Abstract
Midbrain dopaminergic (DAergic) neurons are heterogeneous and composed of functionally distinct cell populations projecting to the basal ganglia, prefrontal cortex and limbic system. Despite their functional significance, the midbrain population of DAergic neurons is sparse, constituting 20–30,000 neurons in mice, and development of novel tools to identify these cells is warranted. Here, we characterize a bacterial artificial chromosome (BAC) mouse line (Dat1-eGFP) from Gene Expression Nervous System Atlas that expresses enhanced green fluorescent protein (eGFP) under control of the dopamine transporter (DAT) promoter. Confocal microscopy analysis of brain sections showed strong eGFP reporter in midbrain regions and striatal terminals that co-localized with the DAergic markers DAT and tyrosine hydroxylase (TH). Thorough quantification of co-localisation of the eGFP reporter signal with DAT and TH in the ventral midbrain showed that a vast majority of eGFP-expressing neurons are DAergic. Importantly, expression profiles also revealed DAergic heterogeneity when comparing substantia nigra (SN) and ventral tegmental area (VTA). Dat1-eGFP mice showed neither change in synaptosomal dopamine uptake nor altered levels of DAT and TH in both striatum and midbrain. We found no behavioral difference between Dat1-eGFP and wildtype suggesting that the strain is not aberrant. Finally, cell populations highly enriched in DAergic neurons can be obtained from postnatal mice by fluorescence-activated cell sorting and the sorted neurons can be cultured in vitro. Our investigation demonstrates that eGFP expression in this mouse line is selective for DAergic neurons, suggesting that the Dat1-eGFP mouse strain constitutes a promising tool for delineating new aspects of dopamine biology.
Keywords: transgenic mice, dopaminergic cultures, enhanced green fluorescent protein, fluorescence-activated cell sorting, ventral midbrain
Introduction
Dopamine (DA) plays a fundamental role in the central nervous system as a modulatory neurotransmitter regulating important brain functions including locomotion, reward, cognition and neuroendocrine functions (Tritsch & Sabatini, 2012). DAergic neurons are primarily localized in the retrorubral field (RRF, A8), the substantia nigra (SN, A9) and ventral tegmental area (VTA, A10) of the midbrain from where they project to different areas of the basal ganglia, prefrontal cortex and limbic system. The adult midbrain DAergic system is composed of molecularly and functionally distinct cell populations that show specific projection targets, gene expression profiles and physiological features (Lammel et al., 2008). Despite their functional significance, the midbrain population of DAergic neurons is sparse, constituting 20–30,000 neurons in mice (Prasad & Richfield, 2010). It has therefore been a major challenge to study processes selectively taking place in these neurons. Little is therefore known about the global gene expression pattern in DAergic neurons under normal physiological and pathophysiological conditions.
The application of transgenic methodologies has enabled selective expression of fluorescent markers in distinct neuronal populations and greatly facilitated visualization and isolation of neuronal subpopulations (Valjent et al., 2009). GENSAT has created a plethora of BAC transgenic mice where eGFP is expressed under control of promoter regions derived from different cell type-specific neuronal markers. BAC mouse strains have proven useful for identification and detailed characterization of neuronal cell types (Gong et al., 2003; Lobo et al., 2006; Surmeier et al., 2007). However, studies have raised concerns regarding the use of BAC mice, since recent reports revealed behavioural abnormalities in both Drd1-eGFP and Drd2-eGFP mouse lines (Ade et al., 2011; Bagetta et al., 2011; Kramer et al., 2011). Another recent investigation showed that transgenic lines targeting midbrain TH positive DAergic neurons show extensive transgene expression in non-DAergic neurons that might confound midbrain circuitry analysis (Lammel et al., 2015). It is clear that novel transgenic mouse lines should be thoroughly characterized in order to reveal putative aberrant phenotypes before being used as tools in research (Kramer et al., 2011).
We have obtained a BAC transgenic mouse strain from GENSAT expressing eGFP under control of the DAT promoter (Dat1-eGFP). DAT, which mediates reuptake of DA from extracellular medium, is specifically expressed in DAergic neurons (Giros et al., 1996; Rickhag et al., 2013a). Previously, transgenic mouse lines have been generated in order to drive reporter expression in DAergic neurons, taking advantage in particular of the TH promotor (Sawamoto et al., 2001; Donaldson et al., 2005; Jomphe et al., 2005; Kelly et al., 2006). TH, the rate-limiting enzyme in DA synthesis, is a well established marker for identification of DAergic neurons. Although TH has been the golden standard for defining DAergic cells, alone it is not enough to define a dopaminergic phenotype since not all dopaminergic cells are TH positive (Bjorklund & Dunnett, 2007). In the present study, we set out to characterize hemizygous Dat1-eGFP mice to assess the applicability of this strain in the study of DA biology.
Materials and methods
Mice
Dat1-eGFP mice were generated by the GENSAT project (Gong et al., 2003) and cryopreserved embryoes were obtained from Mutant Mouse Regional Resource Centre, strain name: Tg(Slc6a3-EGFP)JN119Gsat. Dat1-eGFP mice were bred to C57BL/6 mice and used in the F1 generation. To avoid or minimize effects of passenger genes in the BAC insert and possible genomic gene-disturbances at insertion site, we used hemizygous Dat1-eGFP mice for all studies. WT and hemizygous Dat1-eGFP mice were housed under diurnal light conditions with food and water ad libitum and mice of both genders were used in the experiments. All mice experiments were performed in accordance with guidelines of the Danish Animal Experimentation Inspectorate (permission number: 2012-15-2934-00279). In total, 101 adult mice were used in the study.
Genotyping
To distinguish between eGFP-expressing mice and non-expressing littermates, all mice were genotyped before use. The BAC insert includes the sequence of eGFP and the DAT promoter, and by testing for the presence of a 450 kb product covering a partial sequence of both the eGFP and the DAT promoter region it was possible to verify the presence of the BAC insert in hemizygous mice. DNA was extracted from tail tips from progeny by adding 75 μl lysis buffer (25 mM NaOH, 0.2 mM disodium EDTA, pH 12) and heating to 95 °C for 30 min and then adding 75 μl neutralizing buffer (40 mM Tris HCl, pH 5). DNA was amplified using PCR by following STOCK Tg(Slc6a3-EGFP) 119Gsat#30526-UCD PCR protocol form from Mutant Mouse Regional Resource Center: US Davis and the primer pairs of Slc6a3 (30526) F1 (CATGGAATTTCAGGTGCTTGGACTA) and Gfp R2 (TAGCGGCTCAAGCACTGCA) under the conditions: one cycle at 95°C of 1 minute, one cycle at 94°C of 20 seconds, 35 cycles at 60–50°C of 30 seconds, one cycle at 72°C of 30 seconds, one cycle at 72°C of 5 min. DNA products were run on a 1% agarose gel at 90 V for 35 min. A 100 base pair ladder was used as a marker of size.
Immunohistochemistry
Adult mice (3–4 months) were anaesthetized and transcardially perfused with 4% paraformaldehyde in 0.1 M PBS. Subsequently, coronal sections (40 μm) from prefrontal cortex, striatum, midbrain, amygdala and hypothalamus were used for immunohistochemistry.
Bright-field immunohistochemistry was performed using a standard peroxidase-based method with 3, 3′-diamino-benzidine. Briefly, brain sections were rinsed in PBS followed by preincubation with 5% goat serum in PBS containing 0.3% Triton X-100 and 1% bovine serum albumin. Sections were incubated with rabbit polyclonal eGFP antibody (AB) (1:1000, Abcam, USA) at 4°C overnight. On the second day, sections were incubated with biotinylated goat anti-rabbit IgG for 1h (1:400, DAKO Cytomation A/S, Denmark). Following avidin-biotin-peroxidase complex (Vector laboratories, USA) incubation, peroxidase was visualized using 3, 3′-diamino-benzidine (0.5 mg/ml) and 0.01% H2O2 treatment. After additional washing, sections were mounted, air-dried overnight and finally cover-slipped using Pertex (Histolab, Sweden).
Co-localization studies of eGFP expressing neurons were performed using triple and dual-labeling immunofluorescence (with and without AB enhancement of the eGFP signal). Co-localization was performed using the following primary and secondary antibodies: rabbit polyclonal eGFP (1:1000, Abcam, USA) followed by secondary AB Alexa-488-goat anti-rabbit IgG (1:1000), rat polyclonal DAT (1:1000, MAB369, Millipore, USA) followed by secondary AB Alexa-647 goat anti rat IgG (1:1000), mouse monoclonal TH (1:1000, MAB318, Millipore, USA) followed by secondary AB Alexa-568 goat anti mouse IgG (1:1000). Briefly, sections were incubated over night with or without rabbit anti-eGFP AB and mouse anti-TH and rat anti-DAT primary AB. The following day, sections were rinsed and incubated with secondary antibodies for 1h (Molecular Probes, USA). Additional rinsing was followed by mounting and cover-slipping using Prolong® Gold antifade reagent (Molecular Probes, USA). Detailed information regarding antibodies can be found in Table 1.
Table 1.
Primary antibodies used in this study
| Antibody | Immunogen | Source | Cat. No | Dilution | Reference |
|---|---|---|---|---|---|
| DAT | N-terminus of DAT | Millipore | MAB369 | 1:1000, rat, polyclonal | Miller et al. 1997 |
| GFP | entire GFP | Abcam | ab290 | 1:1000, rabbit, polyclonal | Dawson et al. 2014 |
| TH | native rat TH | Millipore | MAB318 | 1:1000, mouse monoclonal | Ilijic. et al. 2011 |
| GFP | entire GFP | Invitrogen | A-11120 | 1:1000, mouse monoclonal | Landry et al. 2014 |
Synaptosomal DA uptake
Striata from adult mice (3–5 months) were dissected from coronal slices using a brain matrix and homogenized in ice-cold HEPES buffer (4mM) pH 7.4 containing 0.32 M sucrose using a motor-driven teflon pestle. Crude synaptosome fraction (P2) was obtained as previously described (Richards & Zahniser, 2009) and re-suspended in homogenization buffer followed by protein concentration assessment using BCA™ Protein Assay kit (Thermo Scientific Pierce). Synaptosomal preparations were preincubated with uptake buffer containing 25 mM HEPES pH 7.4, 120 mM NaCl, 5 mM KCl, 1.2 mM CaCl2, 1.2 mM MgSO4, 1 mM L-ascorbic acid, 5 mM D-glucose, 1 μM pargyline, 100 nM desipramine and 1 μM cathechol-O-methyl-transferase inhibitor (RO-41-0960) for 10 min at 37°C. Various DA concentrations (0.015625, 0.03125, 0.0625, 0.125, 0.25, 0.5 and 1.0 μM) were added together with 2, 5, 6-[3H]-DA (91.1 Ci/mmol, Perkin Elmer Life Sciences, USA) for assessment of DA uptake. Non-specific uptake was determined in the presence of 100 μM cocaine. DA uptake was performed for 5 min at 37°C and terminated by addition of ice-cold uptake buffer. Synaptosomes were then loaded on glass microfiber filters (GF/C Whatman® GE Healthcare Life Sciences, USA), rinsed 4 × 5 ml in uptake buffer and allowed to air-dry. Scintillation fluid was added and filters were agitated for 1h followed by counting.
Open-field behavior and cocaine-induced hyperlocomotion
Adult mice (3–4 months) were placed in a white pre-washed open-field arena (41×41×60cm) after 30 min of habituation in the same room as the open field testing. All testing was performed at the same time of the day during the active phase, with a minimum of light in the testing room. The activity was recorded with avideocamera for 180 min, and the behaviour analysed post-recording using EthoVision software as previously described (Fernagut et al., 2003; Rickhag et al., 2013b).
Cocaine-induced hyperlocomotion was performed using a previously described setup (Sorensen et al., 2012). Cocaine-induced hyperactivity was assessed by intraperitoneal injection of cocaine (30mg/kg) or saline 3–7 min. before placing the mice in an activity box for 1 hour.
Culture of DAergic neurons
Ventral midbrain DAergic neurons were prepared according to a previously decribed protocol (Rayport et al., 1992). Briefly, the midbrain was located by dissection in 1 day old Dat1-eGFP mice, and digested for 20 min at 37°C in Hank’s Buffered Salt Solution (HBSS), 1% (v/v) HEPES, 1% (v/v) pen-strep, 1% (v/v) pyruvate, 30 mM glucose, and papain 20 U/ml. The tissue was gently triturated to single cells using pipettes with decreasing size, spun for 10 min at 100 g, and seeded in warm SF1C consisting of 50% MEM (Modified Eagle’s Medium), 40% DMEM, and 10% F-12 Ham’s nutrient mixture (all from Invitrogen) supplemented with bovine serum albumin (2.5 mg/ml), D-glucose (0.35%), glutamine (0.5 mM), 1% heat-inactivated calf serum (Invitrogen), kynurenic acid (5 mM), penicillin, streptomycin, liquid catalase (0.05%), and DiPorzio (di Porzio et al., 1980) (containing insulin, transferrin, superoxide dismutase, progesterone, cortisol, Na2SeO3, and T3) Neurons were grown on a glia cell monolayer on glass coverslips for immunocytochemistry or in LabTek chamber slides (Nunc, Thermo Scientific) for live imaging. All animals were dissected, dissociated and seeded separately. After 2 h the cells were treated with 10 ng/ml glial-derived neurotrophic factor (Millipore, USA). 5-fluorodeoxyuridine was added at 2 DIV (days in vitro) to inhibit further growth of glia cells. The DAergic neurons were used at 8 DIV.
Single cell dissociation and FACS
Hemizygous Dat1-eGFP mice at the age of 2–6 days (P2–6) were sacrificed by decapitation and the brain was removed from the cranial cavity followed by micro-dissection of the midbrain under a microscope (Leica M60, Leica Microsystems) in cold dissection media (10% 10 x HBSS, 1% pyruvate, 1% HEPES; 30 mM glucose). The meninges were removed and the midbrain was cut into three pieces. These were placed in oxygenated (5% CO2, 95% O2) papain solution (1 mM cysteine, 1.5 mM CaCl2, 116 mM NaCl, 5.4 mM KCl, 26 mM NaHCO3, 2 mM NaH2PO2*H2O, 1 mM MgSO4*7H2O, 0.5 mM EDTA, 25 mM glucose, 20 U/ml papain, 0.001% phenol red, 0.5 mM kynurenate, 3.75 mM HCl, in tissue culture water) at 37°C for 25 min. The tissue was then transferred to a new tube and washed twice with 3 ml ice-cold artificial cerebrospinal fluid (aCSF) (120 mM NaCl, 5 mM KCl, 30 mM glucose, 2 mM MgCl2*6H2O, 2 mM CaCl2*2H2O, 25 mM HEPES, pH=7.4). Two ml aCSF was added and trituration was performed with flame heated Pasteur pipettes of decreasing tip diameter. The single cell solution was placed for incubation with Hoechst (1:10,000) for 20 min on ice and centrifuged for 10 min at 200 x g for elimination of debris. The supernatant was discarded and the pellet re-suspended in ice-cold aCSF. The single cell solution was filtered with a 70-μm pore diameter cell strainer before cell sorting.
FACS was performed using a MoFlo Astrios (Beckman Coulter, USA) and Summit software. eGFP reporter expression was used as a reporter for DAergic neurons and gate settings were performed using a WT animal as control. Side and forward scatter gating and Hoechst gating were furthermore used to distinguish between debris, and cells of interest. Cells were therefore selected based on size, complexity and granularity using eGFP fluorescence (at 513 nm (500–526 nm)) vs auto fluorescence at 576 nm (563–589 nm) after activation with a 488 nm laser. As a control, 500 eGFP positively sorted cells were viewed immediately with microscopy (Olympus IX70) to ensure correct setting parameters. Collection of cells was performed on dry ice, and all events positive for eGFP and 50,000 events negative for eGFP were separately collected. Samples were kept at −80°C until further use.
Immunoblotting of brain lysates
Adult mice (3–4 months) were decapitated and the brain was removed from the cranial cavity, and placed on dry-ice for immediate freezing. Brains were sectioned using an ice cold brain matrix and striatum and midbrain were dissected. For midbrain sections, only the ventral part of the sections containing the RRF (A8), SN (A9) and VTA (A10) were used. The microdissected tissue were homogenized in lysis buffer (20 mM Tris-HCl, 300 mM NaCl, 2% Triton-X 100, 0.5% NP-40, 2 mM EDTA/EGTA) and mechanically disrupted using a motor-driven teflon pestle (800 rpm, 10 × 5s). Samples were centrifuged at 16.000 x g for 20 min at 4°C. The supernatant was transferred to a new Eppendorf tube and incubated with loading buffer (100mM dithiothreitol, 4 x SDS loading buffer) for 1h at 37°C, followed by protein concentration assessment using a BCA™ protein Assay kit (Thermo Scientific Pierce). Striatal (5 μg protein) and midbrain (10μg protein) lysates were separated by SDS-PAGE (using any kD gels, BioRad, USA) and transferred to Immobilon-P membranes (Millipore, USA). Membranes were blocked in TBS containing 0.05% Tween-20 and 5% dry milk for 1h (RT), followed by incubation overnight (4°C) with either rat anti-DAT AB (1:1000, MAB369, Millipore, USA) or mouse anti-TH AB (1:1000, MAB318, Millipore, USA) in blocking buffer (TBS/0.05% tween-20 with 5% dry milk). This was followed by washing 3 × 5 min in washing buffer (TBS/0.05% tween-20) and subsequently incubated with secondary HRP-coupled goat anti-rat AB (1:1000, Pierce, USA) (for DAT) or HRP-coupled goat anti-mouse AB (1:1000, pierce, USA) for 1h. After additional rinsing in washing buffer (TBS/0.05% tween-20), the blots were visualized by chemiluminescence (ECL-kit, Amersham, UK) using AlphaEase (Alpha Innotech, USA). Membranes were re-probed for β-actin (HRP-coupled anti-β-actin, 1:15000, Sigma) to verify equal loading. Images were processed using the ImageJ software.
Immunoblotting of FACS purified neurons
FACS-collected eGFP-negative (eGFP−)− and eGFP-positive (eGFP+) cells derived from Dat1-eGFP midbrain where lysed in solubilization buffer (25 mM Tris pH=7.5 containing 150 mM NaCl, 1% Triton X-100, 0.2 mM phenylmethanesulphonylflouride, 5 mM N-ethylmaleimide, and, 1mM EDTA supplemented with protease inhibitor (Roche, Switzerland)). Samples were centrifuged at 16.000 x g for 20 min at 4°C. The supernatant was transferred to a new tube and incubated with loading buffer (100mM dithiothreitol, 4 x SDS loading buffer) for 1h at 37°C. An equal amount of cell lysate (amount adjusted according to cell number derived from FACS) was separated by SDS-PAGE (using any kD gels, BioRad, USA) and transferred to Immobilon-P membranes (Millipore, USA). Membranes were blocked in TBS containing 0.05% Tween-20 and 5% dry milk for 1h (RT). Membranes were incubated overnight (4°C) with either rat anti-DAT AB (1:200, MAB369, Millipore, USA) or mouse anti-TH AB (1:200, MAB318, Millipore, USA) in blocking buffer (TBS/0.05% tween-20 with 5% dry milk). This was followed by washing 3×5 min in washing buffer (TBS/0.05% tween-20) and subsequently 1h incubation with secondary HRP-coupled goat anti-rat AB (1:1000, Pierce, USA) (for DAT) or HRP-coupled goat anti-mouse AB (1:1000, pierce, USA) (for TH). After washing three times for 5 min with washing buffer (TBS/0.05% tween-20), the blots were visualized by chemiluminescence (ECL-kit, Amersham, UK) using AlphaEase (Alpha Innotech, USA). Membranes were re-probed for β-actin (HRP-coupled anti-β-actin, 1:5–15000, Sigma) to verify equal loading. Images were processed with ImageJ software.
FACS-purified DAergic neurons in culture
P0-P2 Dat1-eGFP mice were cryoanesthetized and decapitated for brain tissue collection. For older mice, animals were anaesthetized with halothane (Sigma-Aldrich, Canada) and decapitated. Primary cultures of mesencephalic DA neurons were prepared according to a previously described protocol (Fasano et al., 2008). Mesencephalic cells were plated onto a pre-established monolayer of astrocytes. Cultures were prepared at a density of 5,000 cells/ml of FACS-purified mesencephalic DA neurons from Dat1-eGFP mice, as previously reported (Mendez et al., 2008).
Immunocytochemistry on midbrain cultures
DAergic neurons were fixed using 4% paraformaldehyde in PBS for 15 min at room temperature before blocked and permeabilized using 0.2% saponin in blocking buffer (5% goat serum in PBS). and incubated with the following antibodies: rat anti-DAT (1:1000, MAB369, Millipore, USA), mouse anti-eGFP (1:1000, Abcam, USA), andrabbit anti-TH (1:1000, Affinity Bioreagents, USA) for 1 h. After three washes in blocking buffer, samples were incubated with secondary antibodies; donkey anti-rat-Alexa-568 (for DAT), donkey anti-mouse-Alexa-488 (for eGFP), and donkey anti-rabbit-Alexa-647 for TH) (Invitrogen, USA) for 45min. Finally, the samples were washed twice in blocking buffer and PBS, and once in water before mounted with Prolong® Gold antifade reagent (Molecular Probes, USA).
Immunocytochemistry on FACS-purified cell cultures
Cells on coverslips were fixed with 4% paraformaldehyde, permeabilized, and nonspecific binding sites blocked. Coverslips were then incubated overnight with a rat anti-DAT AB (1:1000, Millipore, USA) and a rabbit anti-eGFP (1:1000, Abcam, UK); these were subsequently detected using a rabbit Alexa-488-conjugated secondary AB and a rat Alexa-647-conjugated secondary AB (1:400, Invitrogen, USA). Images of live or fixed cells were acquired using a 40 × water-immersion objective with a 488 and/or 647 nm laser.
Live cell imaging
The cells were incubated with 10 nM of a rhodamine-conjugated cocaine analogue, JHC 1–64, with high affinity for DAT, for 30 min at either 4°C or room temperature. Incubation at 4°C blocks internalization of bound transporter.
Confocal microscopy
Confocal microscopy was performed using a LSM 510 confocal laser-scanning microscope with an oil immersion 63×1.4 numerical aperture objective (Carl Zeiss). An argon-krypton laser was used to excite Alexa-488 dye, and detection of the emitted light was performed using a 505–530 nm bandpass filter. A 543 nm helium-neon laser was employed to excite Alexa-568 dye and JHC 1–64, and fluorescence was recorded using a 560 nm long-pass filter. Alexa-647 dye was excited using a 633 helium-neon laser and fluorescence was recorded using a 650 nm long-pass filter. Images were analyzed using ImageJ software.
Epifluorescence microscopy
Epifluorescence microscopy was performed using a Zeiss axio scan.Z1, with a plan-apochromat 20x/0.8 objective (Carl Zeiss). LED light sources were used for excitation of fluorophores (493nm (eGFP), 577 (Alexa-568), 653 (Alexa-647)), with an exposure time of 100ms except for eGFP-ir for which the exposure time for the 470 nm LED light could be reduced to 7ms. Fluorescence was recorded using a 525/50 nm band pass filter (eGFP), a 600/70nm band pass filter (Alexa-568) and a 690/50 nm bandpass filter (Alexa-647). We used enhanced dept focusing, performing a maximal projection of a 10 image z-stack covering a total of 10μm of the 40μm thick brain slices.
Quantification of co-localization in ventral midbrain region using immunohistochemistry
Quantification was performed on tilescan images generated by epifluoresence microscopy, and areas of quantification were choosen according to Paxinos and Franklin mouse brain atlas (depicted areas are also shown in Fig. 4A, C). Images for all filters were merged using ImageJ software and the outline of each cell was marked in the merged image. The total number of cells detected and marked in this image was set as the total number of detected cells. A stack was created using all channels together with the merged image containing the marked cells, and hereafter each cell marked in the merged image was investigated in all channels for detection of fluorescent signal. A neuron was considered positive if the fluorescent signal were above background signal. Accordingly seven different cell categories were detected; eGFP+/TH+/DAT+, eGFP+/TH+/DAT−, eGFP+/TH−/DAT+, eGFP+/TH−/DAT−, eGFP−/TH+/DAT+, eGFP−/TH+/DAT−, and eGFP−/TH−/DAT+. Of these types, we further made the distinction of three different populations we named; (i) eGFP-labeled DAergic neurons (eGFP+/TH+/DAT+, eGFP+/TH+/DAT−, eGFP+/TH−/DAT+), (ii) non-eGFP labeled DAergic neurons (eGFP−/TH+/DAT+, eGFP−/TH+/DAT− and eGFP−/TH−/DAT+) and (iii) non-DAergic eGFP labeled neurons (eGFP+/TH−/DAT−). For each region the number of cells in each category was divided by the total number of cells detected in that region to give the relative distribution in percentages. The relative distribution in percentages was calculated for each experiment.
Data analysis
Analysis of difference in subgroup populations between eGFP reporter signal and eGFP-ir experiments was performed using non-parametric Kruskal-Wallis test followed by post hoc analysis. Km and Vmax values for DA uptake experiments were derived by fitting Michaelis-Menten kinetics to the background corrected uptake data. Cocaine-induced hyperactivity was analysed using a one-way ANOVA followed by the Bonferroni post-hoc test. Analysis of changes in behavior of Dat1-eGFP mice compared to WT where performed using non-parametric Mann- Whitney test. All analysis was performed using GraphPad Prism 5.0. If not otherwise stated values are shown as means ± SEM.
Results
eGFP-reporter signal in DAergic areas of Dat1-eGFP mice
In order to investigate cellular and regional eGFP distribution in DAergic areas of Dat1-eGFP mice, we performed dual-labelling immunohistochemical analysis for DAT and the rate-limiting enzyme for DA synthesis, TH, in brain sections derived from hemizygous Dat1-eGFP mice. Confocal immunofluorescence microscopy analysis of the stainings revealed a strong eGFP reporter signal in the ventral midbrain (SN and VTA) of adult mice that localized to the same neurons showing DAT immunoreactivity (DAT-ir) and TH immunoreactivity (TH-ir) (Fig. 1A). Non-transgenic WT littermates showed similar DAT-ir and TH-ir distribution but were completely devoid of eGFP reporter expression (data not shown).
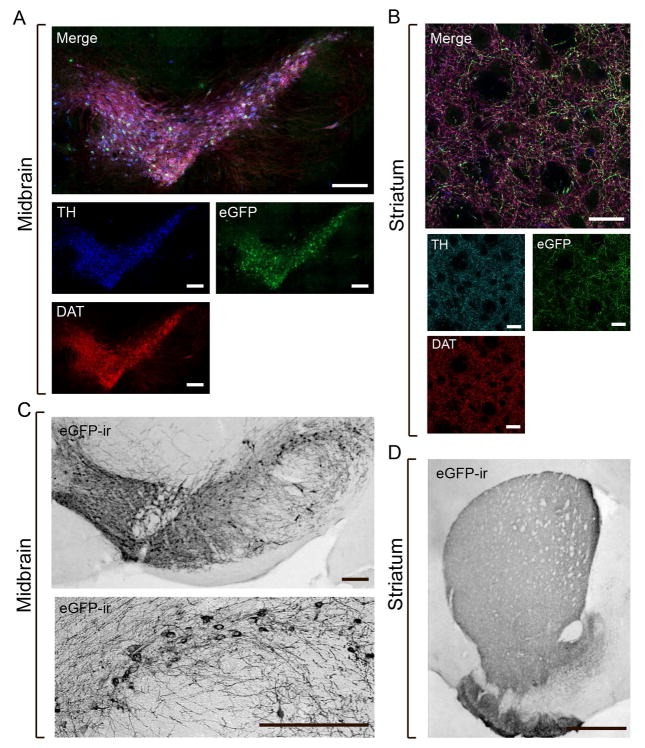
Figure 1. Characterization of eGFP reporter signal in the ventral midbrain and striatum of Dat1-eGFP mice.
Immunohistochemical characterization shows strong eGFP reportersignal in ventral midbrain and striatum of Dat1-eGFP mice that clearly co-localizes with DAergic markers, DAT (red) and TH (blue). Images shown are representative (Dat1-eGFP, n = 3). (A) Epifluorescence tilescan microscopy of the ventral midbrain of Dat1-eGFP mice, showing eGFP reporter signal (green), TH-ir (blue) and DAT-ir (red). Images show substantial but not complete, overlay between eGFP reporter signal and DAT-ir and TH-ir. Scalebar = 200μm. (B) Confocal microscopy of striatum from Dat1-eGFP mice, showing eGFP reporter signal (green), TH-ir (blue) and DAT-ir (red). The eGFP expressing terminals in striatum show a ramified, elaborate network of punctate filaments with significant overlay with the DAergic markers. Scalebar = 20μm. (C–D) eGFP-immunoreactivity (-ir) using immunoperoxidase labelling in ventral midbrain and striatum of hemizygous Dat1-eGFP mice shown using bright-field microscopy. Representative photomicrographs demonstrate dense eGFP-ir in both midbrain perikarya and dendritic processes (C) (scalebar = 200 μm for upper panel; scalebar = 30 μm for lower panel), and in striatal terminals with a dense pattern of labelled axonal fibers in dorsal/ventral striatum and olfactory tubercle (D) (scalebar = 1 mm). Images shown are representative of at least three independent stainings.
Confocal imaging of axonal terminals in the striatum revealed eGFP signal in an extensive network of punctate filaments (Fig. 1B) and, importantly, eGFP distribution showed substantial overlap with DAT-ir and TH-ir. Apart from labelled terminals in the striatal slices, we also observed intervening neuronal perikarya and protruding white matter bundles of the internal capsule (dark areas) (Fig. 1B). In WT mice, we observed no eGFP signal consistent with absence of eGFP expression (data not shown). Use of immunoperoxidase labeling revealed strong eGFP-immunoreactivity (eGFP-ir) in the ventral midbrain as well as dorsal/ventral striatum and olfactory tubercle of the Dat1-eGFP mice (Fig. 1C, D). In the midbrain, the strong eGFP-ir was observed in both perikarya and dendritic processes (Fig. 1C). Extensive eGFP-ir was furthermore observed in striatal terminals with a dense pattern of labelled axonal fibers (Fig. 1D). Altogether, both DAT and TH labelling of showed substantial overlap with the eGFP reporter signal, confirming the DAergic distribution of the eGFP signal in Dat1-eGFP mice.
Midbrain DAergic neurons project to other areas in addition to the dorsal/ventral striatum, including the medial prefrontal cortex and basolateral amygdala. DAergic cell bodies are also localized to the hypothalamus (Lammel et al., 2008). Thus, we investigated the eGFP reporter signal in these additional DAergic areas. However, we were not able to detect any eGFP signal in these areas, except for a few (~5–10) neurons in the hypothalamus near the 3rd ventricle. These neurons also showed strong TH-ir but lacked DAT-ir (data not shown). Mesocortical and basolateral amygdala projections have previously been shown to express sparse immunoreactivity for DAT and to be difficult to detect by immunohistochemistry (Revay et al., 1996; Sesack et al., 1998), which could explain our failure to detect both eGFP signal as well as DAT-ir in these areas. In order to determine whether lack of eGFP signal in these additional DAergic regions might be due to a weak eGFP signal, we enhanced reporter signal in these sections by performing AB labeling of eGFP. However, this did not allow us to detect any reporter signal in these areas (data not shown).
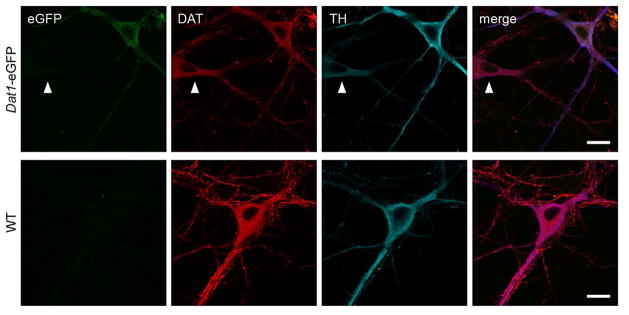
Anatomical characterization of eGFP distribution in RRF (A8), SN (A9) and VTA (A10) in Dat1-eGFP mice
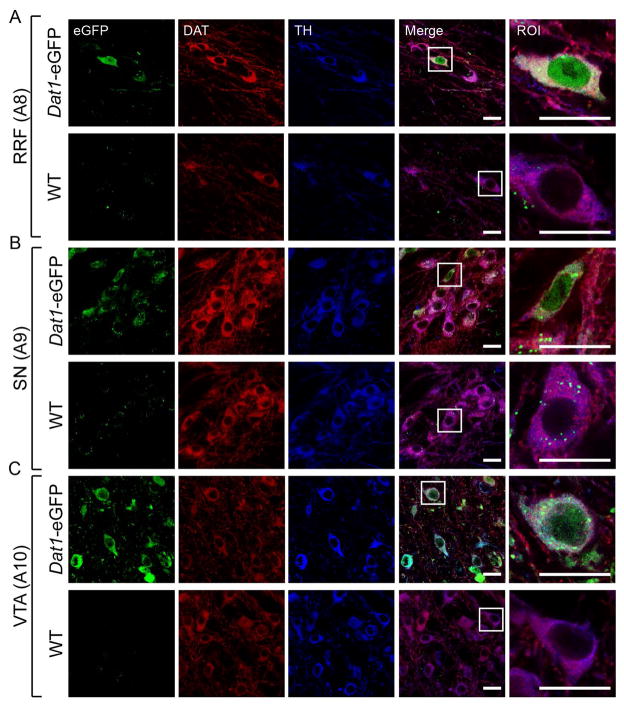
Detailed confocal microscopy analysis of ventral midbrain in hemizygous Dat1-eGFP mice showed strong eGFP signal in RRF, SN and VTA (Fig. 2A, B and C respectively, upper panel). The eGFP signal was particular prominent in the cell body region with diffuse distribution in the entire cell body including the nucleus. DAT-ir and TH-ir showed strong detection in the cytoplasm and especially for DAT-ir, a strong signal in the neuronal processes was observed. For both DAergic markers, we observed substantial overlay with the eGFP signal. Non-transgenic littermates (WT) showed similar DAT-ir and TH-ir distribution, with extensive overlay, but eGFP expression was completely absent (Fig. 2A, B and C lower panels). However, in all regions, a subset of TH-positive or DAT-positive neurons did not contain eGFP signal above background level. We conclude that Dat1-eGFP mice demonstrate extensive, but not complete, overlap of eGFP expression with DAT-ir and TH-ir in the ventral midbrain.
Figure 2. eGFP-expressing neurons from Dat1-eGFP mice demonstrate co-localization with DAergic markers in midbrain subregions.
Confocal images of midbrain sections (40μm) in A8 (A), A9 (B) and A10 (C) from Dat1-eGFP mice (upper panels) and WT (lower panels). Brains sections were immunostained for DAT (red) and TH (blue) as described in Material and Methods. Midbrain neurons show DAT and TH immunofluorescence in the cytoplasm and neuronal processes and extensive overlay is observed with eGFP reporter signal, which is diffusely distributed in the entire cell body including nucleus. Non-transgenic WT littermates show similar DAT and TH distribution but the eGFP signal is completely absent (Fig. 2A–C, lower panels). Scalebar = 20 μm. A region of interest (ROI) from RRF, SN and VTA respectively is depicted to visualize eGFP signal and co-expression with the DAergic markers. Scalebar = 20 μm. Identical confocal laser settings were used for all images, and images shown are representative (Dat1-eGFP: n = 3, WT: n = 1).
Quantification of co-localization between eGFP-immunoreactivity and DAT/TH-immunoreactivity in RRF, SN and VTA
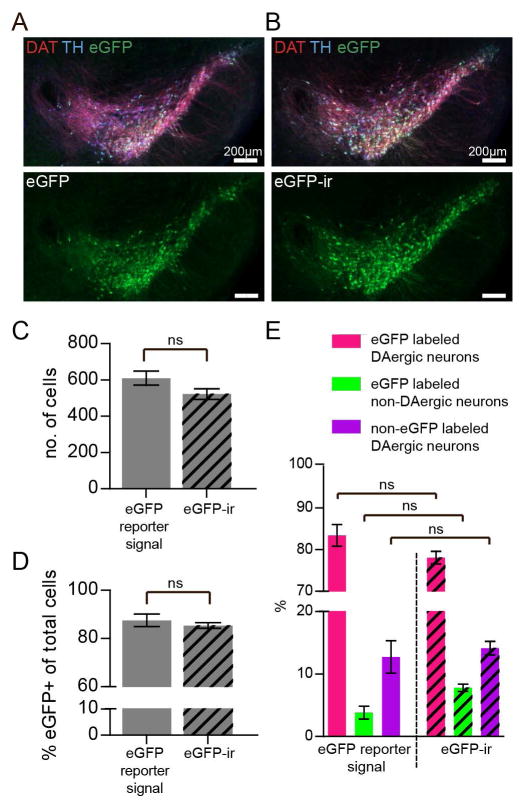
To test the possibility that the apparent absence of eGFP signal in some DAergic neurons was due to weak immunofluorescence at levels close to background, we performed an amplification of the reporter signal by taking advantage of eGFP antibodies (Fig. 3A–E). However, this approach did not increase the total number of detected cells in the midbrain (Fig 3C, eGFP reporter signal = 609.7 ± 38.7, eGFP-ir = 522.3 ± 29.0, non-parametric Mann-Whitney test: (P = 0.40). In addition, there was no change in the percentage of detected eGFP positive cells compared to to the total number of cells (Fig 3D, eGFP reporter signal = 87.5 ± 2.6%, eGFP-ir = 85.4 ± 1.2%; non-parametric Mann-Whitney test: (P = 1.0).
Figure 3. Enhancement of the eGFP signal with antibody does not increase the number of cell detected or the number of eGFP-positive cells in the midbrain of Dat1-eGFP.
(A) Epi-fluorescene images of ventral midbrain sections (40μm) from Dat1-eGFP mice showing the overlay between eGFP reporter signal and DAT-ir and TH-ir. Sections were immunostained for DAT (red) and TH (blue) as decribed in Materials and Methods. Laser exposure times: 493nm/100ms, 668nm/100ms, 577nm/100ms, n = 3. (B) Epi-fluorescene images of sections (40μm) of the ventral midbrain of Dat1-eGFP mice showing the overlay between antibody enhanced eGFP signal (eGFP-ir) (green), DAT-ir (red) and TH-ir (blue) as decribed in Materials and Methods. Laser exposure times: 493nm/7ms, 668nm/100ms, 577nm/100ms, n = 3. (C) Total number of cells detected in merged image for eGFP reporter fluorescence, DAT-ir and TH-ir compared to Antibody (AB) enhanced eGFP fluorescence (eGFP-ir) and DAT-ir and TH-ir. (D) Total number of eGFP positive cells detected in merged image for eGFP reporter signal, DAT-ir and TH-ir compared to eGFP-ir and DAT-ir and TH-ir. (E) Quantification of co-localization of reporter signal or AB enhanced-eGFP signal and DAT-ir and TH-ir in the midbrain of Dat1-eGFP mice based on the three groups; (i) eGFP-labeled DAergic neurons, (ii) non-eGFP labeled DAergic neurons and (iii) eGFP labeled non-DAergic neurons (non-parametric Kruskal-Wallis test and post hoc Dunn’s multiple comparison test: P > 0.05).
We conclude that some of the DAergic neurons in the midbrain did not co-express detectable levels of eGFP, and we therefore set out to investigate and quantify the coexpression of eGFP, DAT and TH in the ventral midbrain of Dat1-eGFP mice. For the quantification, we discriminated between three categories of labelled cells: (i) eGFP-labeled DAergic neurons (eGFP+/DAT+/TH+-neurons, eGFP+/DAT+-neurons or eGFP+/TH+-neurons), (ii) eGFP-labeled non-DAergic neurons (eGFP+/DAT−/TH−- neurons) and (iii) non-eGFP labeled DAergic neurons (DAT+-neurons, TH+-neurons or DAT+/TH+-neurons). See material and methods for further description of labeled cells. We performed this quantification using AB-enhanced eGFP signal, since amplification of the eGFP signal allowed us to decrease the exposure time ~10-fold, thereby decreasing bleaching of the tissue and experimental time. We analyzed whether the quantification would be affected by using AB-enhanced eGFP (eGFP-ir) compared to eGFP reporter signal and found no influence of AB labeling (Fig. 3E, non-parametric Kruskal-Wallis test and post hoc Dunn’s multiple comparison test: P > 0.05).
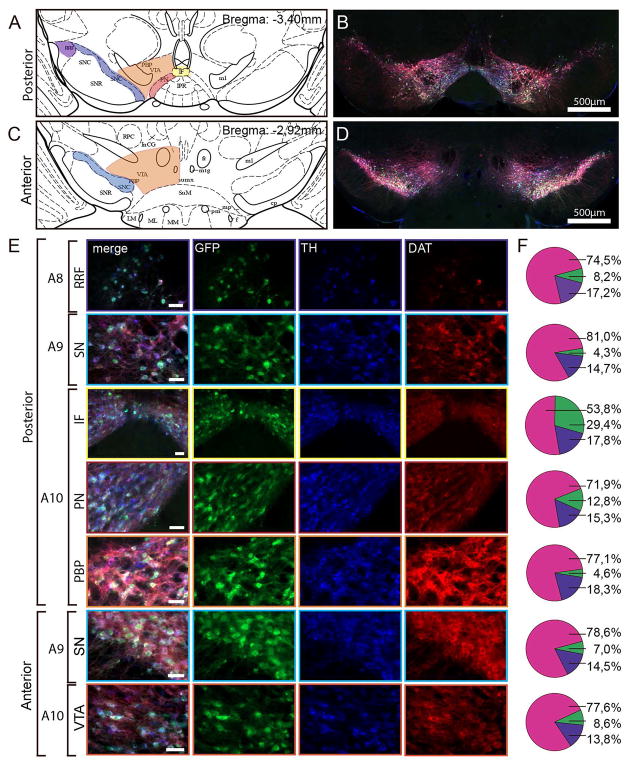
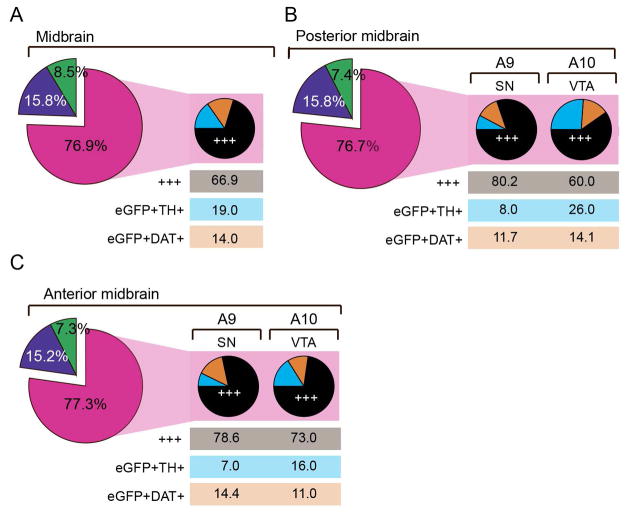
We performed a comprehensive quantification in different midbrain areas (Fig. 4A, C), including RRF (A8), SN (A9) and VTA (A10). In addition, the major subnuclei of the VTA were analyzed including paranigral nucleus (PN), parabrachial nucleus (PBP) and interfascicular nucleus (IF) (Swanson, 1982; Lammel et al., 2008). The quantification was performed as described in Materials and Methods. Our immunohistochemical analysis showed that a vast majority of the total number of neurons detected using the triple labeling for eGFP, DAT and TH was eGFP labeled DAergic neurons (positive for eGFP and either TH or DAT or both) (posterior midbrain; RRF: 74.5 ± 6.3%, SN: 81 ± 1.2%, PBP: 77,1 ± 4.9%, PN: 71,9 ± 5.4% and IF: 53,8 ± 5.1%) (Fig. 4E-F). We observed no difference in distribution of eGFP-labeled DAergic neurons between the different subregions of the midbrain, except for IF in which the percentage of eGFP-labeled DAergic neurons was reduced. Importantly, we observed a minor cell population that was targeted by the eGFP reporter but devoid of detectable levels of DAT or TH (eGFP-labeled non-DAergic neurons) (posterior midbrain; RRF: 8.2 ± 5.3%, SN: 4.3 ± 0.8%, PN: 12.8 ± 2.7%, PBP: 4.6 ± 2.1%, IF: 29.4 ± 5.8%). Another cell population showed undetectable eGFP signal but was still DAergic (non-eGFP labeled DAergic neurons) (posterior midbrain; RRF: 17.2 ± 10.24%, SN: 14.7 ± 1.0%, PN: 15.3 ± 2.7%, PBP: 18.3 ± 3.5%, IF: 17.8 ± 4.1%). A similar pattern was observed for the anterior midbrain, in which a majority of the total number of neurons using triple labeling for eGFP, DAT and TH was eGFP labeled DAergic neurons (SN: 78.6 ± 2.1%, VTA: 77.6 ± 2.6%, Fig. 4E, F). eGFP labeled non-DAergic-neurons and non-eGFP labeled DAergic neurons showed a similar distribution in anterior midbrain as for posterior midbrain (eGFP labeled non-DAergic-neurons, SN: 8.6 ± 1.0%, VTA: 7.0 ± 0.4%, non-eGFP labeled DAergic neurons SN: 14,5 ± 1.7%, VTA: 13.8 ± 1.7%, Fig. 4E, F). We conclude that a similar expression pattern was observed for both anterior and posterior midbrain.
Figure 4. Quantification of co-localization between eGFP-, DAT- and TH-immunofluorescence in the midbrain of Dat1-eGFP mice.
(A) Schematic view of the mouse posterior midbrain at bregma = −3,40mm modified from Paxinos and Franklin (2001). Different midbrain subregions are depictured, including the RRF, SN and VTA subnuclei: IF = interfascicular nucleus, PN = paranigral nucleus, PBP = parabrachial pigmented nucleus, VTA = ventral tegmental area, SNC = substantia nigra pars compacta, SNR = substantia nigra pars reticulata, RRF = retrorubral field. Marked areas represent regions in the posterior midbrain in which co-localization was quantified (purple: RRF, blue: SN, yellow: IF, red: PN, orange: PBP). (B) Fluorescence image of coronal brain slice (40μm) from a representative Dat1-eGFP mouse at bregma −3,4mm showing TH immunofluorescence (blue), eGFP immunofluorescence (green) and DAT immunofluorescence (red). Scalebar = 500μm. Note that eGFP signal was antibody-amplified to obtain stronger fluorescence signal for the quantification analysis. (C) Schematic view of the mouse anterior midbrain at bregma = −2,92mm (modified from Paxinos and Franklin, 2001). VTA = ventral tegmental area, SNC = substantia nigra pars compacta. Marked areas represent regions in the anterior midbrain in which co-localization were quantified (blue: SN, orange: VTA). (D) Fluorescence image of coronal brain slice (40μm) from a Dat1-eGFP mouse at bregma −2,92mm showing TH immunofluorescence (blue), eGFP immunofluorescence (green) and DAT immunofluorescence (red). Scale bar = 500μm. (E) Fluorescence images showing selected ROIs in the following subregions for posterior midbrain; A8 (RRF), A9 (SN), A10 (IF, PN and PBP) and the anterior midbrain; A9 (SN) and A10 (VTA). Scalebars = 50μm. (F) Pie charts illustrate the percentage of co-localization between TH, eGFP and DAT using the following groups: eGFP labeled DAergic neurons (pink) (comprising eGFP+/DAT+/TH+-labeled neurons, eGFP+/DAT+-labeled neurons or eGFP+/TH+-labeled neurons), eGFP labeled non-DAergic neurons (green, comprising eGFP+/DAT−/TH−-labeled neurons) and non-eGFP labeled DAergic neurons (purple, comprising DAT+, TH+ or DAT+/TH+-labeled neurons) (n = 3). Total number of cells counted for quantification: Posterior midbrain; VTA PN = 378, VTA PBP = 437, SN = 499, RRF = 77, IF = 56. Anterior midbrain; SN = 1205, VTA = 556.
For the eGFP labeled DAergic neurons, we further investigated the distribution of the subgroups (eGFP+/DAT+/TH+, eGFP+/DAT+/TH− and eGFP+/DAT−/TH+) in subregions of the posterior and anterior midbrain (Fig. 5A–C). This analysis demonstrated that the majority of this group was triple-labeled when assessing the entire midbrain (eGFP+/DAT+/TH+: 66.9 ± 2.8%, eGFP+/TH+: 19.0 ± 3.1% and eGFP+/DAT+: 14.0 ± 4.8%, Fig. 5A). However, when investigating the distribution of these groups in VTA and SN of the posterior midbrain, we observed a clear shift in distribution of the subpopulations from 80.2 ± 4.2% triple-labeled in SN to 60.0 ± 2.1% in VTA with a similar distribution of eGFP+/DAT+ in both regions but a corresponding increase in eGFP+/TH+ in VTA (eGFP+/TH+: VTA = 26.0 ± 3.0%, SN = 8.1 ± 1.8%) (Fig. 5B). The same tendency was observed in the anterior midbrain (eGFP+/TH+/DAT+; SN = 78.6 ± 4.0%, VTA = 73 ± 3.6%, eGFP+/DAT+; SN = 14.4 ± 3.6%, VTA = 11.0 ± 1.8%, eGFP+/TH+: SN = 7.0 ± 0.4% VTA = 16.0 ± 2.5%). Thus, we observed that for the eGFP labeled DAergic neurons more heterogeneity was observed in the VTA compared to the SN, where the majority of cells was triple labeled.
Figure 5. Subpopulations of eGFP-labeled DAergic neurons.
Piecharts showing the distribution of subpopulations of eGFP-labeled DAergic neurons in the midbrain (A), subregions of the anterior midbrain (B) and subregions of the posterior midbrain (C). Following color scheme is applied for the three subpopulations: (i) eGFP+/DAT+/TH+ (+++, triple positive) = black, (ii) eGFP+/DAT/+TH− (eGFP+DAT+, DAT positive eGFP-labeled neurons) = orange and (iii) eGFP+/DAT−/TH+ (eGFP+TH+, TH positive eGFP-labeled neurons) = cyan. The percentages are weighed according to total number of detected cells in region (large piecharts, no. of total detected cells: midbrain = 3015, anterior midbrain = 1567, posterior midbrain = 1448) or total number of eGFP-labeled DAergic neurons in each subregion (small piecharts; no. of eGFP-labeled DAergic neurons in regions: Midbrain = 2321, anterior midbrain: SN = 761, VTA = 449, posterior midbrain: SN = 398, VTA= 655. Quantifications were performed on brain slices (40μm) from three hemizygous Dat1-eGFP mice (n = 3). Areas of quantification were choosen based on Paxinos and Franklin (2001). Images for quantification were aquired using epi-fluorescence microscopy with enhanced depth focusing (Z-stack covered 10μm) and for all experiments the same laser settings were applied.
In conclusion, our detailed quantification of eGFP-labeling in RRF, SN and the major subnuclei of VTA (PBP, PN and IF) revealed that a vast majority is selectively targeted to DAergic neurons (both for posterior and anterior midbrain more than 75% of the labeled neurons were considered DAergic). Nevertheless, we also identified two minor cell populations that showed either ectopic expression of the eGFP reporter (eGFP labeled non-DAergic) or were DAergic neurons without detectable eGFP expression (non-eGFP labeled DAergic neurons). Our analysis of the eGFP-labeled DAergic neurons revealed DAergic midbrain heterogeneity when comparing SN and VTA, in which cells located in SN showed a higher fraction of triple labeled neurons (eGFP+/DAT+/TH+) while neurons in VTA showed increased number of cells dual-labeled for eGFP and TH (eGFP+/TH+).
Unaltered DAT and TH protein expression and functional DAT-mediated DA uptake in Dat1-eGFP mice
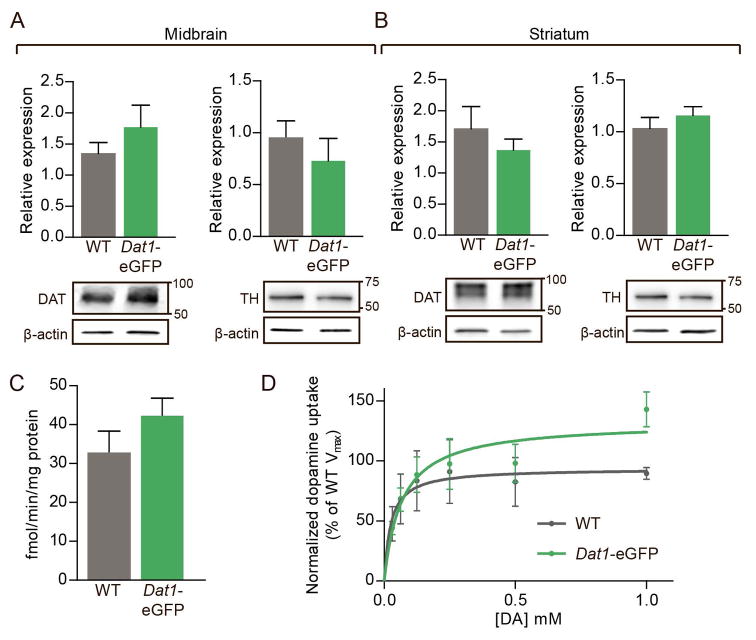
Recent investigations have shown that some BAC transgenic mouse lines display altered behavioral and functional properties compared to their WT littermates (Bagetta et al., 2011; Kramer et al., 2011). To confirm that expression levels of the different DA markers, DAT and TH, are not aberrant in Dat1-eGFP mice compared to WT mice, we performed immunoblotting on striatal and midbrain total tissue lysates. Immunoblotting showed that protein levels for both DAT and TH were not altered in DAergic areas of hemizygous Dat1-eGFP mice when compared to WT (Fig. 6A, B). Note that DAT in striatal extracts eluted as a broad band correponding to a MW of 70–80 kDA, representing fully glycosylated DAT (Rickhag et al., 2013a). In the midbrain extracts, DAT also eluted as a rather broad band but correponding to a slightly lower MW suggesting predominant existence of less glycosylated forms in the perikarya.
Figure 6. DAT and TH protein expression is unaltered in dopaminergic areas from Dat1-eGFP mice concomitant with unaltered DAT-mediated DA transport.
(A) Immunoblotting confirmed that protein levels of both DAT and TH were unaltered in midbrain from Dat1-eGFP mice when compared to WT. Upper panels, densitometric analysis of immunoblots for WT and Dat1-eGFP (n = 4). DAT and TH expression levels were normalized to β-actin. Lower panels, representative immunoblots for DAT and TH. (B) Immunoblotting confirmed that levels of both DAT and TH were unaltered in striatum from Dat1-eGFP mice compared to WT. Upper panels, densitometric analysis of immunoblots for WT and Dat1-eGFP (n = 4). DAT and TH expression levels were normalized to β-actin. Lower panels, representative immunoblots for DAT and TH. (C) Dopamine uptake in striatal synaptosomes from Dat1-eGFP mice (green circles) and WT (black circles). No difference in the Vmax values for synaptosomal DA uptake in Dat1-eGFP and WT mice (n = 4) compared to WT (Vmax for Dat1-eGFP = 42.3±4.5 fmol/min/μg; Vmax for WT = 32.8±5.6 fmol/min/μg, n = 4). (D) Data are shown as normalized saturation curves for DA uptake (normalized to WT littermate controls, n = 4 of triplicate determinations).
To investigate whether hemizygous Dat1-eGFP mice display normal DA transport kinetics compared to non-transgenic WT, we isolated synaptosomes from striatum and assessed DA uptake. Experiments showed that Dat1-eGFP mice have unaltered DA uptake compared to WT littermates (Fig. 6C, D). Dat1-eGFP mice demonstrated similar uptake capacity (Vmax) compared to WT (Vmax for Dat1-eGFP = 42.3 ± 4.5 fmol/min/μg; Vmax for WT = 32.8 ± 5.6 fmol/min/μg, (non-parametric Mann-Whitney test: P = 0.34, n = 4)) with no difference in apparent affinity (Km for Dat1-eGFP = 83 ± 19 nM; Km for WT = 65 ± 39 nM, n = 4).
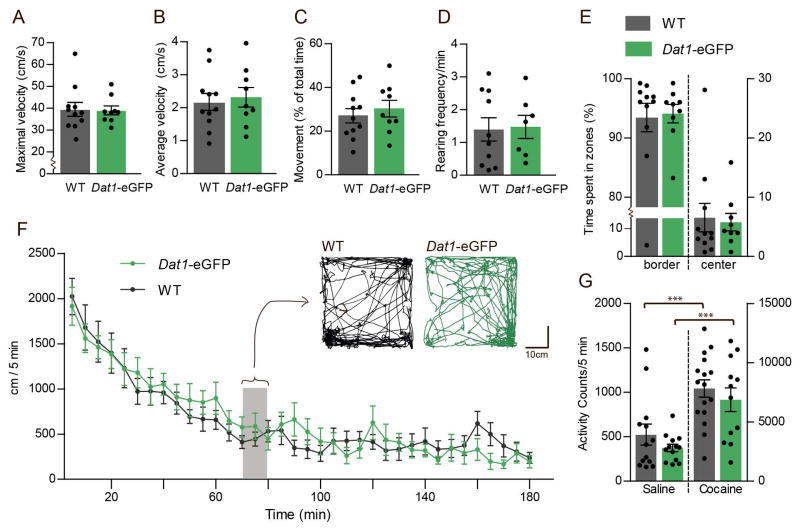
Dat1-eGFP mice demonstrate unaltered open-field behavior and cocaine-induced hyperactivity compared to wildtype mice
To investigate potential behavioural abnormalities in the transgenic Dat1-eGFP mouse strain, we assessed basal locomotion, anxiety-related behavior and cocaine-induced hyperactivity in hemizygous Dat1-eGFP mice and WT controls (Fig. 7). We investigated the basal locomotion of Dat1-eGFP mice compared to WT littermates in an open-field (41×41cm) over a time period of 180 min. We observed no difference in maximal velocity (Fig. 7A, non-parametric Mann-Whitney test: P = 0.82), average velocity (Fig. 7B, non-parametric Mann-Whitney test: P = 0.29) or percentage of movement (Fig. 7C, non-parametric Mann-Whitney test: P = 0.17, n ≥ 9 in all groups). Furthermore, we found that the habituation of Dat1-eGFP mice were similar to that of WT littermates (Fig 7F, n ≥ 9 in all groups). We furthermore investigated exploratory behavior by measuring the rearing frequency (i.e. standing on hindlimbs) of Dat1-eGFP mice compared to WT and found no difference when compared to WT littermates (Fig. 7D, non-parametric Mann-Whitney test: P = 0.29, n ≥ 9 in all groups). We also investigated anxiety-related behavior in the Dat1-eGFP mice by quantification of time spent in zones (border and center) during 180 min in open-field (41×41cm, center zone 21×21cm) and observed no difference based on genotype (Fig. 7E, non-parametric Mann-Whitney test: P = 0.44, n ≥ 9 in all groups). Finally, the WT and Dat1-eGFP mice responded similarly to a cocaine challenge (30mg/kg) when mice were placed in activity boxes and the number of beam brakes was recorded (Fig. 7G, versus saline control, one-way ANOVA followed by posthoc Bonferroni multiple comparison test: F3,50 = 41.84, P < 0.001, n ≥ 12 in all groups). Importantly, saline treated mice from both Dat1-eGFP mice and WT showed few counts in the activity boxes due to habituation prior to the psychostimulant challenge.
Figure 7. Dat1-eGFP mice demonstrate unaltered open-field behavior and cocaine-induced hyperactivity compared to wildtype mice.
Open-field locomotor activity (41×41cm) showing the average maximal velocity (A), average velocity (B), percentage of movement (D) and rearing frequency (C) of the of Dat1-eGFP mice (n = 9) compared to WT mice (n = 11). No difference is observed between the two groups (E) Assessment of anxiety-related behavior in Dat1-eGFP mice compared to WT. Open-field activity showing percentage of time spent in border versus center of the open-field arena. No difference was found between Dat1-eGFP and WT littermates. (F) Basal locomotion of Dat1-eGFP compared to WT littermates in open-field showing habituation (5 min. time bins, 180 min.). No difference was found between Dat1-eGFP and WT littermates. Insert shows representative traces for (10min) Dat1-eGFP (green trace) and WT (black trace) respectively at t = 70–80 min during open field testing (41x41cm). (H) Assessment of locomotor activity for Dat1-eGFP and WT mice upon treatment with the psychostimulant cocaine (30mg/kg) or saline in activity boxes (F3,50 = 41.84, P < 0.001 versus saline control, one-way ANOVA followed by posthoc Bonferroni multiple comparison test, n ≥ 12 in all groups).
In conclusion, we did not observe any apparent functional or behavioural abnormalities in the transgenic mice when assessing DA uptake, basal locomotion, explorative behavior, anxiety-like behavior and cocaine-induced hyperlocomotion.
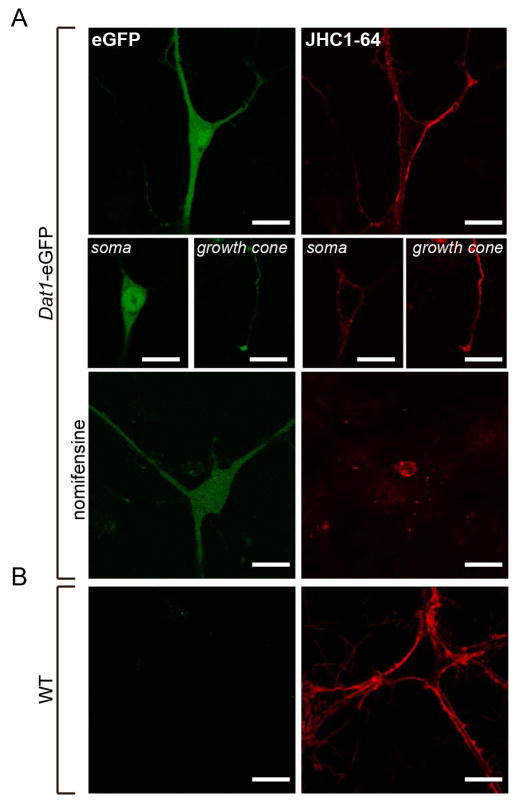
Labeling of live midbrain Dat1-eGFP neurons using the fluorescent cocaine analogue JHC 1–64
Next we generated primary cultures of midbrain neurons from hemizygous Dat1-eGFP mice in order to evaluate the usefulness of this strain to confirm the neurochemical phenotype of living neurons labelled with fluorescent indicators, using the fluorescent cocaine analogue JHC 1–64 as a proof of principle. Individual mice at postnatal day 1 (P1) were dissected and neurons dissociated and seeded as described in Material and Methods. To visualize DAT, we used JHC 1–64, which we previously reported as a suitable and highly specific marker for visualization of DAT plasma membrane expression in live DAergic neurons (Eriksen et al., 2009; Rickhag et al., 2013a). As illustrated in Fig. 8A, a strong eGFP reporter signal was found in the same neurons that displayed distinct JHC 1–64 labeling of surface DAT (Fig. 8A, upper panel). The eGFP reporter signal was particular strong in the perikarya but also visible in most dendritic and axonal regions while JHC 1–64 labelling was most prominent in the dendritic and axonal regions, which are regions where the plasma membrane levels of DAT are most predominant (Fig. 8A, middle panels) (Nirenberg et al., 1996; Eriksen et al., 2009). By applying the specific DAT antagonist nomifensine, we prevented all labeling with JHC 1–64 of eGFP positive neurons, validating the specificity of the signal and the selectivity of the identification of DAT-expressing neurons based on the eGFP reporter signal (Fig. 8A, lower panel). As expected, no eGFP signal was detected in JHC 1–64 labelled neurons in the non-transgenic WT mice (Fig. 8B).
Figure 8. The fluorescent cocaine analogue JHC 1–64 labels eGFP-expressing cells in live midbrain dopaminergic cultures from Dat1-eGFP mice.
(A) Confocal images of cultured midbrain neurons from Dat1-eGFP mice labeled with JHC 1–64. The representative images show the signal corresponding to the somatodendritic compartment, and growth cones (8A, middle panels). Incubation with the DAT specific antagonist nomifensine (100 μM) prior to and during JHC 1–64 incubation completely abolished the binding. The JHC 1–64 signal distributes to the plasma membrane of the somas and to the neuronal extensions as well as a clear signal is found corresponding to the growth cones. Importantly, JHC 1–64 exclusively labeled neurons from Dat1-eGFP mice that also displayed an eGFP signal. The eGFP signal was homogenously distributed in the entire cytoplasm with intense expression in the somatic region. (B) Confocal images of cultured midbrain neurons from non-transgenic WT mice labeled with JHC 1–64. Despite JHC 1–64 labeling of several neurons, we observe no eGFP signal. Scale bar = 20 μm. Images shown are representative of at least three independent experiments.
Finally, taking advantage of these primary cultures, we examined if the profile expression of eGFP and DAergic markers in vivo was maintained in vitro. We immunostained the neurons for both DAT and TH, and performed confocal microscopy analysis on the resulting fixed samples. DAT- and TH- immunofluorescence were observed in the same neurons as those expressing the eGFP reporter, thus validating a DAergic phenotype of eGFP neurons (Fig. 9). Importantly, we observed a small subset of neurons that showed DAT and TH immunofluorescence but that did not express detectable levels of the eGFP reporter signal (Fig. 9, white arrow). This further substantiates our finding from the quantification of co-localization in the intact ventral midbrain that a minor fraction of the DAergic neurons does not show detectable eGFP signal (“non-eGFP labeled DAergic neurons”).
Figure 9. Cultured midbrain dopaminergic neurons from Dat1-eGFP mice show overlap between the immunosignal from eGFP and dopaminergic markers DAT and TH.
Confocal images of fixed neuronal midbrain cultures from Dat1-eGFP mice (upper panels) and WT littermates (lower panels) immunostained for eGFP (green), DAT (red) and TH (blue). The images show overlap between DAergic markers (DAT and TH) and immunosignal of the eGFP reporter. Both DAT and TH signal are present in the soma, axonal network and the dendrites. Note the presence of a non-eGFP expressing DAergic cell, consistent with our previous finding of non-eGFP expressing DAergic neurons in the ventral midbrain of Dat1-eGFP mice (white arrow). Non-transgenic WT mice showed complete lack of eGFP signal. Scale bar = 20 μm. Images shown are representative of at least three independent stainings.
Fluorescence activated cell sorting and culturing of postnatal DAergic neurons from Dat1-eGFP mice
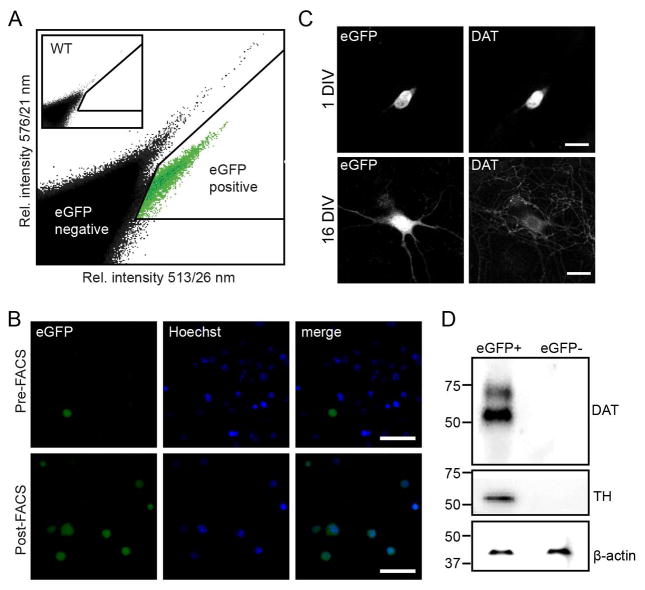
To solve the major challenge of obtaining pure preparations of DAergic neurons, we sought to take advantage of the Dat1-eGFP mice in FACS experiments to purify these neurons. Ventral midbrain DAergic neurons from Dat1-eGFP mice at postnatal day 2–6 (P2–6) were dissociated and subjected to FACS to select eGFP-positive neurons. Pooled midbrain neurons from 1–4 hemizygous Dat1-eGFP mice yielded in the range of 3,000 to 20,000 eGFP+ events. FACS-isolation of Dat1-eGFP neurons convincingly demonstrated enrichment of a population of eGFP+ cells in mice carrying the Dat1-eGFP transgene while controls were completely devoid of eGFP+ signal (Fig. 10A). Purified cells were investigated using epifluorescence microscopy and the majority of all sorted eGFP+ cells showed eGFP expression and were co-labeled for Hoechst, a nuclear staining marker used for identification of viable neurons (Fig. 10B). To confirm the DAergic phenotype of the eGFP+ cells, we performed immunoblotting on FACS-purified neurons for the DAergic markers, DAT and TH. A rat anti-DAT AB recognized two bands, one that most likely corresponded to the fully glycosylated transporter (~70 kDa) and one conceivably detecting core glycosylated immature transporter eluting at 55 kDa (Bjerggaard et al., 2004) (Fig. 10D). The high intensity of the 55 kDa band might reflect that we have specifically isolated DAergic perikarya that predictably would contain more immature transporter in the biosynthetic pathway compared to the entire neuron. Indeed, a similar band is not readily detected when assaying whole brain lysates and lysates from midbrain DAergic neuronal cultures (Rickhag et al., 2013a). A mouse anti-TH AB recognized a band at approximately 55 kDa, corresponding to the TH protein. This analysis of the sorted samples confirms isolation of proper DAergic cells. The eGFP-negative population showed complete absence of DAT and TH proteins (Fig. 10D). FACS-purified neurons were also cultured in vitro to assess post-sorting viability. Images from representative neurons showed that eGFP-ir was restricted to perikarya at 1 DIV and included more dendritic and axonal processes at 16 DIV due to a differentiated cellular morphology. Furthermore, at 1 DIV, 100% of the eGFP+ cells were expressing DAT (21 neurons out of 21 investigated neurons) while at 16 DIV, 91% of eGFP+ cells were expressing DAT (30 neurons out of 33 investigated neurons) (Fig. 10C). Importantly, no DAT+ cells were present in the eGFP− cells. In conclusion, we demonstrated that we can purify postnatal DA neurons on the basis of eGFP signal. Furthermore we show that these neurons are viable, enriched for DAT and TH, and can be cultured for more than two weeks.
Figure 10. Fluorescence activated cell sorting and culturing of sorted postnatal dopaminergic neurons from Dat1-eGFP mice.
(A) FACS-purification of cells isolated from the midbrain of hemizygous Dat1-eGFP mice (P2–6). Representative scatter plot showing the emission intensity of sorted events at 513/26 nm against 576/21 nm after excitation at 488 nm. Events arise from sorting of a single cell suspension of ventral midbrain neurons from transgenic Dat1-eGFP mice. Insert shows a similar sorting from a WT animal. Note the appearance of a population of events at a higher 513/26 nm intensity for the Dat1-eGFP animal. These events are designated eGFP positive events (eGFP+), events outside this gate are designated eGFP negative events (eGFP−). (B) Confocal microscopy of cultured FACS sorted eGFP+ events at 1 DIV (upper panel) and 16 DIV (lower panel), showing eGFP reporter signal and DAT-ir. Sorted eGFP+ cells are viable and can be cultured for more than two weeks, in which period they will protrude extensions. Furthermore, expression of eGFP reporter signal and DAT-ir is detectable from 1 DIV. Scalebars = 50μm. (C) Visualization of FACS-purified cells from the Dat1-eGFP midbrain suspensions confirmed eGFP+ phenotype. Upper panels; epifluorescence microscopy of cells before FACS (pre-FACS), Lower panels; epi-fluorescence microscopy of FACS-purified eGFP+ cells (post-FACS). Hoechst, a nuclear staining marker, was included to discriminate between viable and non-viable cells during the sorting process. Scalebars = 50μm. (D) Immunoblotting confirms a dopaminergic phenotype of the eGFP+ sorted cell population. Representative immunoblots demonstrate enrichment of the dopaminergic markers DAT and TH in cell lysates from the eGFP+ cell population while DA markers are completely absent in the GFP− population (n = 3). Cell lysates from purified eGFP+ and eGFP− cells were analyzed with SDS-PAGE followed by immunoblotting for anti-DAT and anti-TH antibody as described in Material and Methods. DAT and TH is exclusively expressed in lysates from eGFP+ sorted cells. Note that anti-DAT antibody recognizes two bands corresponding to the mature (~70 kDa) and immature (~55 kDa) isoforms of DAT while anti-TH recognizes one band at (~55 kDa). β-actin was used to verify equal loading for respective cell population.
Discussion
To assess the applicability of Dat1-eGFP mice as a novel tool for investigation of the DAergic system, we carried out an extensive immunohistochemical, functional and behavioral analysis of this transgenic mouse strain. Our study demonstrates that basal locomotion, exploratory behavior, anxiety-related behavior and cocaine-induced hyperactivity are not compromised in hemizygous Dat1-eGFP mice when compared to WT. Furthermore, we show that the functional properties of DAT are not compromised in Dat1-eGFP mice and that the bulk expression levels of DAT and TH in striatum and midbrain is unaltered compared to WT. A detailed quantification of eGFP-reporter distribution in RRF, SN and VTA midbrain DAergic cell populations reveals that a vast majority is localized to bona fide DAergic neurons. Furthermore, we successfully FACS-purified DAergic cell populations from postnatal Dat1-eGFP mice, and validated that the eGFP+ sorted cells are enriched for proteins specific for DAergic neurons. We finally show post-sorting viability of the sorted midbrain DAergic neurons, which can be cultured in vitro for more than two weeks.
Phenotypic characterization of the novel transgenic mouse line Dat1-eGFP
GENSAT has created a plethora of BAC transgenic mice with constructs encoding eGFP driven by promoter regions of different cell type-specific neuronal markers. Several of these mouse strains have already been used as tools when addressing biological and physiological questions (Gong et al., 2003; Lobo et al., 2006; Surmeier et al., 2007). Recently, it has been discussed whether BAC transgenic mouse lines display altered behavioural and physiological traits when compared to their WT littermates (Kramer et al., 2011; Nelson et al., 2012). The issue of artificial behavioural traits caused by the random BAC insertion was, to our knowledge, first raised by Kramer and colleagues (Kramer et al., 2011). They reported that a homozygous BAC transgenic mouse expressing eGFP in dopamine D2 receptor positive neurons (Drd2-eGFP mice) displayed increased dopamine D2 receptor mRNA and protein levels accompanied by hyperlocomotion. Furthermore, Bagetta and colleagues demonstrated differences in the pattern of long-term depression in medium spiny neurons from BAC transgenic mice expressing eGFP in dopamine D1 receptor positive neurons (Drd1-eGFP mice) (Bagetta et al., 2011). However, another study concluded that hemizygous Drd1-eGFP and Drd2-eGFP mice are indistinguishable from WT littermates in open-field behaviour and in their acute response to cocaine (Nelson et al., 2012). Recently, Lammel et al (2015) showed that a transgenic mouse line targeting TH-expressing neurons exhibit substantial transgene expression in non-DAergic neurons in the ventral midbrain. The authors argued that ectopic transgene expression reporter substantially confounds interpretation of midbrain circuitry studies, and further highlighting the importance of genuine characterization of transgenic mouse lines that target distinct neuronal cell populations (Lammel et al., 2015). The debate about potential aberrant phenotypes underlines the significance of thoroughly characterizing new transgenic mouse strains before using them as tools in research.
In the present study, we set out to to assess the value of the Dat1-eGFP mice as a tool for studying the DAergic system. The strain was created by introducing a BAC construct covering the genetic sequence of the DAT promoter region and an eGFP protein into the genome of C57BL/6 mice (Heintz, 2004). We investigated DA-mediated behavior to reveal any aberrant phenotype in Dat1-eGFP mice compared to WT by performing open-field behaviour and cocaine-induced hyperactivity in activity boxes. Open-field test has previously been used to assess aspects of anxiety-like behavior (Prut & Belzung, 2003) and also revealed aberrant anxiety-like behavior in Drd2-eGFP mice (Ade et al., 2011). Our investigation of basal locomotion, exploratory behavior, anxiety-related behavior and cocaine-induced hyperlocomotion showed no aberrant phenotype in Dat1-eGFP mice compared to WT. We found no change in DAT-mediated DA uptake in striatal synaptosomes, which is in further agreement with an unaltered DAT function. Furthermore, protein levels of DAT and TH in the midbrain and striatum of Dat1-eGFP mice are unaltered compared to WT. DAT is essential for maintaining DA homeostasis and loss of striatal DAT is known to cause hyperlocomotion and alterations of postsynaptic DA receptor expression (Giros et al., 1996; Rickhag et al., 2013a). We convincingly show preserved DA transporter function and DA-mediated behavior in Dat1-eGFP mice compared to WT ruling out an aberrant DAergic phenotype in this transgenic strain.
Immunohistochemical analysis of targeting efficiency for eGFP reporter in ventral midbrain
In our initial investigation of the reporter eGFP signal in Dat1-eGFP mice, we observed that the eGFP reporter signal co-localized with neurons showing DAT- and TH-ir. However, we also observed that the overlap was not complete and we therefore quantified the degree of co-localisation between eGFP reporter and DAT and TH in ventral midbrain.
TH-ir has been the golden standard for identification of DA neurons for many years but concerns have been raised regarding the use of a single gene marker to define the DAergic phenotype (Bjorklund & Dunnett, 2007; Stuber et al., 2015). Midbrain DAergic neurons are heterogeneous and display differential axonal projection targets, gene expression patterns as well as electrophysiological properties (Lammel et al., 2008). A recent investigation showed that a small subset of rat VTA neurons express enzymes responsible for DA synthesis (TH and L-amino acid decarboxylase (AADC)) but lack detectable expression of vesicular monoamine transporter-2 and DAT, proteins controlling reuptake and vesicular packing of DA (Yamaguchi et al., 2015). Another cell population in the striatum and cortical areas shows TH expression but lack AADC further arguing that presence of TH alone is not always a reliable criteria to identify DA neurons (Weihe et al., 2006). These unconventional features have recently been brought to attention to argue that the very definition of a DAergic phenotype has become complex, and highlights the importance of not only using a single gene marker to identify DAergic neurons (Bjorklund & Dunnett, 2007; Stuber et al., 2015).
In our study, we combined the expression of DAergic markers (TH and DAT) in order to include the heterogeneous DAergic midbrain population, accurately defined based on multiple markers (Bjorklund & Dunnett, 2007). Our immunohistochemical analysis showed that a vast majority of the total number of neurons detected using the triple labeling for eGFP, DAT and TH were eGFP labeled DAergic neurons (positive for eGFP and either TH or DAT or both, Fig. 4). A minor cell population showed expression of eGFP but was devoid of detectable levels of DAT or TH (eGFP-labeled non-DAergic neurons). This population of eGFP-expressing neurons can be considered as ectopically labeled neurons. Previous investigations of transgenic strains have also reported ectopic reporter expression (Sawamoto et al., 2001; Kelly et al., 2006). Recently, Lammel et al (2015) showed substantial transgene expression in non-DAergic VTA neurons of TH-Cre and TH-eGFP mice. The authors only reported the ectopic transgene expression but did not investigate the presence of DAergic neurons lacking eGFP expression (Lammel et al., 2015). Importantly, our investigation reveals that a minor fraction of DAergic neurons is not labeled by the eGFP reporter. Our detailed quantification of the ventral midbrain shows that, although not all DA neurons expressed eGFP all DAergic regions expressed the reporter gene with no subregion specific change in expression. This finding argues that the eGFP reporter is a reliable marker for the identification of DAergic neurons in all subnuclei of the ventral midbrain. Moreover, although we do identify a minor population devoid of eGFP reporter signal, this population is also distributed equally throughout the ventral midbrain subnuclei.
Our analysis of subgroups of eGFP labeled DAergic neurons demonstrate that the majority of this group was triple labeled (positive for eGFP, TH and DAT) when assessing the entire midbrain (Fig. 5). However, when comparing SN to VTA, we observed a shift in the fraction of triple labeled (eGFP+/DAT+/TH+) cells, with fewer cells triple labeled in the VTA compared to SN. Instead, in this subregion of the midbrain, a larger fraction of cells showed dual labeling for eGFP and TH (eGFP+/DAT−/TH+) (Fig. 5B, C). This finding substantiates a previous study showing low DAT mRNA expression in mesocortical projecting DA neurons originating specifically from VTA subnuclei (Lammel et al., 2008). Mesocortical projections have also been shown to express sparse immunoreactivity for DAT and have been difficult to detect by immunohistochemistry (Sesack et al., 1998). Our findings emphasizes DAergic heterogeneity in ventral midbrain since DAergic mesocortical projections is different from mesostriatal projections by showing distinct expression profiles (e.g. triple labeled neurons in SN compared to dual labeled neurons in VTA).
Purification of postnatal DAergic neurons from Dat1-eGFP mice by fluorescence-activated cell sorting
The midbrain area in mice only contains 20–30,000 DAergic neurons and isolation of these neurons has been a major challenge because they only constitute approximately 10–50% of the total neuronal population in this area (Rayport et al., 1992; Eriksen et al., 2009; Prasad & Richfield, 2010). Both the SN and VTA contain cell types expressing other neurotransmitters such as γ-amino butyric acid, glutamate as well as various neuropeptides (Yetnikoff et al., 2014) some of which are co-localized (El Mestikawy et al., 2011). Our investigation demonstrated that viable populations of DAergic neurons can be obtained from early postnatal Dat1-eGFP mice by utilizing FACS. The neurons were isolated from postnatal Dat1-eGFP mice (P2–6) and a DAergic phenotype was confirmed by immunoblotting for DAT and TH.
FACS is a commonly used method for isolation of various cell subtypes. Recently, the method has successfully been applied in the field of neuroscience to isolate neuronal subtypes from brain regions with high cell-diversity, in particular from an embryonic stage (Lobo et al., 2006; Zhou et al., 2009; Okada et al., 2011; Guez-Barber et al., 2012). Lobo et al. showed successful FACS-purification of both juvenile and adult striatal neurons derived from BAC mice labelled with the fluorescent reporter eGFP. Specifically, they showed that two functionally different subsets of striatal neurons isolated by FACS retain cell-type specific transcripts and identified differentially regulated genes. This pivotal approach opened the possibility of gene expression profiling on subsets of neuronal ensembles in the basal ganglia (Lobo et al., 2006).
Isolation of DAergic neurons using FACS has been accomplished previously. In one case, this involved AB labelling and fixation of the cells (Guez-Barber et al., 2012), thus, preventing isolation of viable neurons. Zhou et al. (2009) used knock-in mice expressing DAT fused to eGFP and were able to isolate viable DAergic neurons by FACS. However, the knock-in mouse strain expressing the eGFP-DAT was compromised by an abolished DAT function, which presumably was the cause of a significant locomotor hyperactivity observed in these mice (Zhou et al., 2009). Interestingly, DAergic neurons have also been isolated from mice expressing eGFP or YFP under control of the TH promoter (TH-eGFP and TH-YFP mice) (Sawamoto et al., 2001; Donaldson et al., 2005; Kelly et al., 2006). Moreover, Trudeau and co-workers have utilized TH-eGFP mice for FACS-purification and culture of postnatal DA neurons (Mendez et al., 2008; Fortin et al., 2012).
The present study is, to the best of our knowledge, the first study to purify viable DAergic neurons from postnatal mice not phenotypically aberrant using the expression of eGFP under control of the DAT promoter. Dat1-eGFP mice represent a promising new tool for obtaining viable preparations of pure DAergic neurons using FACS. Indeed, this possibility for high-yield isolation of these neurons now makes numerous applications possible. Thus, mRNA can be isolated from the cells and used for transcriptional analysis of selected protein families by employment of real-time PCR or for full-genome transcriptional profiling using microarray technologies (Lobo et al., 2006; Karsten et al., 2008). Alternatively, despite the relatively small cell yields, it should be possible to also employ advanced proteomics to assess protein expression in such isolated neurons. Altogether, Dat1-eGFP mice hold great promises for novel detailed investigations of the physiology and pathophysiology of the DAergic system.
Acknowledgments
The work was supported by the National Institute of Health Grants P01 DA 12408 (UG), the Lundbeck Foundation (MR), the Danish Medical Research Council, University of Copenhagen BioScaRT Program of Excellence (UG, GS) and the Lundbeck Foundation Center for Biomembranes in Nanomedicine (UG, JE, TRC). The work was also supported by the Canadian Institutes of Health, the Krembil Foundation and Brain Canada (LT). We thank Consiglia Pacelli for contributing to the manuscript. The fluorescent cocaine analogue JHC 1–64 was kindly provided by Dr. Amy Hauck Newman at the National Institute on Drug Abuse, Baltimore, USA. Epifluorescent images were provided by the Core Facility for Integrated Microscopy at University of Copenhagen.
Abbreviations
- ANOVA
analysis of variance
- AB
antibody
- BAC
bacterial artificial chromosome
- DIV
days in vitro
- DA
dopamine
- DAergic
dopaminergic
- DAT
dopamine transporter
- eGFP
enhanced green fluorescent protein
- FACS
fluorescence activated cell sorting
- GENSAT
Gene Expression Nervous System Atlas
- ir
immunoreactivity
- IF
interfascicular nucleus
- PN
paranigral nucleus
- PBP
parabrachial nucleus
- RRF
retrorubral field
- ROI
region of interest
- SN
substantia nigra
- TH
tyrosine hydroxylase
- VTA
ventral tegmental area
- WT
wildtype
References
- Ade KK, Wan Y, Chen M, Gloss B, Calakos N. An Improved BAC Transgenic Fluorescent Reporter Line for Sensitive and Specific Identification of Striatonigral Medium Spiny Neurons. Front Syst Neurosci. 2011;5:32. doi: 10.3389/fnsys.2011.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagetta V, Picconi B, Marinucci S, Sgobio C, Pendolino V, Ghiglieri V, Fusco FR, Giampa C, Calabresi P. Dopamine-dependent long-term depression is expressed in striatal spiny neurons of both direct and indirect pathways: implications for Parkinson’s disease. J Neurosci. 2011;31:12513–12522. doi: 10.1523/JNEUROSCI.2236-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerggaard C, Fog JU, Hastrup H, Madsen K, Loland CJ, Javitch JA, Gether U. Surface targeting of the dopamine transporter involves discrete epitopes in the distal C terminus but does not require canonical PDZ domain interactions. J Neurosci. 2004;24:7024–7036. doi: 10.1523/JNEUROSCI.1863-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund A, Dunnett SB. Dopamine neuron systems in the brain: an update. Trends Neurosci. 2007;30:194–202. doi: 10.1016/j.tins.2007.03.006. [DOI] [PubMed] [Google Scholar]
- di Porzio U, Daguet MC, Glowinski J, Prochiantz A. Effect of striatal cells on in vitro maturation of mesencephalic dopaminergic neurones grown in serum-free conditions. Nature. 1980;288:370–373. doi: 10.1038/288370a0. [DOI] [PubMed] [Google Scholar]
- Donaldson AE, Marshall CE, Yang M, Suon S, Iacovitti L. Purified mouse dopamine neurons thrive and function after transplantation into brain but require novel glial factors for survival in culture. Mol Cell Neurosci. 2005;30:601–610. [PubMed] [Google Scholar]
- El Mestikawy S, Wallen-Mackenzie A, Fortin GM, Descarries L, Trudeau LE. From glutamate co-release to vesicular synergy: vesicular glutamate transporters. Nat Rev Neurosci. 2011;12:204–216. doi: 10.1038/nrn2969. [DOI] [PubMed] [Google Scholar]
- Eriksen J, Rasmussen SG, Rasmussen TN, Vaegter CB, Cha JH, Zou MF, Newman AH, Gether U. Visualization of dopamine transporter trafficking in live neurons by use of fluorescent cocaine analogs. J Neurosci. 2009;29:6794–6808. doi: 10.1523/JNEUROSCI.4177-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano C, Thibault D, Trudeau LE. Culture of postnatal mesencephalic dopamine neurons on an astrocyte monolayer. Curr Protoc Neurosci. 2008;Chapter 3(Unit 3):21. doi: 10.1002/0471142301.ns0321s44. [DOI] [PubMed] [Google Scholar]
- Fernagut PO, Chalon S, Diguet E, Guilloteau D, Tison F, Jaber M. Motor behaviour deficits and their histopathological and functional correlates in the nigrostriatal system of dopamine transporter knockout mice. Neuroscience. 2003;116:1123–1130. doi: 10.1016/s0306-4522(02)00778-9. [DOI] [PubMed] [Google Scholar]
- Fortin GM, Bourque MJ, Mendez JA, Leo D, Nordenankar K, Birgner C, Arvidsson E, Rymar VV, Berube-Carriere N, Claveau AM, Descarries L, Sadikot AF, Wallen-Mackenzie A, Trudeau LE. Glutamate corelease promotes growth and survival of midbrain dopamine neurons. J Neurosci. 2012;32:17477–17491. doi: 10.1523/JNEUROSCI.1939-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giros B, Jaber M, Jones SR, Wightman RM, Caron MG. Hyperlocomotion and indifference to cocaine and amphetamine in mice lacking the dopamine transporter. Nature. 1996;379:606–612. doi: 10.1038/379606a0. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Guez-Barber D, Fanous S, Harvey BK, Zhang Y, Lehrmann E, Becker KG, Picciotto MR, Hope BT. FACS purification of immunolabeled cell types from adult rat brain. J Neurosci Methods. 2012;203:10–18. doi: 10.1016/j.jneumeth.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. Gene expression nervous system atlas (GENSAT) Nat Neurosci. 2004;7:483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- Jomphe C, Bourque MJ, Fortin GD, St-Gelais F, Okano H, Kobayashi K, Trudeau LE. Use of TH-EGFP transgenic mice as a source of identified dopaminergic neurons for physiological studies in postnatal cell culture. J Neurosci Methods. 2005;146:1–12. doi: 10.1016/j.jneumeth.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Karsten SL, Kudo LC, Geschwind DH. Gene expression analysis of neural cells and tissues using DNA microarrays. Curr Protoc Neurosci. 2008;Chapter 4(Unit 4):28. doi: 10.1002/0471142301.ns0428s45. [DOI] [PubMed] [Google Scholar]
- Kelly BB, Hedlund E, Kim C, Ishiguro H, Isacson O, Chikaraishi DM, Kim KS, Feng G. A tyrosine hydroxylase-yellow fluorescent protein knock-in reporter system labeling dopaminergic neurons reveals potential regulatory role for the first intron of the rodent tyrosine hydroxylase gene. Neuroscience. 2006;142:343–354. doi: 10.1016/j.neuroscience.2006.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer PF, Christensen CH, Hazelwood LA, Dobi A, Bock R, Sibley DR, Mateo Y, Alvarez VA. Dopamine D2 receptor overexpression alters behavior and physiology in Drd2-EGFP mice. J Neurosci. 2011;31:126–132. doi: 10.1523/JNEUROSCI.4287-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Hetzel A, Hackel O, Jones I, Liss B, Roeper J. Unique properties of mesoprefrontal neurons within a dual mesocorticolimbic dopamine system. Neuron. 2008;57:760–773. doi: 10.1016/j.neuron.2008.01.022. [DOI] [PubMed] [Google Scholar]
- Lammel S, Steinberg EE, Foldy C, Wall NR, Beier K, Luo L, Malenka RC. Diversity of transgenic mouse models for selective targeting of midbrain dopamine neurons. Neuron. 2015;85:429–438. doi: 10.1016/j.neuron.2014.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Mendez JA, Bourque MJ, Dal Bo G, Bourdeau ML, Danik M, Williams S, Lacaille JC, Trudeau LE. Developmental and target-dependent regulation of vesicular glutamate transporter expression by dopamine neurons. J Neurosci. 2008;28:6309–6318. doi: 10.1523/JNEUROSCI.1331-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AB, Hang GB, Grueter BA, Pascoli V, Luscher C, Malenka RC, Kreitzer AC. A comparison of striatal-dependent behaviors in wild-type and hemizygous Drd1a and Drd2 BAC transgenic mice. J Neurosci. 2012;32:9119–9123. doi: 10.1523/JNEUROSCI.0224-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg MJ, Vaughan RA, Uhl GR, Kuhar MJ, Pickel VM. The dopamine transporter is localized to dendritic and axonal plasma membranes of nigrostriatal dopaminergic neurons. J Neurosci. 1996;16:436–447. doi: 10.1523/JNEUROSCI.16-02-00436.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, Saiwai H, Kumamaru H, Kubota K, Harada A, Yamaguchi M, Iwamoto Y, Ohkawa Y. Flow cytometric sorting of neuronal and glial nuclei from central nervous system tissue. J Cell Physiol. 2011;226:552–558. doi: 10.1002/jcp.22365. [DOI] [PubMed] [Google Scholar]
- Prasad K, Richfield EK. Number and nuclear morphology of TH+ and TH− neurons in the mouse ventral midbrain using epifluorescence stereology. Exp Neurol. 2010;225:328–340. doi: 10.1016/j.expneurol.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Prut L, Belzung C. The open field as a paradigm to measure the effects of drugs on anxiety-like behaviors: a review. Eur J Pharmacol. 2003;463:3–33. doi: 10.1016/s0014-2999(03)01272-x. [DOI] [PubMed] [Google Scholar]
- Rayport S, Sulzer D, Shi WX, Sawasdikosol S, Monaco J, Batson D, Rajendran G. Identified postnatal mesolimbic dopamine neurons in culture: morphology and electrophysiology. J Neurosci. 1992;12:4264–4280. doi: 10.1523/JNEUROSCI.12-11-04264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revay R, Vaughan R, Grant S, Kuhar MJ. Dopamine transporter immunohistochemistry in median eminence, amygdala, and other areas of the rat brain. Synapse. 1996;22:93–99. doi: 10.1002/(SICI)1098-2396(199602)22:2<93::AID-SYN1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Richards TL, Zahniser NR. Rapid substrate-induced down-regulation in function and surface localization of dopamine transporters: rat dorsal striatum versus nucleus accumbens. J Neurochem. 2009;108:1575–1584. doi: 10.1111/j.1471-4159.2009.05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickhag M, Hansen FH, Sorensen G, Strandfelt KN, Andresen B, Gotfryd K, Madsen KL, Vestergaard-Klewe I, Ammendrup-Johnsen I, Eriksen J, Newman AH, Fuchtbauer EM, Gomeza J, Woldbye DP, Wortwein G, Gether U. A C-terminal PDZ domain-binding sequence is required for striatal distribution of the dopamine transporter. Nat Commun. 2013a;4:1580. doi: 10.1038/ncomms2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickhag M, Owens WA, Winkler MT, Strandfelt KN, Rathje M, Sorensen G, Andresen B, Madsen KL, Jorgensen TN, Wortwein G, Woldbye DP, Sitte H, Daws LC, Gether U. Membrane-permeable C-terminal dopamine transporter peptides attenuate amphetamine-evoked dopamine release. J Biol Chem. 2013b;288:27534–27544. doi: 10.1074/jbc.M112.441295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamoto K, Nakao N, Kobayashi K, Matsushita N, Takahashi H, Kakishita K, Yamamoto A, Yoshizaki T, Terashima T, Murakami F, Itakura T, Okano H. Visualization, direct isolation, and transplantation of midbrain dopaminergic neurons. Proc Natl Acad Sci U S A. 2001;98:6423–6428. doi: 10.1073/pnas.111152398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesack SR, Hawrylak VA, Matus C, Guido MA, Levey AI. Dopamine axon varicosities in the prelimbic division of the rat prefrontal cortex exhibit sparse immunoreactivity for the dopamine transporter. J Neurosci. 1998;18:2697–2708. doi: 10.1523/JNEUROSCI.18-07-02697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen G, Jensen M, Weikop P, Dencker D, Christiansen SH, Loland CJ, Bengtsen CH, Petersen JH, Fink-Jensen A, Wortwein G, Woldbye DP. Neuropeptide Y Y5 receptor antagonism attenuates cocaine-induced effects in mice. Psychopharmacology (Berl) 2012;222:565–577. doi: 10.1007/s00213-012-2651-y. [DOI] [PubMed] [Google Scholar]
- Stuber GD, Stamatakis AM, Kantak PA. Considerations when using cre-driver rodent lines for studying ventral tegmental area circuitry. Neuron. 2015;85:439–445. doi: 10.1016/j.neuron.2014.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmeier DJ, Ding J, Day M, Wang Z, Shen W. D1 and D2 dopamine-receptor modulation of striatal glutamatergic signaling in striatal medium spiny neurons. Trends Neurosci. 2007;30:228–235. doi: 10.1016/j.tins.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Swanson LW. The projections of the ventral tegmental area and adjacent regions: a combined fluorescent retrograde tracer and immunofluorescence study in the rat. Brain Res Bull. 1982;9:321–353. doi: 10.1016/0361-9230(82)90145-9. [DOI] [PubMed] [Google Scholar]
- Tritsch NX, Sabatini BL. Dopaminergic modulation of synaptic transmission in cortex and striatum. Neuron. 2012;76:33–50. doi: 10.1016/j.neuron.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valjent E, Bertran-Gonzalez J, Herve D, Fisone G, Girault JA. Looking BAC at striatal signaling: cell-specific analysis in new transgenic mice. Trends Neurosci. 2009;32:538–547. doi: 10.1016/j.tins.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Weihe E, Depboylu C, Schutz B, Schafer MK, Eiden LE. Three types of tyrosine hydroxylase-positive CNS neurons distinguished by dopa decarboxylase and VMAT2 co-expression. Cell Mol Neurobiol. 2006;26:659–678. doi: 10.1007/s10571-006-9053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Qi J, Wang HL, Zhang S, Morales M. Glutamatergic and dopaminergic neurons in the mouse ventral tegmental area. Eur J Neurosci. 2015;41:760–772. doi: 10.1111/ejn.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yetnikoff L, Lavezzi HN, Reichard RA, Zahm DS. An update on the connections of the ventral mesencephalic dopaminergic complex. Neuroscience. 2014 doi: 10.1016/j.neuroscience.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Lee YM, Guy VC, Freed CR. Embryonic stem cells with GFP knocked into the dopamine transporter yield purified dopamine neurons in vitro and from knock-in mice. Stem Cells. 2009;27:2952–2961. doi: 10.1002/stem.216. [DOI] [PubMed] [Google Scholar]