Abstract
We report the first microdeletion (26 kb) of the biotinidase gene (BTD) that involves three of the four exons of the gene. This deletion further exemplifies the importance of performing microarray analysis or other methodologies for a deletion of the BTD gene when the enzymatic activity indicates lower activity than can be attributed to the mutations identified by DNA sequencing.
Keyword: Biotinidase, Biotinidase deficiency, Microdeletion, Deletion, Mutation, Microarray
1. Introduction
Biotinidase (EC 3.5.1.12) is the enzyme responsible for cleaving biocytin and recycling biotin from dietary protein-bound sources [1], [2]. Profound biotinidase deficiency (less than 10% of mean normal serum activity) (OMIM #253260) is an autosomal recessively inherited metabolic disorder [3]. Untreated individuals with profound biotinidase deficiency usually exhibit neurological and cutaneous symptoms with metabolic acidosis and organic aciduria [2], [3]. Symptoms of the disorder can be markedly improved or prevented with pharmacological doses of oral biotin. However, if treatment is delayed, once vision or hearing problems or developmental delays occur, they are usually irreversible [3]. All states in the United States and many countries screen their newborns for the disorder.
The gene encoding biotinidase (BTD) has been isolated and characterized [4], [5] and over 150 mutations causing biotinidase deficiency have been identified [6]. We now report the first microdeletion of BTD that involves three of the four exons of the gene. This deletion further exemplifies the importance of performing microarray analysis or other methodologies for a deletion of BTD when the enzymatic activity indicates lower activity than can be attributed to the mutations identified by DNA sequencing.
2. Material and methods
Microarray analysis was performed by whole genome chromosome Prenatal Reveal SNP microarray (Integrated Genetics, LabCorp Specialty Testing Group).
DNA sequencing of the biotinidase (BTD) gene was performed by PCR amplification using primers and conditions described previously [7]. All exonic and intron-exon boundaries of the BTD gene were sequenced by Prevention Genetics (Marshfield, WI).
3. Case report and results
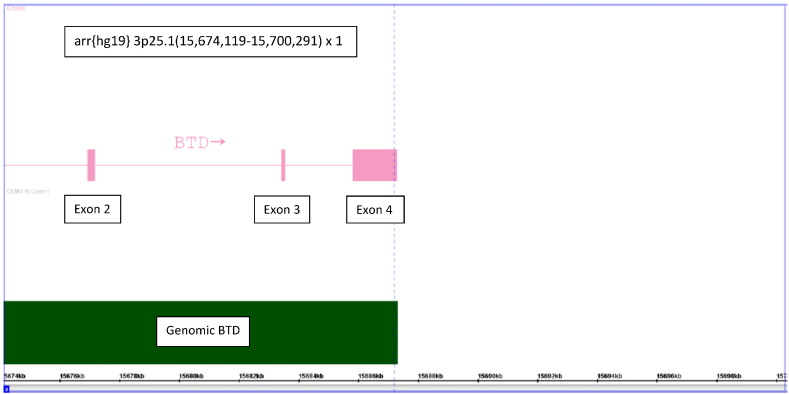
A non-consanguineous couple had prenatal diagnosis by microarray analysis for advanced maternal age. The mother is from India. The results of the microarray analysis revealed a 26 kb interstitial microdeletion of chromosome 3p25.1 → p25.1 (arr {hg19} 3p25.1 (15,674,119–15,700,291) × 1. This heterozygous deletion is involves a major portion of the BTD gene, including exons 2–4 (Fig. 1). Based on this information, it was important to determine if the baby had a mutation on the other allele causing biotinidase deficiency. To determine the likelihood that the baby had such a mutation, the parents had their serum biotinidase activities determined. The father's activity was 5.5 nmol/min/dl (range of normal activity is 5.7 to 8.7 nmol/min/dl) and the mother's activity was 2.9 nmol/min/dl. These results indicated that the father had normal activity and did not have a mutation of BTD and the mother had activity in the heterozygous range. If the microdeletion is inherited, then it is likely inherited from the mother. In fact, microarray analysis of the parents revealed that the mother did have the microdeletion. The fetal DNA was sequenced and did not reveal any other mutations or variants. At birth, the infant did not have biotinidase deficiency on newborn screening; however, the infant did not have serum enzymatic testing to confirm that her biotinidase activity was in the heterozygous range.
Fig. 1.
The 26 kb deletion of the child encompasses exons 2–4 of the BTD gene (pink). The normal genomic BTD gene is shown in green.
4. Discussion
We have previously reported a child with a contiguous gene deletion that involved three genes, including the BTD gene [8]. The child reported here is heterozygous for a microdeletion that only involves the BTD gene. This deletion involves three of the four exons of the BTD gene and is predicted to result in complete loss of biotinidase activity. This was confirmed by finding activity in the heterozygous range in the mother who also has the microdeletion.
The reference laboratory that performed the microarray analysis indicates that they report deletions as small as 50 kb and they may report susceptibility genes when they are associated with clinical presentations that have a clear phenotype. However, many commercial laboratories that perform microarray analyses do not report microdeletions or duplications of less than 200 to 400 kb, unless the alteration involves a gene known to cause a dominant pathogenic disorder. In the instance reported here, the deletion is only 26 kb and an alteration of the involved gene, BTD, which is only pathogenic as an autosomal recessive disorder. Therefore, it is possible that some or most laboratories would have not reported this deletion, unless they were specifically performing testing for biotinidase deficiency.
If, for example, an individual has biotinidase activity in the profoundly biotinidase deficient range and has a missense mutation on one allele and a deletion on the other, sequencing would have only identified the missense mutation. Only microarray analysis could identify the deletion of BTD explaining the reduced enzymatic activity. If this scenario occurs in an asymptomatic, profoundly enzyme deficient infant identified by newborn screening, it is imperative to reconcile the low enzymatic activity with finding only a single mutation.
There are occasions when enzymatic activity is lower than expected from mutation analysis. In these cases, the validity of the enzymatic data is usually questioned. The reduced activity is often attributed to poor sample storage [3]. It is precisely for this reason we have recommended that confirmatory enzymatic activities be performed on the proband, parents and an unrelated control [3], [9]. If there is confidence that the reduced enzymatic is not due to poor sample storage, then it is important to consider the possibility that a microdeletion is present on the second allele to explain the lower enzyme activity.
It is important to consider microarray analysis for a possible deletion in children identified as having enzymatic activity on newborn screening consistent with profound biotinidase deficiency, but only are found to have a single mutation by BTD sequencing. The possibility of a deletion involving part or all of the BTD genes must be considered in those children having enzymatic deficiency that is inconsistent with the results of their mutation analysis.
Acknowledgment
I thank Dr. Sainan Wei for generating the figure. This work was funded in part by the Safra Research Fund at Henry Ford Hospital.
References
- 1.Pispa J. Animal biotinidase. Ann. Med. Exp. Biol. Fenn. 1965;43(Suppl.5):1–39. [PubMed] [Google Scholar]
- 2.Wolf B. Disorders of biotin metabolism. In: Scriver C.R., Beaudet A.L., Sly W.S., Valle D., editors. The Metabolic and Molecular Bases of Inherited Disease. eighth ed. McGraw-Hill; New York: 2001. pp. 3935–3962. [Google Scholar]
- 3.Wolf B. Biotinidase deficiency: If you have to have an inherited metabolic disease, this is the one to have. Genet. Medicine. 2012;14:565–575. doi: 10.1038/gim.2011.6. [DOI] [PubMed] [Google Scholar]
- 4.Cole H., Reynolds T.R., Buck G.B., Lockyer J.M., Denson T., Spence J.E., Hymes J., Wolf B. Human serum biotinidase: cDNA cloning, sequence and characterization. J. Biol. Chem. 1994;269:6566–6570. [PubMed] [Google Scholar]
- 5.Knight H.C., Reynolds T.R., Meyers G.A., Pomponio R.J., Buck G.A., Wolf B. Structure of the human biotinidase gene. Mammal. Genome. 1998;9:327–330. doi: 10.1007/s003359900760. [DOI] [PubMed] [Google Scholar]
- 6.Procter M., Wolf B., Crockett D.K., Mao R. The biotinidase variants registry: a paradigm public database. Genes Genomics Genetics. 2013 doi: 10.1534/g3.113.005835. pii: g3.113.005835v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pomponio R.J., Hymes J., Reynolds T.R., Meyers G.A., Fleischhauer K., Buck G.A., Wolf B. Mutations in the human biotinidase gene that cause profound biotinidase deficiency in symptomatic children: molecular, biochemical and clinical analysis. Pediatr. Res. 1997;42:840–848. doi: 10.1203/00006450-199712000-00020. [DOI] [PubMed] [Google Scholar]
- 8.Senanayake D.N., Jasinge F.A., Pindolia K., Wanigasinghe J., Suchy S.F., Wei S., Jaysna S., Wolf B. First contiguous gene deletion causing biotinidase deficiency; the enzyme deficiency in three Sri Lankan children. Molecul. Genet. Metabol. Rep. 2016;2:81–84. doi: 10.1016/j.ymgmr.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf, B. Clinical issues and frequent questions about biotinidase deficiency. Mol. Genet. Metab. 100, 6-13. 20130. [DOI] [PubMed]