Abstract
Cryptochromes (CRY) are blue-light photoreceptors that mediate various light responses in plants and animals. It has long been demonstrated that Arabidopsis CRY (CRY1 and CRY2) C termini (CCT1 and CCT2) mediate light signaling through direct interaction with COP1. Most recently, CRY1 N terminus (CNT1) has been found to be involved in CRY1 signaling independent of CCT1, and implicated in the inhibition of gibberellin acids (GA)/brassinosteroids (BR)/auxin-responsive gene expression. Here, we performed RNA-Seq assay using transgenic plants expressing CCT1 fused to β-glucuronidase (GUS-CCT1, abbreviated as CCT1), which exhibit a constitutively photomorphogenic phenotype, and compared the results with those obtained previously from cry1cry2 mutant and the transgenic plants expressing CNT1 fused to nuclear localization signal sequence (NLS)-tagged YFP (CNT1-NLS-YFP, abbreviated as CNT1), which display enhanced responsiveness to blue light. We found that 2903 (67.85%) of the CRY-regulated genes are regulated by CCT1 and that 1095 of these CCT1-regulated genes are also regulated by CNT1. After annotating the gene functions, we found that CCT1 is involved in mediating CRY1 regulation of phytohormone-responsive genes, like CNT1, and that about half of the up-regulated genes by GA/BR/auxin are down-regulated by CCT1 and CNT1, consistent with the antagonistic role for CRY1 and these phytohormones in regulating hypocotyl elongation. Physiological studies showed that both CCT1 and CNT1 are likely involved in mediating CRY1 reduction of seedlings sensitivity to GA under blue light. Furthermore, protein expression studies demonstrate that the inhibition of GA promotion of HY5 degradation by CRY1 is likely mediated by CCT1, but not by CNT1. These results give genome-wide transcriptome information concerning the signaling mechanism of CRY1, unraveling possible involvement of its C and N termini in its regulation of response of GA and likely other phytohormones.
Keywords: CRY1, CRY1 C terminus (CCT1), CRY1 N terminus (CNT1), RNA-Seq, gibberellin acids (GA), brassinosteroids (BR), auxin
Introduction
Light not only provides plants with energy to maintain life, but also supplies plants with signal to regulate their developmental processes in the whole life cycle (Chen et al., 2004). Plants have evolved various photoreceptors including blue/UV-A light receptors cryptochromes (CRY1 and CRY2), phototropins (PHOT1 and PHOT2) (Cashmore et al., 1999; Briggs and Christie, 2002), the red/far-red light receptors phytochromes (phyA to phyE) (Quail et al., 1995; Quail, 2002), and the UV-B receptor UVR8 (Rizzini et al., 2011). It is known that CRY1 and CRY2 regulate a variety of physiological processes (Ahmad and Cashmore, 1993; Guo et al., 1998; Somers et al., 1998; Mao et al., 2005; Kang et al., 2009), including seedling photomorphogenesis and photoperiodic flowering, respectively. CRY contains an N-terminal domain related to the photolyase and a C-terminal extension domain (Yang et al., 2000; Sang et al., 2005). The C-termini of CRY1 and CRY2 (CCT1 and CCT2) are known to mediate CRY1 signaling, since transgenic plants expressing CCT1 or CCT2 fused to β-glucuronidase (GUS) show a constitutively photomorphogenic phenotype (Yang et al., 2000). It is demonstrated that the N termini of CRY1 and CRY2 (CNT1 and CNT2) are not only responsible for chromophores binding and blue light perception (Lin et al., 1995; Lin, 2002), but also mediation of their dimerization, which is essential for light activation of their photoreceptor activity (Sang et al., 2005; Yu et al., 2007). Most recently, CNT1 has been shown to mediate CRY1 signaling independent of its C terminus, since CNT1 fused to nuclear localization signal sequence (NLS)-tagged YFP confers enhanced responsiveness to blue light (He et al., 2015).
Several CRY-interacting proteins have been characterized. Of them, the master negative regulator of photomorphogenesis, COP1 (CONSTITUTIVELY PHOTOMORPHOGENIC1) (Osterlund et al., 1999), is the first characterized, with which CRY1 or CRY2 interacts through their C termini (Yang et al., 2000, 2001; Wang et al., 2001). COP1 has E3 ubiquitin ligase activity, and physically interacts with various substrates, such as HY5/HYH, HFR1, LAF1, and CONSTANS, to promote their ubiquitination and degradation to regulate photomorphogenesis and flowering (Holm et al., 2002; Seo et al., 2003; Yang et al., 2005; Liu et al., 2008b). Among them, HY5, a bZIP transcription factor, plays a pivotal role in promoting photomorphogenesis (Osterlund et al., 2000). Recent studies have revealed that CRY1 and CRY2 interact with COP1's E3 ubiquitin ligase enhancer, SPA1 (SUPPRESSOR OFPHYTOCHROME A 1), through their C and N termini, respectively (Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011). The interactions of CRY1 with COP1 and SPA1 result in the dissociation of the COP1-SPA1 complex and eventually the accumulation of HY5 protein (Lian et al., 2011; Liu et al., 2011). CRY2 is also found to interact with a family of bHLH transcription factors, cryptochrome-interacting bHLH (CIBs), through its N terminus, to regulate flowering (Liu et al., 2008a; Kennedy et al., 2010). To date, no CRY1 N terminus-interacting protein has been identified.
Several phytohormones, such as gibberellin acids (GA), brassinosteroids (BR), and auxin, are involved in promoting hypocotyl cell elongation. The biosynthesis mutants of GA, BR, and auxin, as well as of the loss-of-function mutants of GA receptor, GID1, BR receptor, BRI1, and auxin receptors, TIR1/AFBs, show a shortened hypocotyl phenotype during photomorphogenic development (Chory et al., 1991; Clouse et al., 1996; Kim et al., 1996; Szekeres et al., 1996; Richards et al., 2001; Dharmasiri et al., 2005; Ueguchi-Tanaka et al., 2005). The antagonistic regulation of hypocotyl cell elongation by light and these phytohormones indicates that the signaling mediated by light and these phytohormones may antagonize. Indeed, it has been demonstrated that the signaling crosstalk between GA and light is mediated through repression of the transcriptional activity of Phytochrome-Interacting Factors 3 and 4 (PIF3 and PIF4), pivotal negative regulators of photomorphogenesis (Leivar et al., 2008), through direct interactions of DELLA proteins with PIF3 and PIF4 (de Lucas et al., 2008; Feng et al., 2008). In addition, brassinosteroid and light can be converged through the interaction of PIF4 with BZR1, a critical transcription factor acting togerther with BES1 to mediate BR response (Oh et al., 2012). HY5 is also shown to be involved in regulating auxin, GA, and BR signaling through regulating genes involved in phytohormone biosynthesis or signaling (Cluis et al., 2004; Weller et al., 2009; Li and He, 2016). The newly identified branch inhibiting phytohormone, strigolactones (Jiang et al., 2013; Zhou et al., 2013), regulates hypocotyl elongation through promoting HY5 protein accumulation, which is dependent on cryptochrome and phytochrome signaling pathways (Jia et al., 2014).
It has been demonstrated that cryptochromes regulate GA catabolic/metabolic genes to inhibit the biosynthesis of active GA and thus hypocotyl elongation (Zhao et al., 2007). A recent study shows that CNT1 is likely involved in mediating CRY1 repression of GA/BR/auxin-responsive gene expression (He et al., 2015). Given that CNT1 and CCT1 are both involved in mediating CRY1 signaling, it is worth investigating how they are involved in regulating phytohormone-responsive gene expression at the transcriptomic level. In this study, we performed RNA-Seq assay using transgenic plants expressing CCT1 fused to β-glucuronidase (GUS) (GUS-CCT1, abbreviated as CCT1), and compared with the RNA-Seq results we obtained from cry1cry2 and CNT1 seedlings previously (He et al., 2015). We uncovered 6780 CCT1-regulated genes, a portion of which are CRY-regulated GA/BR/auxin-responsive genes, indicating the possible involvement of CCT1 in mediating CRY1 regulation of phytohormone-responsive genes. CCT1 and CNT1 not only co-regulate, but also separately regulate some of the GA/BR/auxin-responsive genes that are regulated by CRY. The results of physiological studies show that both CCT1 and CNT1 are likely involved in mediating CRY1 reduction of seedlings sensitivity to GA under blue light. Protein expression studies indicate that CCT1, but not CNT1, may be involved in mediating CRY1 inhibition of GA-promoted degradation of HY5, indicating possible different mechanisms for CCT1 and CNT1 in mediating CRY1 regulation of GA response. Hence, our study gives new insight into the signaling mechanism of CRY1, providing clue to looking for potential downstream components of CRY1 that may interact with CCT1 or CNT1.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild type (WT) control. The transgenetic line overexpressing CCT1 fused to β-glucuronidase (GUS) (GUS-CCT1, abbreviated as CCT1), transgenic cry1 mutant line #9 overexpressing CNT1-NLS-YFP (abbreviated as CNT1), the seedlings overexpressing MYC-tagged full-length CRY1 (CRY1-ovx), cop1-4, and cry1 mutant were described previously and all in the Col-0 background (Yang et al., 2000; Mao et al., 2005; He et al., 2015). Imbibed seeds were kept for 3 days at 4°C and grown on half-strength Murashige-Skoog (MS) nutrient medium plus 1% sucrose with 0.8% agar at 22°C under white light (100 μmol/m2/s) or 30 μmol/m2/s blue light.
Rna extraction, library preparation, and sequencing
Seeds were germinated on half-strength MS plates plus 1% sucrose and placed at 4°C for 3 days and then transferred to white light for 12 h before placed in darkness for another 4 days. Two biological replicates were prepared for WT, CCT1 and cop1-4 plants. Total RNA were extracted with RNAprep plant kit (TIANGEN) and treated with DNase I (TIANGEN) following the manufactuer's instruction. Quality control was performed with Agilent 2100 Bioanalyzer. The cDNA libraries were constructed using NEBNextUltra™ RNA library Prep Kit and submitted for sequencing using Illumina Hiseq2500. The library construction and sequencing were performed by the Hanyu BioTech in Shanghai (Pu Dong, Shanghai, China).
Processing of RNA-seq data
Raw sequencing reads were processed with FASTX-Toolkit (v0.0.13) to trim adaptor contaminations and filter out low quality reads with default parameters. The processed reads were then mapped to the Arabidopsis TAIR10 genome assembly using Bowtie 2.2.2 with default parameters (Langmead and Salzberg, 2012). The mapped reads for each gene were counted with samtool and then converted into RPKM (Mortazavi et al., 2008). The MARS (MA-plot-based method with Random sampling model) from DEGseq package was used to call significant differentially expressed genes (DEGs) (Wang et al., 2010). When identifying differentially expressed genes between two samples, we considered both fold change and p-value. P-value was calculated for each gene with expression values obtained from RNA-Seq analysis being subjected to two-tailed unpaired Student t-test. Genes with a fold change >2 and p < 0.01 were defined as differentially expressed genes. Venn diagram was generated in Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html). Heat-map was generated with hierarchical clustering analysis by MeV 4.7 software (He et al., 2015).
Function and pathway enrichment analysis
The Gene Ontology (GO) enrichment analysis is based on the GOseq method (Young et al., 2010), which is based on Wallenius non-central hyper-geometric distribution. We identified the significantly enriched GO term of DEGs with FDR (q-value) < 0.05.
For KEGG analysis, we used KEGG pathway as a unit and applied the hypergeometric test to find significant enriched pathways compared to the whole genome background (Kanehisa and Goto, 2000). We identified the significantly enriched KEGG pathway of DEGs with FDR (q-value) < 0.05. Differential gene KEGG enrichment and scatter plot is a graphical display of the results of KEGG enrichment analysis. Rich Factor means the number of genes that are differentially expressed in the pathway entries and the ratio of the total number of genes located in the pathway entries to all of the annotated genes.
Real-time quantitative PCR
We used WT, cry1 mutant, CRY1-ovx, CCT1, and CNT1 seedlings for real-time quantitative PCR. Seedlings were germinated on half-strength MS plates plus 1% sucrose and placed at 4°C for 3 days and then transferred to white light for 12 h before all seedlings placed in darkness for another 4 days. Then half of the seedlings were exposure to 30 μmol/m2/s blue light for 1 h, and another half of the seedlings were continue grown in dark. Total RNAs were isolated with RNAprep Plant kit (TIANGEN) followed by DNase I (TIANGEN) treatment. Then 500 ng sample of total RNA were used to reverse-transcribed to 10 μl cDNA using iScriptcDNA Synthesis kit (Bio-Rad). To validate our expression profile data, we selected genes that showed significant expression changements according to our different aim and performed real-time quantitative PCR. qRT-PCR was described previously (Zhang et al., 2014; He et al., 2015) and ACT2 was used as internal control for qRT-PCR. The used primers are listed in Table S4.
GA and PAC treatment
GA3 and PAC stocks were prepared in ethanol. WT, cry1 mutant, CRY1-ovx, CCT1, and CNT1 seedlings were grown on half-strength MS plates supplementing gradient GA (mock, 0.05, 0.2, 0.5, 1.0 μM), or PAC concentrations (mock, 0.025, 0.05, 0.2, 0.5 μM), or PAC (1 μM) plus GA concentrations (0.0, 0.5, 1.0, 2.5, 5.0, 10.0 μM) at 4°C for 3 days, then placed in white light overnight before transferred into blue light for another 5–6 days. Seedlings were neatly stacked on the agar plates. Then seedlings were photographed and the hypocotyl length was measured with Image J software.
Western blotting
WT, cry1 mutant, CRY1-ovx, CCT1, and CNT1 seedlings were grown in plates treated with gradient GA concentrations (mock, 1, 2.5 μM), PAC (mock, 0.5, 1 μM) at 4°C for 3 days, then placed in white light overnight before transferred into blue light for another 4 days. Seedlings were collected and homogenized in lysis buffer (50 mM Tris-HCl pH7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.2% Trition-X-100, 1 mM Pefabloc, cocktail, 50 μM MG132). The protein were quantified with Bradford assay (Bio-Rad), and subjected to Western blotting analyses with anti-HY5 antibody (Jia et al., 2014), and anti-ACT11 antibody (Abmart), respectively. The cop1-4 mutant served as a positive control.
Accession numbers
All RNA-seq data have been submitted to the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra/) under the accession number SRP067949. The accession numbers of previous microarray or RNA-Seq datasets used in this study are GSE58552 and GSE59763 (He et al., 2015), GSE863 (auxin/mock) (Nemhauser et al., 2006), GSE51772 (BR/mock) (Oh et al., 2014), and GSE22681 (ga1-3/WT) (Cheminant et al., 2011).
Results
Identification of genes regulated by CCT1
It has been reported recently that CRY1 N terminus (CNT1) mediates CRY1 signaling independent of its C terminus (CCT1), and CNT1 influenced about one third of the CRY-regulated genes (He et al., 2015). To gain more information concerning the signaling mechanism of CRY1 mediated by its C terminus at the transcriptomic level, we performed RNA-Seq analysis with WT and the transgenic plants expressing CCT1 fused to β-glucuronidase (GUS) (CCT1) through the Illumina HiSeq2500 system. A total of 22,791,289 and 25,743,501 successful sequencing reads were produced for WT and CCT1 plants, respectively (Table 1). Notably, more than 90.56 and 70.39% of the reads for WT and CCT1 were mapped to the Arabidopsis genome (version 10), and 84.18 and 63.81% reads were uniquely mapped to a single location in WT and CCT1, respectively (Table 1). Furthermore, 6780 differentially expressed genes (DEGs) between WT and CCT1 were identified with a p < 0.01 and fold change >2 (Table S1). In this study, we primarily focused on the DEGs.
Table 1.
Summary of RNA-Seq data from WT and CCT1.
| Map to gene | CCT1_D1 | CCT1_D2 | WT_D1 | WT_D2 | ||||
|---|---|---|---|---|---|---|---|---|
| Reads number | Percentage (%) | Reads number | Percentage (%) | Reads number | Percentage (%) | Reads number | Percentage (%) | |
| Clean reads | 22791286 | 100 | 27312797 | 100 | 25743501 | 100 | 22651750 | 100 |
| Total mapped reads | 16041945 | 70.39 | 18807037 | 68.86 | 23312549 | 90.56 | 20382779 | 89.98 |
| Unique_match | 14543021 | 63.81 | 17086853 | 62.56 | 21670195 | 84.18 | 18921019 | 83.53 |
| Perfect match | 15022315 | 65.91 | 17613248 | 64.49 | 21919460 | 85.15 | 19150500 | 84.54 |
Functional enrichment analysis indicates CCT1 involvement in CRY1 regulation of phytohormone-responsive gene expression
Since both CCT1 and CNT1 mediate CRY1 signaling (Yang et al., 2000; He et al., 2015), and CRY1 N terminus is involved in the regulation of growth-related phytohormone-responsive gene expression, we examined whether CCT1 might also be involved in mediating CRY1 regulation of phytohormone-responsive genes. Firstly, we examined how many of the CRY-regulated genes might be regulated by CCT1. Notably, CCT1 regulated 2903 (67.85%) of the 4278 CRY-regulated genes in the same direction (Figures 1A,B). Venn diagram and Heat-map graph revealed that, of the 2903 CCT1-regulated genes, 1095 were also regulated by CNT1 in the same direction (Figures 1A,B), indicating that the overlapping is statistically significant. These results suggested that CCT1 is involved in mediating CRY1 signaling at the transcriptomic level.
Figure 1.

CCT1, CNT1, and CRY regulate a large number of overlapping genes in the same direction. (A) Venn diagram showing significant overlapping genes between CCT1, CNT1, and CRY datasets. (B) Hierarchical clustering analysis of 1095 overlapping genes as shown in (A). Red and green colors in the heat-maps represent induced and repressed genes, respectively. Scale bar denotes the log2 value of fold change.
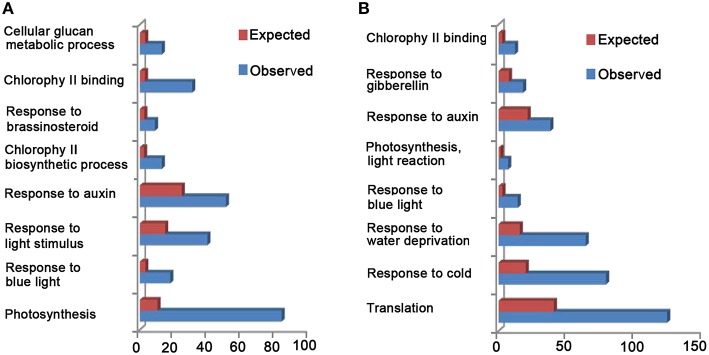
Since CRY is shown to be related to many biological processes, such as “photosynthesis,” “chloroplast organization,” “cell wall organization or biogenesis,” “response to auxin,” “response to GA,” “response to BR,” “response to ABA,” “response to JA,” and “response to SA” (Folta et al., 2003; He et al., 2015), we examined whether CCT1 might be involved in regulating these processes through GO and KEGG analyses, respectively. We found that the DEGs of CCT1 can be classified into 128 GO terms (Table S2), which suggest that CCT1 is associated with several biological processes, such as “photosynthesis,” “response to blue light,” “response to light stimulus,” “response to auxin,” and “response to brassinosteroid” (Figure 2A). KEGG pathway analysis further showed that many DEGs are mapped to “photosynthesis” and “plant hormone signal transduction” pathways (Figure S1A). These results indicated that CCT1 might be involved in mediating CRY1 regulation of phytohormone-responsive gene expression. Since CCT1 and CNT1 co-regulate a large number of genes, which include phytohormone-responsive genes that are regulated by CRY1, we further determined the potential functional relevance of CCT1 and CNT1 through GO and KEGG analyses, respectively. GO analysis revealed that both CCT1 and CNT1 are associated with many biological processes, such as “translation,” “response to cold,” “response to water deprivation,” “response to blue light,” “photosynthesis, light reaction,” “response to auxin,” and “response to gibberellin” (Figure 2B). And KEGG analysis demonstrated that the genes co-regulated by CCT1 and CNT1 mainly participated in “ribosome” pathway (Figure S1B). Taken together, these results indicated that CCT1 alone tends to be involved in mediating CRY1 regulation of photosynthesis, light response, and hormonal regulation similar to CNT1, whereas CCT1 and CNT1 are jointly involved in mediating CRY1 regulation of protein synthesis and stress responses.
Figure 2.
Gene Ontology (GO) analyses of all the DEGs between WT and CCT1, CCT1, and CNT1. (A) GO enrichment analysis of DEGs between WT and CCT1. Expect means the gene number base on all Arabidopsis genes. Observed means the gene number base on the DEGs of WT and CCT1.(B) GO enrichment analysis of DEGs between CCT1 and CNT1. Expect means the gene number base on all Arabidopsis genes. Observed means the gene number base on the DEGs of CCT1 and CNT1.
CCT1 and CNT1 mediate CRY1 regulation of GA/BR/auxin-responsive genes
With the demonstrations that CCT1 is involved in CRY1 regulation of phytohormone-responsive genes, we further confirmed the relationship among CRY1, CCT1, and CNT1 in regulating phytohormone-responsive genes. We compared the CRY1-, CCT1-, and CNT1-regulated genes with the transcriptomic data relative to GA/BR/auxin reported previously (Nemhauser et al., 2006; Cheminant et al., 2011; Oh et al., 2014), respectively. Of the 311 GA-regulated genes, 134 (43.09%) genes were regulated by CRY, and of these 134 genes, 105 (78.35%) and 66 (49.25%) genes were also regulated by CCT1 and CNT1, respectively, and 56 genes were co-regulated by CCT1 and CNT1 (Figure 3A). Similarly, of the 1308 BR-regulated genes, 427 (32.65%) were regulated by CRY, and of these 427 genes, 320 (74.94%), and 228 (53.40%) genes were also regulated by CCT1 and CNT1, respectively, and 183 genes were co-regulated by CCT1 and CNT1 (Figure 3B). Of the 211 auxin-regulated genes, 77 (47.39%) were regulated by CRY, and of these 77 genes, 58 (75.32%) and 46 (59.74%) genes were also regulated by CCT1 and CNT1, respectively, and 36 genes were co-regulated by CCT1 and CNT1 (Figure 3C). These results confirmed that both CCT1 and CNT1 participate in CRY1 regulation of phytohormone-responsive genes.
Figure 3.
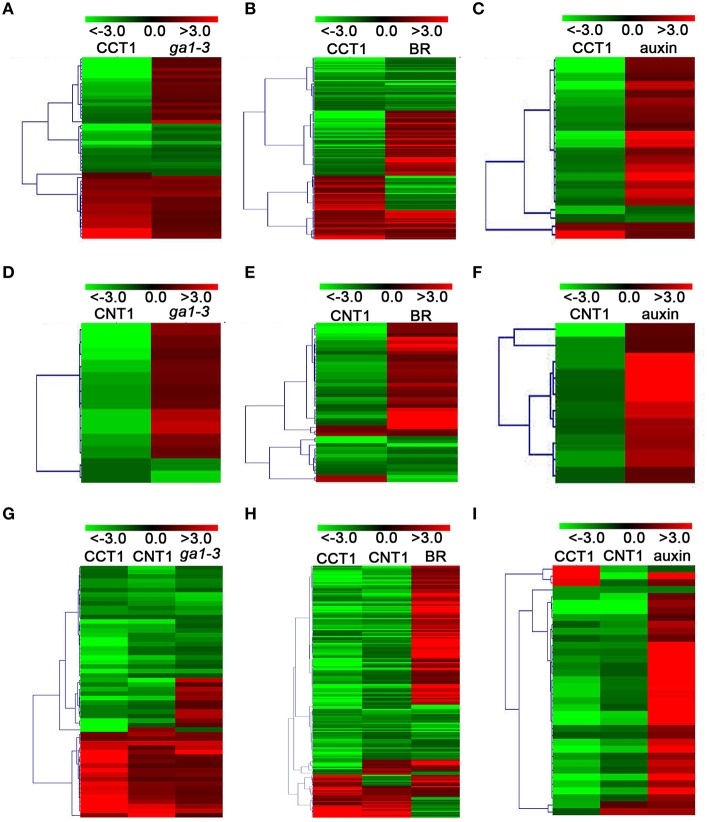
CRY, CCT1, and CNT1 regulate a large number of GA/BR/auxin-responsive genes. (A–C) Venn diagram showing the number of unique and common DEGs among CRY, CCT1, CNT1, and GA (A), BR (B), auxin (C). (D) Hierarchical clustering analysis of 105 CRY/CCT1/ga1-3 overlapping genes as shown in (A). (E) Hierarchical clustering analysis of 320 CRY/CCT1/BR overlapping genes as shown in (B). (F) Hierarchical clustering analysis of 58 CRY/CCT1/auxin overlapping genes as shown in (C). (G) Hierarchical clustering analysis of 66 CRY/CNT1/ga1-3 overlapping genes as shown in (A). (H) Hierarchical clustering analysis of 228 CRY/CNT1/BR overlapping genes as shown in (B). (I) Hierarchical clustering analysis of 46 CRY/CNT1/auxin overlapping genes as shown in (C). Red and green colors in the heat-maps represent induced and repressed genes, respectively. Scale bar denotes the log2 value of fold change.
Heat-map analyses showed that most of the 105 genes co-regulated by CRY, CCT1 and GA were regulated by CRY and CCT1 in the same direction, and that about one third of these genes were up-regulated by GA, but down-regulated by CRY and CCT1 (Figure 3D). Similarly, most of the 320 genes co-regulated by CRY, CCT1, and BR (Figure 3E), and 58 genes co-regulated by CRY, CCT1, and auxin (Figure 3F), were also regulated by CRY and CCT1 in the same direction, respectively, and a number of these genes were up-regulated by BR/auxin, but down-regulated by CRY and CCT1. Moreover, 66 genes co-regulated by CRY, CNT1, and GA (Figure 3G), and 228 genes co-regulated by CRY, CNT1, and BR (Figure 3H), and 46 genes co-regulated by CRY, CNT1, and auxin (Figure 3I), were regulated by CRY and CNT1 in the same direction, and part of these genes were up-regulated by GA/BR/auxin, but down-regulated by CRY and CNT1. These results suggest that the antagonistic regulation of hypocotyl elongation by CRY1 and GA/BR/auxin is likely mediated through CCT1- and CNT1-mediated CRY1 inhibition of genes responsive to GA/BR/auxin.
CCT1 and CNT1 specifically regulate yet co-regulate some of the GA/BR/auxin-responsive genes
To further verify the potential relationship between CCT1 and CNT1 in regulating phytohormone-responsive genes, we analyzed the CCT1- and CNT1-regulated genes that are regulated by CRY in details. We found that, of the 134 GA- and CRY-co-regulated genes, 49 and 10 genes were specifically regulated by CCT1 and CNT1, respectively, and 56 genes were co-regulated by CCT1 and CNT1. Similarly, of the 427 BR- and CRY-co-regulated genes, 137 and 45 genes were specifically regulated by CCT1 and CNT1, respectively, and 183 genes were co-regulated by CCT1 and CNT1. Of the 77 auxin- and CRY-co-regulated genes, 22 and 10 genes were specifically regulated by CCT1 and CNT1, respectively, and 36 genes were co-regulated by CCT1 and CNT1 (Figures 3A–C). The aforementioned genes were indicated by hierarchical clustering analyses, respectively (Figure 4), and heat-map graphs showed that more than half of the GA/BR/auxin up-regulated genes were down-regulated by CCT1 and CNT1, respectively (Figures 4A–F), or both of them (Figures 4G–I). Since CRY1 and GA/BR/auxin act antagonistically to regulate hypocotyl elongation, we further analyzed the genes that are up-regulated by GA/BR/auxin but down-regulated by CRY, CCT1, and CNT1(Table S3), and found that many of these genes have been reported to regulate hypocotyl elongation, including XTHs, SAURs, IAAs, PREs, BBXs, and PARs (Kim et al., 1996; Zhang et al., 2009; Sasidharan et al., 2010; Hao et al., 2012; Spartz et al., 2012; Gangappa and Botto, 2014). Taken together, these results indicate that CCT1 and CNT1 may mediate CRY1 signaling by specifically regulating yet co-regulating some of the phytohormone-responsive genes.
Figure 4.
CCT1 and CNT1 specifically regulate yet co-regulate a large number of the GA/BR/auxin-responsive genes. (A) Hierarchical clustering analysis of 49 CRY/CCT1/ga1-3 overlapping genes as shown in (Figure 3A). (B) Hierarchical clustering analysis of 137 CRY/CCT1/BR overlapping genes as shown in (Figure 3B). (C) Hierarchical clustering analysis of 22 CRY/CCT1/BR overlapping genes as shown in (Figure 3C). (D) Hierarchical clustering analysis of 10 CRY/CNT1/ga1-3 overlapping genes as shown in (Figure 3A). (E) Hierarchical clustering analysis of 45 CRY/CNT1/BR overlapping genes as shown in (Figure 3B). (F) Hierarchical clustering analysis of 10 CRY/CNT1/auxin overlapping genes as shown in (Figure 3C). (G) Hierarchical clustering analysis of 56 CRY/CCT1/CNT1/ga1-3 overlapping genes as shown in (Figure 3A). (H) Hierarchical clustering analysis of 183 CRY/CCT1/CNT1/BR overlapping genes as shown in (Figure 3B). (I) Hierarchical clustering analysis of 36 CRY/CCT1/CNT1/auxin overlapping genes as shown in (Figure 3C). Red and green colors in the heat-maps represent induced and repressed genes, respectively. Scale bar denotes the log2 value of fold change.
Validation of differentially expressed genes by RT-qPCR
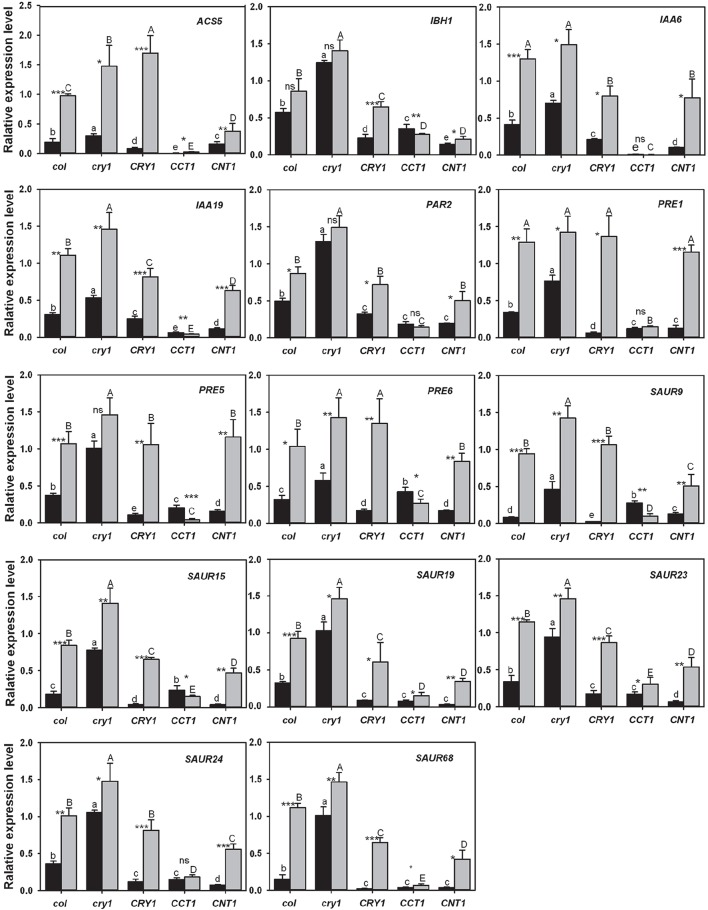
To further confirm the gene expression patterns observed from the transcriptomic analyses, we performed reverse transcription-quantitative PCR (RT-qPCR) analyses with the genes that are relative to hypocotyl elongation and down-regulated by CRY, CCT1, and CNT1 (Table S4), but up-regulated by GA/BR/auxin. We firstly determined whether these genes might be regulated by CRY1-mediated blue light signaling. Indeed, almost all of these genes expressed at significantly higher levels in WT seedlings grown in the dark than in those exposed to blue light (Figure 5). Moreover, upon blue light irradiation, these genes expressed at much higher levels in cry1 mutant than in WT, whereas they expressed at considerably lower levels in CRY1-overexpressing (CRY1-ovx) seedlings than in WT seedlings (Figure 5). We further determined the role for CCT1 or CNT1 in mediating CRY1 regulation of these genes through RT-qPCR, and found that the identified genes specifically regulated by CCT1 indeed expressed at a lower level in CCT1 than in CNT1 seedlings exposed to blue light, such as ACS5. Similarly, the identified CNT1-specific genes expressed at a lower level in CNT1 than in CCT1 seedlings exposed to blue light, such as SAUR15 (Figure 5). Furthermore, all of the identified CCT1- and CNT1-co-regulated genes are indeed down-regulated by CCT1 and CNT1, such as SAUR19 (Figure 5). These results indicated that our RNA-Seq analyses are reliable, and therefore further support the involvement of CCT1 and CNT1 in mediating CRY1 regulation of phytohormone-responsive gene expression.
Figure 5.
qRT–PCR analysis showing the effects of blue light, CRY1, CCT1, and CNT1 on GA/BR/ auxin-responsive gene expression. qRT–PCR analysis showing the effects of blue light, CRY1, CCT1, and CNT1 on the genes that are relative to hypocotyl elongation and are down-regulated by CRY, CCT1, or CNT1, but up-regulated by phytohormones. Data are mean ± SD (n = 3). Asterisks denote significant differences in the gene expression level between the two compared conditions of blue light (black columns) and darkness (gray columns) (*Significant at P < 0.05; **Significant at P < 0.01; ***Significant at P < 0.001, and ns denotes no significant differences at P>0.05) as determined by Student's t test. The lowercase letters (a–e) indicate significant differences among means for the gene expression level of five genotypes of blue-light-grown seedlings, as determined by LSD (least significant difference) at a significance level of 0.05. The uppercase letters (A–E) indicate significant differences among means for the gene expression level of five genotypes seedlings grown in darkness, as determined by LSD (least significant difference) at a significance level of 0.05.
Both CCT1 and CNT1 are involved in mediating CRY1 reduction of the seedlings sensitivity to GA
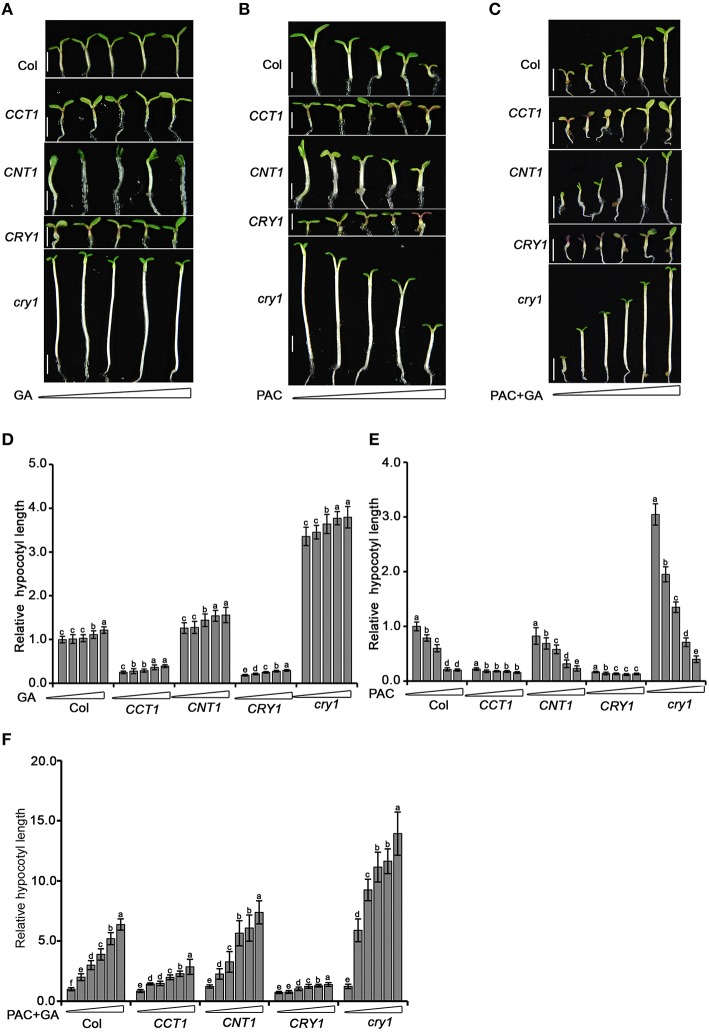
With the demonstration that CCT1 and CNT1 are involved in mediating CRY1 regulation of phytohormone-responsive gene expression, we further explored whether CCT1 and CNT1 are physiologically involved in the regulation of phytohormone response by CRY1. To do this, we tested the hypocotyl elongation response of seedlings to GA and its biosynthesis inhibitor, paclobutrazol (PAC), which are shown to promote and inhibit hypocotyl elongation, respectively (Feng et al., 2008). We found that cry1 mutant shows a long hypocotyl phenotype upon blue light irradiation, but responded only weakly to the increased concentrations of GA compared to WT. However, cry1 mutant showed significantly more sensitive responsiveness to PAC than WT (Figures 6A,B,D,E), indicating that the endogenous GA levels are likely higher in cry1 mutant than in WT (Zhao et al., 2007). In an attempt to minimize the potential GA content difference in WT, cry1 mutant, CRY1-ovx, CCT1, and CNT1 seedlings and obtain more dramatic difference in the responsiveness of these genotypes of plants to GA, we treated these plants firstly with PAC and then with GA, respectively. Indeed, cry1 mutant seedlings exhibited significantly greater responsiveness to GA than WT (Figures 6C,F), confirming the role for CRY1 in repressing GA response (Zhao et al., 2007). However, either CRY1-ovx or CCT1 seedlings hardly responded to GA in either the presence or absence of PAC (Figures 6A–F). Similarly, CNT1 seedlings, which are in the cry1 mutant background, displayed significantly more reduced responsiveness to GA (Figures 6A–F). Taken together, these results indicate that CCT1 and CNT1 are involved in CRY1 inhibition of GA response.
Figure 6.
Hypocotyl elongation analysis of WT, cry1 mutant, CRY1-ovx, CCT1, and CNT1 in response to GA or PAC. (A) Five-day-old Arabidopsis WT, cry1 mutant, CRY1-ovx, CCT1, and CNT1 seedlings grown on half-strength MS plates supplemented gradient concentrations (0, 0.05, 0.2, 0.5, 1 μM) of GA under 30 μmol/m2/s blue light. (B) Five-day-old Arabidopsis WT, cry1 mutant, CRY1-ovx, CCT1, and CNT1 seedlings grown on half-strength MS plates supplemented with gradient concentrations of PAC (0, 0.025, 0.05, 0.2, 0.5 μM) under 30 μmol/m2/s blue light. Means ± SD were obtained from 30 independent plants. (C) Six-day-old Arabidopsis WT, cry1 mutant, CRY1-ovx, CCT1, and CNT1 seedlings grown on half-strength MS plates supplemented with 1 μM PAC plus gradient concentrations of GA (0, 0.5, 1.0, 2.5, 5.0, 10.0 μM) under 30 μmol /m2/s blue light. (D) Hypocotyl length measurement of blue-light-grown seedlings treated with increasing amounts of GA. The concentrations of GA used are 0, 0.05, 0.2, 0.5, and 1 μM (from left to right). (E) Hypocotyl length measurement of blue-light-grown seedlings treated with increasing amounts of PAC. The concentrations of PAC used are 0, 0.025, 0.05, 0.2, and 0.5 μM (from left to right). (F) Hypocotyl length measurement of blue-light-grown seedlings treated with 1 μM PAC plus gradient concentrations of GA. The concentrations of GA used are 0, 0.5, 1.0, 2.5, 5.0, 10.0 μM (from left to right). Seedlings in (D–F) were grown as described in (A–C), respectively. In (D–F), hypocotyl length of untreated wild-type seedlings is set to 100%. Scale bars in (A–C) represent 2.5 mM. The letters (a–f) in (D–F) indicate significant differences among means for the hypocotyl lengths of seedlings treated with the gradient concentration of GA, PAC, PAC plus GA, as determined by LSD (least significant difference) at a significance level of 0.05.
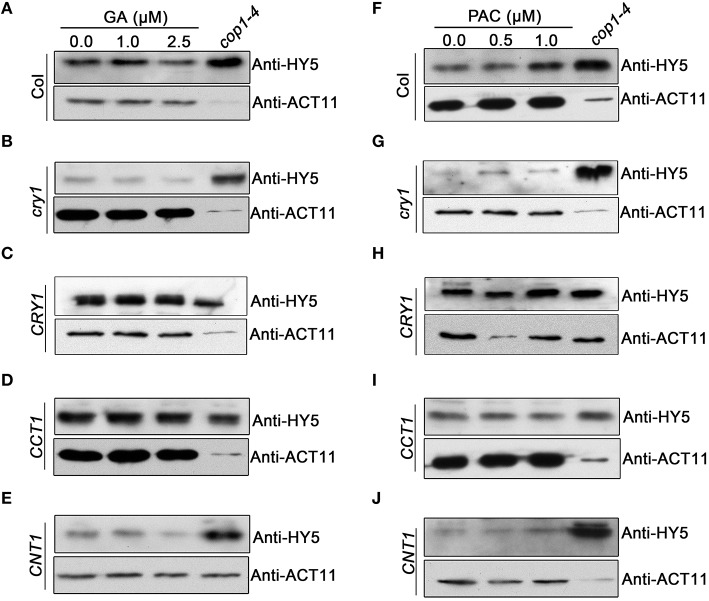
CCT1 mediates CRY1 inhibition of GA-promoted degradation of HY5 protein
HY5 is known to be an important positive regulator of photomorphogenesis, whose protein stability is promoted by light (Osterlund et al., 2000), but reduced by GA (Alabadi et al., 2008). To explore whether CRY1 is involved in regulating GA-promoted degradation of HY5, we performed Western blotting assay using an anti-HY5 antibody to analyze the potential changes of HY5 protein in WT, cry1 mutant, and CRY-ovx seedlings in response to GA and PAC treatments, respectively. We observed that the HY5 protein level in cry1 mutant was very low and reduced much faster compared to WT upon GA application (Figures 7A,B). In contrast, upon PAC application, HY5 accumulates more slowly in cry1 mutant than in WT (Figures 7F,G). Interestingly, HY5 protein level in CRY1-ovx seedlings is basically not affected by GA treatments (Figure 7C). These results indicated that CRY1 is involved in repressing GA-promoted degradation of HY5 protein. We further analyzed HY5 protein levels in CCT1 and CNT1 seedlings treated with GA and PAC, respectively. The results indicate that the dynamic changes of HY5 protein levels in CNT1 seedlings, which are in cry1 mutant background, are basically similar to cry1 mutant (Figures 7E,J), indicating that CNT1 is not likely involved in CRY1 inhibition of GA-promoted HY5 protein degradation. However, GA reduction of HY5 protein levels is inhibited in CCT1 seedlings (Figure 7D), indicating that CCT1 may mediate CRY1 repression of GA-induced HY5 degradation.
Figure 7.
Western blotting assay showing the effects of CRY1, CCT1, and CNT1 on GA or PAC regulation of HY5 protein level. (A–E) HY5 protein levels in blue light-grown WT (A), cry1 mutant (B), CRY1 (C), CCT1 (D) and CNT1 (E) plants, as determined by Western blotting using anti-HY5 antibody. Seedlings were grown for 4 days under blue light on half-strength MS media with agar and gradient GA (0, 1, 2.5 μM) concentrations. (F–J) HY5 protein levels in blue light-grown WT (F), cry1 mutant (G), CRY1 (H), CCT1 (I) and CNT1 (J) plants, as determined by Western blotting using anti-HY5 antibody. Seedlings were grown for 4 days under blue light on half-strength MS media with agar and gradient PAC (0, 0.5, 1 μM) concentrations. Anti-HY5 antibody detection in cop1-4 acted as a positive control. Endogenous actin determined by anti-ACT11 was used as a loading control.
Discussion
Both CCT1 and CNT1 regulate many target genes of CRY1 in genome-wide
The C terminus of CRY1 was shown to mediate CRY1 signaling over 14 years ago (Yang et al., 2000, 2001). Most recently, it has been demonstrated that the N terminus of CRY1 is also involved in CRY1 signaling, which is independent on its C terminus (He et al., 2015). In the present study, we obtained 6780 CCT1-regulated genes through RNA-Seq and compared these genes with those shown to be regulated by CRY and CNT1 previously (He et al., 2015). The results demonstrate the involvement of CCT1 in the regulation of gene expression at the transcriptomic level, which facilitated us to obtain the global “digital” transcriptional information of CCT1. We found that CCT1 influences 2903 (67.85%) of the CRY-regulated genes (Figure 1A), and that CRY-, CCT1-, and CNT1-co-regulated genes are modulated in the same direction (Figure 1B), indicating a close correlation of CCT1 and CNT1 with CRY1 in their regulation of gene expression. Although a previous study analyzed the CCT1 and CCT2 global genome expression profiles using an Arabidopsis cDNA microarray (Wang et al., 2001), its aim was to prove whether transgenic CCT1 and CCT2 seedlings share a similar global genome expression profile to cop1 mutant. In this study, our RNA-Seq analysis not only generated much more target genes than cDNA microarray assay, but revealed the potential common and specific gene targets regulated by CCT1 and CNT1 at the transcriptomic level, and all these genes are also regulated by CRY.
Both CCT1 and CNT1 are involved in mediating CRY1 regulation of phytohormone-responsive genes in genome-wide
GO and KEGG analyses suggest that CCT1 participate in several biology processes, similar to CRY (He et al., 2015), of which the process of “response to many hormones” is one of the most enriched (Figure 2A). Most of overlapping genes regulated by CRY and GA/BR/auxin also were regulated by CCT1 and CNT1 (Figure 3). However, some of the genes regulated by GA/BR/auxin are specifically affected by CCT1 or CNT1 (Figure 4). RT-qPCR assay further confirmed that all the genes that respond to GA/BR/auxin and are related to the regulation of hypocotyl elongation are indeed down-regulated by CRY1, which is likely mediated by CCT1 or CNT1 only or them both (Figure 5). Physiological studies demonstrate that both CCT1 and CNT1 are involved in mediating CRY1 inhibition of GA-enhanced seedlings hypocotyl elongation (Figures 6A–F). Protein expression studies suggested that CCT1 may mediate CRY1 repression of GA-induced HY5 degradation, but not CNT1 (Figures 7A–E). These results, in conjunction with the global genome expression profiles of CRY (He et al., 2015) and CCT1 (this study), indicate that CCT1 and CNT1 may be involved in CRY1 regulation of GA and likely other phytohormones response.
Possible mechanisms of CCT1 and CNT1 in mediating CRY1 regulation of phytohormone-responsive genes
Our transcriptome analysis results show that CCT1/CNT1 specifically regulate many GA/BR/auxin-responsive genes that are regulated by CRY (Figures 3, 4), indicating that CCT1 and CNT1 may adopt different mechanism to mediate CRY1 regulation of phytohormone-responsive genes. Since CCT1 and CCT2 mediate CRY1 and CRY2 signaling through direct interaction with COP1 (Wang et al., 2001; Yang et al., 2001), and CRY1 inhibition of GA-promoted HY5 degradation is mediated by CCT1 (Figure 7), we propose that CCT1-mediated CRY1 regulation of GA response may be mediated through CCT1 regulation of COP1. To test this prediction, we performed RNA-Seq analysis using cop1-4 mutant (Table S5) and obtained 5774 COP1-regulated genes (Table S6). Vern diagram showed that CRY and CCT1 regulate a large number of overlapping genes with COP1 in an opposite direction, which is consistent with their opposite role in the regulation of hypocotyl elongation (Figures S2A,B). Given that COP1 is shown to participate in many phytohormones signaling, including GA/BR/auxin (Alabadi and Blazquez, 2008; Luo et al., 2010; Sassi et al., 2012), these results, in conjunction with the previous microarray data obtained from cop1 mutant (Wang et al., 2001), suggest that CCT1 may regulate phytohormone-responsive genes mainly through regulation of COP1. We also found that CRY and CNT1 regulate a large number of overlapping genes with COP1 in an opposite direction (Figures S3A,B), some of which are GA/BR/auxin-responsive genes (Figures S4A–I). Since CNT1 does not interact with COP1 (Yang et al., 2001), and the enhancement of blue light responsiveness by CNT1 is not mediated through promotion of HY5 accumulation (He et al., 2015), we postulate that CNT1 regulation of phytohormone genes, as well as GA response, may not involve its direct regulation of COP1. However, the downstream components of COP1, including its direct substrates and potentially the components involved in phytohormone signaling, can be directly or indirectly regulated by CNT1, which will be worth exploring in future studies.
It is interesting to note that CCT1 and COP1 do not always regulate the same phytohormone-responsive genes (Figures S4A–C), suggesting that CCT1-mediated CRY1 signaling may not exclusively proceed through COP1. It is shown that light and GA signaling is integrated through direct interaction of DELLA proteins, key negative regulators in GA signaling pathway (Achard et al., 2007), with PIF3 and PIF4, pivotal negative transcriptional factors of photomorphogenesis (Leivar et al., 2008), resulting in inhibition of their DNA-binding activities (de Lucas et al., 2008; Feng et al., 2008). CCT1 may regulate these key factors of light and phytohormone signaling to mediate CRY1 regulation of phytohormone response. In view of the demonstration that CNT2 mediates the interaction of CRY2 with CIBs, bHLH transcription factors, to promote the binding to the promoter of FT to regulate flowering, it will also be worth exploring whether CRY1 interacts with the components in GA/BR/auxin signaling pathways to regulate phytohormone-responsive gene expression and physiological response, through CCT1 or CNT1. Taken together, this study provides clue to further exploring the molecular mechanisms by which CCT1 and CNT1 mediate CRY1 signaling, which may involve regulation of phytohormone-responsive gene expression.
Author contributions
WW and HY designed the project; WW, HL, and ZM performed the experiments; WW analyzed the data; WW and HY wrote the manuscript; all authors reviewed and edited the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Drs. Chun-Ying Kang and Li-Jun Liu for critical reading of this manuscript. This work was supported by grants from the National Natural Science Foundation of China (91217307, 90917014, 31530085, and 30830012 to HY, and 31170266 to HL). China Innovative Research Team, Ministry of Education, and 111 Project (B14016).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00294
References
- Achard P., Liao L., Jiang C., Desnos T., Bartlett J., Fu X., et al. (2007). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143, 1163–1172. 10.1104/pp.106.092254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M., Cashmore A. R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166. 10.1038/366162a0 [DOI] [PubMed] [Google Scholar]
- Alabadi D., Blazquez M. A. (2008). Integration of light and hormone signals. Plant Signal. Behav. 3, 448–449. 10.4161/psb.3.7.5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadi D., Gallego-Bartolome J., Orlando L., Garcia-Carcel L., Rubio V., Martinez C., et al. (2008). Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53, 324–335. 10.1111/j.1365-313X.2007.03346.x [DOI] [PubMed] [Google Scholar]
- Briggs W. R., Christie J. M. (2002). Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci. 7, 204–210. 10.1016/S1360-1385(02)02245-8 [DOI] [PubMed] [Google Scholar]
- Cashmore A. R., Jarillo J. A., Wu Y. J., Liu D. (1999). Cryptochromes: blue light receptors for plants and animals. Science 284, 760–765. 10.1126/science.284.5415.760 [DOI] [PubMed] [Google Scholar]
- Cheminant S., Wild M., Bouvier F., Pelletier S., Renou J. P., Erhardt M., et al. (2011). DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23, 1849–1860. 10.1105/tpc.111.085233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38, 87–117. 10.1146/annurev.genet.38.072902.092259 [DOI] [PubMed] [Google Scholar]
- Chory J., Nagpal P., Peto C. A. (1991). Phenotypic and Genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3, 445–459. 10.1105/tpc.3.5.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S. D., Langford M., McMorris T. C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111, 671–678. 10.1104/pp.111.3.671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluis C. P., Mouchel C. F., Hardtke C. S. (2004). The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 38, 332–347. 10.1111/j.1365-313X.2004.02052.x [DOI] [PubMed] [Google Scholar]
- de Lucas M., Daviere J. M., Rodriguez-Falcon M., Pontin M., Iglesias-Pedraz J. M., Lorrain S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. 10.1038/nature06520 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., et al. (2005). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9, 109–119. 10.1016/j.devcel.2005.05.014 [DOI] [PubMed] [Google Scholar]
- Feng S., Martinez C., Gusmaroli G., Wang Y., Zhou J., Wang F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. 10.1038/nature06448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta K. M., Pontin M. A., Karlin-Neumann G., Bottini R., Spalding E. P. (2003). Genomic and physiological studies of early cryptochrome 1 action demonstrate roles for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J. 36, 203–214. 10.1046/j.1365-313X.2003.01870.x [DOI] [PubMed] [Google Scholar]
- Gangappa S. N., Botto J. F. (2014). The BBX family of plant transcription factors. Trends Plant Sci. 19, 460–470. 10.1016/j.tplants.2014.01.010 [DOI] [PubMed] [Google Scholar]
- Guo H., Yang H., Mockler T. C., Lin C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360–1363. 10.1126/science.279.5355.1360 [DOI] [PubMed] [Google Scholar]
- Hao Y., Oh E., Choi G., Liang Z., Wang Z. Y. (2012). Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 5, 688–697. 10.1093/mp/sss011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S. B., Wang W. X., Zhang J. Y., Xu F., Lian H. L., Li L., et al. (2015). The CNT1 Domain of Arabidopsis CRY1 alone is sufficient to mediate blue light inhibition of hypocotyl elongation. Mol. Plant 8, 822–825. 10.1016/j.molp.2015.02.008 [DOI] [PubMed] [Google Scholar]
- Holm M., Ma L. G., Qu L. J., Deng X. W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16, 1247–1259. 10.1101/gad.969702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K. P., Luo Q., He S. B., Lu X. D., Yang H. Q. (2014). Strigolactone-regulated hypocotyl elongation is dependent on cryptochrome and phytochrome signaling pathways in Arabidopsis. Mol. Plant 7, 528–540. 10.1093/mp/sst093 [DOI] [PubMed] [Google Scholar]
- Jiang L., Liu X., Xiong G., Liu H., Chen F., Wang L., et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405. 10.1038/nature12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C. Y., Lian H. L., Wang F. F., Huang J. R., Yang H. Q. (2009). Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21, 2624–2641. 10.1105/tpc.109.069765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M. J., Hughes R. M., Peteya L. A., Schwartz J. W., Ehlers M. D., Tucker C. L. (2010). Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975. 10.1038/nmeth.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. C., Soh M. C., Kang B. J., Furuya M., Nam H. G. (1996). Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J. 9, 441–456. 10.1046/j.1365-313X.1996.09040441.x [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., et al. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823. 10.1016/j.cub.2008.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. F., He J. X. (2016). BZR1 interacts with HY5 to mediate Brassinosteroid- and Light-Regulated Cotyledon Opening in Arabidopsis in darkness. Mol. Plant 9, 113–125. 10.1016/j.molp.2015.08.014 [DOI] [PubMed] [Google Scholar]
- Lian H. L., He S. B., Zhang Y. C., Zhu D. M., Zhang J. Y., Jia K. P., et al. (2011). Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25, 1023–1028. 10.1101/gad.2025111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. (2002). Blue light receptors and signal transduction. Plant Cell 14Suppl., S207–S225. 10.1105/tpc.000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Robertson D. E., Ahmad M., Raibekas A. A., Jorns M. S., Dutton P. L., et al. (1995). Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269, 968–970. 10.1126/science.7638620 [DOI] [PubMed] [Google Scholar]
- Liu B., Zuo Z., Liu H., Liu X., Lin C. (2011). Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25, 1029–1034. 10.1101/gad.2025011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D., et al. (2008a). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539. 10.1126/science.1163927 [DOI] [PubMed] [Google Scholar]
- Liu L. J., Zhang Y. C., Li Q. H., Sang Y., Mao J., Lian H. L., et al. (2008b). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20, 292–306. 10.1105/tpc.107.057281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X. M., Lin W. H., Zhu S., Zhu J. Y., Sun Y., Fan X. Y., et al. (2010). Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 19, 872–883. 10.1016/j.devcel.2010.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J., Zhang Y. C., Sang Y., Li Q. H., Yang H. Q. (2005). From the cover: a role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc. Natl. Acad. Sci. U.S.A. 102, 12270–12275. 10.1073/pnas.0501011102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Nemhauser J. L., Hong F., Chory J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126, 467–475. 10.1016/j.cell.2006.05.050 [DOI] [PubMed] [Google Scholar]
- Oh E., Zhu J. Y., Bai M. Y., Arenhart R. A., Sun Y., Wang Z. Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 3:e03031. 10.7554/eLife.03031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J. Y., Wang Z. Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–809. 10.1038/ncb2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M. T., Ang L. H., Deng X. W. (1999). The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol. 9, 113–118. 10.1016/S0962-8924(99)01499-3 [DOI] [PubMed] [Google Scholar]
- Osterlund M. T., Hardtke C. S., Wei N., Deng X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. 10.1038/35013076 [DOI] [PubMed] [Google Scholar]
- Quail P. H., Boylan M. T., Parks B. M., Short T. W., Xu Y., Wagner D. (1995). Phytochromes: photosensory perception and signal transduction. Science 268, 675–680. 10.1126/science.7732376 [DOI] [PubMed] [Google Scholar]
- Quail P. H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3, 85–93. 10.1038/nrm728 [DOI] [PubMed] [Google Scholar]
- Richards D. E., King K. E., Ait-Ali T., Harberd N. P. (2001). HOW GIBBERELLIN REGULATES PLANT GROWTH AND DEVELOPMENT: a molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 67–88. 10.1146/annurev.arplant.52.1.67 [DOI] [PubMed] [Google Scholar]
- Rizzini L., Favory J. J., Cloix C., Faggionato D., O'Hara A., Kaiserli E., et al. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103–106. 10.1126/science.1200660 [DOI] [PubMed] [Google Scholar]
- Sang Y., Li Q. H., Rubio V., Zhang Y. C., Mao J., Deng X. W., et al. (2005). N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17, 1569–1584. 10.1105/tpc.104.029645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R., Chinnappa C. C., Staal M., Elzenga J. T., Yokoyama R., Nishitani K., et al. (2010). Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol. 154, 978–990. 10.1104/pp.110.162057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi M., Lu Y., Zhang Y., Wang J., Dhonukshe P., Blilou I., et al. (2012). COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in Arabidopsis. Development 139, 3402–3412. 10.1242/dev.078212 [DOI] [PubMed] [Google Scholar]
- Seo H. S., Yang J. Y., Ishikawa M., Bolle C., Ballesteros M. L., Chua N. H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423, 995–999. 10.1038/nature01696 [DOI] [PubMed] [Google Scholar]
- Somers D. E., Devlin P. F., Kay S. A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282, 1488–1490. 10.1126/science.282.5393.1488 [DOI] [PubMed] [Google Scholar]
- Spartz A. K., Lee S. H., Wenger J. P., Gonzalez N., Itoh H., Inze D., et al. (2012). The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 70, 978–990. 10.1111/j.1365-313X.2012.04946.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M., Nemeth K., Koncz-Kalman Z., Mathur J., Kauschmann A., Altmann T., et al. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. 10.1016/S0092-8674(00)81094-6 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. 10.1038/nature04028 [DOI] [PubMed] [Google Scholar]
- Wang H., Ma L. G., Li J. M., Zhao H. Y., Deng X. W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294, 154–158. 10.1126/science.1063630 [DOI] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Wang X., Zhang X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-Seq data. Bioinformatics 26, 136–138. 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- Weller J. L., Hecht V., Vander Schoor J. K., Davidson S. E., Ross J. J. (2009). Light regulation of gibberellin biosynthesis in pea is mediated through the COP1/HY5 pathway. Plant Cell 21, 800–813. 10.1105/tpc.108.063628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. Q., Tang R. H., Cashmore A. R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13, 2573–2587. 10.1105/tpc.13.12.2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. Q., Wu Y. J., Tang R. H., Liu D., Liu Y., Cashmore A. R. (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103, 815–827. 10.1016/S0092-8674(00)00184-7 [DOI] [PubMed] [Google Scholar]
- Yang J., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., et al. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17, 804–821. 10.1105/tpc.104.030205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K., Oshlack A. (2010). Gene ontology analysis for RNA-Seq: accounting for selection bias. Genome Biol. 11:R14. 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Shalitin D., Liu X., Maymon M., Klejnot J., Yang H., et al. (2007). Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc. Natl. Acad. Sci. U.S.A. 104, 7289–7294. 10.1073/pnas.0701912104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Y., He S. B., Li L., Yang H. Q. (2014). Auxin inhibits stomatal development through MONOPTEROS repression of a mobile peptide gene STOMAGEN in mesophyll. Proc. Natl. Acad. Sci. U.S.A. 111, E3015–E3023. 10.1073/pnas.1400542111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Y., Bai M. Y., Wu J., Zhu J. Y., Wang H., Zhang Z., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21, 3767–3780. 10.1105/tpc.109.070441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Yu X., Foo E., Symons G. M., Lopez J., Bendehakkalu K. T., et al. (2007). A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiol. 145, 106–118. 10.1104/pp.107.099838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Lin Q., Zhu L., Ren Y., Zhou K., Shabek N., et al. (2013). D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410. 10.1038/nature12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z., Liu H., Liu B., Liu X., Lin C. (2011). Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21, 841–847. 10.1016/j.cub.2011.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.