicate and the mean values are presented.
Fermentations of hydrolysates derived from the acid pre-treatment of seaweed using selected yeast strains
Yeast strains which appeared to be more metabolically active by having a redox signal intensity value (RSI) of ≥20 on the seaweed specific sugars and seaweed hydrolysates from the yeast strain screening assay were directly used for bioethanol fermentations. Twenty-five-millilitre fermentations of the seaweed acid hydrolysate produced from section 2.3 were conducted in glass serum bottles (30 mL; Wheaton, USA) using a method adapted from Quain et al. (1985) and Powell et al. (2003). The bottles were made anaerobic by sealing the vessels with rubber septa and a one-way valve was used in order to facilitate the expulsion of CO2 produced during the fermentation process as sugars are converted to ethanol. The fermentation vessels containing seaweed hydrolysate were inoculated with the selected yeast strain at a pitching rate of ca 107 cells/mL. The vessels were incubated at 30 °C (MIR-253 incubator, Sanyo Electric Co., Japan) with magnetic stirring set at 120 rpm, and the progression of the fermentation was monitored by tracking the weight loss of the vessels at frequent intervals resulting from the removal of CO2. The end of the fermentation was indicated by the vessels reaching constant mass. Samples were taken at the end of the fermentation for sugar and ethanol analysis via HPLC. All fermentations were carried out in triplicate.
Analysis of monosaccharides, inhibitors and bioethanol
The monosaccharide concentrations were quantified via HPLC using Dionex ICS-3000 Reagent-Free Ion Chromatography, electrochemical detection using ED 40 and computer controller. The CarboPacTM PA 20 column (3 × 150 mm) was used, and the mobile phase was 10 mM NaOH with a flow rate of 0.5 mL min−1. The injection volume was 10 μL and the column temperature was 30 °C. Authentic standards of monosaccharides (mannitol, fucose, arabinose, galactose, glucose and xylose) with concentrations within range of 1 to 0.0625 g L−1 were used for monosaccharide quantification.
The analysis of inhibitors (5-hydroxy-methyl-furfural (HMF), furoic acid, furfural, vanillic acid, vanillin, ferulic acid and p-coumaric acid) were quantified by HPLC using UV detection at 270 nm (2695 HPLC system and 996 Photodiode Array Detector, Waters, USA) and a Techsphere ODS C18 column (5 μm, 4.6 mm × 250 mm; HPLC Technologies, UK) was used at room temperature. The sample volume was 10 μL, and the mobile phase was a gradient of methanol in 1 % acetic acid at an overall flow rate of 1.0 mL min−1. The methanol concentration was increased from 20 to 50 % over 30 min with a 100 % methanol column cleaning phase and a 9-min re-equilibration period. Data were recorded using Millennium Chromatography software (Waters, USA).
Ethanol yields produced during fermentation were quantified by HPLC using an AS-2055 Intelligent Auto-sampler and a PU-1580 Intelligent HPLC Pump (Jasco, Japan). The Rezex ROA Organic Acid H+ organic acid column (5 μm, 7.8 mm × 300 mm; Phenomenex, UK) was operated at ambient temperature with a mobile phase of 0.005 N H2SO4 was used at a flow rate of 0.5 mL min−1. A Refractive Index cell (RI-2031 Intelligent Refractive Index detector, Jasco, Japan) was used for detection, and the injection volume was 10 μL. Data were acquired using the Azur software package v. 4.6.0.0 (Datalys, France). Prior to HPLC analysis, all samples and standards were filtered using Whatman GD/X syringe filters (GF/C 25 mm filter diameter/1.2 μm pore size; Whatman, UK). All analyses were conducted in triplicate.
Results
Biochemical composition of native UK seaweed species
The gross chemical compositions of the five seaweed species investigated in this study are listed in Table 2. The ash content was relatively high across all species, ranging from 18.8–25.7 % dry weight (d/w), which is substantially higher than the amounts present in most terrestrial plants as seaweeds have the ability to easily absorb inorganic substances from their environment (Yanik et al. 2013). Determination of protein content revealed that protein was within the range of 9.6–26.8 % (dry wt) and the lipid content was between 0.48–3.3 % (dry wt) across all species. Carbohydrate content was measured as the total of the monosaccharides, glucose, galactose, mannitol, fucose, xylose and arabinose, analysed by HPLC following an acid hydrolysis. Amongst the five seaweed species screened in this study, P. palmata had the highest carbohydrate content, 39.4 % ± 1.00 of the dry material, which likely reflects the hydrolysis of agar, a major component located in the extracellular matrix of red seaweed (Noseda et al. 1999). L. digitata had the lowest measured carbohydrate content of 21.7 %.
Table 2.
Proximate composition of seaweed species used in this study
| Composition % (dry weight basis) | |||||
|---|---|---|---|---|---|
| Seaweed species | Ash | Protein | Lipid | Carbohydratea | Moisture |
| L. digitata | 24.3 ± 0.38 | 26.8 ± 0.19 | 1.9 ± 0.09 | 21.7 ± 0.68 | 12.1 ± 0.39 |
| F. serratus | 18.8 ± 0.58 | 9.6 ± 0.72 | 2.8 ± 0.38 | 26.4 ± 0.75 | 10.6 ± 0.06 |
| C. crispus | 19 ± 1.02 | 19.9 ± 0.27 | 0.48 ± 0.25 | 21.8 ± 1.57 | 12.6 ± 0.21 |
| P. palmata | 25.7 ± 0.31 | 22.9 ± 0.16 | 3.3 ± 0.60 | 39.4 ± 1.00 | 5.2 ± 0.01 |
| U. lactuca | 21.5 ± 0.29 | 16.4 ± 0.14 | 1.0 ± 0.23 | 23.8 ± 0.80 | 10.0 ± 0.01 |
Data are the mean ± SD of three measurements
aCarbohydrate was estimated as the sum of monosaccharides arabinose, galactose, glucose, xylose, fucose and mannitol. It is assumed that the unaccounted for dry matter is principally polysaccharide material either nor broken down under the hydrolysis conditions employed or not quantified against authentic standards during HPLC analysis
Analysis of the hydrolysates generated from acid pre-treatment
The composition of hydrolysates derived from the seaweed using a 5 % (w/v) H2SO4 pre-treatment in a benchtop autoclave at 120 °C for 15 min can be seen in Table 3. The range and concentrations of monosaccharides liberated into the hydrolysate differed according to seaweed species. The red species C. crispus and P. palmata and the green seaweed U. lactuca yielded the highest concentrations of glucose, 2.02, 2.99 and 5.30 g L−1, respectively, which most likely resulted from the hydrolysis of cellulose. C. crispus and P. palmata produced the highest yields of galactose (4.24 and 6.84 g L−1, respectively) which would most likely be a product of carrageenan hydrolysis. Brown seaweed species L. digitata and F. serratus only released 1.51 and 0.95 g L−1 of glucose, respectively, but yielded 2.99 and 3.77 g L−1 of fucose and 3.49 and 4.38 g L−1 of mannitol, respectively. Interestingly, P. palmata yielded 3.04 g L−1 of fucose, a sugar which has not previously been reported or identified in this particular red algal species. Hydrolysates derived from seaweed were analysed for presence of known yeast-inhibitory compounds (a range of phenolic compounds, organic acids and furan compounds), which can be formed in the pre-treatment of lignocellulosic biomass. Such compounds are generated during the acid hydrolysis of biomass at high temperatures and can negatively impact on ethanol production during fermentation (Meinita et al. 2012). HMF and furfural were the only compounds present in the hydrolysates (Table 3) but were present at low levels and were not detectable in the hydrolysate of F. serratus. HMF derives from the breakdown of hexose sugars and furfural from the breakdown of pentose sugars. C. crispus generated the highest amount of HMF (2.13 g L−1) which is consistent with its high glucose and galactose contents (C-6 sugars).
Table 3.
Analysis of monosaccharides, yeast-inhibitory compounds and free amino nitrogen (FAN) present in the hydrolysates of L. digitata, F. serratus, C. crispus, P. palmata and U. lactuca after 5 % H2SO4 treatment at 120 °C for 15 min in a benchtop autoclave
| Seaweed species | Liberated monosaccharides (g L−1) | Inhibitors (g L−1) | Others (g L−1) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Arabinose | Fucose | Galactose | Glucose | Mannitol | Xylose | Total | HMF | Furfural | FAN | |
| L. digitata | 0.07 ± 0.01 | 2.29 ± 0.11 | 0.57 ± 0.03 | 1.51 ± 0.08 | 3.49 ± 0.08 | 0.55 ± 0.03 | 8.48 ± 0.34 | 0.02 ± 0.02 | – | 0.24 ± 0.001 |
| F. serratus | 0.08 ± 0.00 | 3.77 ± 0.10 | 0.91 ± 0.03 | 0.95 ± 0.03 | 4.38 ± 0.09 | 1.00 ± 0.03 | 11.09 ± 0.30 | – | – | 0.38 ± 0.004 |
| C. crispus | 0.14 ± 0.01 | 3.04 ± 0.15 | 6.84 ± 0.17 | 2.02 ± 0.07 | 0.01 ± 0.02 | 1.31 ± 0.06 | 13.36 ± 0.47 | 2.13 ± 0.16 | – | 0.36 ± 0.01 |
| P. palmata | 0.05 ± 0.01 | 0.08 ± 0.00 | 4.24 ± 0.21 | 2.99 ± 0.15 | 0.04 ± 0.00 | 8.71 ± 0.42 | 16.13 ± 0.79 | 0.02 ± 0.00 | 0.25 ± 0.01 | 0.38 ± 0.01 |
| U. lactuca | 2.15 ± 0.00 | 0.11 ± 0.02 | 0.78 ± 0.00 | 5.30 ± 0.03 | 0.06 ± 0.00 | 3.52 ± 0.02 | 11.91 ± 0.08 | 0.09 ± 0.00 | 0.05 ± 0.00 | 0.16 ± 0.003 |
Data are the mean ± SD of three measurements
Levels of free amino nitrogen (FAN) are also reported for each of the seaweed hydrolysates (Table 3). This is because amino acids represent a significant nitrogen source required for yeast growth and metabolism during fermentation. FAN levels were within the range 0.16 to 0.38 g L−1. The minimum FAN content normally specified for brewing fermentations is 0.15 g L−1, so the seaweed hydrolysates appeared to contain sufficient quantities of amino acids to avoid the need for supplementary nitrogen sources to aid fermentation.
Phenotypic microarray analysis of yeast strains
Metabolic output of yeast strains grown on synthetic minimal media with different carbon sources
The metabolic outputs (defined here as redox signal intensity) of 22 yeast strains on minimal media containing individual carbon sources: glucose, galactose, mannitol, xylose, fucose and rhamnose (at 6 % w/v) were measured (Fig. 1a–f). These carbon sources were chosen because they are the most abundant in sugar composition across the three taxonomical groups of macroalgae. There was measurable metabolic output for all strains utilising glucose after 50 h of incubation at 30 °C, with considerable variation observed between the strains (2.3–98.7 RSI) (Fig. 1). Some strains, however, exhibited extremely low RSI levels when utilising glucose as a sole carbon source, namely Candida shehatae var lignosa NCYC 2389 and Pichia stipitis TP10, and were as a result removed from further study. The majority of the strains displayed a high metabolic output when utilising galactose (Fig. 1b), whereas the strains exhibited very poor metabolic output when utilising fucose (Fig. 1d). The metabolic outputs of all strains screened here appear to be similar in response when utilising mannitol, xylose and rhamnose (Fig. 1b, e, f).
Fig. 1.
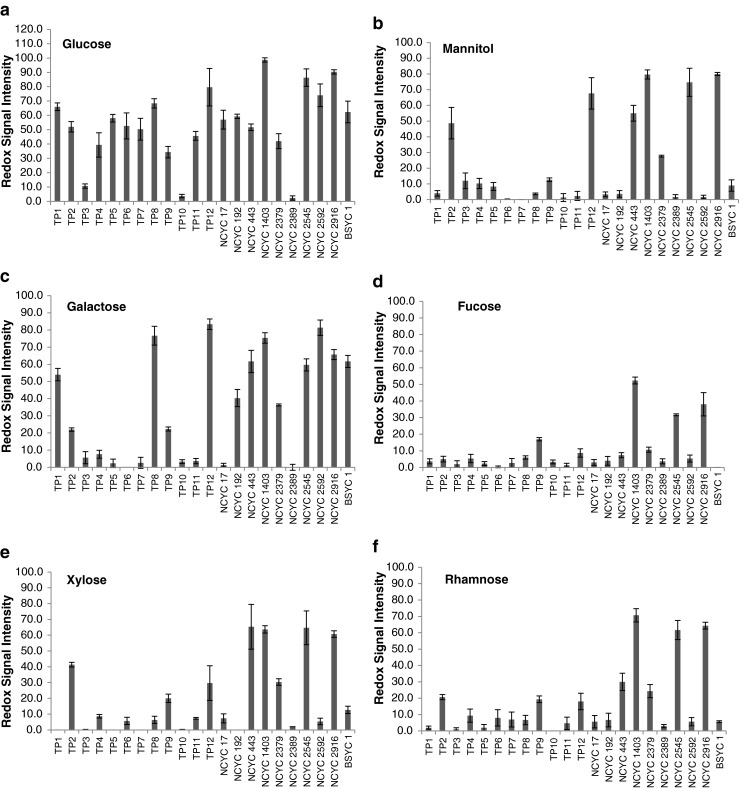
Phenotypic microarray analysis (redox signal intensity) at 50 h for strains on minimal media containing a glucose, b mannitol, c galactose, d fucose, e xylose and f rhamnose at 6 % (w/v) inclusion. Data are the mean ± SD of three replicates. See Table 1 for description of yeast strains
In order to have a better understanding of each yeast strains’ ability to metabolise unusual carbon sources, the PM assay results were adjusted to show each strain’s metabolic output for these sugars in relation to their utilisation of glucose. Interestingly, strains which had relatively high utilisation of glucose (≥79.7) also displayed high metabolic outputs when utilising galactose, mannitol, fucose, xylose and rhamnose. These were: C. succiphila NCYC 1403, P. anomala TP12, C. tenuis NCYC 2545 (Table 4).
Table 4.
Phenotypic microarray results (RSI at 50 h) for each yeast strain investigated in this study on minimal media containing: galactose, mannitol, fucose, xylose and arabinose (6 % v/w) relative to the corresponding glucose value (RSI). Data are the mean ± SD of three replicates. See Table 1 for description of yeast strains
| Monosaccharide (% RSI in comparison to glucose) | ||||||
|---|---|---|---|---|---|---|
| Yeast ID | Glucose (RSI) | Galactose | Mannitol | Fucose | Xylose | Rhamnose |
| TP1 | 66.0 | 81.8 | 6.1 | 5.6 | 0.0 | 3.0 |
| TP2 | 52.0 | 42.3 | 93.6 | 9.6 | 79.5 | 39.7 |
| TP3a | 10.7 | 53.1 | 112.5 | 18.8 | 3.1 | 9.4 |
| TP4 | 39.3 | 19.5 | 26.3 | 13.6 | 22.0 | 23.7 |
| TP5 | 58.0 | 4.0 | 14.4 | 4.0 | 0.0 | 3.4 |
| TP6 | 52.7 | 0.0 | 0.6 | 0.6 | 10.8 | 15.2 |
| TP7 | 50.3 | 5.3 | 0.0 | 5.3 | 0.0 | 13.9 |
| TP8 | 68.3 | 112.2 | 5.4 | 8.8 | 9.3 | 9.8 |
| TP9 | 34.3 | 65.0 | 36.9 | 49.5 | 58.3 | 56.3 |
| TP10a | 3.7 | 90.9 | 36.4 | 90.9 | 9.1 | 0.0 |
| TP11 | 45.7 | 8.0 | 5.1 | 2.9 | 16.1 | 10.2 |
| TP12 | 79.7 | 104.6 | 84.9 | 10.9 | 37.2 | 22.6 |
| NCYC 17 | 57.0 | 2.3 | 5.8 | 5.3 | 12.9 | 9.9 |
| NCYC 192 | 59.3 | 68.0 | 6.2 | 6.7 | 0.0 | 11.2 |
| NCYC 443 | 51.7 | 119.4 | 106.5 | 14.2 | 126.5 | 58.1 |
| NCYC 1403 | 98.7 | 76.4 | 80.7 | 53.0 | 64.5 | 71.6 |
| NCYC 2379 | 42.0 | 86.5 | 65.9 | 25.4 | 72.2 | 57.9 |
| NCYC 2389a | 2.3 | 0.0 | 71.4 | 157.1 | 85.7 | 128.6 |
| NCYC 2545 | 86.3 | 69.1 | 86.5 | 36.7 | 74.9 | 71.4 |
| NCYC 2592 | 74.0 | 109.9 | 2.3 | 7.2 | 7.2 | 7.7 |
| NCYC 2916 | 90.3 | 72.7 | 88.6 | 42.1 | 67.2 | 71.2 |
| BSYC 1 | 62.3 | 98.9 | 14.4 | 0.0 | 20.3 | 9.1 |
aRemoved from study due to low metabolic output on glucose
Metabolic output of yeast strains grown on seaweed hydrolysates
Three yeast strains, C. tenuis NCYC 2545, C. succiphila NCYC 1403 and P. anomala TP12 were able to metabolise the majority of the individual sugars as evaluated using PM analysis. They were thus selected for further experiments to assess their abilities to metabolise the acid hydrolysates generated from five seaweed species (Fig. 2a–e). S. cerevisiae NCYC 2592 was also included in the trial as a reference distiller’s production strain (www.ncyc.co.uk), expected to provide efficient conversion of glucose and galactose to ethanol. Additionally, two S. cerevisiae Y12 and YPS128 strains (Liti et al. 2009) were also included as reference strains. S. cerevisiae Y12 and YPS128 are wild yeast derived from clean lineages that have had no alternations to their genomes from human influences or domestication events (Liti et al. 2009). Assays revealed that all strains cultured on L. digitata, F. serratus and U. lactuca hydrolysates were metabolically active (Fig. 2a, b, e). However, assays with C. tenuis NCYC 2545 revealed a decrease in RIS after 20 h from ca 50–60 RIS to ca 10 RIS in both brown seaweed hydrolysates (Fig. 2a, b). Although this phenomenon has been observed in other microarray assays before, it is unknown why it occurs but could be down to cell lysis and loss of the irreversibly reduced dye (Darren Greetham, personal communication). Due to this result, C. tenuis NCYC 2545 was not included in future work.
Fig. 2.
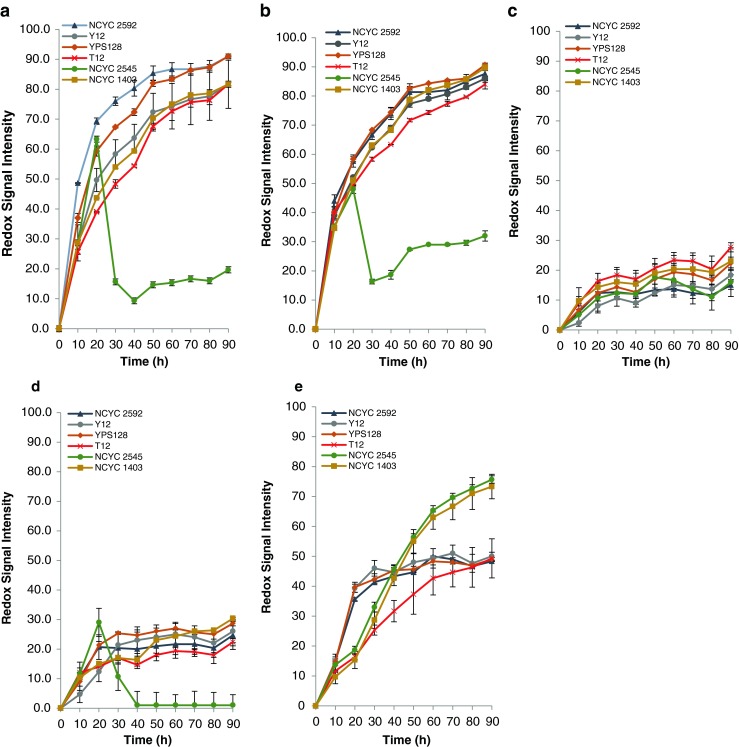
Phenotypic microarray analysis (redox signal intensity) for yeast on a L. digitata hydrolyste, b F. serratus hydrolysate, c C. crispus hydrolysate, d P. palmata hydrolysate, e U. lactuca hydrolysate. Data are the mean ± SD of three replicates. See Table 1 for description of yeast strains
Screening for bioethanol production from seaweed hydrolysates in laboratory-scale fermentations
Trial fermentations were conducted with the yeast strains S. cerevisiae NCYC 2592, C. succiphila NCYC 1403, P. anomala TP12 and S. cerevisiae strains YPS128 and Y12 to determine whether they could produce ethanol as a carbon end point. Hydrolysates derived from the five seaweed species post-acid pre-treatment were neutralised and fermented using each strain individually at 30 °C.
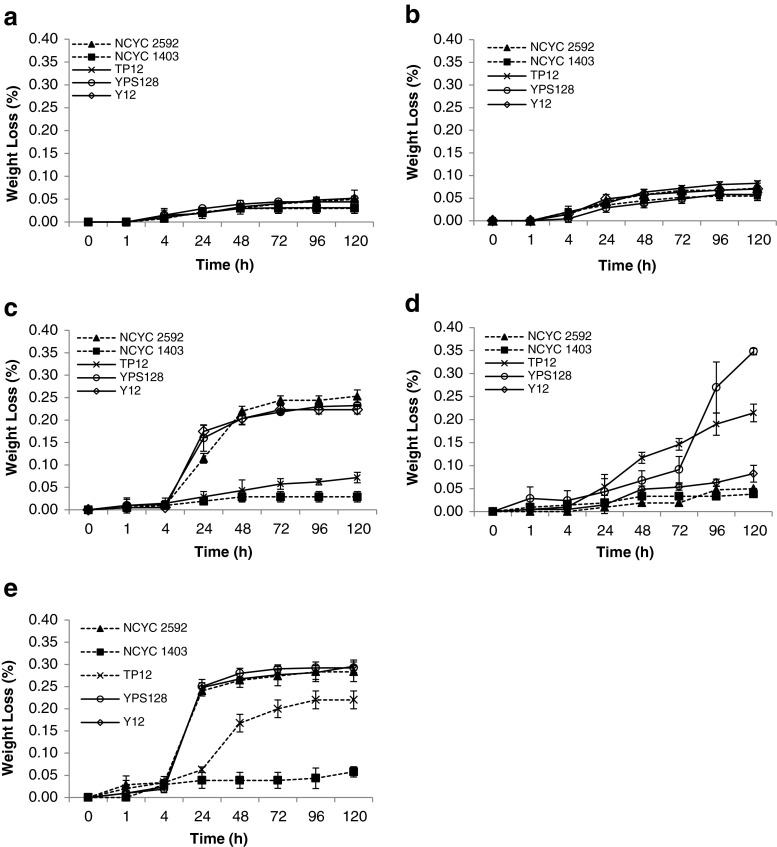
Fermentation progression was monitored as weight loss due to the evolution of carbon dioxide, a metabolic product of the fermentation of any monosaccharide (Fig. 3a–e). There were clear differences in the fermentation progression for each hydrolysate using the strains employed in this trial. Overall, fermentation of the L. digitata and F. serratus hydrolysates by all strains did not perform well as there were only small weight losses after 120 h (Fig. 3a, b). P. palmata and C. crispus fermentations on the contrary progressed better, with P. anomala TP12 performing better on the C. crispus hydrolysate, albeit with a longer lag-phase and longer attenuation time (Fig. 3c, d). S. cerevisiae YPS128 displayed a similar fermentation profile grown on the C. crispus hydrolysate yet S. cerevisiae Y12 did not readily ferment this substrate. All three S. cerevisiae strains demonstrated significant weight loss on the U. lactuca hydrolysate, whereas it took up to 24 h for P. anomala TP12 to start to ferment this hydrolysate (Fig. 3e).
Fig. 3.
Fermentation progression of yeast strains on a F. serratus hydrolysate, b L. digitata hydrolysate, c P. palmata hydrolysate, d C. crispus hydrolysate, e U. lactuca hydrolysate by yeast strains, as indicated by vessel weight loss resulting from the release of carbon dioxide. Data are the mean ± SD of three replicates. See Table 1 for description of yeast strains
Utilisation of monosaccharides pre- and post-fermentation and the mean ethanol yields achieved from each strain on the five hydrolysates was determined. Overall, each strain apart from C. succiphila NCYC 1403 managed to produce ethanol; however, the yields varied between strain and hydrolysate (Table 5). The highest yield of ethanol, ca 13 g L−1, was produced from the fermentation of a hydrolysate derived from C. crispus by S. cerevisiae YPS128, and it appears that this strain was able to consume the majority of the available monosaccharides that were present in the hydrolysate. Interestingly, strains S. cerevisiae Y12 and P. anomala TP12 also consumed the vast majority of monosaccharides, however, were only able to yield ca 4.5 and 3 g L−1 of ethanol, respectively. The highest yields of ethanol produced from the hydrolysates of P. palmata and U. lactuca were ca 7 g L−1, resulting from the use of Saccharomyces spp. Interestingly, the fucose which was present in the C. crispus hydrolysate (at a concentration of ca 3 g L−1) and not in the brown seaweed hydrolysates, was partially consumed by strains S. cerevisiae NCYC 2592, Y12 and P. anomala TP12 during the fermentations. Disappointingly, the five strains hardly yielded any ethanol post-fermentation of hydrolysates derived from the two brown seaweeds (0.00–1.55 g L−1 ethanol).
Table 5.
Utilisation of monosaccharides in hydrolysates of L. digitata, F. serratus, C. crispus, P. palmata and U. lactuca and production of bioethanol by different yeast strains post-fermentation. Data are the mean ± SD of three replicates
| Yeast strain | |||||||
|---|---|---|---|---|---|---|---|
| NCYC 1403 | NCYC 2592 | TP12 | Y12 | YPS128 | |||
| Hydrolysate | Monosaccharide/ethanol (g L−1) | Start | End | End | End | End | End |
| L. digitata | Mannitol | 3.49 ± 0.08 | 3.40 ± 0.07 | 3.46 ± 0.11 | 3.44 ± 0.48 | 3.42 ± 0.48 | 3.48 ± 0.11 |
| Fucose | 2.29 ± 0.11 | 2.25 ± 0.07 | 2.23 ± 0.13 | 2.28 ± 0.24 | 2.30 ± 0.24 | 2.31 ± 0.01 | |
| Arabinose | 0.07 ± 0.01 | 0.06 ± 0.00 | 0.07 ± 0.02 | 0.02 ± 0.05 | 0.08 ± 0.05 | 0.07 ± 0.00 | |
| Galactose | 0.57 ± 0.03 | 0.55 ± 0.04 | 0.02 ± 0.01 | 0.05 ± 0.01 | 0.05 ± 0.01 | 0.00 ± 0.00 | |
| Glucose | 1.51 ± 0.08 | 1.49 ± 0.08 | 0.02 ± 0.01 | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.01 ± 0.00 | |
| Xylose | 0.55 ± 0.03 | 0.56 ± 0.02 | 0.37 ± 0.03 | 0.14 ± 0.10 | 0.14 ± 0.10 | 0.41 ± 0.00 | |
| Ethanol | – | 0.09 ± 0.12 | 1.24 ± 0.12 | 1.26 ± 0.06 | 1.22 ± 0.05 | 1.55 ± 0.17 | |
| F. serratus | Mannitol | 4.38 ± 0.08 | 4.36 ± 0.55 | 4.35 ± 0.56 | 4.32 ± 0.50 | 4.38 ± 0.11 | 4.37 ± 0.13 |
| Fucose | 3.77 ± 0.11 | 3.58 ± 0.40 | 3.58 ± 0.28 | 3.75 ± 0.42 | 3.76 ± 0.05 | 3.77 ± 0.09 | |
| Arabinose | 0.08 ± 0.01 | 0.04 ± 0.01 | 0.04 ± 0.00 | 0.06 ± 0.02 | 0.04 ± 0.00 | 0.04 ± 0.00 | |
| Galactose | 0.91 ± 0.03 | 0.88 ± 0.13 | 0.00 ± 0.00 | 0.00 ± 0.01 | 0.00 ± 0.00 | 0.02 ± 0.00 | |
| Glucose | 0.95 ± 0.08 | 0.89 ± 0.12 | 0.01 ± 0.00 | 0.00 ± 0.00 | 0.02 ± 0.00 | 0.01 ± 0.00 | |
| Xylose | 1.00 ± 0.03 | 0.94 ± 0.13 | 0.82 ± 0.12 | 0.07 ± 0.04 | 0.80 ± 0.02 | 0.97 ± 0.09 | |
| Ethanol | – | 0.00 ± 0.03 | 0.58 ± 0.05 | 0.46 ± 0.04 | 0.78 ± 0.17 | 1.05 ± 0.07 | |
| P. palmata | Mannitol | 0.04 ± 0.02 | 0.02 ± 0.03 | 0.02 ± 0.00 | 0.03 ± 0.02 | 0.01 ± 0.00 | 0.03 ± 0.08 |
| Fucose | 0.08 ± 0.15 | 0.05 ± 0.01 | 0.04 ± 0.00 | 0.05 ± 0.15 | 0.06 ± 0.00 | 0.07 ± 0.04 | |
| Arabinose | 0.05 ± 0.00 | 0.04 ± 0.02 | 0.05 ± 0.00 | 0.03 ± 0.01 | 0.05 ± 0.00 | 0.04 ± 0.00 | |
| Galactose | 4.24 ± 0.17 | 4.22 ± 0.04 | 0.39 ± 0.01 | 4.23 ± 0.17 | 0.40 ± 0.01 | 0.23 ± 0.23 | |
| Glucose | 2.99 ± 0.07 | 2.97 ± 0.02 | 0.00 ± 0.00 | 2.96 ± 0.07 | 0.00 ± 0.00 | 0.00 ± 0.12 | |
| Xylose | 8.71 ± 0.06 | 8.70 ± 0.02 | 8.69 ± 0.14 | 8.70 ± 0.06 | 8.68 ± 0.25 | 6.20 ± 0.00 | |
| Ethanol | – | 0.00 ± 0.01 | 5.21 ± 0.27 | 1.69 ± 0.05 | 6.57 ± 1.20 | 6.08 ± 0.40 | |
| C. crispus | Mannitol | 0.01 ± 0.02 | 0.00 ± 0.03 | 0.01 ± 0.00 | 0.01 ± 0.02 | 0.00 ± 0.04 | 0.01 ± 0.01 |
| Fucose | 3.04 ± 0.15 | 2.35 ± 0.10 | 1.37 ± 0.00 | 0.05 ± 0.00 | 0.25 ± 0.00 | 0.39 ± 0.00 | |
| Arabinose | 0.14 ± 0.01 | 0.11 ± 0.00 | 0.13 ± 0.01 | 0.04 ± 0.00 | 0.11 ± 0.00 | 0.15 ± 0.01 | |
| Galactose | 6.84 ± 0.17 | 6.84 ± 0.20 | 6.63 ± 0.11 | 0.36 ± 0.01 | 0.50 ± 0.03 | 1.32 ± 0.01 | |
| Glucose | 2.02 ± 0.07 | 1.97 ± 0.09 | 0.00 ± 0.10 | 0.00 ± 0.00 | 0.01 ± 0.00 | 0.03 ± 0.04 | |
| Xylose | 1.31 ± 0.06 | 0.79 ± 0.04 | 0.84 ± 0.02 | 0.43 ± 0.13 | 0.51 ± 0.01 | 0.43 ± 0.02 | |
| Ethanol | – | 0.00 ± 0.02 | 2.90 ± 0.02 | 3.26 ± 0.01 | 4.43 ± 0.18 | 12.84 ± 0.70 | |
| U. lactuca | Mannitol | 0.06 ± 0.00 | 0.05 ± 0.05 | 0.06 ± 0.00 | 0.04 ± 0.07 | 0.03 ± 0.01 | 0.06 ± 0.04 |
| Fucose | 0.11 ± 0.02 | 0.09 ± 0.00 | 0.10 ± 0.00 | 0.11 ± 0.00 | 0.01 ± 0.01 | 0.11 ± 0.00 | |
| Arabinose | 2.15 ± 0.00 | 2.15 ± 0.04 | 1.98 ± 0.08 | 2.03 ± 0.14 | 2.15 ± 0.03 | 1.98 ± 0.02 | |
| Galactose | 0.78 ± 0.00 | 0.71 ± 0.01 | 0.70 ± 0.03 | 0.04 ± 0.00 | 0.01 ± 0.00 | 0.04 ± 0.00 | |
| Glucose | 5.30 ± 0.07 | 5.28 ± 0.04 | 0.05 ± 0.00 | 0.00 ± 0.00 | 0.03 ± 0.00 | 0.00 ± 0.00 | |
| Xylose | 3.51 ± 0.02 | 3.49 ± 0.06 | 2.29 ± 0.04 | 2.43 ± 0.04 | 2.71 ± 0.02 | 2.06 ± 0.01 | |
| Ethanol | – | 0.26 ± 0.02 | 6.14 ± 0.22 | 3.49 ± 0.06 | 6.80 ± 0.34 | 7.31 ± 0.50 | |
Discussion
The strains used in this study (mainly Candida and Pichia strains) for the initial screen on various carbon sources were selected based upon the fact that these species of yeast have the ability to ferment C5 sugars such as xylose (present in seaweeds) (Hahn-Hägerdal et al. 2007). Therefore, it was of interest to examine whether these yeasts could utilise the sugars which are only found in seaweeds and whether they would produce ethanol as a result.
In the PM method, changes in colour of a redox sensitive dye are monitored as yeast cells respire on selected media arranged in a 96-well plate format (Bochner et al. 2001). The colour development is a measure of the yeast’s metabolic activity and essentially indicates its ability to metabolise the sugar present in the assay. It is not possible to determine the metabolic products (e.g. ethanol) directly, just metabolic activity. The majority of the strains screened in this study displayed an increase in the rate of metabolic output, defined as redox signal intensity (RSI), in the presence of glucose and galactose (Fig. 1a, c) compared to the other tested monosaccharides (fucose, mannitol, xylose, arabinose and rhamnose). This finding was as expected since these monosaccharides are favourable carbon sources for yeast (Flores et al. 2000). Interestingly, the Candida and Pichia spp examined in this study displayed an increase in metabolic activity, albeit at varying intensities, when utilising mannitol, xylose or rhamnose as sole carbon sources (Fig. 1b, e, f) compared to Saccharomyces, Kluyveromyces and Hanseniaspora spp in this study. Candida spp and Pichia spp have been previously reported to be able to metabolise and ferment pentose sugars (Dien et al. 1996; Jarosz et al. 2008); however, only 1 % of all known xylose utilising yeasts are known to display the potential of fermenting xylose to ethanol (McMillan and Boynton 1994). Pichia angiophorae has already been shown to be able to ferment both mannitol and laminarin in seaweed extracts to ethanol (Horn et al. 2000a). C. tenuis NCYC 2545, C. succiphila NCYC 1403 and P. anomala TP12 exhibited high RSI levels on all monosaccharides analysed in relation to their corresponding glucose values, which implies that they are able to metabolise these sugars (Table 4). What is interesting is that they are the only species which appear to be metabolically active on synthetic media containing fucose, with an RSI ≥ 20 (Fig. 1d). Fucose is a hexose sugar and is the fundamental subunit of the fucoidan polysaccharide found in brown seaweeds (Ale et al. 2011). To date, there has been no evidence to suggest that fucose can be metabolised by yeast and converted into ethanol during a fermentation. Results here revealed that three Candida spp displayed metabolic activity in the presence of fucose as a sole carbon source. This suggests that these strains may possibly contain a fucose utilisation pathway or surface transporter, enabling the uptake of fucose into the cell. A gene has been identified in the C. albicans genome, C2_09280C_B, of which is predicted to be either a membrane transporter, member of the fucose:proton symporter (FHS) family or part of a major facilitator superfamily (MFS), and orthologous genes have been identified in other Candida, Aspergillus and Saccharomyces species (www.candidagenome.org). It would be of great interest to analyse whether the same or orthologous genes associated with fucose uptake or metabolism can also be identified in the Candida spp examined in the present study.
The metabolic activities of the shortlisted strains C. succiphila NCYC 1403, P. anomala TP12, C. tenuis NCYC 2545 and S. cerevisiae NCYC 2592 cultured on red seaweed hydrolysates could not be reliably interpreted using the present methodology. This is because the colour of the hydrolysates interfered with detection of generation of the tetrazolium dye (Fig. 2c, d). This is undoubtedly a limitation on the current application of the PM. If this approach is to be employed in future work, it would be necessary to experiment with the use of other reporter dyes such as DAPI, dihydroethidium or GFP.
This study used hydrolysates generated from the thermo-chemical pre-treatment of different seaweed species. There had been no previous reports of the utilisation of hydrolysates produced from the acid pre-treatment of seaweeds for bioethanol production, until Mutripah et al. (2014) fermented the hydrolysate of the rhodophyte Palmaria palmata with Saccharomyces cerevisiae, where 17.3 mg of ethanol per gramme of seaweed was produced. Typically, the objective of applying an acid (or in some cases alkaline) pre-treatment step is to modify the chemical structure of the biomaterial by removing the more recalcitrant complexes. Hydrolysis of polysaccharides into their monosaccharide constituents is typically a further step that is sequential to the acid pre-treatment, by the addition of selective enzymes (Meinita et al. 2012); however, addition of enzyme is expensive and it is necessary to apply the correct enzyme blend to suit the particular species of seaweed that is being processed. The liquid fraction generated from the acid pre-treatment step is usually discarded. Analysis of the hydrolysates derived from a pre-treatment of seaweed indicates that they contained a cocktail of available monosaccharides which could be fermented to ethanol, without the need to apply an enzyme hydrolysis step, and contained low levels of yeast-inhibitory compounds which should not unduly influence yeast performance during fermentation (Table 5). According to the compositional analysis undertaken in the study, P. palmata had the highest carbohydrate content (39.4 %) and actually yielded the highest concentration of total sugars in its hydrolysate post-pre-treatment (ca 16.1 g L−1). Although it appears that this particular species could be an ideal candidate for bioethanol production, its abundance in UK waters is not as high as that of certain brown kelp species, such as L. hyperborea and A. nodosum (Wilkinson 1995). L. digitata had the lowest measured carbohydrate content of 21.7 % (dry wt), which could in part be due to the low levels of phaeophycean polysaccharides, in particular laminarin and fucoidan, as seaweeds were harvested in May for this study. A study by Adams et al. (2011) revealed that the most suitable month to harvest L. digitata for bioethanol production was July, as this is when carbohydrate yields are highest. Further, some specialised monosaccharides such as mannuronic and guluronic acids from the polysaccharide alginate might have been released but were not quantified using the HPLC method applied. Alginate is extremely difficult to hydrolyse and under the conditions utilised in this study is unlikely to have liberated significant quantities of these particular monosaccharides. The main cell wall constituents that make up brown seaweeds are typically cellulose microfibrils which are embedded in an amorphous matrix of alginate and fucoidan polysaccharides (Kloareg et al. 1986). Mannitol is often found as a terminating side chain on laminarin, a polymer of β-(1-3)-D-glucose that can be easily extracted from brown seaweed (Horn et al. 2000a) and the fucose yields have resulted from the breakdown of fucoidan in the brown seaweed hydrolysates.
Saccharomyces cerevisiae spp are traditionally used in fermentations for the production of commercial bioethanol, as strains can adequately consume hexose sugars and resist any osmotic and ethanol toxicity stresses which may arise during the process (Zhao and Bai 2009). However, their inability to utilise pentose sugars, despite having the relevant pathways in their genomes (Hahn-Hägerdal et al. 2007), limits the full potential of their use in the production of bioethanol.
The yeast strains selected for fermentation trials performed significantly better on the red seaweed hydrolysates than the other hydrolysates produced from the brown and green seaweeds, with the exception of C. succiphila NCYC 1403, which failed to produce any ethanol. Candida spp are obligate aerobes and are Crabtree-negative microorganisms, implying that they respire and produce biomass independent of the sugar concentration as long as oxygen is available (Fredlund et al. 2004). This explains the extremely low levels of ethanol produced in the anaerobic environment encountered during a fermentation. Despite the fact that strain P. anomala TP12 is also a Crabtree-negative yeast, it demonstrated the ability to produce bioethanol in an anoxic environment from all five seaweed hydrolysates, with the highest ethanol yield of ca 3.5 g L−1 being produced from the green U. lactuca hydrolysate. There are reports that have revealed this strain to be ethanol-producing and potentially resistant to high ethanol concentrations (Passoth et al. 2013; Tao et al. 2011). Interestingly, a study by Ruyters et al. (2015) revealed that P. anomala could be a candidate strain for ethanol production from lignocellulosic hydrolysates, as ethanol yields were similar to those with S. cerevisiae and P. anomala displayed tolerance to high levels of ethanol. The group also found that P. anomala needed a longer fermentation time, which is in agreement with the results obtained from this study when P. anomala TP12 was inoculated into the seaweed hydrolysates (Fig. 3). Although the oxygen-sensitive nature of this strain will undoubtedly limit its fermentation performance, it appears that P. anomala TP12 and possibly other P. anomala strains are compatible not only with lignocellulosic hydrolysates but also with seaweed.
The relatively poor fermentation performance of the five strains on the brown seaweed hydrolysates resulted in the lowest ethanol yields (<2 g L−1 ethanol for all strains). This is due to the low levels of fermentable sugars in the hydrolysate, namely glucose and galactose, which together amounted to 2.08 and 1.86 g L−1 in the L. digitata and F. serratus hydrolysates, respectively, principally as a result of harvesting in May. In order to make bioethanol production commercially viable from brown seaweed species, harvesting at the appropriate time period of the year for maximum sugar quantities and hydrolysis with enzymes will most likely be required, as acid hydrolysis alone was not sufficient to generate an adequate yield of fermentable sugars. Polysaccharides such as cellulose would therefore need to be targeted by selective cellulase enzymes in order to release glucose in high yields.
Wild yeast strains S. cerevisiae Y12 and YPS128, which were included in this study as reference strains due to the fact that their genomes have remained unaltered from human influences and domestication events, were both able to consume the vast majority of monosaccharides (including pentose sugars) that were present in the hydrolysate of C. crispus. However, S. cerevisiae Y12 only generated 4.43 g L−1 of ethanol, whereas S. cerevisiae YPS128 produced the highest yield of ethanol in this study of 12.84 g L−1 on the C. crispus feedstock. S. cerevisiae Y12 has been reported to be extremely sensitive to the presence of inhibitory compounds which arise as a result of the thermo-chemical pre-treatment of biomass (Wimalasena et al. 2014). A haploid segregant population derived from these yeasts is available, and it would be interesting to screen for variance in utilisation of these sugars in that population, as research has revealed that populations of haploid segregants have a varied response to any given environment (Greetham et al. 2014b). Hydrolysate from the pre-treatment of C. crispus was shown to have the highest yield of HMF at ca 2 g L−1, which could explain S. cerevisiae Y12’s poor fermentation progress in this instance. Conversely, S. cerevisiae YPS128 displays tolerance to inhibitory compounds (Wimalasena et al. 2014), and such phenotypes are highly desirable for bioethanol production. P. anomala TP12 was also able to consume the majority of the monosaccharides present in the hydrolysate of C. crispus; however, only 3.26 g L−1 of ethanol was generated. It is thus likely possible that the sugars were converted to other carbon end point products in addition to ethanol. In order to enhance the performance of strains S. cerevisiae Y12 and P. anomala TP12 during the fermentation of C. crispus hydrolysate, it would be worthwhile attempting to detoxify the hydrolysate by selective removal of inhibitory compounds. Many methods have been developed in order to remove toxic compounds from hydrolysates, as reviewed by Mussatto and Roberto (2004).
Interestingly, only the fucose which was present in the C. crispus hydrolysate (at a concentration of ca 3 g L−1) was partially consumed by strains S. cerevisiae NCYC 2592, Y12 and P. anomala TP12 during fermentation (Table 4). This calls into question the correct assignment of the HPLC peak to fucose in this instance. This monosaccharide is typically only found in phaeophytes, not rhodophytes. It may be concluded that the C. crispus hydrolysate contained a monosaccharide which demonstrated chromatographic similarity to the fucose standard under experimental conditions. Likewise, it eluted within the same retention window as the peak assigned to fucose in brown seaweed hydrolysates. However, LC-MS studies would be required to further confirm this identification.
In conclusion, in this study, we have demonstrated a novel application of the PM assay to evaluate the ability of yeast strains to metabolise seaweed-derived monosaccharides (individually) as well as entire hydrolysates of five UK seaweed species. The most suitable yeast strains to utilise sugars derived from macroalgae and convert them into bioethanol were S. cerevisiae spp with the highest concentration of ethanol (ca 13 g L−1) resulting from the fermentation of the acid hydrolysate of C. crispus by S. cerevisiae YPS128. Shortlisted stain P. anomala TP12 was able to utilise the majority of the monosaccharides present in the hydrolysate of C. crispus including fucose, generating ca 3.5 g L−1 ethanol, which was similar to yields of ethanol produced by S. cerevisiae strains NCYC 2592 and Y12 from the same hydrolysate. This reveals that P. anomala TP12 could be a possible candidate strain for further research. The present work was focused on strain selection and did not seek to optimise ethanol yields from the fermentation of the seaweed hydrolysates. This would be required in order to make the industrial production of ethanol from seaweed economically viable. Industrial production of ethanol from seaweeds would require the generation of more efficient strains that can ferment the majority of sugars contained in seaweed polysaccharides. Although ethanol was the only product screened for in this study, other high value, carbon end point products such as xylitol, succinic and acetic acids could also be screened for and sourced from the growth of similar or other microorganisms on seaweed hydrolysates.
Acknowledgments
The authors gratefully acknowledge the financial support of the Biotechnology and Biological Science Research Council (BBSRC) and CRODA International plc for funding this research and Miss E. Kostas’s studentship. We also thank Drs D. Greetham and T. Wimalasena for their help and input with the phenotypic microarray (Biolog) screen and Dr T. Phister and Prof. E. Louis for providing yeast strains.
References
- Adams JM, Gallagher JA, Donnison IS. Fermentation study on Saccharina latissima for bioethanol production considering variable pre-treatments. J Appl Phycol. 2009;21:569–574. doi: 10.1007/s10811-008-9384-7. [DOI] [Google Scholar]
- Adams JM, Toop TA, Donnison IS, Gallagher JA. Seasonal variation in Laminaria digitata and its impact on biochemical conversion routes to biofuels. Bioresour Technol. 2011;102:9976–9984. doi: 10.1016/j.biortech.2011.08.032. [DOI] [PubMed] [Google Scholar]
- Ale MT, Mikkelsen JD, Meyer AS. Important determinants for fucoidan bioactivity: a critical review of structure-function relations and extraction methods for fucose-containing sulfated polysaccharides from brown seaweeds. Mar Drugs. 2011;9:2106–2130. doi: 10.3390/md9102106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR, Gadzinski P, Panomitros E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothast RJ, Schlicher MA. Biotechnological processes for conversion of corn into ethanol. Appl Biochem Biotechnol. 2005;67:19–25. doi: 10.1007/s00253-004-1819-8. [DOI] [PubMed] [Google Scholar]
- Dien BS, Kurtzman CG, Saha BC, Bothast RJ (1996) Screening for L-arabinose fermenting yeasts. Appl Biochem Biotechnol 57–58:233–242 [PubMed]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- Enquist-Newman M, Faust AME, Bravo DD, Santos CNS, Raisner RM, Hanel A, Sarvabhowman P, Le C, Regitsky DD, Cooper SR, Peereboom L, Clark A, Martinez Y, Goldsmith J, Cho MY, Donohoue PD, Luo L, Lamberson B, Tamrakar B, Kim EJ, Villari JL, Gill A, Tripathi SA, Karamchedu P, Paredes CJ, Rajgarhia V, Kotlar HK, Bailey RB, Miller DJ, Ohler NL, Swimmer C, Yoshikuni Y. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature. 2014;505:239–243. doi: 10.1038/nature12771. [DOI] [PubMed] [Google Scholar]
- Flores CL, Rodríguez C, Petit T, Gancedo C. Carbohydrate and energy‐yielding metabolism in non‐conventional yeasts. FEMS Microbiol Rev. 2000;24:507–529. doi: 10.1111/j.1574-6976.2000.tb00553.x. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Stanley GHS. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Fredlund E, Druvefors UÄ, Olstorpe MN, Passoth V, Schnürer J. Influence of ethyl acetate production and ploidy on the anti‐mould activity of Pichia anomala. FEMS Microbiol Lett. 2004;238:133–137. doi: 10.1016/j.femsle.2004.07.027. [DOI] [PubMed] [Google Scholar]
- Greetham D, Wimalasena T, Kerruish DWM, Brindley S, Ibbett RN, Linforth RL, Tucker G, Phister TG, Smart KA. Development of a phenotypic assay for characterisation of ethanologenic yeast strain sensitivity to inhibitors released from lignocellulosic feedstocks. J Ind Microbiol Biotechnol. 2014;41:931–945. doi: 10.1007/s10295-014-1431-6. [DOI] [PubMed] [Google Scholar]
- Greetham D, Wimalasena TT, Leung K, Marvin ME, Chandelia Y, Hart AJ, Phister TG, Tucker GA, Louis EJ, Smart KA. The genetic basis of variation in clean lineages of Saccharomyces cerevisiae in response to stresses encountered during bioethanol production. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0103233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn-Hägerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. Towards industrial pentose-fermenting yeast strains. Appl Biochem Biotechnol. 2007;74:937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- Horn SJ, Aasen IM, Ostgaard K. Ethanol production from seaweed extract. J Ind Microbiol Biotechnol. 2000;25:249–254. doi: 10.1038/sj.jim.7000065. [DOI] [Google Scholar]
- Horn SJ, Aasen IM, Ostgaard K. Production of ethanol from mannitol by Zymobacter palmae. J Ind Microbiol Biotechnol. 2000;24:51–57. doi: 10.1038/sj.jim.2900771. [DOI] [Google Scholar]
- Jarosz A, Kordowska-Wiater M, Targonski Z. Biotransformation of L-arabinose to arabitol by yeasts from genera Pichia and Candida. Bio Technologia. 2008;1:177–188. [Google Scholar]
- Jeong TS, Um BH, Kim JS, Oh KK. Optimizing dilute-acid pretreatment of rapeseed straw for extraction of hemicellulose. Appl Biochem Biotechnol. 2010;161:22–33. doi: 10.1007/s12010-009-8898-z. [DOI] [PubMed] [Google Scholar]
- Jung KA, Lim SR, Kim Y, Park JM. Potentials of macroalgae as feedstocks for biorefinery. Bioresour Technol. 2013;135:182–190. doi: 10.1016/j.biortech.2012.10.025. [DOI] [PubMed] [Google Scholar]
- Karakashev D, Thomsen AB, Angelidaki I. Anaerobic biotechnological approaches for production of liquid energy carriers from biomass. Biotechnol Lett. 2007;29:1005–1012. doi: 10.1007/s10529-007-9360-3. [DOI] [PubMed] [Google Scholar]
- Kloareg B, Demarty M, Mabeau S. Polyanionic characteristics of purified sulphated homofucans from brown algae. Int J Bio Macromol. 1986;8:380–386. doi: 10.1016/0141-8130(86)90060-7. [DOI] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Robert IN, Burt A, Koufopanos V, Tsai IJ, Bergman CM, Bensasson D, O’Kelly MJT, van Oudenaarden A, Barton DBH, Bailes E, Nguyen AN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XB, Zhang YM, Angelidaki I. Optimization of H2SO4-catalyzed hydrothermal pretreatment of rapeseed straw for bioconversion to ethanol: focusing on pretreatment at high solids content. Bioresour Technol. 2009;100:3048–3053. doi: 10.1016/j.biortech.2009.01.008. [DOI] [PubMed] [Google Scholar]
- McMillan JD, Boynton BL. Arabinose utilization by xylose-fermenting yeasts and fungi. Appl Biochem Biotechnol. 1994;45:569–584. doi: 10.1007/BF02941831. [DOI] [PubMed] [Google Scholar]
- Meinita MDN, Kang JY, Jeong GT, Koo HM, Park SM, Hong YK. Bioethanol production from the acid hydrolysate of the carrageenophyte Kappaphycus alvarezii (cottonii) J Appl Phycol. 2012;24:857–862. doi: 10.1007/s10811-011-9705-0. [DOI] [Google Scholar]
- Mukherjee V, Steensels J, Lievens B, Van de Voorde I, Verplaetse A, Aerts G, Willems KA, Thevelein JM, Verstrepen KJ, Ruyters S. Phenotypic evaluation of natural and industrial Saccharomyces yeasts for different traits desirable in industrial bioethanol production. Appl Microbiol Biotechnol. 2014;98:9483–9498. doi: 10.1007/s00253-014-6090-z. [DOI] [PubMed] [Google Scholar]
- Mussatto, Roberto Alternatives for detoxification of diluted-acid lignocellulosic hydrolysates for use in fermentative processes: a review. Bioresour Technol. 2004;93:1–10. doi: 10.1016/j.biortech.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Mutripah S, Meinita MDN, Kang JY, Jeong GT, Susanto AB, Prabowo RE, Hong YK. Bioethanol production from the hydrolysate of Palmaria palmata using sulfuric acid and fermentation with brewer’s yeast. J Appl Phycol. 2014;26:687–693. doi: 10.1007/s10811-013-0068-6. [DOI] [Google Scholar]
- Noseda MD, Tulio S, Duarte ME. Polysaccharides from the red seaweed Bostrychia montagnei: chemical characterization. J Appl Phycol. 1999;11:35–40. doi: 10.1023/A:1008098931931. [DOI] [Google Scholar]
- Passoth V, Tabassum MR, Nair HA, Olstorpe M, Tuikova I, Stahlberg J. Enhanced ethanol production from wheat straw by integrated storage and pre-treatment (ISP) Enzyme Microb Technol. 2013;52:105–110. doi: 10.1016/j.enzmictec.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Powell CD, Quain DE, Smart KA. The impact of brewing yeast cell age on fermentation performance, attenuation and flocculation. FEMS Yeast Res. 2003;3:149–157. doi: 10.1016/S1567-1356(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Quain DE, Box WG, Walton EF. An inexpensive and simple small-scale laboratory fermenter. Lab Pract. 1985;34:84. [Google Scholar]
- Roesijadi G, Jones S, Snowden-Swan L, Zhu Y. Macroalgae as a biomass feedstock: a preliminary analysis, PNNL 19944. Richland: Pacific Northwest National Laboratory; 2010. [Google Scholar]
- Ruyters S, Mukherjee V, Verstrepen KJ, Thielevein JM, Willems KA, Lievens B. Assessing the potential of wild yeasts for bioethanol production. J Ind Microbiol Biotechnol. 2015;42:39–48. doi: 10.1007/s10295-014-1544-y. [DOI] [PubMed] [Google Scholar]
- Santos M, Jiménez J, Bartolomé B, Gómez-Cordovés C, del Nozal MJ. Variability of brewer’s spent grain within a brewery. Food Chem. 2003;80:17–21. doi: 10.1016/S0308-8146(02)00229-7. [DOI] [Google Scholar]
- Singh A, Nigam PS, Murphy JD. Renewable fuels from algae: an answer to debatable land based fuels. Bioresour Technol. 2011;102:10–16. doi: 10.1016/j.biortech.2010.06.032. [DOI] [PubMed] [Google Scholar]
- Taherzadeh MJ, Karimi K. Pretreatment of lignocellulosic wastes to improve ethanol and biogas production: a review. Int J Mol Sci. 2008;9:1621–1651. doi: 10.3390/ijms9091621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao N, Gao Y, Liu Y. Isolation and characterization of a Pichia anomala strain: a promising candidate for bioethanol production. Braz J Microbiol. 2011;42:668–675. doi: 10.1590/S1517-83822011000200031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CNS, Kim PB, Cooper SR, Raisner RM, Herman A, Sivitz AB, Lakshamanaswamy A, Kashiyama Y, Baker D, Yoshikuni Y. An engineered microbial platform for direct biofuel production from brown macroalgae. Science. 2012;335:308–313. doi: 10.1126/science.1214547. [DOI] [PubMed] [Google Scholar]
- Wilkinson M (1995) Information review on the impact of kelp harvesting. Scottish Natural Heritage
- Wimalasena TT, Greetham D, Marvin ME, Liti G, Chandelia Y, Hart AJ, Louis EJ, Phister TG, Tucker GA, Smart KA. Phenotypic characterisation of Saccharomyces spp. yeast for tolerance to stresses encountered during fermentation of lignocellulosic residues to produce bioethanol. Microb Cell Fact. 2014;13:47. doi: 10.1186/1475-2859-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa M, Nakamura K, Ariga O, Nakasaki K. Production of high concentrations of bioethanol from seaweeds that contain easily hydrolyzable polysaccharides. Process Biochem. 2011;46:2111–2116. doi: 10.1016/j.procbio.2011.08.001. [DOI] [Google Scholar]
- Yanik J, Stahl R, Troeger N, Sinag A. Pyrolysis of algal biomass. J Anal Appl Pyrolysis. 2013;103:134–141. doi: 10.1016/j.jaap.2012.08.016. [DOI] [Google Scholar]
- Zhao X, Bai F. Mechanisms of yeast stress tolerance and its manipulation for efficient fuel ethanol production. J Biotechnol. 2009;144:23–30. doi: 10.1016/j.jbiotec.2009.05.001. [DOI] [PubMed] [Google Scholar]