Abstract
We examined the phylogenetic relationships between species and genera within the caddisfly subfamily Drusinae (Trichoptera: Limnephilidae) using sequence data from two mitochondrial loci (cytochrome oxidase 1, large subunit rRNA) and one nuclear gene (wingless). Sequence data were analysed for 28 species from five genera from the subfamily. We analysed individual and combined data sets using a Bayesian Markov Chain Monte Carlo and a Maximum Parsimony approach and compared the performance of each partition for resolving phylogenetic relationships at this level. In terms of resolution and phylogenetic utility wingless outperformed the two mitochondrial gene partitions.
Using both Shimodaira-Hasegawa and expected likelihood weights tests we tested several hypotheses of relationships previously inferred based on adult morphological characters. The data did not support the generic concept, or many previously proposed species groupings, based on adult morphology. In contrast, the molecular data correlated with the morphology and feeding ecology of larvae. Using Bayesian ancestral character state reconstructions we inferred the evolution of feeding ecology and relevant larval morphological characters. Our analyses showed that within the subfamily Drusinae two derived feeding types evolved. One of these – grazing epilithic algae – is otherwise unusual in the Limnephilidae and may have promoted the high degree of diversity in the Drusinae.
Keywords: ancestral character state reconstruction, wingless, mtCOI, 16s, larval morphology
Introduction
Trichoptera (caddisflies), is the 7th largest insect order with over 13,000 described species, and the largest order of insects whose members are almost exclusively aquatic. It is the sister taxon to Lepidoptera, the moths and butterflies, but among other characters differs in that most larvae are aquatic. In contrast to the majority of other insects the larval stage of caddisflies is the most conspicuous and familiar to the non-entomologist because of the intricate portable cases and delicate silken nets the larvae construct (Wiggins, 2004). Like Lepidoptera caterpillars, Trichoptera larvae produce silk from the labium, and it is probably due to the diverse ways in which silk is used to exploit various aquatic niches that the order owes its evolutionary success (Mackay and Wiggins, 1978). The larvae of Trichoptera are found in all types of freshwater and even brackish aquatic habitats, but are especially abundant in rivers and streams. Caddisflies exhibit a wide range of feeding ecology. This includes shredding of leaf litter detritus, gathering fine organic particles, sucking algal cells, scraping periphyton off exposed surfaces, filtering the water of suspended food, preying on other aquatic invertebrates, or feeding on living green plants or algae. Through these diverse feeding strategies, caddisflies are fundamental participants in nutrient dynamics and energy flow in aquatic ecosystems (Resh and Rosenberg, 1984; Wallace and Webster, 1996). Despite their ecological importance and the diversity of feeding types, little is known about the evolution of their feeding ecology.
Biological monitoring of water quality depends heavily on caddisflies, especially in North America, Europe and Australia (Wright et al., 1984, 2000; Smith et al., 1999; Barbour and Yoder, 2000; Graf et al., 2002; Hering et al., 2006). The different sensitivity of caddisfly species is widely used for monitoring pollutants and other types of environmental disturbance (Rosenberg and Resh, 1993; Dohet, 2002), making caddisflies primary indicator taxa in monitoring water quality together with mayflies and stoneflies (Buffagni et al., 2006; Moog et al., 2004).
Despite the large number of species known, their unique and diverse life histories, and the important role caddisflies play in stream assessment, our knowledge about the evolutionary history of the group is limited (Morse, 1997). Few molecular phylogenies exist for caddisflies (Morse, 1997), and most of these studies have focussed on resolving the deeper level relationships between suborders or families (Kjer et al., 2001, 2002; Geerts et al., 2001; Dreesmann and Wichard, 2002). Few investigations have examined within family relationships (Myers and Sperling, 2002; Geraci et al., 2005) or intraspecific populations structure (Myers et al., 2001; Wilcock et al., 2001, 2003; Pauls et al., 2006). Thus, while the deep relationships and intraspecific population structure are becoming better understood, there is a significant lack of molecular based studies looking at interspecific relationships and genus level diversification in caddisflies.
The Drusinae Banks, 1916 is a subfamily of the Limnephilidae Kolenati, 1848 (Trichoptera). The group is restricted in its range to Eurasian mountain ranges from the Caucasus in the East to the Iberian Peninsula in the south-west. Most Drusinae are highland insects with a preference for cold running water. Despite its small range and the relatively narrow ecological niche, the group is highly diversified with 87 species known to date. Three quarters of the Drusinae are endemics limited to a single or very few mountain ranges, making the group an ideal model for studying recent evolution, diversification and speciation.
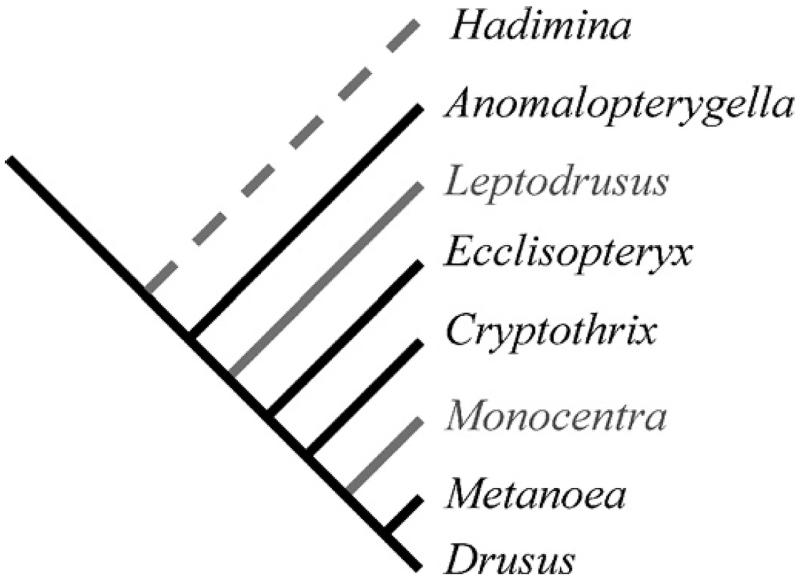
The last major treatment of the group was conducted by Schmid (1956). In his seminal work he described and characterised seven genera and six species groups within genus Drusus Stephens, 1837 based on adult morphology of the 42 Drusinae taxa known at the time. Based on character distribution in the group, Schmid (1956) proposed a phylogeny of the subfamily (Fig. 1). Since then a new genus and many new species have been described, more than doubling the total number of Drusinae known to 87 (Sipahiler, 2002; Malicky, 2005). Marinkovic-Gospodnetic (1976), Kumanski (1988), and Sipahiler (1999) have provided further summaries of species groupings, taking into account some newer species descriptions. Also, several recent studies have focussed on describing larval stages of Drusinae and identifying a variety of feeding strategies within the subfamily (Graf et al., 2005; Waringer et al., 2007a, 2007b, 2007c). Taken together these studies provide an ideal basis for testing hypotheses on relationships within and between genera and the trait evolution in these organisms.
Fig. 1.
Phylogenetic classification proposed by Schmid (1956) and Sipahiler (2002). Dotted branch shows inferred basal position of Hadimia following Sipahiler (2002). Grey branches indicate monotypic genera we could not sample for this study.
Our study has three main objectives. First, we want to provide the first multi-gene molecular phylogeny in caddisflies at the level of genera and species. Using a molecular phylogeny we will examine the evolutionary relationships within the Limnephilidae subfamily Drusinae and test existing hypotheses on the generic concept and specific relationships. We will also examine the phylogenetic utility of adult and larval morphology. Second, we want to reconstruct the evolution of feeding types in the subfamily using a coupled Bayesian/Maximum Likelihood approach (Pagel and Lutzoni, 2002) that allows more realistic reconstructions than Maximum Parsimony based methods. Third, we want to compare and evaluate the utility of three gene regions for reconstructing evolutionary relationships within caddisfly families and within and between genera to provide a basis for future phylogenetic studies of caddisflies. One of these regions (mtCOI) was previously used for studying intraspecific population structure and within family relationships. To date, 16S rRNA has not been tested in an extensive framework, and the third gene (wingless) has not been used in previous studies of caddisflies.
2. Materials and Methods
2. 1 Taxon sampling
A data matrix containing 53 specimens from 28 species of Drusinae was constructed with sequences from mitochondrial cytochrome oxidase I (mtCOI, 498 bp), 16S rRNA (mtLSU, 506 bp), and nuclear wingless (nuWG, 472bp) genes (Table 1). The data for this study were generated at the Research Institute Senckenberg (RIS), at the Pritzker Laboratory for Molecular Systematics and Evolution at The Field Museum (FM-PL) and by the Nano+Bio Zentrum Kaiserslautern University (NBZ). Five of the currently eight recognised genera in the Drusinae (Schmid, 1956; Sipahiler, 2002; Table 2) were included. We have not been able to get fresh material of the monotypic genera Hadimia, Leptodrusus, and Monocentra. Three species of one other Limnephilidae subfamily (Chaetopterygini, Stenophylacini) were included as outgroups.
Table 1.
Material used in this study. Localities are given with country code, mountain range, locality and collection date. Stage/Sex refers to L: larva; M: adult male; F: adult female. GenBank Accession codes are given for each taxon for each gene region used in the study.
| Taxon | Locality | Stage/Sex | mtCOI | mtLSU | nuWG | Collector |
|---|---|---|---|---|---|---|
| Anomalopterygella chauviniana | D, Spessart, Bieber ab. Rossbach, 13.07.2004 | L | EU215079 | EU215174 | EU215121 | Pauls & Sundermann |
| Chaetopterygopsis maclachlani | D, Black Forest, Brotenaubach, 04.05.2003 | L | EU215081 | EU215176 | EU215123 | Pauls |
| Chaetopteryx rugulosa | AT, Fischbacher Alps, Stiftingstalbach, 12.10.2006 | M | EU215083 | EU215178 | EU215125 | Graf |
| Consorophylax consors | CH, Alps, Furkapass, 15.10.2006 | M | EU215080 | EU215175 | EU215122 | Graf |
| Cryptothrix nebulicola | I, Bergamask Alps, San Marco Pass, 14.08.2000 | M | EU215082 | EU215177 | EU215124 | Graf |
| Drusus alpinus | CH, Alps, Furkapass, Sidelen tributary, 17.07.2004 | M | EU215084 | EU215179 | EU215126 | Lubini, Pauls & Sundermann |
| Drusus alpinus | CH, Alps, St. Gotthardt Pass, 21.07.2006 | M | EU215085 | EU215180 | EU215127 | Graf |
| Drusus annulatus | SK, Muranska Planina, Havranik tributary, 22.05.2003 | L | EU215086 | EU215181 | EU215128 | Blanar & Pauls |
| Drusus annulatus | D, Black Forest, Brotenaubach, 11.05.2006 | M | EU215087 | EU215182 | EU215129 | Sundermann |
| Drusus balcanicus | BG, Balkan Range, Zavodna River, 24.08.2003 | M | EU215088 | EU215183 | EU215130 | Beskov, Kumanski & Pauls |
| Drusus biguttatus | AT, Nockberge, St.Oswald Stream, 27.07.2006 | M | EU215089 | EU215184 | EU215131 | Graf, Pauls & Schmidt-Kloiber |
| Drusus botosaneanui | BG, Pirin Mts, Demyanishka River, 19.08.2003 | M | EU215090 | EU215185 | EU215132 | Kumanski & Pauls |
| Drusus brunneus | RO, Caliman Mts, Toplita, Lomas river, 29.07.2003 | L | EU215091 | EU215186 | EU215133 | Pauls & Ujvarosi |
| Drusus brunneus | RO, Hăşmaşu Mare Mts., Voşlăbeni, Sugó Cave | M | EU215092 | - | EU215134 | Balint |
| Drusus brunneus | RO, Apuseni Mts, Buscat springs, 03.08.2003 | L | - | EU215187 | - | Pauls & Ujvarosi |
| Drusus chrysotus | AT, Soboth, Krumbach tributary, 18.05.2002 | L | AY954395 | EU215188 | EU215135 | Graf & Pauls |
| Drusus chrysotus | AT, Soboth, Krumbach tributary, 18.05.2002 | L | - | - | EU215136 | Graf & Pauls |
| Drusus chrysotus | AT, Saualpe, Springs near Ladinger Hütte, 30.06.2006 | M | EU143739 | EU215189 | - | Graf, Pauls & Schmidt-Kloiber |
| Drusus destitutus | AT, Soboth, Krumbach tributary, 18.05.2002 | L | EU143738 | EU215193 | EU215140 | Graf & Pauls |
| Drusus destitutus | AT, Saualpe, Springs near Ladinger Hütte, 30.06.2006 | M | EU215096 | EU215194 | EU215141 | Graf, Pauls & Schmidt-Kloiber |
| Drusus discolor | RO, Retezat Mts, Rausor Valley, 08.08.2003 | L | EU215095 | EU215192 | EU215139 | Pauls & Ujvarosi |
| Drusus discolor | BG, Pirin Mts, Demyanishka River, 19.08.2003 | M | EU215093 | EU215190 | EU215137 | Beskov, Kumanski & Pauls |
| Drusus discolor | RO, Bucegi Mts, Pietra Alba, 05.08.2003 | F | EU215094 | EU215191 | EU215138 | Pauls & Ujvarosi |
| Drusus discophorus pallidus | BG, Pirin Mts, Banderishka River, 18.08.2003 | M | EU215097 | EU215195 | EU215142 | Beskov, Kumanski & Pauls |
| Drusus discophorus pallidus | BG, Pirin Mts, Banderishka River, 18.08.2003 | M | EU215098 | EU215196 | EU215143 | Beskov, Kumanski & Pauls |
| Drusus franzi | AT, Saualpe, 29.5.2006 | M | - | EU215197 | - | Graf |
| Drusus franzi | AT, Saualpe, 29.5.2006 | M | EU215099 | - | EU215144 | Graf |
| Drusus franzi | AT, Koralpe, Weinebene, 27.5.2006 | M | EU215100 | EU215198 | EU215145 | Graf |
| Drusus melanchaetes | CH, Alps, Meienreuss tributary, Sustenpass, 18.07.2004 | M | EU143740 | EU215199 | EU215146 | Lubini, Pauls & Sundermann |
| Drusus mixtus | CH, Jura, Dou springs near Cormoret, 17.04.2006 | L | EU215101 | EU215200 | EU215147 | Stucki |
| Drusus monticola | AT, Soboth, Krumbach tributary, 18.05.2002 | L | EF464556 | EU215201 | EU215148 | Graf & Pauls |
| Drusus monticola | AT, Saualpe, Springs near Ladinger Hütte, 15.6.2006 | F | EF464560 | EU215202 | EU215149 | Graf |
| Drusus muelleri | CH, Alps, Meienreuss tributary, Sustenpass, 18.07.2004 | M | AY954400 | EU215203 | EU215150 | Lubini, Pauls & Sundermann |
| Drusus muelleri | CH, Alps, Furkapass, Springs of Mutt tributary, 17.07.2004 | M | AY954398 | EU215204 | EU215151 | Lubini, Pauls & Sundermann |
| Drusus muelleri | CH, Alps, Grimselsee Zulauf, 18.07.2004 | M | AY954401 | EU215205 | EU215152 | Lubini, Pauls & Sundermann |
| Drusus nigrescens | CH, Alps, Furkapass, Springs of Mutt tributary, 17.07.2004 | M | EF464562 | EU215206 | EU215153 | Lubini, Pauls & Sundermann |
| Drusus nigrescens | CH, Alps, Furkapass 21.7.2006 | M | EF464565 | EU215207 | EU215154 | Graf |
| Drusus rectus | F, Pyrenees, Breche de Roland, 13.7.1999 | M | EU215105 | - | EU215158 | Lorenz |
| Drusus rectus | F, Pryenees, Cirque de Govarine, 14.07.1999 | F | - | EU215211 | - | Lorenz |
| Drusus romanicus | RO, Apuseni Mts, Buscat springs, 03.08.2003 | M | EU215102 | EU215208 | EU215155 | Pauls & Ujvarosi |
| Drusus romanicus | RO, Retezat Mts, Rausor Valley, 08.08.2003 | M | EU215103 | EU215209 | EU215156 | Pauls & Ujvarosi |
| Drusus romanicus | BG, Pirin Mts, Banderishka River, 18.08.2003 | M | EU215104 | EU215210 | - | Beskov, Kumanski & Pauls |
| Drusus romanicus | BG, Pirin Mts, Banderishka River, 18.08.2003 | M | - | - | EU215157 | Beskov, Kumanski & Pauls |
| Drusus trifidus | AT, Ennstaler Alps, Gesäuse, 02.07.2006 | M | EU215108 | EU215214 | EU215161 | Graf |
| Drusus trifidus | AT, Ennstaler Alps, Gesäuse, 02.07.2006 | M | EU215109 | EU215215 | EU215162 | Graf |
| Ecclisopteryx asterix | AT, Soboth, Krumbach tributary, 18.05.2002 | L | EU215111 | EU215217 | EU215164 | Graf & Pauls |
| Ecclisopteryx asterix | AT, Karawanken, Babniakgraben, 22.7.2006 | L | EU215110 | EU215216 | EU215163 | Graf |
| Ecclisopteryx dalecarlica | RO, Ţarcău, Poiana Mărului | F | EU215106 | EU215212 | EU215159 | Balint |
| Ecclisopteryx dalecarlica | RO, Ţarcău, Poiana Mărului | F | EU215107 | EU215213 | EU215160 | Balint |
| Ecclisopteryx dalecarlica | D, Spessart, Jossa below Sahlensee, 10.03.2003 | L | EU215112 | EU215218 | EU215165 | Lohse |
| Ecclisopteryx dalecarlica | D, Spessart, Jossa below Sahlensee, 10.03.2003 | L | EU215113 | EU215219 | EU215166 | Lohse |
| Ecclisopteryx guttulata | AT, Jogland, Lafnitz tributary, 24.05.2002 | M | EU215114 | EU215220 | EU215167 | Graf & Pauls |
| Ecclisopteryx madida | RO, Bucegi Mts, Pietra Alba, 05.08.2003 | M | EU215115 | EU215221 | EU215168 | Pauls & Ujvarosi |
| Ecclisopteryx madida | AT, Nockberge, St.Oswald Stream, 12.08.2006 | M | EU215116 | EU215222 | - | Graf |
| Ecclisopteryx madida | AT, Nockberge, St.Oswald Stream, 17.08.2006 | M | - | - | EU215169 | Graf |
| Ecclisopteryx malickyi | IT, Alto-Adige, Campo Rosso, 15.10.2006, | F | EU215117 | EU215223 | EU215170 | Graf |
| Metanoea flavipennis | CH, Val Münstair, 20.07.2006 | M | EU215118 | EU215224 | EU215171 | Graf |
| Metanoea rhaetica | AT, Nockberge, St.Oswald Stream, 01.07.2003 | M | EU215119 | EU215225 | - | Graf |
| Metanoea rhaetica | AT, Saualpe, Springs north of Offener Hütte, 30.06.2006 | M | - | - | EU215172 | Graf, Pauls & Schmidt-Kloiber |
| Metanoea rhaetica | AT, Nockberge, St.Oswald Stream, 01.07.2006 | M | EU215120 | EU215226 | EU215173 | Graf, Pauls & Schmidt-Kloiber |
Table 2.
Morphological characters of Drusinae genera. Sources: Rambur (1842), McLachlan (1880), Schmid (1956), Sipahiler (2002).
| Genus | Distinctive characters and selected synapomorphies in adults |
|---|---|
| Anomalopterygella |
|
| Cryptothrix |
|
| Drusus |
|
| Ecclisopteryx |
|
| Hadimia |
|
| Leptodrusus |
|
| Metanoea |
|
| Monocentra |
|
Most of the sequence data used in this study was generated from adult male specimens. Females or larvae were only used a) when sequences were also available from adult males, or b) in cases where no adult males were available, only for those taxa where female or larval stages are clearly recognised and easily delimited from other species (e.g. Drusus chrysotus larvae). The material for this study was collected by the authors and several other colleagues (Table 1) using water nets, sweeping nets or light traps. The nomenclature follows Malicky (2005).
2.2 Molecular Techniques
Whole genomic DNA was extracted from the abdomen or 2 legs from adults or larvae using the DNEasy Tissue or QIAmp Micro Kits (both Qiagen) following the manufacturer’s protocol. Cleared genitalia, remaining legs, head, thorax and wings were kept as specimen vouchers.
PCR mixes and procedures varied for each target region. PCR primers and procedures for mtCOI are described in Pauls et al. (2006). PCR primers were LR-J-12887 (5′-CGCCTGTTTATCAAAAACAT-3′) and LR-N-13398 (5′-CCGGTCTGAACTCAGATCACGT-3′) (both Simon et al., 1994) for mtLSU, and Wingnut1a (5′-GAAATGCGNCARGARTGYAA-3′) and Wingnut3 (5′-ACYTCRCARCACCARTGRAA-3′) (Goldstein, unpublished) for nuclear wingless. PCR mixes for mtLSU (New England Biolabs) contained 2.5μl 10x standard PCR buffer, 0.2μM dNTPs, 0.8μM of each PCR primer, 1mM MgCl, 5μg BSA, 1U Taq-polymerase and 4μl undiluted DNA in 25μl. The amplification program included 35 cycles of 95 °C for 45s, 52 °C for 45s, and 72 °C for 80s. PCR mixes for nuclear wingless (Roche) contained 2.5μl 10x standard PCR buffer, 0.2μM dNTPs, 1.6μM of each PCR primer, 1.5 mM MgCl, 2.5μl BSA (New England Biolabs), 0.4μl Taq-polymerase and 4μl undiluted DNA in 25μl. The amplification program included 35 cycles of 95 °C for 45s, 60°C for 45s, and 72 °C for 90s.
Purified PCR products were sequenced using the PCR primers on a ABI 3730XL capillary sequencer (Applied Biosystems) at FM-PL or an ABI 3100 at NBZ. Sequences were edited in Seqman II 4.0 (DNAStar).
2.3 Phylogenetic analysis
Sequences were aligned using Clustal W (Thompson et al., 1994) as implemented in BioEdit (Hall, 1999) and manually edited. All individual and combined data sets were analysed using Bayesian (B/MCMC) and maximum parsimony (MP) methods. Analyses were performed in Paup* 4.0b10 (Swofford, 2001) and MrBayes 3.1 (Ronquist and Huelsenbeck, 2003) with gaps treated as missing data.
For all MP analyses, a heuristic search with 100 random taxon addition replicates was conducted with TBR branch swapping and MULTREES option in effect, MAXTREES set to autoincrease, equally weighted characters and gaps treated as missing data. Robustness of individual branches was estimated by maximum parsimony bootstrap proportions (BP) (Felsenstein, 1985) following Sung et al. (2007). Nonparametric bootstrap support values were obtained with 100 bootstrap replicates, each with five replicates of random sequence addition, TBR branch swapping, MULTREES off and with a maximum of two trees saved per replicate. To assess homoplasy levels, consistency index (CI), retention index (RI), and rescaled consistency (RC) index (Farris, 1989) were calculated from each parsimony search.
Bayesian phylogenetic analyses were performed using the Markov chain Monte Carlo method (B/MCMC) and the model selected for each partition using Modeltest version 3.5 (Posada and Crandall, 1998) for single gene and combined data set analyses. Two parallel analyses with 12 chains each were run for 2*106 generations for single gene and two-gene partition data sets and 5*106 generations for the three gene combined data set. Trees were sampled every 100th generation. The first 1*106 generations were discarded as burn-in. We plotted the log-likelihood scores of sample points against generation time using TRACER 1.0 (http://evolve.zoo.ox.ac.uk/software.html?idDtracer) to ensure that stationarity was achieved after the first 1*106 generations by checking whether the log-likelihood values of the sample points reached a stable equilibrium plateau. From the remaining trees a majority-rule consensus tree with average branch lengths was calculated using the sumt option of MrBayes. Posterior probabilities were obtained for each clade.
We used a Bayesian approach to examine the heterogeneity in phylogenetic signal among the data partitions (Buckley et al., 2002). For the separate genes and the concatenated analyses, the set of topologies reaching 0.95 posterior probabilities were estimated. The combined analysis topology was then examined for conflict with the 0.95 posterior intervals of the single gene analyses. If no conflict was evident, it was assumed that the two data sets were congruent and could be combined.
2.4 Hypothesis Testing
We used the Shimodaira-Hasegawa (1999) (SH) test and expected likelihood weights test (ELW) (Strimmer and Rambaut, 2002) to evaluate whether our data are sufficient to reject alternative topologies using the combined data set. Such topologies, which may not be significantly worse than the obtained topology, might be present in suboptimal trees not sampled or not present in the 50%-majority rule consensus tree of the MCMC sampling. The following hypotheses were tested if they were not supported by the phylogenetic topology: 1) monophyly of the genera Ecclisopteryx (H1), Metanoea (H2), and Drusus (H3); 2) species groupings within Drusus (H4-H10), which primarily reflect adult genital morphology based on Schmid (1956), Marinkovic-Gospodnetic (1976), Kumanski (1988), and Sipahiler (1999) (Table 4, 5); 3) the generic concept proposed by Schmid (1956) (H11-H14) (Fig. 1, Table 4, 5). The SH and ELW tests were performed using Garli (Zwickl, 2006) and Tree-PUZZLE 5.2 (Schmidt et al., 2002) with the combined data set. Unconstrained and constrained maximum likelihood (ML) analyses were performed using Garli employing the GTR+I+G nucleotide substitution model. A pair of trees including the best tree agreeing with each of the null hypotheses, i.e. the constrained ML tree, and the unconstrained ML tree were compared in SH and ELW tests using the “user tree evaluation” option with accurate parameter estimation assuming the GTR model in Tree-PUZZLE 5.2 for each hypothesis (H1-H14).
Table 4.
Summary of interspecific nodes identified in single gene and combined analyses. Shown are B/MCMC posterior probabilities (before slash) and parsimony bootstrap support values (after slash). Nodes not present or unresolved in a certain data set are indicated by “-”.
| Clade | Species included in analyses | Hypothesis | mtCOI | mtLSU | wg | mtCOI+mtLSU | mtCOI+wg | mtLSU+wg | mtCOI+mtLSU+wg |
|---|---|---|---|---|---|---|---|---|---|
| Taxonomic groupings | |||||||||
| Drusinae | 1.00 / 77 | 1.00 / 100 | 1.00 / 100 | 1.00 / 99 | 1.00 / 100 | 1.00 / 100 | 1.00 / 100 | ||
| Chaetopterygini | - / 50 | 0.97 / 65 | 1.00 / 100 | 0.74 / - | 0.98 / 83 | 1.00 / 100 | 1.00 / 83 | ||
| Ecclisopteryx dalecarlica+E. guttulata | H1 | 1.00 / 100 | 0.53 / - | 0.68 / - | 1.00 / 93 | 1.00 / 91 | 0.86 / 61 | 1.00 / 93 | |
| Ecclisopteryx dalecarlica+E. guttulata+E. madida | H1 | - / 57 | 0.58 / 53 | 0.68 / - | 0.88 / 84 | - / - | 0.80 / 59 | - / 64 | |
| Ecclisopteryx asterix+E. malickyi | H1 | - / - | - / - | 0.98 / 66 | - / - 1 | 1.00 / - | 0.97 / 69 | 1.00 / - | |
| Metanoea spp. | rhaetica | H2 | 0.71 / - | - / - | 0.95 / 71 | - / - | 1.00 / 69 | 0.74 / 64 | 1.00 / 80 |
| flavipennis | |||||||||
| Drusus spp. | all | H3 | - / - | - / - | - / - | - / - | - / - | - / - | - / - |
| D. discolor-group | discolor | H4 | - / - | - / - | - / - | - / - | - / - | - / - | - / - |
| chrysotus | |||||||||
| destitutus | |||||||||
| D. muelleri-group | muelleri | H5 | - / - | - / - | - / - | - / - | - / - | - / - | - / - |
| romanicus | |||||||||
| D. annulatus-group | annulatus | H6 | - / - | - / - | - / - | - / - | - / - | - / - | - / - |
| rectus | |||||||||
| botosaneanui | |||||||||
| D. mixtus-group | mixtus | H7 | - / - | - / - | - / - | - / - | - / - | - / - | - / - |
| biguttatus | |||||||||
| brunneus | |||||||||
| trifidus | |||||||||
| D. alpinus-group | alpinus | H8 | - / - | 0.56 / - | 1.00 / 100 | - / - | 1.00 / 87 | 1.00 / 100 | 1.00 / 94 |
| franzi | |||||||||
| D. discophorus-group | d. pallidus | H9 | 1.00 / 100 | 1.00 / 89 | 1.00 / 100 | 1.00 / 100 | 1.00 / 100 | 1.00 / 100 | 1.00 / 100 |
| balcanicus | |||||||||
| D. bosnicus-group | monticola | H10 | 1.00 / 100 | 0.71 / - | 1.00 / 90 | 1.00 / 100 | 1.00 / 100 | 1.00 / 98 | 1.00 / 100 |
| nigrescens | |||||||||
| Ecclisopteryx+Cryptothrix+Drusus+Metanoea | all | H11 | - / - | - / - | - / - | - / - | - / - | - / - | - / - |
| Cryptothrix+Drusus+Metanoea | all | H12 | - / - | - / - | - / - | - / - | - / - | - / - | - / - |
| Drusus+Metanoea | all | H13 | - / - | - / - | - / - | - / - | - / - | - / - | - / - |
| Metanoea outside Drusus | all | H14 | - / - | - / - | - / - | - / - | - / - | - / - | - / - |
| Feeding ecology gropuings | |||||||||
| Filtering Carnivores | - / - | - / - | - / - | - / - | 0.91 / 55 | - / - | 0.88 / 75 | ||
| Filtering Carnivores within genus Drusus | - / - | - / - | 0.75 / 82 | - / 52 | 0.99 / 55 | 0.64 / 73 | 0.99 / 86 | ||
| Omnivorous generalists | - / - | 0.56 / - | 1.00 / 100 | - / - | 1.00 / 87 | 1.00 / 100 | 1.00 / 94 | ||
| Epilithic grazers | - / - | - / - | 1.00 / 100 | - / - | 1.00 / 94 | 1.00 / 100 | 1.00 / 100 |
Table 5.
Alternative topology hypotheses tested with ELW-Test and SH-Test.
| Hypothesis | ELW | SH | |||
|---|---|---|---|---|---|
| result | c | result | p | ||
| monophyly of… | 0.185 | 0.979 | |||
| H1 | Ecclisopteryx | rejected | <0.01 | rejected | <0.01 |
| H2 | Metanoea | ||||
| H3 | Drusus | rejected | <0.01 | rejected | <0.01 |
| H4 | D. discolor-group | rejected | <0.01 | rejected | <0.01 |
| H5 | D. muelleri-group | rejected | 0.015 | 0.518 | |
| H6 | D. annulatus-group | rejected | <0.01 | 0.531 | |
| H7 | D. mixtus-group | rejected | <0.01 | 0.178 | |
| H8 | D. alpinus-group | 0.199 | 0.995 | ||
| H9 | D. discophorus-group | 0.199 | 0.995 | ||
| H10 | D. bosnicus-group | 0.199 | 1.0 | ||
| H11 | Ecclisopteryx + Cryptothrix + Drusus + Metanoea | rejected | <0.01 | 0.073 | |
| H12 | Cryptothrix + Drusus + Metanoea | rejected | 0.019 | rejected | <0.01 |
| H13 | Drusus + Metanoea | rejected | <0.01 | rejected | <0.01 |
| H14 | Metanoea outside Drusus | rejected | <0.01 | rejected | <0.01 |
2.5 Ancestral Character State Reconstructions
We used BayesMultiState (Pagel et al., 2004) as implemented in BayesTraits (http://www.evolution.rdg.ac.uk) to reconstruct how traits relevant to the feeding ecology of the Drusinae evolved on the phylogenetic inference. This Bayesian approach estimates ML rates of character change and ancestral character states and incorporates uncertainty about the process of character change and the phylogeny by using a Bayesian tree sampling (Huelsenbeck et al., 2000; Pagel et al., 2004). Ancestral state posterior probabilities for a given node were estimated by multiplying the mean ancestral character state probability at that node across all trees by the portion of the trees in which that node was found (Pagel et al., 2004). The analysis was performed on 2000 trees taken every 1000th generation from the last 1*106 generations from both of the combined-data B/MCMC runs to ensure independence of successive trees.
3. Results
3.1 Molecular data and utility of individual gene partitions
To evaluate the relationships within the Drusinae 151 new sequences were obtained for this study (45 mtCOI, 53 mtLSU, 53 nuWG) (Table 1). We summarised sequence and tree characteristics for single-gene and combined data sets in Table 3. Variability and number of parsimony informative characters ranged between 21.74-34.74% (110-173 sites) and was highest in mtCOI, followed by nuWG and mtLSU respectively (Table 3). No significantly supported conflicts were observed between the three partitions when comparing the 95% majority rule consensus trees of single gene analyses. This is consistent with the hypothesis that all data partitions evolved along the same underlying topology. We thus combined data partitions to three two-gene and one three-gene data sets.
Table 3.
Comparison of performance of data partitions under maximum parsimony and in a Bayesian framework
| mtCOI | mtLSU | wg | mtCOI+mtLSU | mtCOI+wg | mtLSU+wg | mtCOI+mtLSU+wg | |
|---|---|---|---|---|---|---|---|
| Total Characters | 498 | 506 | 472 | 1004 | 970 | 978 | 1476 |
| Variable (%) | 173 (34.74) | 110 (21.74) | 145 (30.72) | 283 (28.19) | 328 (32.78) | 255 (26.07) | 428 (29.0) |
| Maximum Parsimony | |||||||
| Uninformative characters | 12 | 55 | 41 | 67 | 53 | 96 | 108 |
| Informative characters (%) | 161 (32.33) | 55 (10.87) | 104 (22.03) | 216 (21.51) | 265 (27.32) | 159 (16.26) | 320 (21.68) |
| Consistency Index CI | 0.2819 | 0.6543 | 0.6822 | 0.3373 | 0.3553 | 0.6528 | 0.3911 |
| Retention Index RI | 0.6246 | 0.7833 | 0.9085 | 0.6384 | 0.7035 | 0.8671 | 0.7078 |
| Rescaled consistency index RC | 0.1761 | 0.5125 | 0.6197 | 0.2153 | 0.2499 | 0.5660 | 0.2768 |
| Tree length | 965 | 188 | 258 | 1171 | 1261 | 458 | 1460 |
| No mp trees | 16 | 416 | 64 | 16 | 25 | 128 | 2 |
| Polyspecific nodes bootstrap ≥70 | 6 | 2 | 11 | 7 | 9 | 11 | 13 |
| Bayesian/MCMC | |||||||
| Selected model under hLRT | GTR+I+G | K81uf+I+G | TrN+G | GTR+I+G/K81uf+I+G | GTR+I+G/TrN+G | K81uf+I+G/TrN+G | GTR+I+G/K81uf+I+G/TrN+G |
| Polyspecific nodes pp ≥0.95 | 6 | 4 | 14 | 9 | 16 | 15 | 18 |
| Average distance between runs | 0.007708 | 0.026674 | 0.007544 | 0.016278 | 0.004448 | 0.011423 | 0.002051 |
| Log likelihood | −4635.113 | −1912.363 | −2346.681 | −6627.26 | −7158,902 | −4417.382 | −9156.271 |
| Tree length | 6.369 | 6.734 | 9.43 | 2.995 | 3.252 | 6.224 | 2.303 |
| Model parameter rate of change A/C | 0.0192 | 0.02339 | 0.04757 | 0.02699 | 0.03718 | 0.03693 | 0.03958 |
| Model parameter rate of change A/G | 0.448 | 0.731 | 0.312 | 0.412 | 0.238 | 0.381 | 0.273 |
| Model parameter rate of change A/T | 0.03424 | 0.03627 | 0.07971 | 0.04833 | 0.07508 | 0.102 | 0.06585 |
| Model parameter rate of change G/C | 0.03807 | 0.0803 | 0.07486 | 0.03345 | 0.04837 | 0.114 | 0.06314 |
| Model parameter rate of change C/T | 0.449 | 0.113 | 0.413 | 0.463 | 0.584 | 0.315 | 0.538 |
| Model parameter rate of change G/T | 0.01256 | 0.01591 | 0.07246 | 0.01619 | 0.01704 | 0.05166 | 0.0203 |
| Stationary nucleotide frequency of A | 0.355 | 0.411 | 0.234 | 0.356 | 0.279 | 0.322 | 0.32 |
| Stationary nucleotide frequency of C | 0.203 | 0.076 | 0.323 | 0.165 | 0.253 | 0.2 | 0.195 |
| Stationary nucleotide frequency of G | 0.082 | 0.097 | 0.268 | 0.104 | 0.189 | 0.193 | 0.164 |
| Stationary nucleotide frequency of T | 0.361 | 0.416 | 0.175 | 0.374 | 0.279 | 0.284 | 0.321 |
| Shape parameter α for gamma distribution of rate variation. | 0.911 | 0.129 | 0.119 | 0.169 | 0.499 | 0.144 | 0.229 |
| Proportion of invariable sites | 0.579 | 0.783 | 0.312 | 0.146 | 0.476 | 0.324 | 0.291 |
We summarised maximum parsimony (MP) data and indices in Table 3. In single gene analyses, levels of homoplasy were highest in mtCOI and lowest in nuWG. Combined gene partition analyses exhibited lowest levels of homoplasy, when mtCOI was not included (RC: 0.57). In analyses of the combined three-gene data set RC was relatively low (0.28). The number of most parsimonious trees found in single gene analyses decreased with number of variable sites. Only two shortest trees were found in analyses of the combined three-gene data set. The number of polyspecific clades (i.e. clades with two or more species) supported by bootstrap values ≥70, was highest for nuWG (11) and lowest for mtLSU (2) and increased in combined data sets. The highest number of highly supported clades was found in analyses of the combined three-gene data set (13).
We summarised likelihood parameters in the seven B/MCMC samples in Table 3. G/C content was highest in the nuWG partition (59.1%), while the mitochondrial genes showed a strong bias toward A/T (GC-content 29.1% for mtCOI, 17.3% for mtLSU). The gamma shape parameter α was similar in mtLSU (0.129) and nuWG (0.119), but much higher in mtCOI (0.911). The number of clades with more than one species that receive posterior probabilities ≥ 95% was highest in nuWG for single gene analyses (14) and lowest in mtLSU (4). In combined analyses, the lowest number of supported clades was found when combining the two mitochondrial genes (9), the highest number (18) was reached after combining all three partitions. After 2*106 generations, the average standard deviation of split frequencies between parallel runs was considerably higher in mtLSU than in nuWG or mtCOI or whenever mtLSU was combined with one of the other partitions. In analyses of the combined three-gene data set, however the deviation was relatively low (0.005881 after 2*106 generations).
3.2 Phylogenetic reconstructions
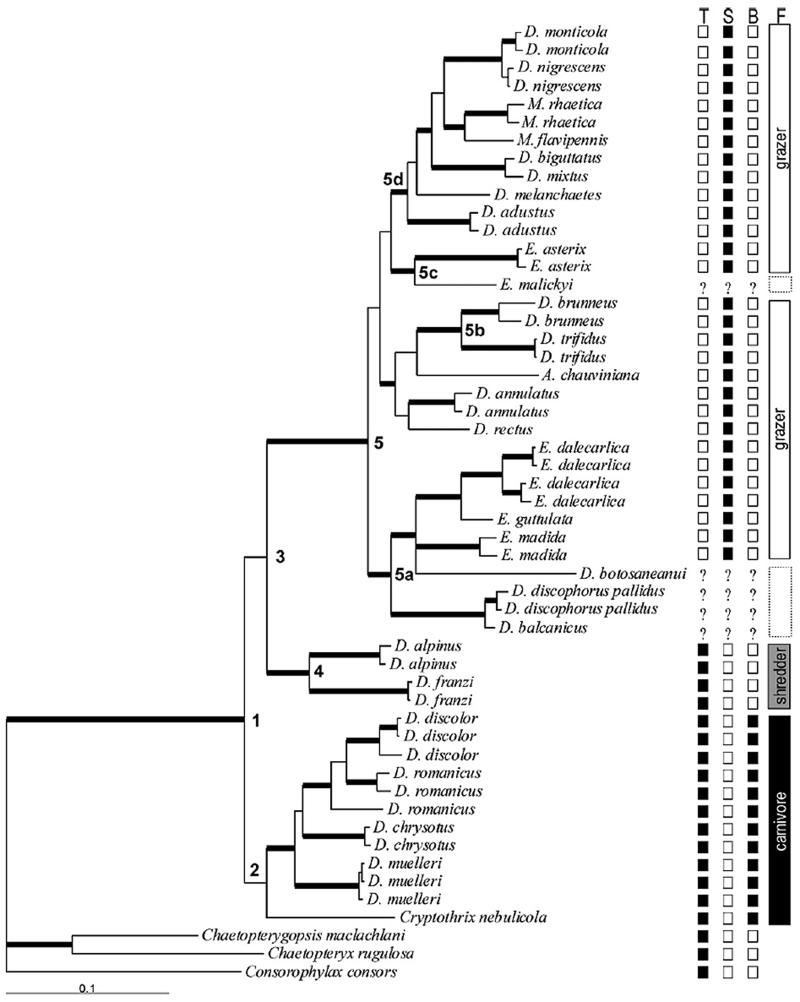
Phylogenetic relationships were investigated using single-gene, two-gene and a three gene region data set (mtCOI+mtLSU+nuWG). The summary of support for individual clades using individual gene partitions and the four possible combinations is given in Table 4. The combined three-gene data set had an aligned length of 1476bp with 428 variable positions (Table 3). Parsimony analysis of the three-gene data set yielded two most parsimonious trees (1460 steps, CI=0.39, RI=0.71, RC=0.28, Table 3). The two trees only differed in the order of specimens within Drusus muelleri. The topology of the most parsimonious trees differed slightly from the 50% majority-rule consensus tree obtained from the B/MCMC tree sampling in the position of clades within the epilithic grazers, but the clades themselves are the same (Fig. 2 and 3). There were no differences in the monophyly of and relationships within genera, species groups, or feeding ecology.
Fig. 2.
B/MCMC inference for 53 Drusinae taxa based on 1468 base pairs from three gene regions (mtCOI, mtLSU nuWG). Bold branch ends indicate posterior probabilities ≥ 0.95. Numbers on the nodes indicate clades referred to in the text. Shown is the 50% majority rule consensus tree from 80002 trees samples in two parallel runs with 11 heated and 1 cold chain each. Tree space was searched for 5000000 generations. Boxes on the right indicate mandible with teeth (T), spoon-shaped mandible without teeth (S), presence of filtering bristles (B) and feeding type (F). “?” indicates that the character state is unknown, because the larva remains unknown. Dotted feeding type boxes indicate that the feeding type is only predicted, as the the larva remains unknown.
Fig. 3.
One of two most parsimonious trees recovered in the MP analysis. Above nodes are bootstrap values above 70%. Bold numbers indicate the nodes referred to in the text. Other annotations as in Fig. 2.
Both the MP and B/MCMC trees from the three-gene combined analysis support monophyly of the subfamily Drusinae (pp=1.0; bs=100%) (Clade 1, Figs. 2, 3). All species where more than one specimens were analysed are monophyletic, although not always significantly supported (D. romanicus and D. nigrescens with pp below 0.95). Within the Drusinae, both topologies show a basal clade with Cryptothrix nebulicola, Drusus muelleri, D. romanicus, D. chrysotus and D. discolor (Clade 2, Figs. 2, 3). This clade, however, lacks strong support. Within this clade, C. nebulicola is basal to a highly supported clade (pp=0.99, bs=86) comprising members of the genus Drusus. Within this clade, D. muelleri and D. chrysotus are basal to D. discolor and D. romanicus.
The remaining species fall into two major clades. One clade (Clade 4, Figs. 2, 3). includes Drusus alpinus and D. franzi (pp=1.0, bs=100). Clade 4 is sister to a clade comprising the remaining Drusinae (Clade 5, Figs. 2, 3). Clade 5 falls into four supported smaller clades in the B/MCMC analysis, all of which are also recovered in the MP analysis, but not supported (Fig. 2, 3). Clade 5a (pp=1.0, bs=n.a.) comprises two well-resolved sister groups with Drusus balcanicus and D. discophorus pallidus (pp=1.0, bs=100%) and D. botosaneanui, Ecclisopteryx dalecarlica, E. guttulata and E. madida (pp=0.97, bs=58%), respectively. Clade 5b (pp=0.98, bs=n.a.) groups D. trifidus and D. brunneus. Sister to this clade is Anomalopterygella chauviniana but the relationship is not supported by high pp or bs scores. Drusus annulatus, and D. rectus form an unsupported clade which is basal to clades 5a and 5b in the MP topology and sister to 5b in the B/MCMC inference. The position of Anomalopterygella chauviniana, Drusus annulatus, and D. rectus within clade 5 remains uncertain, since their phylogenetic positions are not significantly supported. Clade 5c groups the other two Ecclisopteryx species, E. asterix and E.malickyi (pp=1.0, bs=n.a.). Clade 5d (pp=1.0, bs=n.a.) groups the remaining Drusus species. Within this clade, D. destitutus is basal to D. melanchaetes, a clade with D. monticola and D. nigrescens (pp=1.0, bs=100), the sister taxa D. mixtus and D. biguttatus (pp=1.0, bs=100), and genus Metanoea (pp=1.0, bs=80).
The current generic concept is not supported in our analysis. The genera Anomalopterygella, Cryptothrix, Ecclisopteryx and Metanoea are nested in Drusus. Ecclisopteryx is polyphyletic and falls into two separate well-supported clades. Within Drusus three groups distinguished by adult genital morphology were supported in our study: the Drusus alpinus-group, D. discophorus-group, and D. bosnicus-group. The remaining species groups within Drusus are not supported in our study (D. discolor-group, D. muelleri-group, D. annulatus-group, D. mixtus-group).
3.3 Hypothesis testing
The results of the SH and ELW tests for probabilities of alternative topologies are shown in Table 5. Monophyly of the genera Drusus (H3) and Ecclisopteryx (H1) is significantly rejected in both, as was the placement of Metanoea outside Drusus (H14). The original classification proposed by Schmid 1956 (Fig. 1) is rejected with both tests (H12, H13). The basal position of Anomalopterygella (H11) is not rejected significantly by both tests (SH, p=0.073). Within Drusus, monophyly of the discolor-group (H4) is significantly rejected in both tests. Following the ELW test the muelleri-group (H5), annulatus-group (H6), and mixtus-group (H7) are significantly rejected. However, in the SH test these three groups cannot be significantly rejected with the data at hand. Hypotheses H2 and H8-H10 could not be rejected in ELW or SH tests based on the data at hand.
3.4 Feeding ecology and ancestral character state reconstruction
With respect to larval mouthpart anatomy, three distinct species groupings exist in known Drusinae larvae: 1) carnivorous filterers, with teeth around the mandible edges, modifications of head capsules, additional spines on the legs, and long filtering bristles at the first abdominal sternum; 2) omnivorous generalists, with shredder mandibles with teeth around the edges but lacking additional spines and long bristles; 3) epilithic grazers, with spoon-shaped mandibles without teeth along the edges and additional fine setae on faces of femora (Waringer et al., 2007a). These morphologies and feeding types were found to be characteristic for each of the three major clades (Clades 2, 4, and 5) found in the Drusinae (Fig. 2, 3).
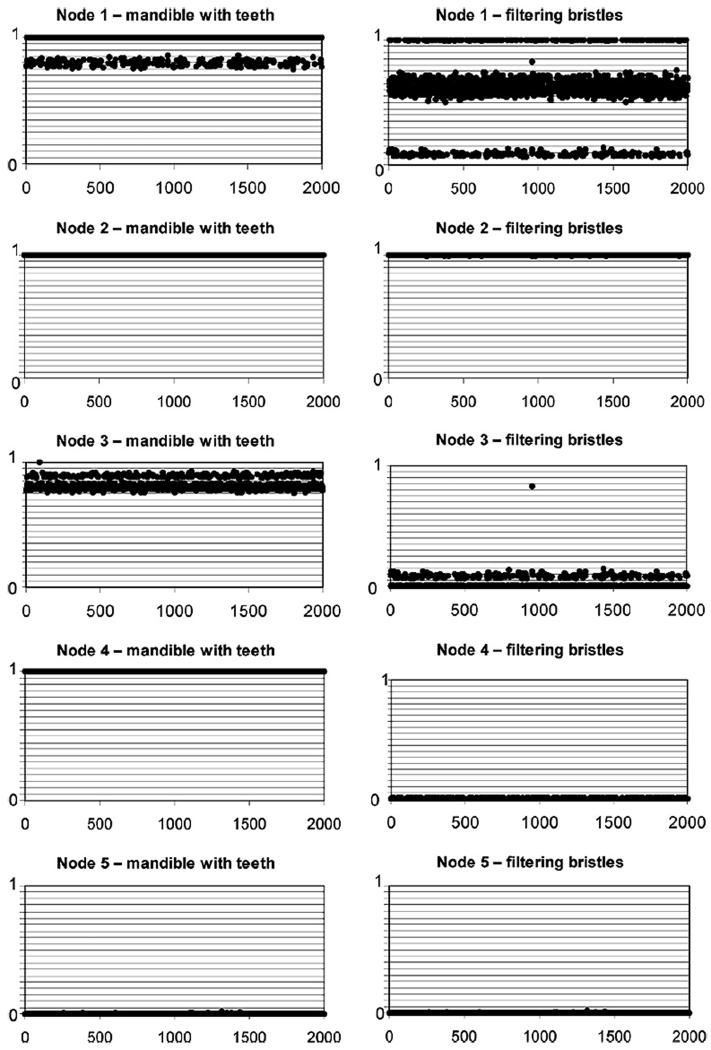
We reconstructed ancestral character states for the 5 nodes described in Section 3.2, to investigate the evolution of mandible type and presence of filtering bristles within the Drusinae. Fig. 4 shows the results of this ancestral character reconstruction analysis for the five nodes over the 2000 examined trees. Only the probabilities of a single character state are shown, since the two alternative states are complementary. Shown in the plots are presence of teeth on the mandible edge (left) and presence of filtering bristles on the thorax and legs (right). The mean p-values over all 2000 examined trees (ØML) are given for each node. At node 1, which includes all Drusinae, the analysis suggested significantly the presence of a mandible with teeth (ØMLT=0.969). The presence or absence of filtering bristles is not resolved (ØMLB=0.593). At node 2, the analysis infers that both teeth on mandibles (ØMLT>0.999) and filtering bristles (ØMLB>0.999) are present. At node 3 the state of teeth on mandibles is not resolved by the analysis (ØMLT=0.812), while filtering bristles are evaluated to be significantly absent (ØMLT=0.014). At node 4 filtering bristles are inferred absent (ØMLB=0.003), but the analysis significantly suggests that there are teeth on the mandible edges (ØMLT>0.999). At node 5, the analysis significantly infers that both teeth on mandibles and filtering bristles are absent (both ØML<0.001).
Figure 4.
Ancestral character reconstruction of mandible type and presence of filtering bristles on thorax and legs for five specific clades (as shown in Figures 2 and 3). The probability of presence of teeth on mandible edges (left column) and presence of filtering bristles on first abdominal sternum and legs (right column) for each node was reconstructed on 2000 trees generated by Markov chain Monte Carlo sampling.
4. Discussion
4.1 Phylogenetic relationships in Drusinae
The phylogenetic relationships recovered in our study do not support the current generic classification, which is based on adult morphology (Schmid, 1956). Our study supports monophyly of the subfamily, with respect to outgroups from subfamily Limnephilinae. Two currently recognised genera, Drusus and Ecclisopteryx, are not monophyletic. Drusus is polyphyletic with Anomalopterygella, Ecclisopteryx and Metanoea nested within. Schmid (1956) considered Anomalopterygella as the basal taxon in the Drusinae. Our phylogeny does not support this position and ELW tests rejects it. Anomalopterygella clusters in Drusus (sister to D. brunneus and D. trifidus) but its placement within the group is not supported by our statistic tests. Its position within Drusus is somewhat surprising considering the large number of characteristic and distinctive features of A. chauviniana (Table 2).
In our analyses Ecclisopteryx falls into two well-supported clades within Drusus. Monophyly of the genus is also rejected with our hypothesis testing approaches. In one clade we find E. asterix and E. malickyi. The second clade of Ecclisopteryx groups E. dalecarlica, E. guttulata, and E. madida with Drusus balcanicus, D. botosaneanui, and D. discophorus pallidus. This relationship is well-supported by the Bayesian inference, and was resolved in the maximum parsimony tree, but not recovered in the bootstrap analysis. Schmid (1956) stated that the morphological differences between Ecclisopteryx and Drusus are limited to the genital armature, but consistent and stable enough to support two genera. In contrast, our study suggests that Ecclisopteryx and Drusus are not true evolutionary units.
Monophyly of the genus Metanoea is supported in our analyses. However, the genus is nested within and not sister to Drusus. Schmid (1956) accepted Metanoea but raised doubts of its distinction based on the known differential characters (Table 2). In this study we could only include two of the five recognised species in the genus. Further investigations including more species of Metanoea are needed to fully resolve its status.
Four of the seven species groupings proposed by Schmid (1956), Marinkovic-Gospodnetic (1976), Kumanski (1988), and Sipahiler (1999) we tested are not supported by our analyses. For those species-groups that are observed in our topologies, we were only able to sample two sister species. It seems that very close adult similarities result from close relationships and could represent natural evolutionary units. Some of these species pairs comprise more or less vicariant species. D. alpinus and D. franzi are endemics of the western-central and south-eastern Alps respectively. D. nigrescens is a local endemic of the western central Alps, while D. monticola mainly occurs further east. Both species pairs may be examples of refugial lineage divergence of a common ancestor which was forced to retreat to south-eastern and south-western refugia during the early or middle Pleistocene (Hewitt, 2004; Pauls et al., 2006). The divergence between the sister taxa is less strong in D. nigrescens and D. monticola. This could result from a later split of the lineage or recurring introgression in the two species that do occasionally occur in the same region.
4.2 Larval Morphology and Feeding Type Evolution
4.2.1 Larval morphology and phylogenetic grouping
With exception of a few species groups, the three major clades within Drusinae (Clades 2, 4, 5) do not correspond to the adult genitalia-based generic and species-group classifications proposed by Schmid (1956), Marinkovic-Gospodnetic (1976) or Kumanski (1988). Instead, they correspond to the three distinct species groupings based on mouthpart anatomy and feeding ecology (Waringer et al., 2007a) (Fig. 2, 3). The larvae of the subfamily Drusinae all have single filament abdominal gills, a fully sclerotised pronotum and mesonotum, and build a cylindrical, slightly curved and slightly conical sand case (Szczesny, 1978; Ulmer, 1909; Waringer et al., 2003). In Cryptothrix nebulicola, Drusus chrysotus, D. discolor, D. muelleri and D. romanicus, mandibles with teeth around edges are present; this, together with additional setae on the legs and with long filtering bristles at the first abdominal sternum, identifies this group as carnivorous filterers. Gut content analysis of several species within this group (D. discolor, D. romanicus, D. muelleri) confirms their carnivorous filtering feeding ecology (Bohle, 1983; Graf and Pauls, unpublished data). These species also share a unique synapomorphy: all species in this group have concave cavities or major indentations in the head capsule. In all other species of Drusinae, the head capsule is rounded without cavities or indentations. In our analyses these species group together in clade 2. Although the inclusion of C. nebulicola in this group is not significant in the Bayesian inference (pp<0.95), the relatively high bootstrap values support inclusion (bs = 74%). Also all other Drusinae belong to one of two other significantly supported clades (pp>0.95; bs≥94%).
A second larval feeding type (omnivorous generalist shredders) was recently identified using DNA-based associations with adult specimens (Graf et al., in preparation; Waringer et al., 2007a). D. franzi and D. alpinus have mandibles with teeth around edges, but additional filtering spines on legs and bristles on the first abdominal sternum are lacking, characterising them as omnivorous generalist shredders. These two species are sister taxa in the highly supported clade 4 (pp>0.95, bs=94%). To date only Drusus alpinus and D. franzi are known to have the morphological characteristics of omnivorous generalist shredders as described above.
Of the remaining 21 species in our analyses 17 have a spoon-shaped mandible without teeth and additional setae and bristles are lacking (Szcesny, 1978; Waringer et al., 2000, 2007b, 2007c). This identifies these species as grazers, which feed mainly on epilithic algae. For the four other species included in our analyses (D. balcanicus, D. botosaneanui, D. discophorus pallidus, E. malickyi), the larval stages are unknown. However, based on their position in the phylogeny, we predict that these larvae are epilithic grazers with a spoon-shaped mandible.
4.2.2 Feeding type evolution
Based on our study, two alternative scenarions of the evolution of feeding ecology are possible: 1. progression from ancestral omnivorous shredders to both filtering carnivores and epilithic grazers or 2. evolution from filtering carnivores to omnivorous shredders and epilithic grazers. The first alternative is more likely based on the fact that all other Limnephilids are known to be shredders, but the latter alternative cannot be ruled out with the data at hand. The mandible in the shredders D. alpinus and D. franzi is of the ancestral type with teeth along the edges (Graf et al., in preparation). Based on our ancestral character state reconstructions the mandible with teeth appears to be the ancestral state, which is maintained in the carnivorous filterers and omnivore generalist shredders (Node 1-4). The spoon-shaped grazer mandible seems to be derived (Node 5), having reduced or lost the teeth on the mandible edge. The acquisition of filtering bristles seems to be a derived character unique to a single clade in our study (Node 2).
With few exceptions, all Limnephilidae are shredders (Graf et al., 2002). Other feeding types are only found in the Drusinae and sporadically among other genera (Melampophylax and Micropterna). Melampophylax mucoreus, M. nepos and Micropterna testacea, for example, are Limnephilinae grazers with spoon-shaped mandibles. Whether the feeding type evolved only once or independently several times within Limnephilidae requires further phylogenetic analysis with a larger sampling of Limnephlidae taxa. The evolution of feeding types in the Drusinae follows the ontogeny of individuals. Nielsen (1942) studied the larval development of Ecclisopteryx guttulata and observed that in first instar larvae both mandibles have a ventral tooth. Additionally, two or three dorsal teeth are present on the left and right mandible, respectively. From the second instar larvae onward, the mandibles are spoon-shaped without any teeth on the mandible edges.
Most of the extant Drusinae species whose larvae are known are grazers or carnivorous filterers. Weaver and Morse (1986) hypothesised generally for caddisflies that feeding specialisation may have opened opportunities to colonise new ecological niches and could have promoted diversification in these organisms significantly. Considering the high number of derived grazers, such changes in feeding ecology may be responsible for much of the diversification within Drusinae. Dietary shifts have also been made responsible for high levels of diversity in other groups including beetles (e.g. Leschen and Buckley, 2007; Farrel, 1998) and fish (e.g. Horstkotte and Strecker, 2006).
4.2.3 Importance of larval morphology for phylogenetic studies
Larval features correspond well with our phylogenetic results. In Drusinae larval morphology outperforms adult characters as phylogenetic discriminators. While most classifications of caddisfly species within genera are based on adult characters (Schmid, 1956; Flint, 1989; Holzenthal and Andersen, 2007), the use of larval characters in systematics and caddisfly phylogeny has been recognised for some time (Scott, 1975; Wiggins, 1981; Weaver and Morse, 1986; Kjer et al., 2001, 2002; De Moor, 2002; Kjaerndsen, 2004). In previous studies larval and adult characters have been incorporated into a joint matrix for phylogenetic studies. Studies using a molecular phylogeny to examine utility of adult and larval characters, however, are lacking. In their subordinal molecular phylogeny of Trichoptera, Kjer et al. (2001, 2002) were unable to fully resolve the basal relationships in Trichoptera, which traditionally consists of three suborders and infraorders based on larval morphology and behaviour: Annulipalpia, Integripalpia, and Spicipalpia (Martynov, 1924; Weaver, 1984).
Our study explicitly shows that at the level of genera and species groupings adult genital morphology does not recover the same relationships we find with an independent, molecular phylogeny, while our species are all recovered as monophyletic entities. While adult genital morphology clearly delimits individual species of Drusinae, relationships between these species are better understood if we also incorporate larval characteristics. From our results it appears that larval features and characters may be more useful in resolving evolutionary relationships between species within families or subfamilies than previously recognised. Utility of larval characters or characters of immature, life stages is widely recognised in other insect groups including beetles (Michat, 2006, Solodovnikov, 2007) and butterflies and moths (Hebert et al., 2004, Wagner et al., 2006). Additional lower-level molecular studies on caddisfly families and genera are needed to better judge the use of adult and larval characters in Trichoptera.
4.3 Utility of mtLSU, mtCOI and nuWG for lower level phylogenetic reconstruction of Drusinae
All three gene partitions we used are suitable for phylogenetic inference at the level examined in this study. Based on CI, RI, variability and resolution, nuWG performs best as a single marker for resolving relationships among species within this subfamily. Both mitochondrial genes are also useful, but variability and resolution are limited in mtLSU, while problems of homoplasy seem to limit the utility of mtCOI. Despite shortcomings in individual partitions, the best resolution is obtained using a combined data set, a result that is consistent with those observed in other studies of insects (Kjer et al., 2001, 2002; Hughes and Vogler, 2004; Nazari et al., 2007). Previous molecular studies on caddisflies have used a common set of molecular markers including various fragments of mitochondrial DNA (COI, 16S rRNA) and nuclear loci including (EF1- α, rRNA) (Kjer et al., 2001, 2002; Myers et al., 2001; Myers and Sperling, 2002; Geraci et al., 2005; Leese, 2004; Pauls et al., 2006). The utility of some of these genes for studying subordinal relationships in caddisflies was rigorously tested by Kjer et al. (2001). These authors concluded that at subordinal levels, rRNA data was very useful, while taken alone, both mtCOI and EF1-α are of little use for resolving subordinal phylogenetic relationships due to saturation and homoplasy issues. However, combined with other data sets, they still proved useful in finding the best estimate of phylogenetic hypotheses (Kjer et al., 2001), especially at lower phylogenetic levels. For example, Geraci et al. (2005) used EF1-α in combination with mtCOI and nu rRNA, to examine relationships between subfamilies in Hydropsychidae, but its utility was not evaluated. Based on our results, mtCOI is valuable in combination with other genes as it increases resolution. The present study is the first that uses and evaluates the utility of mtLSU and nuWG in a larger phylogenetic context in caddisflies. Both genes, especially nuWG, appear to be useful in multi-gene analyses which aim to resolve phylogenetic relationships at lower taxonomic rank in caddisflies.
Acknowledgements
We wish to thank all those colleagues mentioned in Table 1 for providing us with specimens for this study. We thank Kathrin Theißinger and Mag. Philipp Wenzl for assistance in the field and lab, Imke Schmitt for providing us with analytical support, and two anonymous reviewers for their helpful comments. Special thanks go to Paul Z. Goldstein for providing us with unpublished primer sequences fur wingless. Sequencing at the Pritzker Laboratory for Molecular Systematics and Evolution was financially supported through the Pritzker Foundation. Funding support for field work was provided through a fellowship to SUP through the German Academic Exchange Service (DAAD). This paper is part of the outcomes of a project dealing with larval taxonomy of Central European Drusinae (project number P18073-B03, PI: J.Waringer) funded by the Austrian Science Fund (FWF).
References
- Barbour MT, Yoder CO. The multimetric approach to bioassessment, as used in the United States of America. Freshwater Biological Association; Ambleside: 2000. [Google Scholar]
- Bohle HW. Drift-catching and feeding behaviour of the larvae of Drusus discolor (Trichoptera: Limnephilidae) Arch. Hydrobiol. 1983;97:455–470. [Google Scholar]
- Buckley TR, Arensburger P, Simon C, Chambers G. Combined data, Bayesian phylogenetics, and the origin of the New Zealand Cicada genera. Syst. Biol. 2002;51:4–18. doi: 10.1080/106351502753475844. [DOI] [PubMed] [Google Scholar]
- Buffagni A, Erba S, Cazzola M, Murray-Bligh J, Soszka H, Genoni P. The STAR common metrics approach to the WFD intercalibration process: Full application for small, lowland rivers in three European countries. Hydrobiologia. 2006;566:379–399. [Google Scholar]
- Bull JJ, Huelsenbeck JP, Cunningham CW, Swofford DL, Waddell PJ. Partitioning and combining data in phylogenetic analysis. Syst. Biol. 1993;42:384–397. [Google Scholar]
- DeMoor FC. An assessment of the global distribution of Leptocerinae (Trichoptera) and use of larval characters for determining phylogenetic relationships. Nova. Suppl. Entomol. 2002;15:293–308. [Google Scholar]
- Dohet A. Are caddisflies an ideal group for the biological assessment of water quality in streams? Nova. Suppl. Entomol. 2002;15:507–520. [Google Scholar]
- Dreesmann DC, Wichard W. The basal phylogenetical relationships of Trichoptera - a molecular approach. Nova. Suppl. Entomol. 2002;15:309–316. [Google Scholar]
- Farrell BD. “Inordinate fondness” explained: Why are there so many beetles? Science. 1998;281(5376):555–559. doi: 10.1126/science.281.5376.555. [DOI] [PubMed] [Google Scholar]
- Farris JS. The retention index and the rescaled consistency index. Cladistics. 1989;5:417–419. doi: 10.1111/j.1096-0031.1989.tb00573.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Flint OS., Jr Studies of neotropical caddisflies, XXXIX: The genus Smicridea in the chilean subregion (Trichoptera: Hydropsychidae) Smithsonian Contributions to Zoology. 1989;472:1–45. [Google Scholar]
- Geerts R, Löfsted C, Menken SBJ. Molecular phylogeny of Trichoptera. Proc. Exper. Appl- Entomol. 2001;12:165–167. [Google Scholar]
- Geraci C, Kjer KM, Morse J, Blahnik RJ. Phylogenetic relationships of Hydropsychidae subfamilies based on morphology and DNA sequence data. Proc. 11th Int. Symp. Trichoptera.2005. pp. 131–136. [Google Scholar]
- Graf W, Grasser U, Waringer J. Trichoptera. In: Moog O, editor. Fauna Aquatica Austriaca, Lieferung 2002. Wasserwirtschaftskataster, Bundesministerium für Land- und Forstwirtschaft; Vienna: 2002. pp. 1–10. [Google Scholar]
- Graf W, Lubini V, Pauls S. Larval description of Drusus muelleri McLachlan, 1868 (Trichoptera : Limnephilidae) with some notes on its ecology and systematic position within the genus Drusus. Ann. Limnol. - Int. J. Lim. 2005;41:93–98. [Google Scholar]
- Graf W, Waringer J, Pauls SU. A new morpho-ethological feeding group within larval Drusinae (Trichoptera: Limnephilidae): the alpinus-group sensu Schmid, 1956 including larval descriptions of Drusus franzi Schmid, 1956 and Drusus alpinus Meyer-Dür, 1875. in preparation. [PMC free article] [PubMed]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. P. Natl. Acad. Sci. USA. 2004;101:14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering D, Johnson RK, Kramm S, Schmutz S, Szoszkiewicz K, Verdonschot PFM. Assessment of European streams with diatoms, macrophytes, macroinvertebrates and fish: a comparative metric-based analysis of organism response to stress. Freshwater Biol. 2006;51:1757–1785. [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Phil. Trans. R. Soc. Lond. B. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzenthal RW, Andersen T. Review of the caddisfly genus Tagalopsyche with the description of new species and a related new genus (Trichoptera: Leptoceridae: Mystacidini) Zootaxa. 2007;1483:1–32. [Google Scholar]
- Horstkotte J, Strecker U. Trophic differentiation in the phylogenetically young Cyprinodon species flock (Cyprinodontidae, Teleostei) from Laguna Chichancanab (Mexico) Biol. J. Linn. Soc. 2005;85:125–134. [Google Scholar]
- Huelsenbeck JP, Nielsen R, Bollback J. Stachastic mapping of morphological characters. Syst. Biol. 2007;52:131–158. doi: 10.1080/10635150390192780. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Rannala B, Masly JP. Accomodating phylogenetic uncertainty in evolutionary studies. Science. 2000;288:2349–2350. doi: 10.1126/science.288.5475.2349. [DOI] [PubMed] [Google Scholar]
- Hughes J, Vogler AP. The phylogeny of acorn weevils (genus Curculio) from mitochondrial and nuclear DNA sequences: the problem of incomplete data. Mol. Phylogenet. Evol. 2004;32:601–615. doi: 10.1016/j.ympev.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Kjaerandsen J. A revision of the Afrotropical genus Dhatrichia (Trichoptera, Hydroptilidae) Zool. Scr. 2004;33:131–185. [Google Scholar]
- Kjer KM, Blahnik RJ, Holzenthal RW. Phylogeny of Trichoptera (Caddisflies): Charaterization of signal and noise within multiple datasets. Syst. Biol. 2001;50:781–816. doi: 10.1080/106351501753462812. [DOI] [PubMed] [Google Scholar]
- Kjer KM, Blahnik RJ, Holzenthal RW. Phylogeny of caddisflies (Insecta, Trichoptera) Zool. Scr. 2002;31:83–91. [Google Scholar]
- Kumanski K. Trichoptera, Integripalpia. Fauna Bulgarica. 1988;19:1–354. [Google Scholar]
- Leese F. Diploma Thesis. Biological Faculty Ruhr-Universität; Bochum: 2004. Molecular genetic, chemotaxonomic, and autecological investigations of European Sericostomatidae (Insecta: Trichoptera) pp. 1–137. [Google Scholar]
- Leschen RAB, Buckley TR. Multistate characters and diet shifts: Evolution of Erotylidae (Coleoptera) Syst. Biol. 2007;56:97–112. doi: 10.1080/10635150701211844. [DOI] [PubMed] [Google Scholar]
- Mackay RJ, Wiggins GB. Ecological diversity in Trichoptera. Annu. Rev. Entomol. 1979;24:185–208. [Google Scholar]
- Malicky H. Ein kommentiertes Verzeichnis der Köcherfliegen (Trichoptera) Europas und des Mediterrangebietes. Linzer biol. Beitr. 2005;37:533–596. [Google Scholar]
- Marinkovic-Gospodnetic M. The differentiation of Drusus species of the group bosnicus. Proc. 1st Int. Symp. Trichoptera.1976. pp. 77–85. [Google Scholar]
- Martynov AV. Trichoptera. In: Bogdanova-Kat’kova NN, editor. Prakticheskaya entomologiya. Vol. 5. Leningrad; 1924. p. 384. [Google Scholar]
- Michat MC. The phylogenetic position of Hydrovatus Motschulsky: evidence from larval morphology of H-caraibus Sharp (Coleoptera : Dytiscidae : Hydroporinae) Insect Syst. Evol. 2006;37:419–432. [Google Scholar]
- Moog O, Graf W, Janecek B, Ofenböck T. Inventory of sensitive taxa of Austrian rivers and streams, Part III E. In: Moog O, editor. Fauna Aquatica Austriaca, Ergänzungen 2004. Wasserwirtschaftskataster, Bundesministerium für Land- und Forstwirtschaft; Vienna: 2004. pp. 1–4. [Google Scholar]
- Moretti GP, Pirisinu Q. Morphological characteristics of the immature stages in Leptodrusus budtzi Ulm. Series Entomologica. 1981;20:231–236. [Google Scholar]
- Morse JC. Phylogeny of Trichoptera. Annu. Rev. Entomol. 1997;42:427–450. doi: 10.1146/annurev.ento.42.1.427. [DOI] [PubMed] [Google Scholar]
- Myers MJ, Sperling FAH. Preliminary evaluation of subgeneric designations within the caddisfly genus Lepidostoma (RAMBUR) (Trichoptera: Lepidostomatidae) based on mtDNA sequences. Nova. Suppl. Entomol. 2002;15:187–194. [Google Scholar]
- Myers MJ, Sperling FAH, Resh VH. Dispersal of two species of Trichoptera from desert springs: Conservation implications for isolated vs connected populations. J. Insect Conservat. 2001;5:207–215. [Google Scholar]
- Nazari V, Zakharov EV, Sperling FAH. Phylogeny, historical biogeography, and taxonomic ranking of Parnassiinae (Lepidoptera, Papilionidae) based on morphology and seven genes. Mol. Phylogenet. Evol. 2007;42:131–156. doi: 10.1016/j.ympev.2006.06.022. [DOI] [PubMed] [Google Scholar]
- Nielsen A. Über Entwicklung und Biologie der Trichopteren mit besonderer Berücksichtigung der Quelltrichopteren Himmerlands. Arch. Hydrobiol. Suppl. 1942;17:255–626. [Google Scholar]
- Pagel M, Lutzoni F. Accounting for phylogenetic uncertainty in comparative studies of evolution and adaptation. In: Lässig M, Valleriani A, editors. Biological evolution and statistical physics. Springer; Berlin: 2002. pp. 148–161. [Google Scholar]
- Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- Pauls SU, Lumbsch HT, Haase P. Phylogeography of the montane caddisfly Drusus discolor: evidence for multiple refugia and periglacial survival. Mol. Ecol. 2006;15:2153–2169. doi: 10.1111/j.1365-294X.2006.02916.x. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Resh VR, Rosenberg DM. The ecology of aquatic insects. Praeger Scientific; New York: 1984. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rosenberg DM, Resh VH. Freshwater biomonitoring and benthic macroinvertebrates. Chapman& Hall; New York: 1993. [Google Scholar]
- Schmid F. La sous-famille des Drusinae (Trichoptera, Limnophilidae) Mem. Inst. Roy. Sci. Nat. Belg. 1956;2(Serie 55):1–92. [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Scott KMF. The value of larval stages in systematic studies of the Trichoptera, with particular reference to the Hydropsychidae from Africa south of the Sahara. Proceedings of the 1st congress of the Entomological Society of southern Africa; Stellenbosch. 1975. pp. 41–52. [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999;16:1114–1116. [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994;87:651–701. [Google Scholar]
- Sipahiler F. Distribution of Drusinae (Limnephilidae) species in Turkey. Proc. 9th Int. Symp. Trichoptera.1999. pp. 329–336. [Google Scholar]
- Sipahiler F. Hadimina torosensis, new genus and new species of Drusinae from southern Turkey (Trichoptera: Limnephilidae) Nova. Suppl. Entomol. 2002;15:239–248. [Google Scholar]
- Smith MJ, Kay WR, Edward DHD, Papas PJ, Richardson KS, Simpson JC, Pinder AM, Gale DJ, Horwitz PHJ, Davis JA, Yung FH, Norris RH, Halse SA. AusRivAS: using macroinvertebrates to assess ecological condition of rivers in Western Australia. Freshwater Biol. 1999;41:269–282. [Google Scholar]
- Solodovnikov AY. Larval chaetotaxy of Coleoptera (Insecta) as a tool for evolutionary research and systematics: less confusion, more clarity. J. Zool. Syst. Evol. Res. 2007;45:120–127. [Google Scholar]
- Strimmer K, Rambaut A. Inferring confidence sets of possibly misspecified gene trees. Proc. R. Soc. Lond. Ser. B. 2002;269:137–142. doi: 10.1098/rspb.2001.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung GH, Sung JM, Hywel-Jones NL, Spatafora JW. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007;44:1204–1223. doi: 10.1016/j.ympev.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods) Sinauer Associates; Sunderland: 2001. [Google Scholar]
- Szczesny B. Larvae of the subfamily Drusinae (Insecta: Trichoptera) from the Polish part of the Carpathian Mts. Acta Hydrobiol. 1978;20:35–53. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer G. Trichoptera. In: Brauer A, editor. Süsswasserfauna Deutschlands. Gustav Fischer; Jena: 1909. pp. 1–326. [Google Scholar]
- Wagner DL, Hossler EW, Hossler FE. Not a tiger but a dagger: The larva of Comachara cadburyi and reassignment of the genus to Acronictinae (Lepidoptera : Noctuidae) Ann. Entomol. Soc. Am. 2006;99:638–647. [Google Scholar]
- Wallace JB, Webster JR. The role of macroinvertebrates in stream ecosystem function. Annu. Rev. Entomol. 1996;41:115–139. doi: 10.1146/annurev.en.41.010196.000555. [DOI] [PubMed] [Google Scholar]
- Waringer J. The larva of Metanoea rhaetica Schmid, 1955 (Trichoptera: Limnephilidae: Drusinae) from a small Austrian mountain brook. Aquat. Insect. 1985;7:243–248. [Google Scholar]
- Waringer J, Graf W. Atlas der Österreichischen Köcherfliegenlarven. Facultas; Vienna: 1997. [Google Scholar]
- Waringer J, Graf W. Ergänzungen und Berichtigungen zum “Atlas der österreichischen Köcherfliegenlarven unter Einschluß der angrenzenden Gebiete“. Facultas; Vienna: 2004. Beilage zum 2. unveränderten Nachdruck. [Google Scholar]
- Waringer J, Graf W, Maier K-J. The larva of Metanoea flavipennis Pictet, 1834 (Trichoptera: Limnephilidae: Dusinae) Aquat. Insect. 2000;22:66–70. [Google Scholar]
- Waringer J, Graf W, Pauls SU. Functional feeding ecology in Central European species of subfamily Drusinae (Insecta: Trichoptera) Lauterbornia. 2007a (in press) [PMC free article] [PubMed] [Google Scholar]
- Waringer J, Graf W, Pauls SU, Lubini V. The Larva of Drusus nigrescens Meyer-Dür, 1875 (Trichoptera: Limnephilidae: Drusinae) with notes on its ecology, genetic differentiation and systematic position. Ann. Limnol. - Int. J. Lim. 2007b doi: 10.1051/limn:2007010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waringer J, Graf W, Pauls SU, Vicentini H, Lubini V. DNA based association and description of the larval stage of Drusus melanchaetes McLachlan, 1876 (Trichoptera: Limnephilidae: Drusinae) with notes on ecology and zoogeography. Limnologica. 2007c doi: 10.1016/j.limno.2007.09.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver JS., III. Evolution and classification of Trichoptera, part 1: groundplan of Trichoptera. Series Entomologica. 1984;30:413–419. [Google Scholar]
- Weaver JS, III, Morse JC. Evolution of feeding and case-making behaviour in Trichoptera. J. N. Am. Benthol. Soc. 1986;5:150–158. [Google Scholar]
- Wiggins MF. Considerations on the relevance of immature stages to the systematics of Trichoptera. Series Entomologica. 1981;20:395–407. [Google Scholar]
- Wiggins MF. Caddisflies the underwater architects. Toronto Univ. Press; Toronto Buffalo London: 2004. [Google Scholar]
- Wilcock HR, Hildrew AG, Nichols RA. Genetic differentiation of a European caddisfly: past and present gene flow among fragmented larval habitats. Mol. Ecol. 2001;10:1821–1834. doi: 10.1046/j.0962-1083.2001.01310.x. [DOI] [PubMed] [Google Scholar]
- Wilcock HR, Nichols RA, Hildrew AG. Genetic population structure and neighbourhood population size estimates of the caddisfly Plectrocnemia conspersa. Freshwater Biol. 2003;48:1813–1824. [Google Scholar]
- Wright JF, Moss D, Armitage PD, Furse MT. A preliminary classification of running-water sites in Great Britain based on macroinvertebrate species and the prediction of community type using environmental data. Freshwater Biol. 1984;14:221–256. [Google Scholar]
- Wright JF, Sutcliffe DW, Furse MT. Assessing the biological quality of fresh waters: RIVPACS and other techniques. Freshwater Biological Association; Ambleside: 2000. [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. The University of Texas at Austin; 2006. Ph.D. dissertation. [Google Scholar]