Abstract
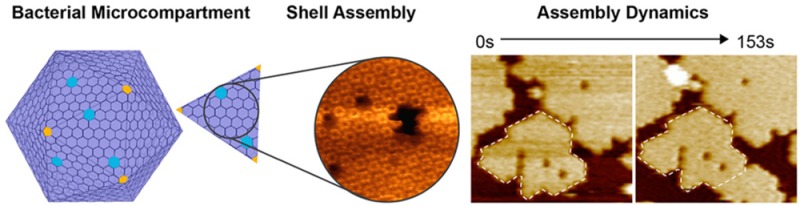
Bacterial microcompartments (BMCs) are proteinaceous organelles widespread among bacterial phyla. They compartmentalize enzymes within a selectively permeable shell and play important roles in CO2 fixation, pathogenesis, and microbial ecology. Here, we combine X-ray crystallography and high-speed atomic force microscopy to characterize, at molecular resolution, the structure and dynamics of BMC shell facet assembly. Our results show that preformed hexamers assemble into uniformly oriented shell layers, a single hexamer thick. We also observe the dynamic process of shell facet assembly. Shell hexamers can dissociate from and incorporate into assembled sheets, indicating a flexible intermolecular interaction. Furthermore, we demonstrate that the self-assembly and dynamics of shell proteins are governed by specific contacts at the interfaces of shell proteins. Our study provides novel insights into the formation, interactions, and dynamics of BMC shell facets, which are essential for the design and engineering of self-assembled biological nanoreactors and scaffolds based on BMC architectures.
Keywords: Bacterial microcompartment, high-speed atomic force microscopy, protein dynamics, protein interaction, self-assembly
Bacterial microcompartments (BMCs) are organelles that are widespread among bacterial phyla.1−5 They are composed of a protein shell that surrounds an enzymatic core. BMCs are proposed to assemble from the inside out,4,6 first forming the metabolic core around which an apparently icosahedral shell self-assembles. To date, models for BMC shells are based on crystal structures of individual shell proteins with the assumption of icosahedral symmetry.7 The facets of the shell are predominantly composed of a homohexameric (BMC-H) shell protein,1,8 suggested to be either a single or a double layer.1,7,9,10 Empty shells and various architectures including tubes and “swiss rolls” can be visualized by transmission electron microscopy (TEM) after heterologous expression of shell proteins in Escherichia coli (E. coli).11 However, how shell proteins self-assemble into higher order structures is an open question. Here we use a combination of X-ray crystallography and high-speed atomic force microscopy (HS-AFM) to characterize a molecular sheet of BMC shell hexamers and visualize for the first time the dynamics of shell facet self-assembly.
Hexamers formed by BMC-H proteins are typically the major component of BMC shell facets. The genome of the halophilic myxobacterium Haliangium ochraceum (HO) encodes only one BMC-H protein.12 Thin-sectioned TEM samples of E. coli recombinantly expressing HO BMC-H show that there are no polyhedral structures, presumably because shell proteins responsible for curvature, or the capping of an icosahedral shell, are lacking. Instead, the protein appears to form rolled up sheets in vivo (Supporting Information Figure S1). The formation of rolls of sheets could be a consequence of being limited to, essentially confined to, the cytoplasmic space. When the HO BMC-H protein is purified from E. coli, it spontaneously forms extended two-dimensional arrays,12 constituting a proxy for a BMC shell facet.
To characterize the building block of these higher order assemblies we crystallized the HO BMC-H protein and determined its structure at a resolution of 1.8 Å by molecular replacement (Supporting Information Table S1). Eight monomers (four partial hexamers) form the asymmetric unit of the P3 space group. The crystallographic 3-fold axis generates cyclic hexamers (Supporting Information Figure S2). Each monomer consists of the α/β fold characteristic of BMC-H monomers (Pfam00936 domain, Figure 1A). The closest structural homologue is a C-terminal deletion mutant of the carboxysome shell protein CcmK2 (PDB: 3DNC) with a root-mean-square deviation (RMSD) of 0.7 Å over 84 (out of 90) aligned Cα atoms. The lateral edges of the HO BMC-H hexamer are 37 Å long and it has an overall thickness of approximately 36 Å including the C-terminal extensions (Figure 1B). The hexamers have a distinct sidedness, concave and convex (Figure 1B), and pack into uniformly oriented layers in the crystal (Figure 1C). The lateral interface between two hexamers buries 307 Å2 per hexamer. A total of 12 nonbonded contacts are made among 8 residues per interface. Residues K28 and R78 of two separate monomers on adjoining hexamers make relatively large contributions to the lateral interface. The layers are stacked in both a concave–concave and offset convex–convex fashion in the crystal (Supporting Information Figure S2). The concave–concave interface primarily consists of interactions made by the N- and C- termini of the proteins. Convex–convex interfaces are mediated by hydrogen bonds and two salt bridges between charged residues (R66 and R62 to E65) on the surface of the protein.
Figure 1.
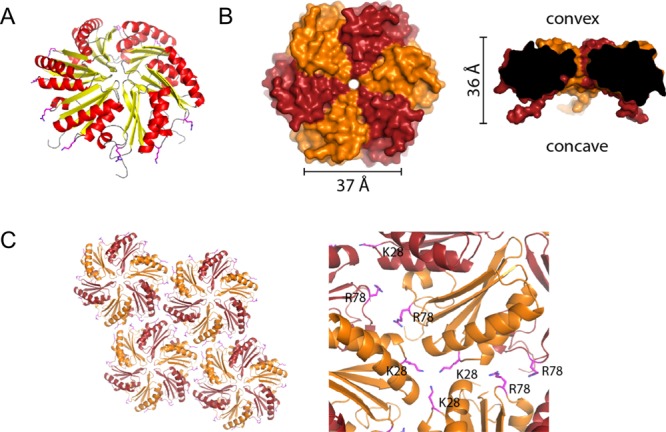
Crystal structure of the HO BMC-H. (A) HO BMC-H adopts the characteristic α/β BMC-H fold (strands in yellow, helices in red). Edge residues K28 and R78 are shown as sticks. (B) Surface top and center sliced view with edge and thickness dimensions. The hexamer has a pronounced sidedness with distinct convex (top) and concave (bottom) surface. Protomer chains are alternatingly colored dark red and orange. (C) The hexamers pack into uniformly oriented layers in the crystal. Close-up view on the right shows residues K28 and R78 interacting at the interfaces (shown as magenta sticks).
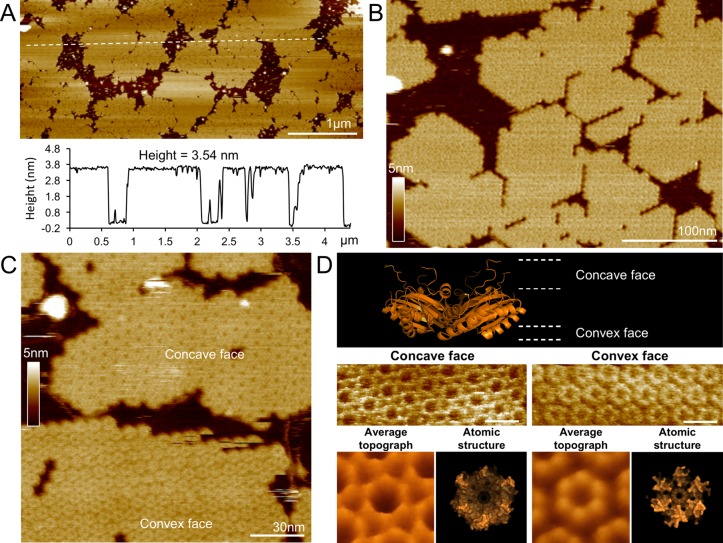
Sheet formation by the HO BMC-H was characterized by atomic force microscopy (AFM) in solution, which has the advantage of characterizing protein organization and assembly under near physiological conditions (in buffer, under ambient temperature and pressure).13−17 The proteins were observed to form two-dimensional patches with varying sizes on the surface of AFM substrate (Figure 2A). Cross-sectional analysis illustrates that the HO BMC-H forms flat protein sheets of ∼3.5 nm thick (Figure 2A), consistent with the thickness of a single hexamer in the crystal structure. This indicates that the sheets are composed of a single layer of hexamers. Higher-magnification AFM images of the protein sheets, captured at 17 seconds per frame (s/frame), further show the regular angles and straight edges of the hexamers within the extended assemblies (Figure 2B, Supporting Information Figure S3). By minimizing the AFM scanning force, we were able to resolve the molecular details of protein organization within these sheets. Two distinct surface morphologies of the hexamer patches could be distinguished in AFM topographs based on the relative sizes of the central depression, measured as the distance between the protruding regions of a single hexameric on each surface (Figure 2C,D). These correspond to the concave and convex faces of the hexamers observed in the crystal structure (Figure 1B). The AFM results show that the shell layers are composed of uniformly oriented BMC-H proteins, which were also observed in the crystal packing.
Figure 2.
AFM analysis of the HO BMC-H sheet. (A) Cross-section analysis of hexamers adsorbed to the mica surface indicates a thickness of 3.54 nm, corresponding to a single protein layer. (B) Higher magnification reveals the organization of hexamers as seen by straight edges and regular 120° angles (see Supporting Information Figure S3). (C) Two distinct surfaces morphologies can be observed between patches of hexamers. (D) The relative sidedness of the sheet (convex versus concave face) accounts for the differing surface morphologies and can be distinguished by the perceived size of the central depression during AFM scanning. The concave face has a depression diameter of 52.8 Å whereas the convex face has a diameter of 47.1 Å measured by AFM cross-section analysis, compared with 51.6 and 45.7 Å, respectively, based on the crystal structure.
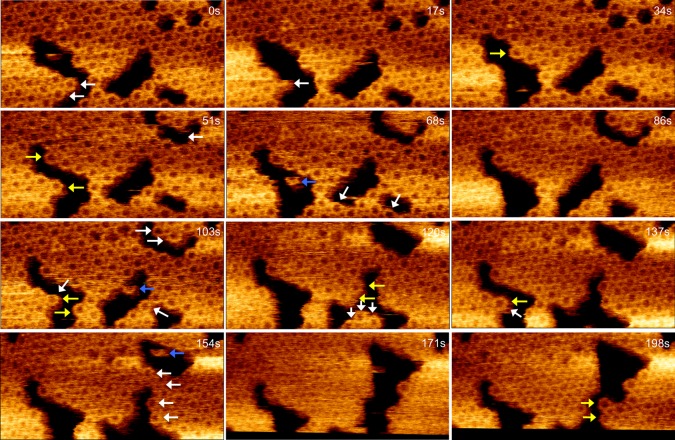
To investigate how BMC-H proteins self-assemble into facets, we observed the dynamics of HO hexamer sheets using HS-AFM (17 s/frame). HS-AFM has evolved into a powerful tool for exploring the structure and dynamics of biomolecular systems.18,19 As shown in Figure 3, the lattice of the BMC-H hexamers is clearly identified, demonstrating the ability of HS-AFM to image individual hexamers. Two hexamers (depicted by white arrows at 0 s) were observed to dissociate from the sheet at 17 s. Subsequently, another hexamer dissociated (white arrow shown at 17 s) and one new hexamer assemble into the protein sheet at 34 s (yellow arrow). Such translational motions of hexamers were continually observed during the time sequence (Figure 3, Supporting Information Movie S1) and higher scanning speed (2 s/frame, Supporting Information Figure S4, Supporting Information Movie S2), revealing the dynamic nature of self-assembly of the shell proteins and flexible interactions between proteins in the shell. These results demonstrate that sheets are formed by the incorporation of preassembled hexamers.
Figure 3.
Time-lapse AFM imaging reveals the dynamics of HO BMC-H sheet formation. Hexamers are both removed from (white arrows) and incorporated into the sheet (yellow arrows) during the course of scanning. Blue arrows depict hexamers not associated with the sheet that are translocating across the mica surface. Twelve aligned AFM images (100 × 47.7 nm) were captured at 17 s per frame from a 20 min AFM movie. Scale bar: 10 nm. See Supporting Information Movie S1.
Time-lapse AFM images further revealed that individual hexamers could migrate along the edges of the sheets, suggesting dynamic association of newly recruited hexamers with existing hexamers located at the periphery of the sheet (Supporting Information Figure S5, Supporting Information Movie S3). Hexamer motion was also observed within the center of sheets, however, these were fewer in number (Supporting Information Figure S6, Supporting Information Movie S4). Together, the motion events captured at both the periphery and center of the sheets reveal its overall dynamic nature and we infer that establishing an increasing number of lateral interactions increases the stability of incorporation of a hexamer in the facet. Nevertheless, fully surrounded hexamers are able to leave the facet, albeit much less frequently than those only partially encompassed. In the cell, this could allow a damaged hexamer to be replaced within an existing shell without the major reconstruction of entire facets.
We further demonstrated that the inclusion of additional hexamers into existing sheets occurs only when the docking hexamer is in the same orientation as the sheet (Supporting Information Figure S7). Hexamers with concave face up could assemble together but could not associate with the hexamers with convex face down. This is most likely ascribed to the particular contacts at the protein interfaces and, moreover, that the same surface orientation of shell proteins is prerequisite for the generation of BMC shells.
The formation of new shell edges and large patches from individual hexamers was characterized in detail (Supporting Information Figures S8, S9, and S10; Supporting Information Movies S5, S6, and S7). New hexamers were observed joining an existing sheet at regular intervals. Furthermore, growth of the sheets could be induced by adding more proteins during the course of AFM imaging (Supporting Information Figure S11). In addition to the motions of individual hexamers, we were also able to capture the dynamics of entire patches. We found patches could disassociate from one sheet, travel, and then assemble with another patch (Supporting Information Figure S12, Supporting Information Movie S8). Higher-speed AFM imaging (1 s/frame) was also used to explore the dynamics of BMC-H protein assemblies (Supporting Information Movie S9). The translational motions of single hexamers and larger patches were observed. Whether there is rotational dynamics of these hexamers in addition to the translational dynamics observed here requires a much higher scanning speed.
Distinct electrostatic properties of the concave and convex faces of the hexamers evident from the crystal structure likely account for the different rates of shell protein attachment to the mica surface (Supporting Information Figure S13). When the concave face (which is relatively nonpolar) of the hexamer is exposed to the AFM probe and the relatively polar convex surface is attached to the negatively charged mica substrate, the sheets are more stable than sheets in the opposite orientation (i.e., convex up, concave on mica).
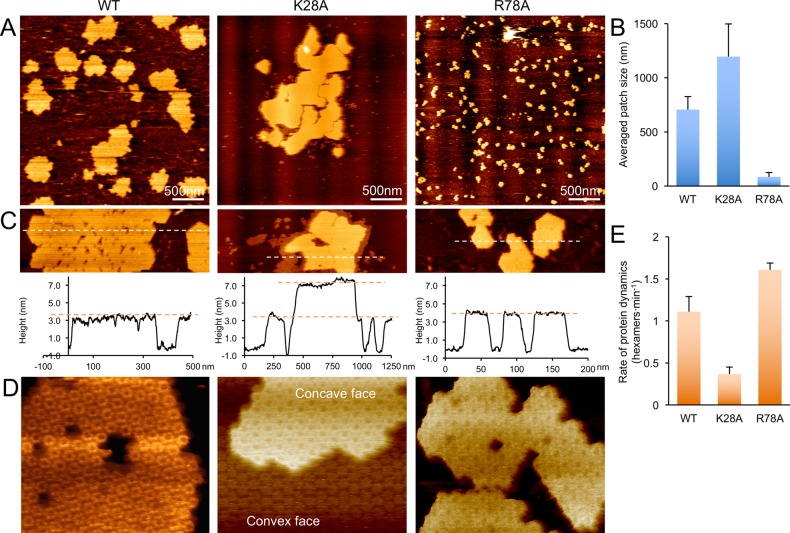
To begin to dissect the structural determinants governing the self-assembly of shell proteins, we made single-point mutations of residues at the lateral interface of adjoining hexamers. Residues K28 and R78 are strongly conserved residues in BMC-H proteins and they are located at the junctions between adjacent hexamers in the layers observed in the crystal packing (Figure 1A,C). AFM images (Figure 4A,B) show that K28A mutant typically forms larger patches (∼1200 nm in diameter), whereas the patches of R78A mutant are much smaller (85 nm in diameter) than those formed by the wild-type (WT) protein (705 nm in diameter). Unexpectedly, the K28A sheets tend to appear as stacked layers, whereas the patches formed by WT or R78A proteins are only single layers (Figure 4C). Molecular-resolution AFM image indicates that the double K28A layer is formed by two protein sheets with convex surfaces making contact with each other (Figure 4D, Supporting Information Figure S14). In addition, the protein dynamics is modified by the K28A and R78A mutations (Figure 4E, Supporting Information Figure S15). The results indicate that K28 and R78 residues of the HO BMC-H protein play key roles in governing the assembly of HO shell hexamers and that the strength of lateral hexamer interactions within the sheets can be modulated without complete disruption. In addition, we found that subtle changes (e.g., a single-point mutation) can promote stacking of the shell protein hexamers.
Figure 4.
Characterization of the impact on assembly of point mutations of the HO BMC-H protein. (A) AFM images of patches of WT, K28A, and R78A BMC-H protein. (B) Quantitative analysis shows the average patch size of the R78A mutant (85 nm) is smaller than the WT (705 nm) while that of the K28A mutant (1196 nm) is larger than the WT. (C) Cross-section analysis shows the thickness of sheets formed by the WT and R78A mutant to correspond to that of a single protein layer (3.5 nm). The K28A mutant sheet has thickness consistent with a double layer (7.0 nm). (D) The double layer of the K28A mutant is formed by convex–convex contacts. Scale bar: 10 nm. (E) Normalized rates of protein dynamics in the WT, K28A, and R78A hexamer sheets. The dynamics features of these assemblies are variable: R78A proteins in the self-assembled patches present higher translational dynamics than WT proteins, whereas the K28A sheets appear relatively stable during AFM imaging. Measurements were made based on a series of AFM images taken at multiple distinct regions (n = 12, Supporting Information Figure S15). The data were normalized to correct for differences in frame capture time, the scan area which have diverse ratios of protein to mica and the scan size, relative to the WT dynamic events. Error bars represent standard deviation.
It is worth noting that with the minimized AFM scanning force, the association and dissociation events can be recorded simultaneously in the same scanning area (Figure 3, Supporting Information Movie S1), at the edges (Supporting Information Figure S5, Supporting Information Movie S3), or centers (Supporting Information Figure S6, Supporting Information Movie S4) of hexamer sheets. Moreover, a difference in protein dynamics was observed between concave up and concave down facets in the same view (Supporting Information Figure S13) and between wild-type, K28A, and R78A mutant hexamers (Figure 4E, Supporting Information Figure S15). These results provide evidence that the hexamer dynamics as captured by high-speed AFM is not biased by the AFM tip scanning.
Recently, the dynamic aggregation of the trp-RNA binding attenuation protein (TRAP) cages, which are self-assembled by the TRAP ring-shaped proteins, has been explored using HS-AFM imaging.20 In the presence of dithiothreitol, the disassembly of TRAP cages was monitored. Interestingly, in contrast to the dynamic assembly of BMC shell proteins on mica surface visualized in this study, the TRAP cage formation could not occur on mica, likely due to the different capacity for interactions between proteins under study and the substrate. Nevertheless, both studies, TRAP cages20 and BMC-H hexamer sheets (in this work), demonstrate the dynamics of the self-assembly of large protein complexes.
In summary, our data provide molecular insights into the formation, interactions, and dynamics of BMC shell facets. We show that preassembled shell hexamers form single layer sheets of uniform orientation. In addition to the arrangement of hexameric proteins, we find that individual hexamers can dissociate from and incorporate into assembled sheets, indicating an overall flexible intermolecular interaction. We also show that specific contacts at the interfaces of neighboring proteins influence the dynamic features of shell proteins, and thereby the self-assembly of the shell facets. The design and construction of synthetic BMCs have attracted intense interest for the bioengineering of nanoreactors and molecular scaffolds.4,21 Understanding the details of self-assembly of BMC shell proteins is a prerequisite for control and engineering of BMC-based architectures with the aim of building designed nanoreactors and molecular scaffolds.
Acknowledgments
L.-N.L. acknowledges the Royal Society University Research Fellowship (UF120411), the Royal Society Research Grant for URF (RG130442), the Biotechnology and Biological Sciences Research Council Grant (BB/M024202/1) and the Biotechnology and Biological Sciences Research Council ALERT 2014 Grant (BB/M012441/1). C.A.K acknowledges the support from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID) Grant (1R01AI114975-01). The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy (DE-AC02-05CH11231). We thank Dr. Alex Winkel from JPK and Dr Alexander Dulebo from Bruker for technical assistance with high-speed AFM imaging. We thank the Centre for Advanced Microscopy at Michigan State University for assistance with TEM.
Glossary
Abbreviations
- AFM
atomic force microscopy
- BMCs
bacterial microcompartments
- BMC-H
bacterial microcompartment hexamer
- HO
Haliangium ochraceum
- HS-AFM
high-speed atomic force microscopy
- s/frame
seconds per frame
- TEM
transmission electron microscopy
- TRAP
trp-RNA binding attenuation protein
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.5b04259.
Materials and methods, crystallization and structure determination, AFM imaging, and Image analysis. (PDF)
These are accompanied by additional supporting table and figures providing further crystallization and AFM data. HS-AFM movies showing the assembly dynamics of HO hexamers are also provided. (ZIP)
Author Contributions
L.-N.L., M.F., and S.B. conducted AFM imaging and data analysis. C.A. and B.P. purified the proteins and B.P. crystallized the wild-type protein. C.A. prepared the mutants and performed visualization using TEM. M.S. determined and analyzed the structure and wrote the manuscript. C.A.K. and L.-N.L. designed the study, analyzed the data and wrote the manuscript.
M.S. and M.F. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Kerfeld C. A.; Sawaya M. R.; Tanaka S.; Nguyen C. V.; Phillips M.; Beeby M.; Yeates T. O. Science 2005, 309 (5736), 936–938. 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- Axen S. D.; Erbilgin O.; Kerfeld C. A. PLoS Comput. Biol. 2014, 10 (10), 20. 10.1371/journal.pcbi.1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld C. A.; Heinhorst S.; Cannon G. C., Bacterial microcompartments. In Annual Review Microbiology; Gottesman S., Harwood C. S., Eds. Annual Reviews: Palo Alto, 2010; Vol. 64, pp 391–408. [DOI] [PubMed] [Google Scholar]
- Kerfeld C. A.; Erbilgin O. Trends Microbiol. 2015, 23 (1), 22–34. 10.1016/j.tim.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Chowdhury C.; Sinha S.; Chun S.; Yeates T. O.; Bobik T. A. Microbiol. Mol. Biol. Rev. 2014, 78 (3), 438–468. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron J. C.; Wilson S. C.; Bernstein S. L.; Kerfeld C. A. Cell 2013, 155 (5), 1131–1140. 10.1016/j.cell.2013.10.044. [DOI] [PubMed] [Google Scholar]
- Tanaka S.; Kerfeld C. A.; Sawaya M. R.; Cai F.; Heinhorst S.; Cannon G. C.; Yeates T. O. Science 2008, 319 (5866), 1083–6. 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- Tanaka S.; Sawaya M. R.; Yeates T. O. Science 2010, 327 (5961), 81–4. 10.1126/science.1179513. [DOI] [PubMed] [Google Scholar]
- Tsai Y.; Sawaya M. R.; Cannon G. C.; Cai F.; Williams E. B.; Heinhorst S.; Kerfeld C. A.; Yeates T. O. PLoS Biol. 2007, 5 (6), 1345–1354. 10.1371/journal.pbio.0050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samborska B.; Kimber M. S. Structure 2012, 20 (8), 1353–62. 10.1016/j.str.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Parsons J. B.; Frank S.; Bhella D.; Liang M.; Prentice M. B.; Mulvihill D. P.; Warren M. J. Mol. Cell 2010, 38 (2), 305–15. 10.1016/j.molcel.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Lassila J. K.; Bernstein S. L.; Kinney J. N.; Axen S. D.; Kerfeld C. A. J. Mol. Biol. 2014, 426 (11), 2217–2228. 10.1016/j.jmb.2014.02.025. [DOI] [PubMed] [Google Scholar]
- Liu L. N.; Scheuring S. Trends Plant Sci. 2013, 18 (5), 277–286. 10.1016/j.tplants.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Scheuring S.; Nevo R.; Liu L. N.; Mangenot S.; Charuvi D.; Boudier T.; Prima V.; Hubert P.; Sturgis J. N.; Reich Z. Biochim. Biophys. Acta, Bioenerg. 2014, 1837 (8), 1263–1270. 10.1016/j.bbabio.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Liu L. N.; Sturgis J. N.; Scheuring S. J. Struct. Biol. 2011, 173 (1), 138–45. 10.1016/j.jsb.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Liu L. N.; Duquesne K.; Oesterhelt F.; Sturgis J. N.; Scheuring S. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (23), 9455–9. 10.1073/pnas.1004205108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. N.; Duquesne K.; Sturgis J. N.; Scheuring S. J. Mol. Biol. 2009, 393 (1), 27–35. 10.1016/j.jmb.2009.07.044. [DOI] [PubMed] [Google Scholar]
- Ando T.; Uchihashi T.; Kodera N. Annu. Rev. Biophys. 2013, 42, 393–414. 10.1146/annurev-biophys-083012-130324. [DOI] [PubMed] [Google Scholar]
- Casuso I.; Rico F.; Scheuring S. Curr. Opin. Chem. Biol. 2011, 15 (5), 704–9. 10.1016/j.cbpa.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Imamura M.; Uchihashi T.; Ando T.; Leifert A.; Simon U.; Malay A. D.; Heddle J. G. Nano Lett. 2015, 15 (2), 1331–1335. 10.1021/nl5045617. [DOI] [PubMed] [Google Scholar]
- Frank S.; Lawrence A. D.; Prentice M. B.; Warren M. J. J. Biotechnol. 2013, 163 (2), 273–279. 10.1016/j.jbiotec.2012.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.