Abstract
Genome-wide association studies (GWAS) implicate the CYP17A1 gene in human blood pressure regulation although the causative polymorphisms are as yet unknown. We sought to identify common polymorphisms likely to explain this association. We sequenced the CYP17A1 locus in 60 normotensive individuals and observed 24 previously identified single nucleotide polymorphisms with minor allele frequency >0.05. From these, we selected for further study 7 polymorphisms located up to 2 kilobases upstream of the CYP17A1 transcription start site. In vitro reporter gene assays identified three of these (rs138009835, rs2150927 and rs2486758) as having significant functional effects. We then analysed association between the 7 polymorphisms and urinary steroid metabolites in a hypertensive cohort (n=232). Significant associations included that of rs138009835 with aldosterone metabolite excretion; rs2150927 associated with the ratio of tetrahydrodeoxycorticosterone to tetrahydrodeoxycortisol, which we employed as an index of 17α-hydroxylation. Linkage analysis showed rs138009835 to be the only one of the 7 polymorphisms in strong linkage disequilibrium with the blood pressure-associated polymorphisms identified in previous studies.
In conclusion, we have identified, characterised and investigated common polymorphisms at the CYP17A1 locus that have functional effects on gene transcription in vitro and associate with corticosteroid phenotype in vivo. Of these, rs138009835 – which we associate with changes in aldosterone level – is in strong linkage disequilibrium with polymorphisms linked by genome-wide association studies to blood pressure regulation. This finding clearly has implications for the development of high blood pressure in a large proportion of the population and justifies further investigation of rs138009835 and its effects.
Keywords: Steroid 17-alpha-Hydroxylase; Hypertension; Aldosterone; Polymorphism, Single Nucleotide; Genome-Wide Association Study; Blood pressure
Introduction
Hypertension is a major risk factor for cardiovascular and cerebrovascular disease. Risk of both increases with blood pressure, even within the normal range.1 Blood pressure has a substantial heritable component and, while polymorphic variations in a small number of genes have been shown to associate with blood pressure levels, much of this genetic component remains unidentified. Recently, the International Consortium for Blood Pressure genome-wide association study (GWAS), employing 200,000 individuals of European descent, identified 16 novel loci as being significantly associated with systolic blood pressure (SBP) and/or diastolic blood pressure (DBP),2 and also confirmed association at 12 loci reported by the CHARGE3 and Global BPgen4 consortia. Of these 28 loci, one on chromosome 10 encompasses a cluster of five genes forming a high linkage disequilibrium (LD) block spanning 347kb.5 This locus was flagged by all three GWAS; the most associated variant was rs1004467, located in intron 3 of CYP17A1, in the CHARGE study, while the most associated variant in the other two studies was rs11191548, which lies some 250kb distant from CYP17A1 at the intergenic region between CNNM2 and NT5C2 and is in high linkage disequilibrium (LD) with rs1004467 (r2=0.91 in CEU). In each of the studies, this locus was associated with a potential difference in SBP of ~1.1mm Hg. Subsequent replication of the GWAS findings in East Asian populations has increased the potential global impact of the CYP17A1 locus.6-9 A recent study has also associated rs11191548 with left ventricular mass index (LVMI) in hypertensive patients, which is suggestive of a role in cardiac hypertrophy.10 Rare genetic variants at CYP17A1 are already known to cause hypertension and so this locus represents a rare concurrence of blood pressure candidate gene studies with GWAS evidence.
The CYP17A1 gene is located on chromosome 10q24 and encodes a dual-function cytochrome P450 enzyme expressed primarily in the adrenal cortex, ovarian thecal cells and testes. In the adrenal zona fasciculata, this CYP17A1 enzyme functions as a 17α-hydroxylase, converting pregnenolone and progesterone to 17α-hydroxypregnenolone and 17α-hydroxyprogesterone, respectively.11 In the zona reticularis, it acts as a 17,20-lyase on these hydroxylated products, cleaving the C17,20 bond to produce dehydroepiandrosterone (DHEA) and androstenedione. The co-factor cytochrome b5 is an important determinant of the balance between these two enzymatic functions.12 Recently, the substantial heritability of 17α-hydroxylase activity in the general population has been demonstrated, as has its inverse correlation with adult blood pressure.13 Deficient 17α-hydroxylation has been known for almost 50 years to result in hypertension, with mutation of the CYP17A1 gene now estimated to account for approximately 1% of congenital adrenal hyperplasia (CAH) cases.14,15 Causative mutations usually occur in exons or at intronic splice sites, resulting in defective androgen production – which disrupts sexual development and maturation – and a markedly reduced ability to synthesise cortisol. The consequent sustained surge in adrenocorticotropic hormone (ACTH) production stimulates very high secretion of the weak mineralocorticoid, 11-deoxycorticosterone (DOC), causing hypertension, hypokalaemia and suppression of aldosterone synthesis. Simultaneously, high levels of corticosterone compensate for cortisol deficiency. A valuable biochemical indicator is therefore an abnormal excess of 17-deoxycorticosteroids (i.e. DOC and corticosterone) and lowered 17α-hydroxycorticosteroids (e.g. 17-OH-pregenenolone and 17-OH-progesterone).
We hypothesised that common CYP17A1 polymorphisms exist, which influence blood pressure and account for the significant association identified at this locus. In this study, therefore, we took the crucial next step of identifying common polymorphic variations within CYP17A1 and investigating their potential effects on biochemical function. We first characterised polymorphic frequency and distribution in normotensive individuals and investigated the effects of selected polymorphisms on gene transcription in vitro. We then tested for association between these candidate polymorphisms and intermediate corticosteroid phenotype in a large hypertensive cohort. As a result, we demonstrate that common CYP17A1 polymorphisms significantly alter gene function and correlate with changes in intermediate corticosteroid phenotype – including a significant effect on aldosterone levels – that could account for the strong association of this locus with population blood pressure variation.
Methods
Polymorphism discovery in normotensive subjects
Genetic polymorphisms at the CYP17A1 locus were identified by direct sequencing of genomic DNA from 60 West of Scotland volunteers (27 males, 33 females) recruited for a prior study.16 Participants were in good health and taking no antihypertensive medication. Median age was 51 years (interquartile range: 32–67years); median weight was 70kg (IQ range: 61–76kg); median SBP 126mm Hg (IQ range: 116–136mm Hg); median DBP was 77 mm Hg (IQ range: 70–84mm Hg). Ethical approval was granted by the West Glasgow Ethics Committee and written informed consent was obtained from all participants. Exons, introns and 3’UTR were each amplified separately by PCR using the Thermo-Start Taq DNA Polymerase PCR Enzyme Kit (Thermo Fisher Scientific, U.K.), while the upstream region was amplified using the Expand High Fidelity PCR System (Roche Diagnostics Ltd, Burgess Hill, U.K.), each according to standard kit protocol. Automated sequencing of PCR products was performed using BigDye Terminator v3.1 Cycle Sequencing chemistry (Life Technologies, U.K.) and the ABI 3730 DNA analyser (Life Technologies, U.K.). For further details, see Tables S2 & S3. Investigations were carried out in accordance with the principles of the Declaration of Helsinki. LD patterns were generated using Haploview software.17
Site-directed mutagenesis of CYP17A1 luciferase reporter vector
To create the pGL3-17 Control Vector, 2898bp of human genomic DNA immediately upstream of the CYP17A1 coding region was inserted into the empty pGL3-Basic vector (Promega UK Ltd., Southampton, U.K.), thereby fusing this sequence to a firefly luciferase reporter gene. To investigate polymorphic effects on CYP17A1 promoter activity, the pGL3-17 Control Vector was then mutated separately at each of 7 SNP positions using the QuikChange Site-Directed Mutagenesis standard kit protocol (Agilent Technologies U.K. Ltd, U.K.) and specific primers (Eurofins MWG Operon, Germany; see Table S4). The resulting 7 vectors were each used to transform JM109 competent cells (100μl; Promega, U.S.A) by heat shock and purified using the QIAprep Spin Miniprep kit (QIAGEN, Crawley, U.K.). Direct sequencing of the entire insert and flanking regions confirmed correct incorporation of the desired polymorphism in each of the 7 plasmids and the absence of additional unintended mutations.
Luciferase reporter gene assays
H295R cells (a kind gift from Professor William E. Rainey, University of Michigan) were grown in DMEM/F12 medium supplemented with 2.5% Ultroser G serum (Pall Bioscience, France), 1% insulin-transferrin-selenium (ITS) (BD Biosciences, U.K.) and 1% penicillin/streptomycin (1IU penicillin, 100μg/ml streptomycin; Life Technologies, U.K.) at 37°C, 5% CO2, and transfected with the pGL3-17 Control vector or one of its 7 mutated derivative vectors using siPORT™ NeoFX™ transfection agent (Life Technologies, U.K.) according to the manufacturer’s protocol, at a final cell density of 8×104 cells/well. A pGL4.73 (renilla luciferase; Promega, U.S.A) construct was co-transfected at a ratio of 50:1 to control for transfection efficiency. After 24 hours, transfectant was removed and replaced with complete medium, or complete medium containing 1mM dibutyryl cAMP, for 24 hours. The Dual Luciferase Reporter Assay system (Promega, U.S.A) was used to measure firefly and renilla luciferase activity in cell lysates containing 1x Passive Lysis Buffer, according to the manufacturer’s instructions, on a Lumat LB 9507 tube luminometer (Berthold Technologies, U.K.).
Genotype/phenotype associations in the hypertensive BRIGHT cohort
The MRC BRitish Genetics of HyperTension (BRIGHT) cohort is a large multicentre study, with Caucasian and British ancestry confirmed to grand-parental level for all participants. Data presented here relate to 232 unrelated and successfully genotyped hypertensive individuals drawn randomly from the BRIGHT sibling-pairs group (Table 1) for whom 24-hour urinary corticosteroid metabolite measurements, previously generated by gas chromatography/mass spectrometry (GC/MS),18 were available. Recruitment to the study required blood pressure values of 150/100 mmHg or higher, based on one reading, or 145/90 mmHg, as a mean of three readings, with onset of hypertension diagnosed before age 60 years in at least one sibling; subjects with BMI >30 were excluded.19 Ethical approval for the study was granted by the local ethics committees of the participating centres and fully informed written consent of the subjects was obtained. The 2.4kb upstream region was amplified using the Expand High Fidelity PCR System (Roche Diagnostics Ltd.), according to standard kit protocol. Automated sequencing of PCR products was performed using BigDye Terminator v3.1 Cycle Sequencing chemistry and the ABI 3730 DNA analyser (both Life Technologies). For further details, see Table S5. Measurement of urinary corticosteroid metabolites by gas chromatography/mass spectrometry (GC/MS) was conducted as previously described.18,20
Table 1.
Demographic and urinary corticosteroid data (median and interquartile ranges) for the BRIGHT study subgroup. Males and females were compared by the non-parametric Mann-Whitney test. SBP: systolic blood pressure; DBP: diastolic blood pressure; BMI: body mass index; WHR: waist:hip ratio; THB: tetrahydrocorticosterone; aTHB: allotetrahydrocorticosterone; THA: tetrahydro-11-dehydrocorticosterone; THF: tetrahydrocortisol; aTHF: allotetrahydrocortisol; THE: tetrahydrocortisone; DHEA: dehydroepiandrosterone; Aetio: aetiocholanolone; Andro: androsterone; THAldo: tetrahydroaldosterone.
| Urinary Steroid Metabolite (μg/24h) | All Subjects (n=232) | Males (n=106) | Females (n=126) | p-value (Male vs Female) |
|---|---|---|---|---|
| Age (years) | 63 (56-69) | 63 (56-68) | 64 (56-69) | 0.47 |
| SBP (mm Hg) | 157 (153-190) | 157 (151.25-187) | 181.5 (153-191) | 0.07 |
| DBP (mm Hg) | 103 (98-110) | 103 (98-110) | 102 (98-109.5) | 0.43 |
| BMI (kg/m2) | 27 (25-30) | 28 (25-30.75) | 27 (25-30) | 0.26 |
| WHR | 0.88 (0.81-0.93) | 0.93 (0.90-0.97) | 0.82 (0.78-0.86) | <0.001 |
| Corticosterone (Total B: THB+aTHB+THA) | 103 (61–188) | 140 (79–228) | 86.5 (54–156) | <0.001 |
| Cortisol (Total F: THF+aTHF+THE) | 1467 (759–2559) | 2081 (1130–3526) | 1118 (661–1989) | <0.001 |
| Androgens (DHEA+Aetio+Andro) Aetio+Andro) | 613 (322–1227) | 1008 (503–1852) | 447 (225–815) | <0.001 |
| Aldosterone (THAldo) | 3 (1–5) | 3 (2–6) | 2 (1–4) | 0.002 |
Statistics
In vitro experiments were performed in quadruplicate on four independent occasions (analysed as n=4), with data presented as the mean and standard error of the mean (SEM). pGL3-Basic and pGL3-17 Control construct activity (Figure 2A) was compared using one-way analysis of variance (ANOVA) and Bonferroni’s post-hoc tests on log-transformed values; mutated luciferase vector activity (Figure 2B and 2C) was analysed by one-sample Student’s t-test on log-transformed values. For analysis of the BRIGHT subjects, demographic and steroid excretion data were not normally distributed and were therefore analysed by the non-parametric Mann-Whitney U test. Comparisons of genotype with steroid excretion were conducted using the dominant model, where heterozygote and minor allele homozygote data are grouped and compared against major allele homozygote data. In all instances, 95% confidence intervals were generated and a P-value of <0.05 was set as the threshold for significance.
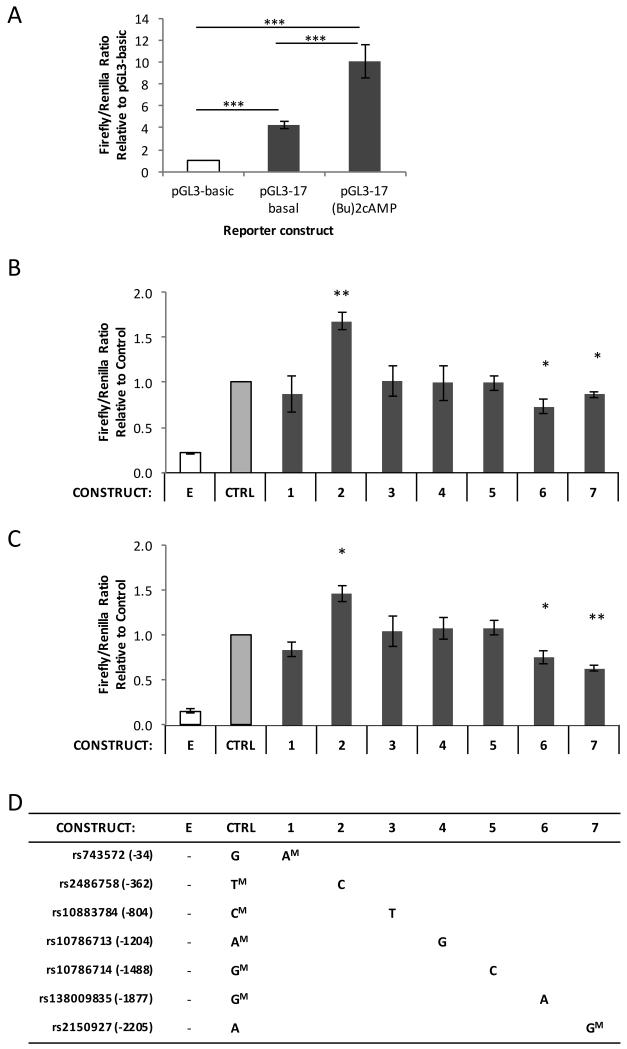
Figure 2.
(A): H295R cells were transfected with pGL3-Basic or pGL3-17 Control reporter constructs and grown 24 hours post-transfection under basal or 1mM dibutyryl cAMP-stimulated conditions. The transcriptional activity of each reporter construct is displayed as a proportion of pGL3-Basic (empty vector) activity (normalised to 1). (B,C): H295R cells were transfected with reporter constructs varying in sequence from the pGL3-17 Control vector (‘CTRL’) at a single base, then grown for 24 hours post-transfection under either basal (B) or 1mM dibutyryl cAMP-stimulated (C) conditions. The transcriptional activity of each reporter construct and of a pGL3-Basic empty construct (‘E’) is displayed as a proportion of pGL3-Control activity (normalised to 1). (D): Table of reporter construct genotypes; major alleles are indicated by a superscript ‘M’. Vector alleles match the control unless otherwise indicated. All data are expressed as the mean (+/−SEM) of four independent experiments (n=4), each performed in quadruplicate.
Results
Polymorphism Distribution Across the CYP17A1 Locus
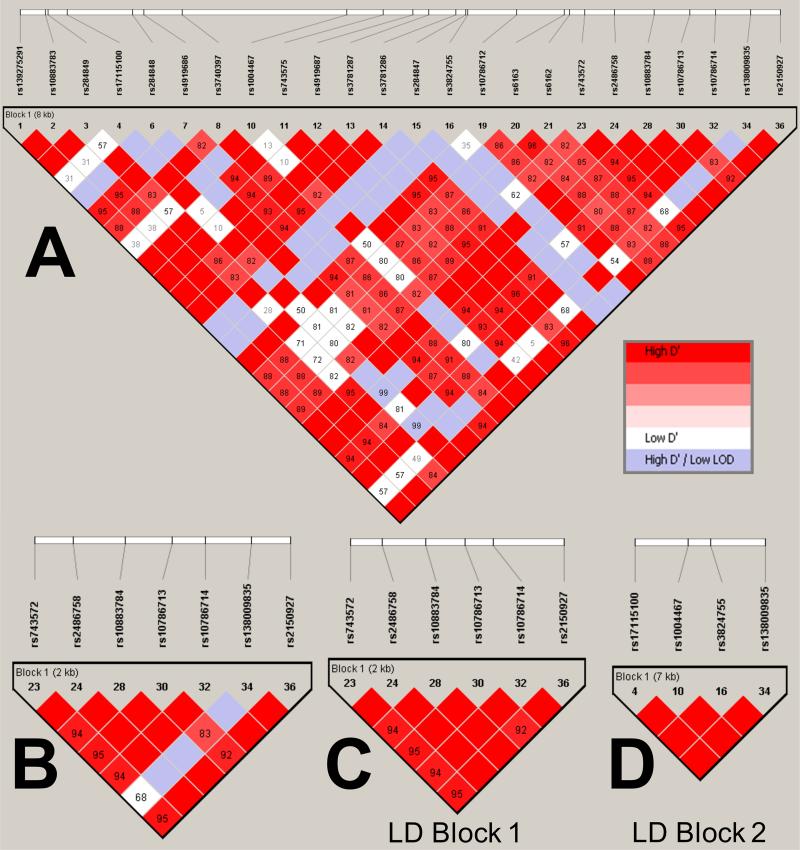
The entire CYP17A1 locus, including exons, introns and a 2kb region upstream of the transcription start site, was sequenced using genomic DNA sourced from 60 healthy Caucasian volunteers, identifying 36 polymorphisms with minor allele frequencies (MAF) ranging from 0.008 to 0.322 (Table S1). All polymorphisms within the coding region were synonymous and none was observed at intronic splice sites. Pairwise LD was analysed between the 24 most common polymorphisms (all MAF>0.05) using Haploview software (Figure 1A). We focused on seven common polymorphisms located upstream of the CYP17A1 coding region with the potential to influence gene transcription (Figure 1B). Analysis of pairwise LD between these SNPs has identified two distinct polymorphic blocks in this area, which we termed LD Block 1 and 2. LD Block 1 is comprised of six of the seven SNPs, which display a high degree of pairwise LD with one another (Figure 1C). LD Block 2 includes the remaining seventh SNP, rs138009835, which is less strongly linked to the other six but is in high LD with three further variants that span the CYP17A1 locus and include rs1004467, the intron 3 SNP associated with blood pressure in the CHARGE GWAS.3
Figure 1.
Pairwise linkage disequilibrium (LD) plots, generated using default settings in Haploview software, showing common polymorphisms (MAF>0.05) at the CYP17A1 locus in a cohort of 60 normotensive volunteers. The various LD plots show: (A) variants found across the entire human CYP17A1 locus; (B): the seven variants subjected to subsequent detailed analysis in vitro and in the BRIGHT study; (C): the six highly-linked variants comprising LD Block 1; (D) the four highly-linked variants comprising LD Block 2. D’ values (%) are displayed for each pair of SNPs in the intersecting square; squares not displaying a figure are D’=100%. Red squares indicate high D’ and a LOD score ≥2; blue squares indicate high D’ and LOD score <2; white squares indicate low D’ (≤80%) and LOD score ≥2.
Effect of Common Polymorphisms on CYP17A1 Transcription
The impact of the 7 upstream polymorphisms on CYP17A1 gene transcription was assessed in vitro through transfection of the H295R adrenocortical cell line with reporter constructs containing 2.9kb of the CYP17A1 5′ upstream region fused to a firefly luciferase reporter gene. Reporter gene activity was measured under basal conditions and following 24-hour stimulation with dibutyryl cAMP to mimic the intracellular activation of cAMP by ACTH. Dibutyryl cAMP caused a two- to threefold increase in the activity of the pGL3-17 Control Vector relative to basal (Figure 2A). Site-directed mutagenesis of this vector generated 7 further plasmids, each varying from the control sequence only at a single polymorphic base. Under basal conditions, 3 polymorphisms resulted in differential transcriptional activity: the minor C allele at position −362 (rs2486758) significantly increased transcriptional activity relative to the control T allele, while the minor A allele at position −1877 (rs138009835) and the major G allele at position −2205 (rs2150927) each reduced activity relative to their control forms (Figure 2B). The polymorphisms at the other four sites had no significant effect on transcription.
Incubation of H295R cells with dibutyryl cAMP stimulated transcription of all constructs relative to basal conditions. The same three polymorphisms resulted in significantly different transcriptional activity under these stimulated conditions as they had under basal (Figure 2C). While the magnitude of change relative to the control vector was similar under basal and stimulated conditions for the −362 (rs2486758) and −1877 (rs138009835) vectors, the differences between the alternative forms of −2205 (rs2150927) vector were more pronounced under stimulation.
Association of CYP17A1 polymorphisms with steroid phenotype in a hypertensive population
A subset of 232 hypertensive BRIGHT study subjects (106 males and 126 females) for whom urinary corticosteroid excretion data were available20 was genotyped for the 7 polymorphisms. Age, SBP, DBP and BMI did not vary significantly with genotype (Table S5) or with sex (Table 1), although levels of tetrahydroaldosterone (THAldo), total corticosterone metabolites (TotalB), total cortisol metabolites (TotalF) and total androgen metabolites (TotalAndrogen) were all significantly lower in females (all p<0.01; Table 1).
In order to provide sufficient power to identify significant associations between corticosteroid biosynthesis and CYP17A1 genotype, data were analysed using a dominant model, grouping heterozygotes with minor allele homozygotes for comparison against major allele homozygotes (Table 2). None of the 7 SNPs departed significantly from Hardy-Weinberg equilibrium and stratification of the study subjects by genotype revealed no association with key demographic characteristics, including blood pressure and BMI (data not shown). All significant associations identified between SNP genotype and steroid phenotype are detailed below.
Table 2.
Genotype data for selected single nucleotide polymorphisms upstream of CYP17A1. Genotypes were generated from a subset (n=232) of the hypertensive BRIGHT population. Major alleles are listed first and the given bases are from the forward strand sequence. The CYP17A1 location is relative to the first codon. HWE = Hardy Weinberg equilibrium.
| SNP | ID | CYP17A1 Location | Chromosomal Location | Alleles | Minor Allele Frequency | HWE (p) | % Genotyped |
|---|---|---|---|---|---|---|---|
| rs743572 | 1 | −34 | 10:104597152 | A/G | 0.420 | 0.9276 | 96.9 |
| rs2486758 | 2 | −362 | 10:104597480 | T/C | 0.183 | 0.9940 | 96.9 |
| rs10883784 | 3 | −804 | 10:104597922 | C/T | 0.304 | 0.7485 | 100.0 |
| rs10786713 | 4 | −1204 | 10:104598322 | A/G | 0.417 | 0.9121 | 99.6 |
| rs10786714 | 5 | −1488 | 10:104598606 | G/C | 0.304 | 0.6991 | 99.6 |
| rs138009835 | 6 | −1877 | 10:104598995 | G/A | 0.109 | 1.0000 | 100.0 |
| rs2150927 | 7 | −2205 | 10:104599323 | G/A | 0.413 | 0.9276 | 99.1 |
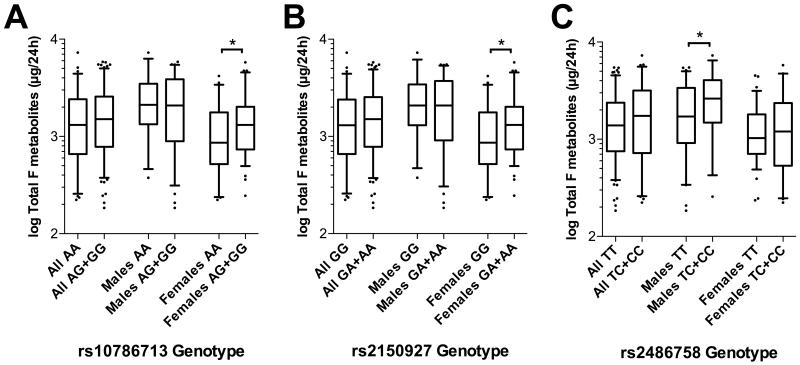
While none of the 7 SNPs associated with significant differences in TotalB, TotalAndrogen or TotalF metabolites when the study population was analysed as a whole, females homozygous for the major alleles at rs10786713 and at rs2150927 (A and G, respectively) had significantly lower levels of TotalF metabolites (each p<0.05) when analysed separately, as did males homozygous for the major T allele at rs2486758 (p<0.05 relative to heterozygotes only; the cohort contained no males homozygous for the minor allele at this locus; Figure 3).
Figure 3.
Box-whisker plots of 24-hour Total F metabolites in the BRIGHT study subgroup (N=232), stratified by (A) rs10786713 genotype, (B) rs2150927 genotype and (C) rs2486758 genotype. Total F (cortisol) metabolites is the sum of THF, aTHF and THE. Plots show the median within the interquartile range box, with whiskers extending to the 5th and 95th percentiles; data points beyond the whiskers are displayed as dots. Groups were compared by Mann-Whitney nonparametric test; *p<0.05.
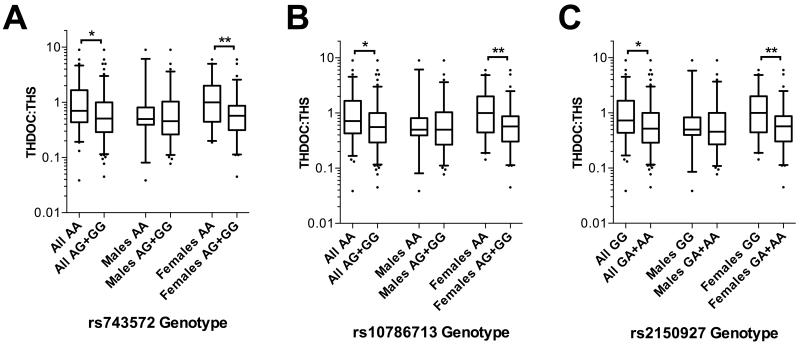
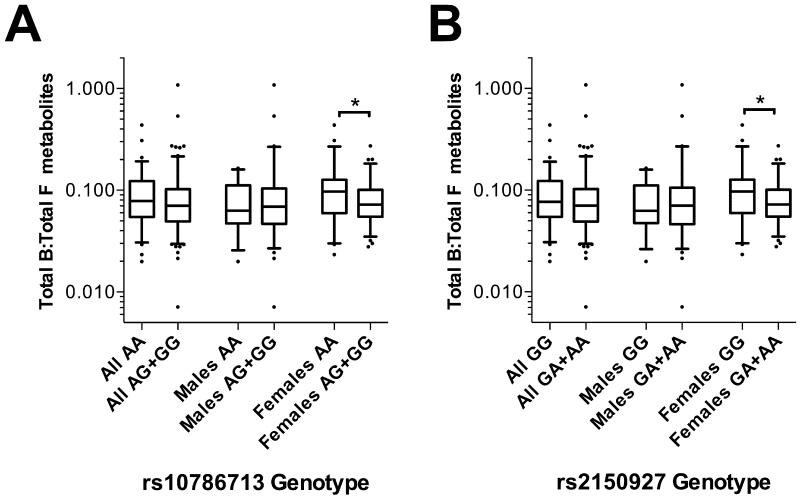
The ratios of tetrahydrodeoxycorticosterone to tetrahydrodeoxycortisol (THDOC:THS) and TotalB:TotalF serve as indices of 17α-hydroxylase activity. Subjects homozygous for the major alleles of rs743572, rs10786713 and rs2150927 all associated with a significantly higher THDOC:THS ratio (each p<0.05), suggesting less efficient 17α-hydroxylation (Figure 4). For each of these SNPs, this effect was more pronounced in females than in the total population (each p<0.01) and was not significant in males alone, when analysed separately (all p>0.05). Female subjects homozygous for the major forms of two of these SNPs – rs10786713 A and rs2150927 G – had a higher TotalB:TotalF ratio (p<0.05), again indicating less efficient hydroxylation; no such difference was detectable in males alone, or in the total cohort (Figure 5).
Figure 4.
Box-whisker plots of 24-hour THDOC:THS ratio in the BRIGHT study subgroup (N=232), stratified by (A) rs743572 genotype, (B) rs10786713 genotype and (C) rs2150927 genotype. Plots show the median within the interquartile range box, with whiskers extending to the 5th and 95th percentiles; data points beyond the whiskers are displayed as dots. Groups were compared by Mann-Whitney nonparametric test; *p<0.05, **p<0.01.
Figure 5.
Box-whisker plots of 24-hour Total B: Total F metabolites ratio in the BRIGHT study subgroup (N=232), stratified by (A) rs10786713 genotype and (B) rs2150927 genotype. Total B (corticosterone) metabolites is the sum of THB, aTHB and THA; Total F (cortisol) metabolites is the sum of THF, aTHF and THE. Plots show the median within the interquartile range box, with whiskers extending to the 5th and 95th percentiles; data points beyond the whiskers are displayed as dots. Groups were compared by Mann-Whitney nonparametric test; *p<0.05.
Ratios of THS:DHEA and TotalF:TotalAndrogen, selected as indices of 17,20 lyase activity showed no significant association with any of the 7 SNPs (data not shown).
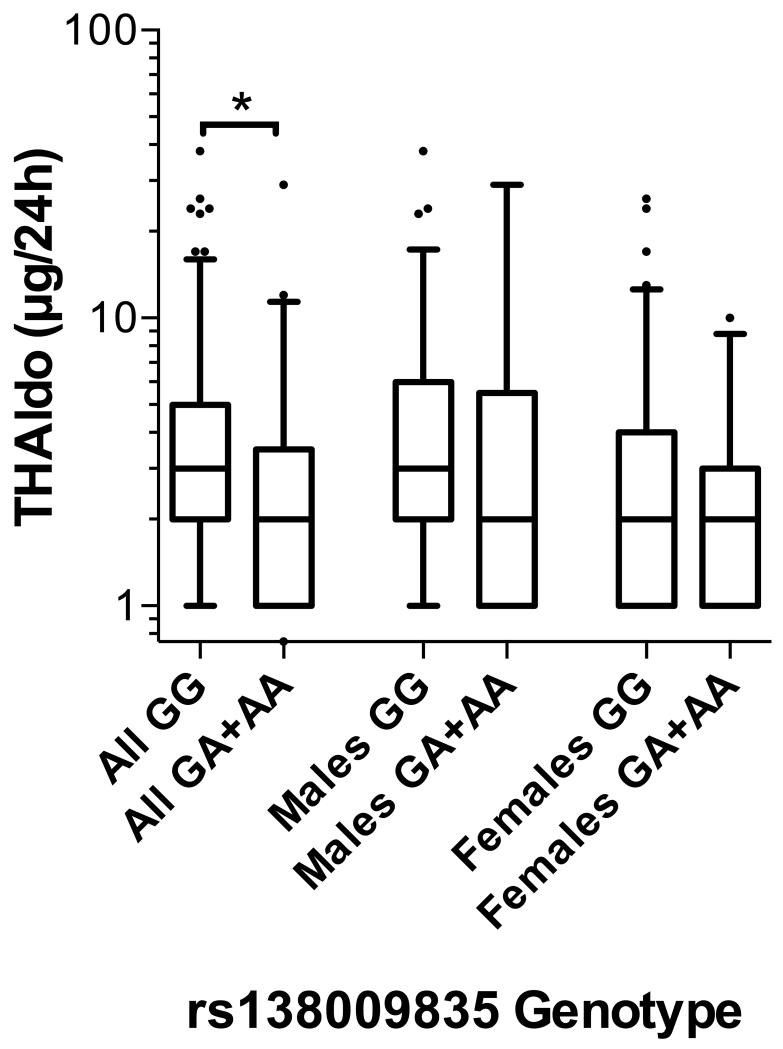
Major allele homozygotes for rs138009835 (GG genotype) had significantly higher levels of the urinary aldosterone metabolite THAldo (p<0.05) in comparison to combined heterozygote and minor allele subjects (Figure 6). In separate analyses of males and females, there was no significant difference in THaldo between these allele groups (p=0.14 and p=0.34, respectively).
Figure 6.
Box-whisker plot of 24-hour THaldo excretion rate in the BRIGHT study subgroup (N=232), stratified by rs138009835 genotype. Plot shows the median within the interquartile range box, with whiskers extending to the 5th and 95th percentiles; data points beyond the whiskers are displayed as dots. Groups were compared by Mann-Whitney nonparametric test; *p<0.05.
Discussion
Our systematic sequencing of a normotensive population identified 24 common (MAF>0.05) polymorphisms spanning the CYP17A1 locus. None is located within the coding region of CYP17A1 or at positions liable to disrupt mRNA splicing. Seven common SNPs located up to 2.2 kilobases from the transcription start site form two distinct and independent linkage blocks. Three of these SNPs disrupt in vitro CYP17A1 transcription: two from LD Block 1 (rs2486758 and rs2150927) and one from LD Block 2 (rs138009835). Previous reporter construct studies found that the 227 base pairs lying immediately upstream of the human CYP17A1 transcription start site (TSS) account for ~60–80% of basal transcriptional activity at that locus21; none of our seven common SNPs lies in that area (rs743572 lies between the TSS and the start codon of CYP17A1). Therefore, a great deal of the basal – not to mention cAMP-activated – transcriptional activity can be attributed to the region further upstream, where 6 of the common SNPs are found. Our own previous studies show that transcriptional activity of steroidogenic genes can be significantly altered by functional polymorphisms lying some 1500 to 2000 bases upstream of the TSS, which alter transcription factor binding affinity.16,18 We propose that the blood pressure association identified by GWAS at this locus is therefore most likely to result from one or more of the common SNPs that significantly alter in vitro transcription: rs2486758, rs138009835 and rs2150927.
Our analysis associates the major forms of the two functional polymorphisms found in LD Block 1, rs2486758 and rs2150927, with gender-dependent changes in corticosteroid excretion rate profiles in hypertensive individuals. (The LD Block 1 SNP, rs10786713, also shows the same association, although this had no functional effect in vitro.) These changes in phenotype are consistent with altered 17α-hydroxylase efficiency; the similarity of these effects together with the close linkage of these SNPs suggests a common underlying factor. There is no associated effect on aldosterone excretion rate. Conversely, the sole transcriptionally functional LD Block 2 SNP, rs138009835, shows no association with apparent 17α-hydroxylase efficiency, but is significantly associated with changes in aldosterone excretion rate. The influence of the LD Block 1 polymorphisms on steroid profile is consistent with their effects on CYP17A1 transcription in vitro: the major alleles rs2486758 T and rs2150927 G each reduce transcription relative to their alternative forms and associate with less efficient hydroxylation in vivo, as reflected in lower total cortisol metabolites for both SNPs as well as higher TotalB:TotalF and THDOC:THS ratios for rs2150927. The rs10786713 A allele has similar associations in vivo but no significant effect on transcription, implying that its association with steroid profile is the result of linkage to one or other of the functional Block 1 polymorphisms. On the basis of the observed steroid ratios, 17-lyase efficiency is unaffected by any of the seven analysed SNPs.
The altered 17α-hydroxycorticosteroid:17-deoxycorticosteroid ratios in subjects carrying the major alleles at selected SNPs imply that they will have higher ACTH drives in order to maintain normal cortisol levels. There is a clear gender difference in the steroid effects of Block 1 polymorphisms, with women tending to have the more altered intermediate phenotype. This may be related to the fact that the women in this cohort have a tendency to higher blood pressure relative to men, although this does not achieve significance (p=0.07; Table 1). Nevertheless, if impaired 17α-hydroxylase function results in increased blood pressure, it seems legitimate to conclude that such an effect is due – at least in part – to sustained changes in adrenal steroid profile. In classical 17α-hydroxylase deficiency, massive DOC excess causes an easily recognisable mineralocorticoid hypertension. Whether the small differences in the proportion of DOC in our population – even persisting over the course of a lifetime – could account for the small but significant blood pressure effects identified by GWAS is debatable, although the potency of DOC relative to aldosterone remains the subject of discussion.22 Alternatively, corticosterone could be responsible. In our study group, women carrying the major alleles at rs10786713 and rs2150927 had higher corticosterone:cortisol ratios. Previously, Soro et al found levels of corticosterone to be higher in subjects with hypertension.23 This may be related to the easier access corticosterone has to the brain when compared to cortisol;24 its concentration relative to cortisol in cerebrospinal fluid is much higher than in plasma25 and it may be preferentially retained in specific regions of the brain,26 where it occupies both MR and GR. Recently, Morris has argued that corticosterone is not merely a minor glucocorticoid subsidiary to cortisol but has distinct properties (e.g. higher mineralocorticoid activity and lower susceptibility to 11β-hydroxysteroid dehydrogenase types 1 and 2) that might cause it and its 5α-metabolites to affect blood pressure significantly.27
Of the 7 analysed SNPs, only rs138009835 is found in LD Block 2, and is therefore strongly linked to the blood pressure GWAS variants at this locus. It shows no association with our chosen indices of steroid 17α-hydroxylation efficiency in vivo but its major G allele – which causes increased CYP17A1 transcription in vitro – does associate with higher levels of aldosterone in a non-gender-dependent manner, unrelated to the effects of the LD Block 1 polymorphisms. Given that the zona glomerulosa does not express CYP17A1 and has no obvious direct interaction with the zona fasciculata, the influence of this SNP on aldosterone levels is not open to a simple explanation. Regardless of the precise mechanism, we demonstrate here that the rs138009835 G allele associates with increased CYP17A1 transcription in vitro and with raised aldosterone levels in vivo. Given its strong linkage (via LD Block 2) to the A allele of rs1004467 – itself significantly associated with increased blood pressure – and the critical role of aldosterone in blood pressure homeostasis, this finding clearly warrants further investigation.
This study had limitations and was not designed with the intention of detecting direct associations of CYP17A1 genotype with blood pressure. Given that the BRIGHT study subjects were all hypertensive and on various forms of antihypertensive therapy, the lack of association between blood pressure and any of the 7 SNPs – including rs138009835 – is therefore unsurprising. It is possible that antihypertensive treatments influenced steroid excretion, although previous analysis of the 512 BRIGHT subjects from which this subset was drawn found no evidence that these drugs systematically affected excretion rates of cortisol, aldosterone or androgens.20 Finally, this study did not adjust for multiple testing. As such there is a danger that some of the results deemed statistically significant may be false positives. However, for an exploratory study such as this, it is recognised that adjusting for multiple testing increases the chance that real differences will be missed and may not be advisable.28 Further investigation in a different study population is now recommended in order to confirm these findings.
Perspectives
Thorough analysis of the CYP17A1 locus in a control human population reveals a high degree of genetic variation, including two distinct LD blocks each containing common upstream SNPs. Several of these SNPs significantly affect in vitro gene expression and associate with in vivo steroid intermediate phenotype in a hypertensive population. Although this study was not designed with the intention of directly analysing blood pressure effects, our identification of the functional rs138009835 SNP might account for the significant blood pressure influence at this locus reported by multiple GWAS. The processes by which such alterations in CYP17A1 transcriptional regulation influence steroid profile and, ultimately, blood pressure require further investigation. If subsequent studies confirm CYP17A1 as a significant factor in population blood pressure variation, it has the potential to serve as a prominent target in the treatment and control of human hypertension.
Supplementary Material
Novelty and Significance.
What Is New?
Previous genome-wide association studies have linked the CYP17A1 locus with blood pressure variation.
This study presents the first evidence of common functional CYP17A1 gene polymorphisms. Of these, the transcriptional, phenotypic and linkage characteristics of the rs138009835 polymorphism suggest it could underlie the blood pressure associations identified at this locus.
What Is Relevant?
Genome-wide association studies have identified several loci as being significantly associated with blood pressure. However, very few of the molecular mechanisms underlying these associations have been identified.
This study provides functional evidence that could explain the known blood pressure associations identified at this region of human chromosome 10. The identification of blood pressure-related pathways affected by this locus is potentially of high clinical benefit.
This study also adds to the substantial evidence base highlighting the importance of adrenal steroid biosynthesis in the development of hypertension.
Summary
We report that common genetic polymorphisms at the CYP17A1 locus alter its expression in vitro and associate with changes in steroid level in vivo. One such variant, rs138009835, is in strong linkage disequilibrium with polymorphisms previously identified by genome-wide association studies as having significant influence on blood pressure. These associations of rs138009835 with transcriptional activity and intermediate steroid phenotype provide a plausible mechanism to explain the known blood pressure associations at this locus.
Acknowledgments
Sources of Funding: LAD was supported by a College of Medical, Veterinary and Life Sciences Medical Research Council Doctoral Training Grant Scholarship; FM and EMF were supported by Medical Research Council Fellowships; MJC and PBM acknowledge the National Institute for Health Research Cardiovascular Biomedical Research Unit at Barts; MJC is a National Institute of Health Research Senior Investigator.
Footnotes
Disclosures: MJC is Chief Scientist for Genomics England, a UK Government company.
References
- 1.Lewington S, Clarke R, Qizilbash N, Peto R, Collins R, Prospective Studies Collaboration Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360:1903–1913. doi: 10.1016/s0140-6736(02)11911-8. [DOI] [PubMed] [Google Scholar]
- 2.International Consortium for Blood Pressure Genome-Wide Association Studies. Ehret GB, Munroe PB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy D, Ehret GB, Rice K, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomez-Rubio P, Meza-Montenegro MM, Cantu-Soto E, Klimecki WT. Genetic association between intronic variants in AS3MT and arsenic methylation efficiency is focused on a large linkage disequilibrium cluster in chromosome 10. J Appl Toxicol. 2010;30:260–270. doi: 10.1002/jat.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Y, Lai X, Chen B, Xu Y, Huang B, Chen Z, Zhu S, Yao J, Jiang Q, Huang H, Wen J, Chen G. Genetic variations in CYP17A1, CACNB2 and PLEKHA7 are associated with blood pressure and/or hypertension in She ethnic minority of China. Atherosclerosis. 2011;219:709–714. doi: 10.1016/j.atherosclerosis.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Liu C, Li H, Qi Q, Lu L, Gan W, Loos RJ, Lin X. Common variants in or near FGF5, CYP17A1 and MTHFR genes are associated with blood pressure and hypertension in Chinese Hans. J Hypertens. 2011;29:70–75. doi: 10.1097/HJH.0b013e32833f60ab. [DOI] [PubMed] [Google Scholar]
- 8.Xi B, Shen Y, Reilly KH, Wang X, Mi J. Recapitulation of four hypertension susceptibility genes (CSK, CYP17A1, MTHFR, and FGF5) in East Asians. Metab Clin Exp. 2013;62:196–203. doi: 10.1016/j.metabol.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Hong K-W, Jin H-S, Lim J-E, Kim S, Go MJ, Oh B. Recapitulation of two genomewide association studies on blood pressure and essential hypertension in the Korean population. J Hum Genet. 2010;55:336–341. doi: 10.1038/jhg.2010.31. [DOI] [PubMed] [Google Scholar]
- 10.Huber M, Lezius S, Reibis R, Treszl A, Kujawinska D, Jakob S, Wegscheider K, Völler H, Kreutz R. A Single Nucleotide Polymorphism near the CYP17A1 Gene Is Associated with Left Ventricular Mass in Hypertensive Patients under Pharmacotherapy. Int J Mol Sci. 2015;16:17456–17468. doi: 10.3390/ijms160817456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller WL, Auchus RJ. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr Rev. 2011;32:81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller WL. Minireview: regulation of steroidogenesis by electron transfer. Endocrinology. 2005;146:2544–2550. doi: 10.1210/en.2005-0096. [DOI] [PubMed] [Google Scholar]
- 13.Ackermann D, Pruijm M, Ponte B, et al. CYP17A1 Enzyme Activity Is Linked to Ambulatory Blood Pressure in a Family-Based Population Study. Am J Hypertens. 2015 doi: 10.1093/ajh/hpv138. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biglieri EG, Herron MA, Brust N. 17-hydroxylation deficiency in man. J Clin Invest. 1966;45:1946–1954. doi: 10.1172/JCI105499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krone N, Arlt W. Genetics of congenital adrenal hyperplasia. Best Pract Res Clin Endocrinol Metab. 2009;23:181–192. doi: 10.1016/j.beem.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McManus F, Sands W, Diver L, MacKenzie SM, Fraser R, Davies E, Connell JM. APEX1 regulation of aldosterone synthase gene transcription is disrupted by a common polymorphism in humans. Circ Res. 2012;111:212–219. doi: 10.1161/CIRCRESAHA.111.262931. [DOI] [PubMed] [Google Scholar]
- 17.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 18.Barr M, MacKenzie SM, Friel EC, et al. Polymorphic variation in the 11beta-hydroxylase gene associates with reduced 11-hydroxylase efficiency. Hypertension. 2007;49:113–119. doi: 10.1161/01.HYP.0000249904.93940.7a. [DOI] [PubMed] [Google Scholar]
- 19.Caulfield M, Munroe P, Pembroke J, Samani N, Dominiczak A, Brown M, Benjamin N, Webster J, Ratcliffe P, O'Shea S, Papp J, Taylor E, Dobson R, Knight J, Newhouse S, Hooper J, Lee W, Brain N, Clayton D, Lathrop GM, Farrall M, Connell J, MRC British Genetics of Hypertension Study Genome-wide mapping of human loci for essential hypertension. Lancet. 2003;361:2118–2123. doi: 10.1016/S0140-6736(03)13722-1. [DOI] [PubMed] [Google Scholar]
- 20.Freel EM, Ingram M, Friel EC, Fraser R, Brown M, Samani NJ, Caulfield M, Munroe P, Farrall M, Webster J, Clayton D, Dominiczak AF, Davies E, Connell JMC. Phenotypic consequences of variation across the aldosterone synthase and 11-beta hydroxylase locus in a hypertensive cohort: data from the MRC BRIGHT Study. Clin Endocrinol (Oxf) 2007;67:832–838. doi: 10.1111/j.1365-2265.2007.02971.x. [DOI] [PubMed] [Google Scholar]
- 21.Lin CJ, Martens JW, Miller WL. NF-1C, Sp1, and Sp3 are essential for transcription of the human gene for P450c17 (steroid 17alpha-hydroxylase/17,20 lyase) in human adrenal NCI-H295A cells. Mol Endocrinol. 2001;15:1277–1293. doi: 10.1210/mend.15.8.0679. [DOI] [PubMed] [Google Scholar]
- 22.Sharma KK, Lindqvist A, Zhou XJ, Auchus RJ, Penning TM, Andersson S. Deoxycorticosterone inactivation by AKR1C3 in human mineralocorticoid target tissues. Mol Cell Endocrinol. 2006;248:79–86. doi: 10.1016/j.mce.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 23.Soro A, Ingram MC, Tonolo G, Glorioso N, Fraser R. Mildly raised corticosterone excretion rates in patients with essential hypertension. J Hum Hypertens. 1995;9:391–393. [PubMed] [Google Scholar]
- 24.Karssen AM, Meijer OC, van der Sandt IC, Lucassen PJ, de Lange EC, de Boer AG, de Kloet ER. Multidrug resistance P-glycoprotein hampers the access of cortisol but not of corticosterone to mouse and human brain. Endocrinology. 2001;142:2686–2694. doi: 10.1210/endo.142.6.8213. [DOI] [PubMed] [Google Scholar]
- 25.Raubenheimer PJ, Young EA, Andrew R, Seckl JR. The role of corticosterone in human hypothalamic-pituitary-adrenal axis feedback. Clin Endocrinol (Oxf) 2006;65:22–26. doi: 10.1111/j.1365-2265.2006.02540.x. [DOI] [PubMed] [Google Scholar]
- 26.McEwen BS, Weiss JM, Schwartz LS. Selective retention of corticosterone by limbic structures in rat brain. Nature. 1968;220:911–912. doi: 10.1038/220911a0. [DOI] [PubMed] [Google Scholar]
- 27.Morris DJ. Why do humans have two glucocorticoids: A question of intestinal fortitude. Steroids. 2015;102:32–38. doi: 10.1016/j.steroids.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 28.Saville DJ. Multiple Comparison Procedures: The Practical Solution. The American Statistician. 1990;44:174–180. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.