Abstract
Shigella is one of the most important waterborne and foodborne pathogens around the world. Emergence of antibiotic-resistant Shigella has made the development of alternatives to conventional antibiotics necessary. In this study, a virulent Myoviridae bacteriophage, pSs-1 was isolated from environmental water in South Korea and showed infectivity to S. flexneri as well as S. sonnei strains. One-step growth analysis showed that pSs-1 has a short latent period (25 min) and a large burst size (97 PFU/cell). According to the genomic analysis, pSs-1 contains 164,999 bp of genome with a G + C content of 35.54% and it is considered as a member of the T4-like bacteriophage group. These results showed that pSs-1 may have potential as a biocontrol agent instead of conventional antibiotics for shigellosis.
Shigella is one of the most important waterborne and foodborne bacterial pathogens in the world1. It is usually related to the ingestion of contaminated water and food1. Shigella is human-adapted Escherichia coli and causes dysentery, spreading efficiently via low-dose fecal-oral transmission route1. It has been known that the majority shigellosis cases occur in developing countries and most of the patients are children under 5 years of age including infants1. However, numerous shigellosis cases are reported every year by military personnel and travelers in developed countries2.
The genus Shigella includes four species: S. dysenteriae, S. flexneri, S. boydii, and S. sonnei3. S. flexneri is the most commonly associated with shigellosis outbreaks in developing countries and causes approximately 2 estimated million cases per year worldwide4,5. S. sonnei has been the predominant agent responsible for dysentery in developed countries but is an emerging problem in developing areas6. The combination of increased incidence and excessive antimicrobial resistance among globally disseminated Shigella populations indicates that the development of effective control method will be increasingly important for long-term prevention of dysentery and associated morbidity and mortality7.
Bacteriophage (phage) can lyse a bacterial cell with acute specificity, which allows for the treatment of a targeted bacterial infection without the disruption of natural host microflora8. Phage therapy has been proposed for the treatment of human bacterial infections since phages were discovered in 1915 and 19179. After a short period of phage therapy development, the developmental focus of antimicrobial therapy shifted from phage therapy to chemotherapy9. On the other hand, phage research has continued in Eastern Europe and the former Soviet Union, enabling phage therapy clinically10. However, more research on phage therapy has been recommended recently because of increasing risk of drug-resistant bacteria, the limited choice of effective treatments, and the declining development of novel antibiotics9. Our current study presented the isolation and characterization of a virulent Myoviridae phage, designated as pSs-1. The biological properties of pSs-1 were evaluated, and it showed its efficient bacteriolytic activity against S. flexneri as well as S. sonnei. Finally, its genome was completely sequenced and analyzed comparatively with its related phages. The main aim of this study was to examine the potential of phage as a biocontrol agent that can be used to control the contaminated water with Shigella.
Results and Discussion
Isolation and characterization of phage
In previous study, Jun and colleagues reported two virulent Shigella phages, a Siphoviridae phage pSf-1 infecting S. flexneri11 and a Podoviridae phage pSb-1 infecting S. boydii12. pSf-1 was able to infect all of the S. flexneri and most of S. sonnei strains, forming clear plaques11; pSb-1 was able to infect all of the S. boydii strains and formed clear plaques12. Although the virulent Shigella phages, pSf-1 and pSb-1 showed the potential usefulness against shigellosis as reported previously, the two phages were considered to present restricted effectiveness because S. sonnei ATCC® 11060 was not infected by any of two phages11,12. This study aimed to isolate a virulent phage infecting S. sonnei strains in order to make the best combination of various phages for use in a phage cocktail as previously noted that the isolation of S. sonnei phage was needed12.
Previous research in our laboratory led to the isolation of S. sonnei phage as a first priority rather than the isolation of different phages infecting other Shigella species. A virulent Shigella phage, pSs-1 infecting S. sonnei and S. flexneri was isolated from the Hongjecheon stream in Seoul in April 2012. From the isolated phages, pSs-1 was selected for further studies depending on the clarity of plaque and the phage titer after a single propagation.
The morphology of pSs-1 places it in the family Myoviridae according to the classification system of Ackermann13 (Fig. 1C). The tail length and width were 120 ± 7 nm (mean ± SD) (n = 10) and 18 ± 2 nm (n = 10), respectively, and the head diameter was 98 ± 4 nm (n = 10). pSs-1 inhibited all of the S. sonnei and S. flexneri strains, producing clear plaques on all of the strains except one S. flexneri strain, ATCC® 12022 (turbid plaque). However, no S. boydii strain was infected by pSs-1. The high EOP value was obtained with S. flexneri ATCC® 29903 (Fig. 1B) although no strain had a higher value than the indicator host strain, S. sonnei ATCC® 25931 (Table 1). Furthermore, pSs-1 was not able to infect Escherichia coli strains used in this study. The result of one-step growth analysis revealed that pSs-1 had a short latent period (25 min) and a large burst size (97 PFU/cell) (Fig. 2).
Figure 1. Phage plaques formed in double-layer agar plates and electron micrograph of negatively stained phage pSs-1.
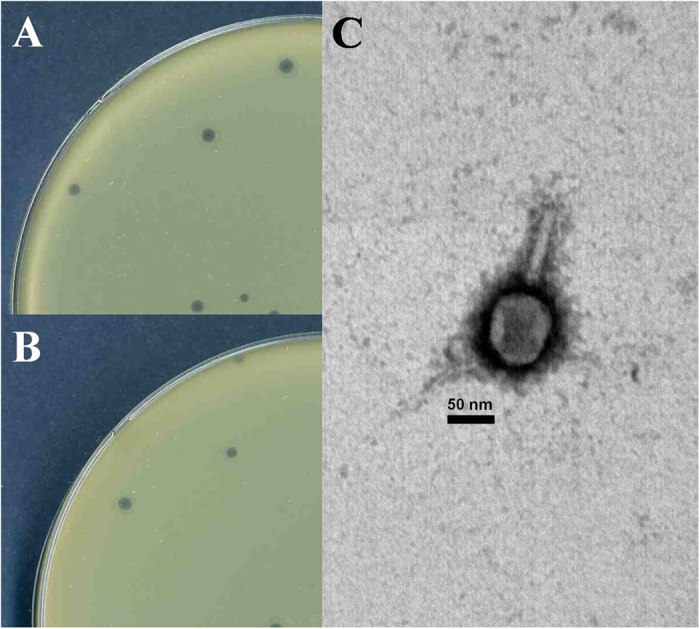
(A) phage plaques formed in agar plates with S. sonnei ATCC® 25931, (B) phage plaques formed in agar plates with S. flexneri ATCC® 29903, and (C) electron micrograph of pSs-1. The bar corresponds to 50 nm.
Table 1. Host range and EOPs of phage pSs-1 against all the bacterial strains used in this study.
| Bacterial species (n) | Strain | Infectivitya | EOPsb |
|---|---|---|---|
| Shigella sonnei (4) | ATCC® c 25931 | ++ | (1.00) |
| ATCC® 29930 | ++ | (0.77 ± 0.07) | |
| ATCC® 11060 | ++ | (0.73 ± 0.08) | |
| ATCC® 9290 | ++ | (0.65 ± 0.05) | |
| Shigella flexneri (3) | ATCC® 29903 | ++ | (0.91 ± 0.05) |
| ATCC® 11836 | ++ | (0.42 ± 0.03) | |
| ATCC® 12022 | + | (0.17 ± 0.07) | |
| Shigella boydii (2) | ATCC® 35966 | − | − |
| ATCC® 8700 | − | − | |
| Escherichia coli (2) | ATCC® 25922 | − | − |
| DH10Bd | − | − |
a++, clear plaque; +, turbid plaque; −, no plaque.
bThe EOP (efficiency of plating) values were shown as the mean of observations at three different occasions.
cpurchased from the American Type Culture Collection.
dpurchased from Invitrogen.
Figure 2. One-step growth curve of pSs-1.
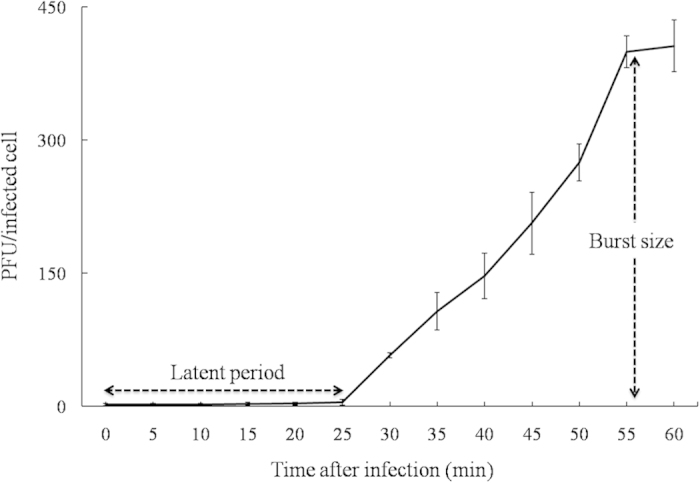
The error bars indicate standard deviations.
The bacteriolytic effect of pSs-1 was tested on early phase cultures of S. sonnei ATCC® 25931 (Fig. 3A) and S. flexneri ATCC® 29903 (Fig. 3B). The OD600 values of the uninfected control culture (MOI: 0) continued to increase during the incubation period. In contrast, the growth of bacteria infected by pSs-1 was retarded at MOIs of 0.1, 1, and 10; the bacterial growth was inhibited most effectively at an MOI of 10. pSs-1 lysed S. sonnei (3 h after incubation) more readily than S. flexneri (12 h after incubation).
Figure 3. Time course of host cell lysis effect of pSs-1 against S. sonnei ATCC® 25931.
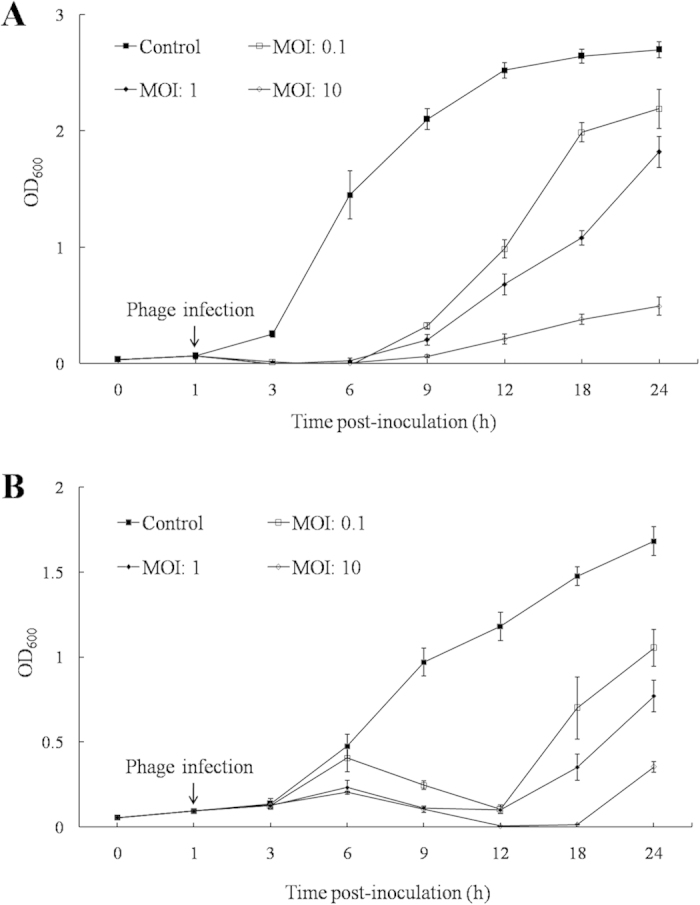
(A) and S. flexneri ATCC® 29903 (B). Early exponential phase cultures of S. sonnei ATCC® 25931 and S. flexneri ATCC® 29903 were co-cultured with pSs-1 at MOIs of 0, 0.1, 1, and 10. The results are shown as mean + standard deviations from triplicate experiments.
The effective bacteriolytic activity and high EOP value of pSs-1 against S. sonnei and S. flexneri strains indicated that pSs-1 could be used for control of both S. sonnei and S. flexneri although pSs-1 was proved to use only S. sonnei strain as a host bacterium; the propagation trials of pSs-1 using S. flexneri strains have not resulted in high titer for further study. These results conclude that the combination of pSf-1, pSb-1, and pSs-1 can inhibit all of the Shigella strains used in this study, increasing the possibility for Shigella control. The bacteriolytic activity of pSs-1 was found to be active for 1 h over the temperature range of 4–50 °C or the pH range of 5.0–9.0 toward S. sonnei and S. flexneri strains although significant reduction of its activity was observed at 50 °C (data not shown). This result emphasized that pSs-1 could be used in various natural environments, especially poor sanitation surroundings.
Comparative genomic analysis of phage
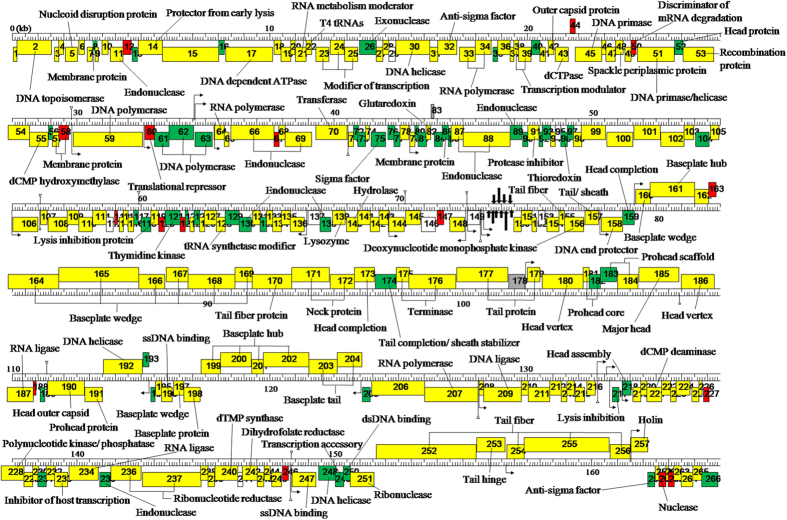
According to the genome sequencing results, the genome size of pSs-1 is 164,999 bp, with a 35.54% total G + C content. The genomic sequence of the Shigella phage pSs-1 which was described in this study has been deposited in the GenBank database under the accession number KM501444. The predominant start codon was ATG; 10 ORFs such as ORF28, ORF51, ORF66, ORF67, ORF84, ORF112, ORF150, ORF178, ORF189, and ORF238 started with an uncommon start codon (GTG). ORFs with a length of at least 34 amino acids were selected. A total of 26 promoters, 14 transcriptional terminator regions, and 266 ORFs were predicted in the genome. However, only 121 ORFs (45.49%) were determined to be functional based on gene predictions and annotation of the genome. Concrete gene information such as positions, directions, sizes, molecular weights, and putative functions of each pSs-1 ORFs are shown in Supplementary Table S1. A total of 121 ORFs were determined to be functional. The functional analysis indicates that pSs-1 has similar functional system to those of T4-like phages. pSs-1 was proved to contain similar host lysis system to Shfl2 (virulent phage against S. flexneri) although orf217 showed higher similarity to SP18 (virulent phage against S. sonnei). Also, the predicted ORFs of phage structural genes were widely scattered across the entire genome although they were mostly located between orf155 and orf204 (74.46%, 35 ORFs in total).
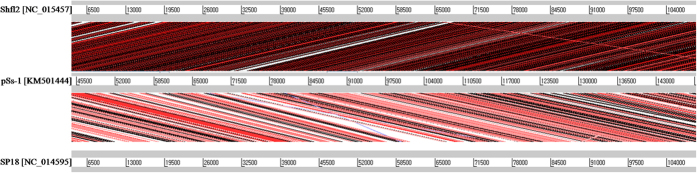
The comparative genome analysis of pSs-1 with T4-like phages, such as Shfl2 and SP18 revealed that pSs-1 had approximately 97% nucleotide sequence identity with Shfl2 and 186 homologous ORFs among its 266 ORFs (Fig. 4). In addition, pSs-1 showed approximately 93% nucleotide sequence identity with SP18 and 54 homologous ORFs (Fig. 4). According to the ACT comparison results, the pSs-1 genome revealed a higher degree of similarity to phage Shfl2 than phage SP18; the results showed the forward matches in the order of the genome of Shfl2 and SP18 (Fig. 5).
Figure 4. Functional genome map of phage pSs-1.
Hypothetical functions of encoded proteins were determined by comparison of amino acid sequences to the non-redundant databank using BLASTP. The + and − stranded ORFs were colored as grey and white, respectively. The CoreGenes between pSs-1 and Shfl2, between pSs-1 and SP18, and between pSs-1 and T4 were colored as yellow, green, and red, respectively. The predicted tRNA was indicated with thick arrow. Putative promoters and terminators were indicated by a bent arrow and inverted triangle on a vertical line, respectively.
Figure 5. Genome comparison of pSs-1 to its related phages (Shfl2 and SP18) using Artemis Comparison Tool (ACT).
Translated BLAST (tblastx, score cutoff: 40) was used to align translated genome sequences of phages. The blue and red lines represent the reverse and forward matches, respectively, and color intensity is proportional to the sequence homology. Nucleotide base-pairs were indicated between grey lines for each phage genome.
The high bacteriolytic activity of pSs-1 against both S. sonnei and S. flexneri may be attributed to the high nucleotide identity with Shfl2 and SP1814. All of the ORFs in pSs-1 exhibited homology to sequences of T4-like phages, such as Shfl2, SP18, T4, T6, AR1, RB14, RB32, RB51, and vB_EcoM_ACG-C40, reported in the GenBank database. T4-like phages, one of the best-characterized groups of phages present common characteristics: (i) morphology of the Myoviridae family; (ii) similar host range, the family Enterobacteriacae; (iii) large genome size in the range between 160 kbp and 250 kbp; (iv) similar G + C content, ranging from 35% to 43%15,16,17,18. Based on these results, pSs-1 is considered as a member of the T4-like phage group since it belongs to the Myoviridae family, infects Shigella species, contains relatively large genome (164,999 bp), and possesses G + C content of 35.54%. More than 200 T4-like phages have been examined and about 90% of T4-like phages grow on Escherichia coli or other enterobacteria, especially its close relatives such as Klebsiella and Shigella17,18.
A total of 10 tRNA genes (cove score 41.18–78.18) were identified19, which is more than the average number of tRNAs in T4-like phage group. The exceptionally large number of tRNAs (24 tRNAs) in Aeh1 may be attributed to its significantly large genome (233,234 bp) compared to those of the other T4-like phages17. Likewise, the genome of KVP40 (244,834 bp) encodes a large number of tRNAs20. Although the exact function of tRNA in phage is still not clear, it demonstrates that it may contribute to short latent period and large burst size of pSs-1 since tRNA in phage is known to influence its reproduction in the host, and facilitate the improvement of propagation and the reduction of latent period21. The genome of pSs-1 did not contain lysogeny genes and all of the ORFs had nothing in common with pathogenic factors.
In summary, it is considered as a universal problem as a lot of shigellosis cases are reported in developed countries as well as developing countries, although shigellosis had been regarded as a problem only in developing countries. In developing countries especially where poor hygiene standards occur, a safe year-round supply of drinking water remains a problem because the effective water treatment facility is often beyond their financial capacity22. The successful protection using phage against shigellosis was reported with animal experiments and the safety of phage administration through drinking water was reported with phage safety test in humans8,23,24,25. Shigella phage has potential uses to control or eradicate epidemic shigellosis in frequently affected area as phage is known to be cost-effective26. In developed countries where there are increasing concerns about antibiotic resistance, phages can be alternatives to conventional antibiotics. Phage research has been conducted continuously in Eastern Europe and Former Soviet Union countries, with leadership of the G. Eliava Institute of Bacteriophages, Microbiology and Virology in Tbilisi, Georgia. For many years, the Eliava Institute had developed and produced phages for the treatment and prophylaxis of bacterial infections including intestinal infections27. In a series of clinical trials, the therapeutic effectiveness of phages against infectious diseases was evaluated and no harmful effects were reported27. As the prevalence of a particular species of Shigella is generally considered to differ in various geographical areas5, a large-scale future screenings using Shigella environmental isolates from different geographical areas are required. This study may serve as a momentum to enhance the international collaborative research aiming control of waterborne infections, particularly shigellosis, on different continents.
Methods
Sampling, phage isolation and purification
Altogether 82 environmental water samples were collected from 5 different sampling sites, such as river and stream in South Korea between October 2011 and April 2012. Phage isolation was performed using a standard enrichment method using an indicator bacterium (S. sonnei ATCC® 25931)11. All of the bacterial strains used in this study were cultured at 37 °C (Table 1). The conventional double-layer agar method was used for the examination of phage activity28. A single plaque was collected from the plate with a sterile Pasteur pipette and used to inoculate TSB containing 2 ml of log-phase S. sonnei ATCC® 25931. This single plaque isolation procedure was repeated three successive times to obtain purified phages11. A Shigella phage, pSs-1, formed clear plaques in S. sonnei ATCC® 25931 (Fig. 1A) and was selected for further studies.
Phage morphology, host range and efficiency of plating (EOP)
In our study, every assay was performed in triplicate except electron microscopy. For the electron microscopy analysis, the phage suspension (7.8 × 108 PFU/ml) was concentrated and purified using continuous CsCl density gradient ultra-centrifugation11. The purified phages (1.0 × 1011 PFU/ml, 6 ml of total volume) after the concentration/ purification using Polyethylene Glycol (PEG) and CsCl gradient centrifugation were negatively stained with 2% uranyl acetate. Electron micrographs were taken using a Zeiss TEM EM902 (Zeiss, Germany) at an accelerating voltage of 80 kV. The phage size was determined from at least 10 measurements29.
To evaluate the host range of pSs-1, its infectivity was tested on the bacterial strains used in this study. The presence of plaque formation and the number of plaques were determined after 24 h of incubation, and the EOP values were quantified by calculating the ratio of the PFU obtained with each phage-susceptible strain to the PFU obtained with the indicator strain.
One-step growth and host cell lysis
Burst size and latent period of pSs-1 were determined by the one-step growth analysis as previously described30. Samples (100 μl) were collected at 5 min intervals and the titers were determined by the double-layer agar method. To evaluate the bacteriolytic activity of pSs-1 against S. sonnei ATCC® 25931 and S. flexneri ATCC® 29903, the absorbance (OD600 nm) was examined in order to determine the change of viable bacteria. After 1 h of the early-exponential phase bacterial incubation, the culture was divided into four 10 ml samples, which were co-cultured with phage suspensions at different multiplicities of infection (MOIs): 0, 0.1, 1, and 10. The preparations were incubated at 37 °C with shaking at 250 rpm. Bacteria not inoculated with pSs-1 (MOI: 0) were used as a control.
Phage genome sequence analysis
The phage DNA was sequenced by GenoTech (Seoul, Korea) using Sanger sequencing and a Next Generation Sequencing System (NGS: Ion PGM 314 sequencer). The full length genome sequence was obtained by sequence assembly using CLC Genomics Workbench v.6.0.5. Contig gaps were filled by additional PCR and primer walking. Potential open reading frames (ORFs) that may encode gene products were predicted using GLIMMER and GeneMarkS, respectively31,32. The putative functions of the ORFs were analyzed by BLASTP searches at the National Center for Biotechnology Information (NCBI). Putative promoter regions were predicted using the Neural Network Promoter Prediction tool of the Berkeley Drosophila Genome Project (minimum promoter score: 0.9)33. Rho-independent transcription terminators were identified using Finding Terminators program (energy threshold value: −11)34. Transmembrane domains and signal sequence regions were predicted with the TMHMM program, ver. 2.0, and the SignalP 3.0 program, respectively35. The phage’s genome map was drawn using DNA Master19. The genome of pSs-1 was subjected to pairwise analysis using the Artemis Comparison Tool (ACT)36 with its close homologs, phage Shfl2 [GenBank accession number: NC_015457] and SP18 [GenBank accession number: NC_014595]. The protein sequence similarities of the phages were analyzed using CoreGenes3.037.
Additional Information
Accession code: The genomic sequence employed in the present study is available at 316 GenBank (No: KM501444).
How to cite this article: Jun, J. W. et al. Bacteriophage application to control the contaminated water with Shigella. Sci. Rep. 6, 22636; doi: 10.1038/srep22636 (2016).
Supplementary Material
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2014R1A2A1A11050093), and under the framework of international cooperation program managed by National Research Foundation of Korea (2015K2A1A2070234).
Footnotes
The authors declare no competing financial interests.
Author Contributions J.W.J., B.C.L. and S.C.P. conceived and designed the experiments; J.W.J. performed the experiments; S.S.G., H.J.K., S.K.Y. and C.C. took part in the design of the study; J.Y.C. contributed materials; J.W.J. and S.C.P. wrote the paper.
References
- Niyogi S. K. Shigellosis. J. Microbiol. 43, 133–143 (2005). [PubMed] [Google Scholar]
- [WHO] World Health Organization Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae type 1 (WHO Document Production Services, Geneva, Switzerland, 2005). [Google Scholar]
- Ranjbar R., Mammina C., Pourshafie M. R. & Soltan-Dallal M. M. Characterization of endemic Shigella boydii strains isolated in Iran by serotyping, antimicrobial resistance, plasmid profile, ribotyping and pulsed-field gel electrophoresis. BMC Res. Notes 1, 74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison G. E. & Verma N. K. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 8, 17–23 (2000). [DOI] [PubMed] [Google Scholar]
- Zafar A., Sabir N. & Bhutta Z. A. Frequency of isolation of Shigella serogroups/serotypes and their antimicrobial susceptibility pattern in children from slum areas in Karachi. J. Pak. Med. Assoc . 55, 184–188 (2005). [PubMed] [Google Scholar]
- Ashkenazi S. et al. Recent trends in the epidemiology of Shigella species in Israel. Clin. Infect. Dis. 17, 897–899 (1993). [DOI] [PubMed] [Google Scholar]
- Holt K. E. et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat. Genet. 44, 1056–1059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housby J. N. & Mann N. H. Phage therapy. Drug Discov. Today 14, 536–540 (2009). [DOI] [PubMed] [Google Scholar]
- Hagens S. & Loessner M. J. Bacteriophage for biocontrol of foodborne pathogens: calculations and considerations. Curr. Pharm. Biotechnol. 11, 58–68 (2010). [DOI] [PubMed] [Google Scholar]
- Pirnay J. P. et al. The phage therapy paradigm: prêt-à-porter or sur-mesure? Pharm. Res. 28, 934–937 (2011). [DOI] [PubMed] [Google Scholar]
- Jun J. W. et al. Characterization and complete genome sequence of the Shigella bacteriophage pSf-1. Res. Microbiol. 164, 979–986 (2013). [DOI] [PubMed] [Google Scholar]
- Jun J. W., Yun S. K., Kim H. J., Chai J. Y. & Park S. C. Characterization and complete genome sequence of a novel N4-like bacteriophage, pSb-1 infecting Shigella boydii. Res. Microbiol. 165, 671–678 (2014). [DOI] [PubMed] [Google Scholar]
- Ackermann H. W. 5500 Phages examined in the electron microscope. Arch. Virol. 152, 227–243 (2007). [DOI] [PubMed] [Google Scholar]
- Kim K. H., Chang H. W., Nam Y. D., Roh S. W. & Bae J. W. Phenotypic characterization and genomic analysis of the Shigella sonnei bacteriophage SP18. J. Microbiol. 48, 213–222 (2010). [DOI] [PubMed] [Google Scholar]
- Ackermann H. W. & Krisch H. M. A catalogue of T4-type bacteriophages. Arch. Virol. 142, 2329–2345 (1997). [DOI] [PubMed] [Google Scholar]
- Petrov V. M. et al. Plasticity of the gene functions for DNA replication in the T4-like phages. J. Mol. Biol. 361, 46–68 (2006). [DOI] [PubMed] [Google Scholar]
- Comeau A. M., Bertrand C., Letarov A., Tétart F. & Krisch H. M. Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology 362, 384–396 (2007). [DOI] [PubMed] [Google Scholar]
- Liao W. C. et al. T4-like genome organization of the Escherichia coli O157:H7 lytic phage AR1. J. Virol. 85, 6567–6578 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. R. et al. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 187, 1091–1104 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E. S. et al. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185, 5220–5233 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Bechet M., Vergassola M. & Rocha E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 17, 1486–1495 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuigan K. G., Joyce T. M. & Conroy R. M. Solar disinfection: use of sunlight to decontaminate drinking water in developing countries. J. Med. Microbiol. 48, 785–787 (1999). [DOI] [PubMed] [Google Scholar]
- Dubos R. J., Straus J. H. & Pierce C. The multiplication of bacteriophage in vivo and its protective effect against an experimental infection with Shigella dysenteriae. J. Exp. Med. 78, 161–168 (1943). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruttin A. & Brűssow H. Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy. Antimicrob. Agents Chemother. 49, 2874–2878 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulakvelidze A. & Kutter E. Bacteriophage therapy in humans In Bacteriophages: biology and applications (eds Kutter E. & Sulakvelidze A.) 381–436 (CRC Press, Boca Raton, FL, 2005). [Google Scholar]
- Miedzybrodzki R., Fortuna W., Weber-Dabrowska B. & Górski A. Phage therapy of staphylococcal infections (including MRSA) may be less expensive than antibiotic treatment. Postepy Hig. Med. Dosw. 61, 461–465 (2007). [PubMed] [Google Scholar]
- Chanishvili N. A Literature Review of the Practical Application of Bacteriophages Research (Nova Science Publishers, New York, USA, 2012). [Google Scholar]
- Adams M. H. Bacteriophages (Wiley-Interscience Publishers, New York, USA, 1959). [Google Scholar]
- Ackermann H. W. & Heldal M. Basic electron microscopy of aquatic viruses In Manual of aquatic virus ecology (eds Wilhelm S. W., Weinbauer M. G. & Suttle C. A.) 182–192 (American Society of Limnology and Oceanography, Waco, 2010). [Google Scholar]
- Ellis E. L. & Delbruck M. The growth of bacteriophage. J. Gen. Physiol. 22, 365–384 (1939). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher A. L., Harmon D., Kasif S., White O. & Salzberg S. L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27, 4636–4641 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J., Lomsadze A. & Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese M. G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 26, 51–56 (2001). [DOI] [PubMed] [Google Scholar]
- Lesnik E. A. et al. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 29, 3583–3594 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento A. L. T. O. et al. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186, 2164–2172 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T. et al. ACT: the artemis comparison tool. Bioinfomatics 21, 3422–3423 (2005). [DOI] [PubMed] [Google Scholar]
- Zafar N., Mazumder R. & Seto D. CoreGenes: a computational tool for identifying and cataloging core genes in a set of small genomes. BMC Bioinformatics 3, 12 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.