Abstract
Dairy goat and sheep farms suffer severe economic losses due to intramammary infections, with Staphylococcus aureus representing the main cause of clinical mastitis in small ruminants. In addition, S. aureus contamination of goat and sheep milk may cause staphylococcal food poisoning, as many traditional caprine and ovine milk products are not subjected to pasteurization. Data on virulence and antimicrobial resistance genes, as well as on the clonality of S. aureus detected in goat and sheep milk is scarce. Therefore, it was the aim of this study to determine (i) spa types and clonal complexes (CC) and (ii) virulence and resistance gene profiles of S. aureus isolated from goat and sheep milk. A total of 162 milk samples from sheep and goats presenting signs of an intramammary infection and 104 bulk milk samples were collected. While low prevalence rates of S. aureus was detected on single animal level, 46% of the bulk tank milk samples from small ruminants were positive for S. aureus. All isolates were spa typed and CC and virulence and resistance gene patterns were determined using a DNA microarray. Data from 49 S. aureus isolates was included in the statistical analysis and the construction of a SplitsTree. The analyzed isolates could be assigned to eleven CC, with the large majority of goat and sheep isolates being assigned to CC130 and CC133. The findings of this study suggest that S. aureus shows pronounced adaptation to small ruminants in general, but not to sheep or goats in particular. Although some common characteristics among S. aureus from caprine, ovine, and bovine milk samples were observed, S. aureus from small ruminants seem to form a distinct population. As 67% of the detected S. aureus strains exhibited at least one enterotoxin gene, many caprine, or ovine raw milk products may be contaminated with low levels of enterotoxigenic S. aureus, stressing the importance of strict maintenance of the cold chain.
Keywords: Staphylococcus aureus, sheep, goat, clonality, enterotoxin genes, virulence gene profile, mastitis
Introduction
Being one of the predominant causes of food poisoning worldwide, Staphylococcus aureus is of particular concern to the dairy industry (Oliver et al., 2009). Dairy sheep and goat farms also suffer severe economic losses due to staphylococcal intramammary infections, with S. aureus being the main cause of clinical mastitis in small ruminants (Bergonier et al., 2003). However, identification of affected animals can be challenging, as in contrast to cattle, high somatic cell counts and positive results in the California mastitis test are not necessarily reliable indicators of intramammary infections among small ruminants.
Over the last decade, the production of caprine and ovine milk in Switzerland has been increasing, with 14,000 registered small ruminant farms and a total population of approximately 490,000 heads in 2014 (Swiss Federal Statistical Office). S. aureus is one of the most commonly found pathogens in raw caprine and ovine milk (Marogna et al., 2012) and has been detected in over 30% of the examined raw milk of Swiss dairy goat and sheep farms (Muehlherr et al., 2003). As goat and sheep milk are often used for traditional, unpasteurized products such as raw milk cheeses, they represent a potential source of staphylococcal food poisoning (SFP).
The Centers for Disease Control estimate a total number of 240,000 SFP cases per year in the US (Scallan et al., 2011). In the EU, the number of SFP outbreaks is rising, with 386 SFP outbreaks reported in 2014 (Anonymous, 2015). SFP patients present with violent vomiting and diarrhea upon ingestion of staphylococcal enterotoxins pre-formed by S. aureus in food (Tranter, 1990). Many different staphylococcal enterotoxins and enterotoxin-like superantigens have been described (Dinges et al., 2000). There is evidence demonstrating emetic activity in humans for all classical enterotoxins SEA-SEE (Dinges et al., 2000) and recently also for some newly described enterotoxins (Jørgensen et al., 2005; Johler et al., 2015).
While the population structure and the genomic characteristics of S. aureus from bovine milk are very well described, similar data on S. aureus isolated from small ruminants is scarce (Scherrer et al., 2004; Concepción Porrero et al., 2012; Gharsa et al., 2012; Linage et al., 2012; Eriksson et al., 2013; Smith et al., 2014). Data on virulence and antimicrobial resistance genes, as well as on the clonality of S. aureus detected in goat and sheep milk is crucial to determine potential routes of transmission, to improve management strategies of affected herds, and to develop effective therapeutic interventions. Therefore, it was the aim of this study to determine clonal complexes (CC) and virulence and resistance gene profiles of S. aureus isolated from goat and sheep milk.
Materials and Methods
Bacterial Isolation and DNA Extraction
In this study, 162 milk samples of goats (n = 31) and sheep (n = 131) exhibiting one or several signs of mastitis (increased somatic cell counts, positive California mastitis test, decreased milk yield), as well as 104 raw bulk milk samples were collected from dairy farms in Switzerland (goat farms: n = 57; sheep farms: n = 47) from March to October 2015. EN ISO 6888-2 was followed for isolation of coagulase-positive staphylococci. One single colony of each different morphology exhibiting an opaque fibrin halo on rabbit plasma fibrinogen agar (Oxoid, Basel, Switzerland) was subcultured. The subcultures were grown on 5% sheep blood agar at 37°C overnight. Chromosomal DNA extraction was performed using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions.
Staphaurex Latex Agglutination Test
All S. aureus isolates were subjected to the Staphaurex latex agglutination test (Oxoid, Basel, Switzerland) following the manufacturer’s instructions. This assay targets microbial surface components recognizing adhesive matrix molecules (SpA, ClfA, FnBPA, and FnBPB) and frequently yields false-negative results in bovine S. aureus (Stutz et al., 2011; Moser et al., 2013).
DNA Microarray, SplitsTree Analysis, and Comparison to Bovine Isolates
DNA microarray analysis was performed using Staphytype genotyping kit 2.0 (Alere, Jena, Germany) following the manufacturer’s instructions. The DNA microarray used in this study determines the presence or absence of over 300 different genes and allelic variants, and allows for assignment of CC (Monecke et al., 2008). All presumptive S. aureus isolates were further characterized by DNA microarray profiling, which also served as a tool for species confirmation. The DNA microarray hybridization results of isolates from goats and sheep were compared to those of isolates from an unrelated collection of 78 bovine S. aureus strains that were obtained in a comprehensive study investigating mastitis isolates from cows in Switzerland (Moser et al., 2013). The resistance and virulence gene profiles of the caprine, ovine, and bovine isolates were visualized using SplitsTree41 as previously described (Wattinger et al., 2012).
spa Typing
spa typing, a high resolution single-locus typing technique in S. aureus, was performed as previously described (Johler et al., 2011). Briefly, PCR amplicons of the polymorphic X region of the spa gene were purified using the GenElute PCR Purification Kit (Sigma–Aldrich, St. Louis, MO, USA), and were subsequently sequenced and assigned to spa types2.
Inclusion Criteria
Stringent inclusion criteria were employed to avoid bias over-representation of strains isolated from both single animals and bulk milk samples of the same dairy farm: only one S. aureus isolate was considered for construction of the SplitsTree and statistical analysis, if the analyzed isolates exhibited the same spa type and ≤3 different hybridization results in the DNA microarray profiling. Two single animal isolates from sheep were therefore excluded from the study, resulting in 49 S. aureus isolates taken into consideration for further analyses.
Statistical Analysis
Statistically significant differences in the distribution of virulence and resistance genes between the bovine, caprine, and ovine isolates were assessed by either Chi squared test or Fisher’s exact test (in case n < 5) using SPSS 23.0 (IBM Corp., Armonk, NY, USA).
Results
A total of 162 milk samples (goats: n = 31; sheep: n = 131) of animals presenting signs of an intramammary infection and 104 bulk milk samples (goat farms: n = 57; sheep farms: n = 47) were collected. On the level of single animals, none of the goat milk samples and 2% (n = 3) of the sheep milk samples were positive for S. aureus. On the level of bulk milk samples, 60% (n = 34) of goat bulk milk samples and 30% (n = 14) of sheep bulk milk samples were positive for S. aureus, which equals an overall prevalence of 46% among the examined bulk milk samples of small ruminants.
S. aureus from small ruminants were compared to bovine mastitis isolates from the study of Moser et al. (2013), with results being presented in Figures 1 and 2, as well as in Table 1. The distribution of CC among caprine, ovine, and bovine strains is depicted in Figure 1, and a SplitsTree comparing gene profiles of caprine, ovine and bovine strains is shown in Figure 2. In general, bovine isolates and isolates from small ruminants represent distinct populations, with CC130 and CC133 exclusively associated with small ruminants. Six main SplitsTree clusters, corresponding to CC CC8, CC97, CC130, CC133, CC151, and CC479, were identified. Isolates not associated with one of the main SplitsTree clusters were assigned to CC1, CC5, CC7, CC9, CC30, CC101, and CC398.
FIGURE 1.
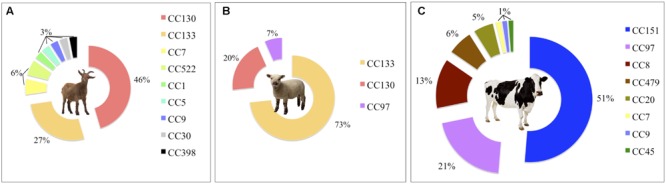
Distribution of clonal complexes (CC) among Staphylococcus aureus isolated from the milk of different animal species: (A) goat (n = 34, pink), (B) sheep (n = 15, blue), (C) cows (n = 78, green).
FIGURE 2.
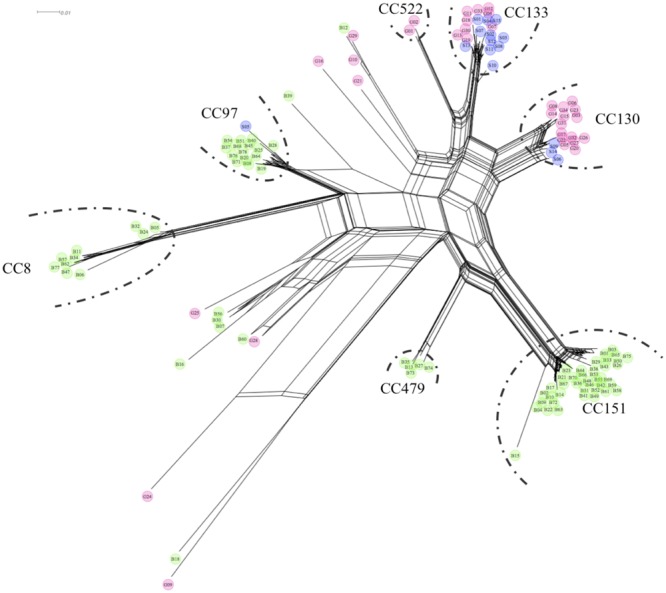
The SplitsTree illustrates similarities between virulence and resistance gene profiles of goat (G), sheep (S), and bovine (B) S. aureus isolates. The isolates grouped into clusters mainly based on assignment to CC. Isolates from small ruminants form a distinct population and were mainly found in the clusters of CC130 and CC133. Bovine isolates were predominant in clusters CC8, CC97, CC151, CC479.
Table 1.
Prevalence rates of selected virulence and resistance genes detected among Staphylococcus aureus strains isolated from goat (G), sheep (S), and bovine (B) milk samples.
| Group | Gene/Probe | Function | G (n = 34) | S (n = 15) | B (n = 78) |
|---|---|---|---|---|---|
| agr | agrI | Accessory gene regulator, type 1 | 44∗S | 80∗G,B | 41∗S |
| agrII | Accessory gene regulator, type 2 | 6∗B | 0∗B | 59∗G,S | |
| agrIII | Accessory gene regulator, type 3 | 50∗S,B | 20∗G,B | 0∗G,S | |
| Capsule | cap5 | Capsule type 5 | 9∗B | 7∗B | 38∗G |
| cap8 | Capsule type 8 | 91∗B | 93∗B | 62∗G | |
| Enterotoxins | sea | Enterotoxin A | 3 | 0 | 10 |
| sea (320E) | Enterotoxin A, allelic variant 320E | 0 | 0 | 9 | |
| sea (N315) | Enterotoxin A, allelic variant N315 | 9 | 0 | 1 | |
| seb | Enterotoxin B | 0 | 0 | 0 | |
| sec | Enterotoxin C | 50∗B | 67∗B | 15∗G | |
| sed | Enterotoxin D | 0∗B | 0 | 13∗G | |
| see | Enterotoxin E | 0 | 0 | 0 | |
| seg | Enterotoxin G | 9∗B | 0∗B | 65∗G,S | |
| sei | Enterotoxin I | 9∗B | 0∗B | 65∗G,S | |
| sek | Enterotoxin K | 0 | 0 | 0 | |
| sel | Enterotoxin L | 50∗B | 67∗B | 15∗G,S | |
| selm | Enterotoxin-like protein M | 9∗B | 0∗B | 64∗G,S | |
| seln | Enterotoxin-like protein N | 9∗B | 0∗B | 65∗G,S | |
| selo | Enterotoxin-like protein O | 9∗B | 0∗B | 65∗G,S | |
| seq | Enterotoxin Q | 0 | 0 | 0 | |
| selu | Enterotoxin-like protein U | 9∗B | 0∗B | 65∗G,S | |
| egc | Enterotoxin Gene Cluster (seg/sei/selm/seln/selo/selu) | 9∗B | 0∗B | 65∗G,S | |
| Other superantigens | tst1 (“bovine” allele) | Toxic Shock Syndrome Toxin, allele from strain RF122 | 35∗B | 40∗B | 8∗G,S |
| etA/B/D | Exfoliative toxins A, B, and D | 0 | 0 | 0 | |
| pvl | Panton Valentine Leukocidin | 0 | 0 | 0 | |
| Resistance1 | tetK | Tetracycline | 12∗B | 0 | 0∗G |
| fosB | Metallothiol Transferase | 38∗S | 73∗G | 19 | |
| Misc | sdrD | Sialoprotein-binding protein D | 97∗B | 100∗B | 37∗G,S |
| splE | Serine protease E | 59∗S,B | 27∗G | 35∗G | |
| lukM/lukF-PV (P83) | Bovine leukocidin | 21∗S,B | 93∗G | 72∗G | |
| Q7A4X2 | Hypothetical protein | 47∗S,B | 80∗G | 86∗G | |
| ssl06/set21 | Staphylococcal superantigen-like protein 6 | 41∗S,B | 73∗G,B | 21∗G,S | |
| ssl10/set4 | Staphylococcal superantigen-like protein 10 | 68∗S | 93∗G | 73 |
A comprehensive list of the prevalence of all genes detected by DNA microarray is provided as a supplement. 1None of the isolates harbored the resistance genes mecA, lnuA, msrA, mefA, mphC, vatA/B, vgaA/B, aacA-aphD, aadD, aphA3, sat, dfrS1, far1, Q6GD50, mupA, cat, fexA, cfr, vanA/Z, qacA/C.∗The distribution of the respective gene differed significantly between strains from the stated sources (G, goat, S, sheep, B, bovine) with p ≤ 0.05.
An overview of all CC and spa types detected is provided in Table 2. The isolates analyzed could be assigned to eleven different CC, of which only two (CC130 and CC133) were common among both caprine (71%) and ovine (93%) isolates. A total of 22 different spa types were detected. The most prevalent spa types were t1773 among the caprine and t1166 among the ovine S. aureus isolates, to which 26 and 27% of the analyzed isolates could be assigned, respectively. Three new spa types were detected: t15248, t15249, and t15404. While 51% of the bovine S. aureus isolates led to false-negative results in the Staphaurex latex agglutination test, all isolates from the milk of small ruminants tested in this study yielded positive results and were thus correctly identified as S. aureus by the Staphaurex latex agglutination test.
Table 2.
Clonal complexes (CC) and spa types of the S. aureus isolates from goat and sheep milk.
| Origin | CC | n | spa type (n) | Isolate ID |
|---|---|---|---|---|
| Goat (n = 34) | CC1 | 1 | t127 (1) | G16 |
| CC5 | 1 | t002 (1) | G25 | |
| CC7 | 2 | t091 (2) | G10, G29 | |
| CC9 | 1 | t899 (1) | G28 | |
| CC30 | 1 | t012 (1) | G09 | |
| CC101 | 1 | t056 (1) | G21 | |
| CC130 | 15 | t1773 (9) | G03, G05, G14, G17, G20, G22, G26, G27, G32 | |
| t11826 (2) | G08, G15 | |||
| t15248 (4) | G06, G23, G31, G34 | |||
| CC133 | 9 | t544 (1) | G18 | |
| t1166 (2) | G12, G19 | |||
| t2678 (3) | G04, G07, G33 | |||
| t3583 (1) | G11 | |||
| t4735 (1) | G13 | |||
| t15249 (1) | G30 | |||
| CC398 | 1 | t4475 (1) | G24 | |
| CC522 | 2 | t1534 (1) | G02 | |
| t5428 (1) | G01 | |||
| Sheep (n = 15) | CC97 | 1 | t267 (1) | S05 |
| CC130 | 3 | t1773 (1) | S09 | |
| t11826 (1) | S14 | |||
| t15404 (1) | S06 | |||
| CC133 | 11 | t998 (1) | S04 | |
| t1166 (4) | S07, S08, S11, S15 | |||
| t2678 (2) | S10, S12 | |||
| t3583 (1) | S01 | |||
| t4735 (2) | S02, S03 | |||
| t6060 (1) | S13 |
An overview of the prevalence of the most important virulence and resistance genes detected by DNA microarray is provided in Table 1. The supplementary files include a comprehensive list of the prevalence rates of all genes detected (Supplementary Table S1) and a complete overview of all hybridization results (Supplementary Table S2). Overall, 67% of all isolates harbored at least one enterotoxin gene. The most prevalent enterotoxin genes were sec and sel, which were present in 55% of the isolates from small ruminants. The sea gene was found exclusively among caprine isolates. None of the genes encoding exfoliative toxins or Panton–Valentine leukocidin were detected. Virulence genes associated with the toxic shock syndrome were found in 27 isolates.
Seven isolates harbored genes conferring penicillin resistance (blaZ/I/R). Genes conferring tetracycline resistance were found only among the caprine isolates. All isolates harbored sdrM, which encodes a multidrug efflux pump. None of the caprine and ovine isolates harbored genes conferring resistance to methicillin, aminoglycosides, streptogramin A, virginiamycin A, glycopeptides, and vancomycin.
Discussion
The prevalence of S. aureus in caprine and ovine bulk tank milk samples varies depending of the country. Muehlherr et al. (2003) detected S. aureus in 32% of the caprine and 33% of ovine bulk tank milk samples in Switzerland, while Linage et al. (2012) and Álvarez-Suárez et al. (2015) detected coagulase positive staphylococci in 66% of caprine and 15% of ovine bulk tank milk samples in Spain. Considering the very low prevalence of S. aureus detected among the analyzed milk samples of single animals in this study, the overall detected prevalence of S. aureus in the bulk milk samples examined was high. This suggests that the prevalence of S. aureus as a subclinical agent of mastitis in small ruminant herds in Switzerland may have been underestimated. This is of particular relevance, as SFP has been associated with raw milk from small ruminants (Giezendanner et al., 2009) and as traditional goat and sheep raw milk cheeses are popular.
Most of the isolates characterized in this study were assigned to CC130 and CC133, suggesting that these lineages may represent the major CC among caprine and ovine S. aureus isolates in Switzerland. These results are consistent with the findings of previous studies suggesting that predominance of either CC130/CC133 or CC522 in S. aureus isolated from milk of small ruminants is associated with geographical, breed- and infection-related aspects (Concepción Porrero et al., 2012; Eriksson et al., 2013; Shepheard et al., 2013; Smith et al., 2014). Only few CC (CC7, CC9, CC97) were detected among strains of both small ruminants and cows.
Even though S. aureus isolates originating from caprine and ovine hosts have been spa typed in several recent studies (Concepción Porrero et al., 2012; Eriksson et al., 2013; Smith et al., 2014; Bar-Gal et al., 2015), three new spa types were detected among the isolates in this study. This suggests that to date, data on the population structure of S. aureus isolates originating from small ruminants is still very limited. The agr types and cap genes detected in this study are consistent with the findings of previous studies investigating S. aureus from small ruminants (Alves et al., 2009; Vautor et al., 2009; Bar-Gal et al., 2015).
Most of the isolates analyzed from small ruminants in this study were lacking antibiotic resistance genes. Resistance gene profiles from caprine and ovine strains in this study were not significantly different from those of bovine isolates (Moser et al., 2013). Only the presence of tetK in 12% of the caprine isolates was significantly higher compared to ovine and bovine isolates (p = 0.007). Overall, the prevalence of blaZ/I/R (14%), tetK (8%), tetM (2%), ermA/B/C (2%) detected was lower than the prevalence detected when analyzing S. aureus from small ruminants milk or nasal swabs in recent studies from the Middle-East and Africa (Gharsa et al., 2012; Bar-Gal et al., 2015; Jamali et al., 2015). The prevalence of antibiotic resistance genes detected was surprisingly high, considering that herd management differs vastly in small ruminants and cattle, with culling being preferred to antimicrobial treatment in small ruminants.
All isolates harboring tst1 also harbored the genes sec and sel, and were assigned exclusively to CC130 and CC133. These genes are located on the ovine pathogenicity island SaPIov1 (Guinane et al., 2010), and have been previously reported in isolates originating from small ruminants (Smyth et al., 2005; Gharsa et al., 2012). Consistent with findings among S. aureus from sheep and goats in Israel (Bar-Gal et al., 2015), in this study, the prevalence of tst1, sec, and sel was significantly higher among small ruminant isolates than among bovine isolates (p < 0.003), which in contrast are more likely to harbor egc genes (p = 0.000). In this study, the detected overall prevalence of 67% of S. aureus carrying at least one enterotoxin gene was similar to 65% reported by Scherrer et al. (2004).
Many genes encoding virulence factors were present at similar rates in caprine, ovine and bovine isolates. This included genes encoding hemolysins (hla, hlb, hld), adhesion factors (clfA, clfB, ebps, fib, fnbA, vwb), hyaluronate lyase (hysA1/A2), immunodominant antigen B (isaB), transferrin binding protein (isdA) and serine proteases (splA, sspa). In several studies, these virulence factors have been reported to play a role in mastitis in cattle (Viana et al., 2010; Ote et al., 2011; Wolf et al., 2011). While many genes were equally distributed among small ruminant and bovine isolates, statistically higher prevalence rates of cap8, sdrD, sec, sel, tst1, ssl06, edinB, and Imrp (RF122) among S. aureus from small ruminants were observed. As for genes associated with biofilm formation (icaA/C/D), very high prevalence rates have been previously reported in isolates originating from small ruminants in particular (Bar-Gal et al., 2015) and from ruminants in general (Snel et al., 2014; Prenafeta et al., 2014).
Comparison of goat and sheep isolates tested in this study showed that caprine and ovine S. aureus exhibited highly similar virulence and resistance gene patterns. However, some species-specific patterns were observed. Higher prevalence rates of splE among the caprine (p = 0.038) and of lukM (p = 0.000) among the ovine isolates was observed. Simultaneous presence of splE and sdrD, which was detected in four ovine and 19 caprine isolates in this study, has been associated with gangrenous mastitis in small ruminants (Vautor et al., 2009). In contrast, lukM was associated with high leukotoxic activity against bovine polymorphonuclear leukocytes (Rainard et al., 2003) and was hypothesized to play a central role in mastitis in ruminants (Barrio et al., 2006). In addition, significant differences in the prevalence of genes ssl06/set21, ssl10/set4, and Q7A4X2 in caprine compared to ovine isolates were observed. While genes encoding for superantigen-like proteins (ssl), have been associated with immunoevasion by interfering with the toll-like receptor system (Zecconi and Scali, 2013), Q7A4X2 may be involved in biofilm formation (Snel et al., 2014). These findings suggest that the virulence genes detected, and especially lukM, sdrD, and splE, represent important virulence factors for S. aureus strains causing mastitis in small ruminants.
Finally, the performance of the Staphaurex latex agglutination test for identification of S. aureus from small ruminants was assessed, as this test was reported to yield false-negative results in 51% of all bovine S. aureus strains tested (Moser et al., 2013). The results of this study show that the Staphaurex latex agglutination test system is a highly reliable diagnostic tool for identification of S. aureus isolates from caprine and ovine milk samples.
Conclusion
The findings of this study suggest that S. aureus shows pronounced adaptation to small ruminants in general, but not to sheep or goats in particular. Comparing S. aureus from caprine, ovine and bovine milk samples collected in the same country, some common virulence genes were observed, but the results indicate that S. aureus from small ruminants may form a distinct population. Further studies covering an extensive strain collection of S. aureus from small ruminants collected at various geographical locations are needed to ensure that this finding can be extrapolated to S. aureus in general. Although low prevalence rates of S. aureus on the level of single animals exhibiting signs of mastitis was detected, 46% of the bulk tank milk samples from small ruminants were positive for S. aureus. This suggests that S. aureus may pose problems for animal and consumer health, in particular, as many products made from the milk of small ruminants are consumed raw.
Author Contributions
SJ and RS conceived and designed the study. AM carried out the laboratory work. AM and SJ analyzed and interpreted the data. AM and SJ wrote the manuscript. All authors critically revised and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank all cooperation partners from the small ruminant dairy industry (Beratungs- und Gesundheitsdienst für Kleinwiederkäuer; Schweizerische Milchschafzucht Genossenschaft, Schweizerischer Ziegenzuchtv-erband) for their kind support in the collection of samples and for helpful discussions regarding small ruminant husbandry in Switzerland.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00319
References
- Álvarez-Suárez M.-E., Otero A., García-López M.-L., Santos J. A. (2015). Microbiological examination of bulk tank goat’s milk in the Castilla y León region in Northern Spain. J. Food Prot. 78 2227–2232. 10.4315/0362-028X.JFP-15-133 [DOI] [PubMed] [Google Scholar]
- Alves P. D. D., McCulloch J. A., Even S., Le Maréchal C., Thierry A., Grosset N., et al. (2009). Molecular characterisation of Staphylococcus aureus strains isolated from small and large ruminants reveals a host rather than tissue specificity. Vet. Microbiol. 137 190–195. 10.1016/j.vetmic.2008.12.014 [DOI] [PubMed] [Google Scholar]
- Anonymous (2015). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 13 4329 10.2903/j.efsa.2014.4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Gal G. K., Blum S. E., Hadas L., Ehricht R., Monecke S., Leitner G. (2015). Host-specificity of Staphylococcus aureus causing intramammary infections in dairy animals assessed by genotyping and virulence genes. Vet. Microbiol. 176 143–154. 10.1016/j.vetmic.2015.01.007 [DOI] [PubMed] [Google Scholar]
- Barrio M. B., Rainard P., Prévost G. (2006). LukM/LukF’-PV is the most active Staphylococcus aureus leukotoxin on bovine neutrophils. Microbes Infect. 8 2068–2074. 10.1016/j.micinf.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Bergonier D., De Crémoux R., Rupp R., Lagriffoul G., Berthelot X. (2003). Mastitis of dairy small ruminants. Vet. Res. 34 689–716. 10.1051/vetres:2003030 [DOI] [PubMed] [Google Scholar]
- Concepción Porrero M., Hasman H., Vela A. I., Fernández-Garayzábal J. F., Domínguez L., Aarestrup F. M. (2012). Clonal diversity of Staphylococcus aureus originating from the small ruminants goats and sheep. Vet. Microbiol. 156 157–161. 10.1016/j.vetmic.2011.10.015 [DOI] [PubMed] [Google Scholar]
- Dinges M. M., Orwin P. M., Schlievert P. M. (2000). Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13 16–34. 10.1128/CMR.13.1.16-34.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J., Espinosa-Gongora C., Stamphøj I., Larsen A. R., Guardabassi L. (2013). Carriage frequency, diversity and methicillin resistance of Staphylococcus aureus in Danish small ruminants. Vet. Microbiol. 163 110–115. 10.1016/j.vetmic.2012.12.006 [DOI] [PubMed] [Google Scholar]
- Gharsa H., Ben Slama K., Lozano C., Gómez-Sanz E., Klibi N., Ben Sallem R., et al. (2012). Prevalence, antibiotic resistance, virulence traits and genetic lineages of Staphylococcus aureus in healthy sheep in Tunisia. Vet. Microbiol. 156 367–373. 10.1016/j.vetmic.2011.11.009 [DOI] [PubMed] [Google Scholar]
- Giezendanner N., Meyer B., Gort M., Müller P., Zweifel C. (2009). [Rohmilch-assoziierte Staphylococcus aureus Intoxikationen bei Kindern]. J. Food Saf. Food Qual. 151 329–331. 10.1024/0036-7281.151.7.329 [DOI] [PubMed] [Google Scholar]
- Guinane C. M., Ben Zakour N. L., Tormo-Mas M. A., Weinert L. A., Lowder B. V., Cartwright R. A., et al. (2010). Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol. Evol. 2 454–466. 10.1093/gbe/evq031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamali H., Paydar M., Radmehr B., Ismail S., Dadrasnia A. (2015). Prevalence and antimicrobial resistance of Staphylococcus aureus isolated from raw milk and dairy products. Food Control 54 383–388. 10.1016/j.foodcont.2015.02.013 [DOI] [Google Scholar]
- Johler S., Giannini P., Jermini M., Hummerjohann J., Baumgartner A., Stephan R. (2015). Further evidence for staphylococcal food poisoining outbreaks caused by egc-encoded enterotoxins. Toxins 7 997–1004. 10.3390/toxins7030997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johler S., Layer F., Stephan R. (2011). Comparison of virulence and antibiotic resistance genes of food poisoning outbreak isolates of Staphylococcus aureus with isolates obtained form bovine mastitis milk and pig carcasses. J. Food Prot. 74 1852–1859. 10.4315/0362-028X.JFP-11-192 [DOI] [PubMed] [Google Scholar]
- Jørgensen H. J., Mathisen T., Lovseth A., Omoe K., Qvale K. S., Loncarevic S. (2005). An outbreak of staphylococcal food poisoning caused by enterotoxin H in mashed potato made with raw milk. FEMS Microbiol. Lett. 252 267–272. 10.1016/j.femsle.2005.09.005 [DOI] [PubMed] [Google Scholar]
- Linage B., Rodriguez-Calleja J. M., Otero A., Garcia-Lopez M. L., Santos J. A. (2012). Characterization of coagulase-positive staphylococci isolated from tank and silo ewe milk. J. Dairy Sci. 95 1639–1644. 10.3168/jds.2011-4734 [DOI] [PubMed] [Google Scholar]
- Marogna G., Pilo C., Vidili A., Tola S., Schianchi G., Leori S. G. (2012). Comparison of clinical findings, microbiological results, and farming parameters in goat herds affected by recurrent infectious mastitis. Small Rumin. Res. 102 74–83. 10.1016/j.smallrumres.2011.08.013 [DOI] [Google Scholar]
- Monecke S., Slickers P., Ehricht R. (2008). Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol. Med. Microbiol. 53 237–251. 10.1111/j.1574-695X.2008.00426.x [DOI] [PubMed] [Google Scholar]
- Moser A., Stephan R., Corti S., Johler S. (2013). Comparison of genomic and antimicrobial resistance features of latex agglutination test-positive and latex agglutination test-negative Staphylococcus aureus isolates causing bovine mastitis. J. Dairy Sci. 96 329–334. 10.3168/jds.2012-5944 [DOI] [PubMed] [Google Scholar]
- Muehlherr J. E., Zweifel C., Corti S., Blanco J. E., Stephan R. (2003). Microbiological quality of raw goat’s and ewe’s bulk-tank milk in Switzerland. J. Dairy Sci. 86 3849–3856. 10.3168/jds.S0022-0302(03)73992-7 [DOI] [PubMed] [Google Scholar]
- Oliver S., Boor K., Murphy S. C., Murinda S. E. (2009). Food safety hazards associated with consumption of raw milk. Foodborne Pathog. Dis. 6 793–806. 10.4315/0362-028X.JFP-11-192 [DOI] [PubMed] [Google Scholar]
- Ote I., Taminiau B., Duprez J.-N., Dizier I., Mainil J. G. (2011). Genotypic characterization by polymerase chain reaction of Staphylococcus aureus isolates associated with bovine mastitis. Vet. Microbiol. 153 285–292. 10.1016/j.vetmic.2011.05.042 [DOI] [PubMed] [Google Scholar]
- Prenafeta A., Sitjà M., Holmes M. A., Paterson G. K. (2014). Short communication: biofilm production characterization of mecA and mecC methicillin-resistant Staphylococcus aureus isolated from bovine milk in Great Britain. J. Dairy Sci. 97 4838–4841. 10.3168/jds.2014-7986 [DOI] [PubMed] [Google Scholar]
- Rainard P., Corrales J.-C., Barrio M. B., Cochard T., Poutrel B. (2003). Leucotoxic activities of Staphylococcus aureus strains isolated from cows, ewes, and goats with mastitis: importance of LukM/LukF’-PV leukotoxin. Clin. Diagn. Lab. Immunol. 10 272–277. 10.1128/CDLI.10.2.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M.-A., Roy S. L., et al. (2011). Foodborne illness acquired in the United States – major pathogens. Emerg. Infect. Dis. 17 7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer D., Corti S., Muehlherr J. E., Zweifel C., Stephan R. (2004). Phenotypic and genotypic characteristics of Staphylococcus aureus isolates from raw bulk-tank milk samples of goats and sheep. Vet. Microbiol. 101 101–107. 10.1016/j.vetmic.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Shepheard M. A., Fleming V. M., Connor T. R., Corander J., Feil E. J., Fraser C., et al. (2013). Historical zoonoses and other changes in host tropism of Staphylococcus aureus, identified by phylogenetic analysis of a population dataset. PLoS ONE 8:e62369 10.1371/journal.pone.0062369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. M., Needs P. F., Manley G., Green L. E. (2014). Global distribution and diversity of ovine-associated Staphylococcus aureus. Infect. Genet. Evol. 22 208–215. 10.1016/j.meegid.2013.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D. S., Hartigan P. J., Meaney W. J., Fitzgerald J. R., Deobald C. F., Bohach G. A., et al. (2005). Superantigen genes encoded by the egc cluster and SaPlbov are predominant among Staphylococcus aureus isolates from cows, goats, sheep, rabbits and poultry. J. Med. Microbiol. 54 401–411. 10.1099/jmm.0.45863-0 [DOI] [PubMed] [Google Scholar]
- Snel G. G. M., Malvisi M., Pilla R., Piccinini R. (2014). Evaluation of biofilm formation using milk in a flow cell model and microarray characterization of Staphylococcus aureus strains from bovine mastitis. Vet. Microbiol. 174 489–495. 10.1016/j.vetmic.2014.09.020 [DOI] [PubMed] [Google Scholar]
- Stutz K., Stephan R., Tasara T. (2011). SpA, ClfA, and FnbA genetic variations lead to Staphaurex test-negative phenotypes in bovine mastitis Staphylococcus aureus isolates. J. Clin. Microbiol. 49 638–646. 10.1128/JCM.01148-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranter H. S. (1990). Foodborne staphylococcal illness. Lancet 336 1044–1046. 10.1016/0140-6736(90)92500-H [DOI] [PubMed] [Google Scholar]
- Vautor E., Cockfield J., Le Marechal C., Le Loir Y., Chevalier M., Robinson D. A., et al. (2009). Difference in virulence between Staphylococcus aureus isolates causing gangrenous mastitis versus subclinical mastitis in a dairy sheep flock. Vet. Res. 40 56 10.1051/vetres/2009039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana D., Blanco J., Tormo-Más M. Á, Selva L., Guinane C. M., Baselga R., et al. (2010). Adaptation of Staphylococcus aureus to ruminant and equine hosts involves SaPI-carried variants of von Willebrand factor-binding protein. Mol. Microbiol. 77 1583–1594. 10.1111/j.1365-2958.2010.07312.x [DOI] [PubMed] [Google Scholar]
- Wattinger L., Stephan R., Layer F., Johler S. (2012). Comparison of Staphylococcus aureus isolates associated with food intoxication with isolates from human nasal carriers and human infections. Eur. J. Clin. Microbiol. Infect. Dis. 31 455–464. 10.1007/s10096-011-1330-y [DOI] [PubMed] [Google Scholar]
- Wolf C., Kusch H., Monecke S., Albrecht D., Holtfreter S., von Eiff C., et al. (2011). Genomic and proteomic characterization of Staphylococcus aureus mastitis isolates of bovine origin. Proteomics 11 2491–2502. 10.1002/pmic.201000698 [DOI] [PubMed] [Google Scholar]
- Zecconi A., Scali F. (2013). Staphylococcus aureus virulence factors in evasion from innate immune defenses in human and animal diseases. Immunol. Lett. 150 12–22. 10.1016/j.imlet.2013.01.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


