Abstract
Salvia miltiorrhiza Bunge is highly valued in traditional Chinese medicine for its roots and rhizomes. Its bioactive diterpenoid tanshinones have been reported to have many pharmaceutical activities, including antibacterial, anti-inflammatory, and anticancer properties. Previous studies found four different diterpenoid biosynthetic pathways from the universal diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP) in S. miltiorrhiza. Here, we describe the functional characterization of ent-copalyl diphosphate synthase (SmCPSent), kaurene synthase (SmKS) and kaurene oxidase (SmKO) in the gibberellin (GA) biosynthetic pathway. SmCPSent catalyzes the cyclization of GGPP to ent-copalyl diphosphate (ent-CPP), which is converted to ent-kaurene by SmKS. Then, SmKO catalyzes the three-step oxidation of ent-kaurene to ent-kaurenoic acid. Our results show that the fused enzyme SmKS-SmCPSent increases ent-kaurene production by several fold compared with separate expression of SmCPSent and SmKS in yeast strains. In this study, we clarify the GA biosynthetic pathway from GGPP to ent-kaurenoic acid and provide a foundation for further characterization of the subsequent enzymes involved in this pathway. These insights may allow for better growth and the improved accumulation of bioactive tanshinones in S. miltiorrhiza through the regulation of the expression of these genes during developmental processes.
Salvia miltiorrhiza Bunge has been widely used in China (and to a lesser extent in Japan, the United States, and European countries) for the treatment of cardiovascular and cerebrovascular diseases. This medicinal herb exhibits anti-inflammatory, antioxidant and radical scavenging effects1,2. Tanshinone I, tanshinone IIA, cryptotanshinone and dihydrotanshinone I are the major diterpene quinones of the lipophilic constituents in Danshen and are responsible for much of its anti-inflammatory, antioxidant, antitumor and a variety of other activities3,4,5. Because these monomeric compounds have significant pharmacological activities, Danshen preparations are more frequently used in the clinic.
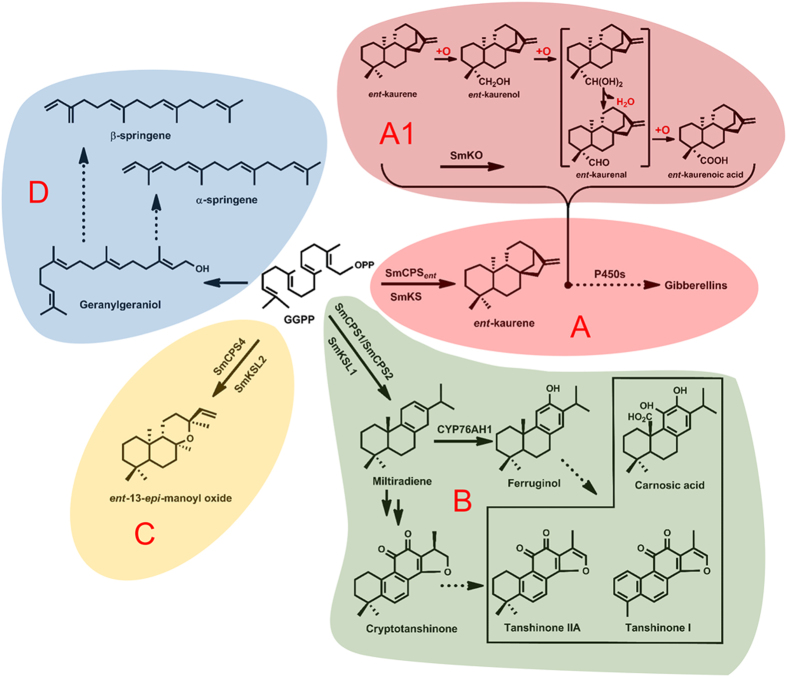
To accommodate the increasing need for clinical applications, researchers have deeply investigated the diterpenoid biosynthetic pathway to obtain the bioactive tanshinones directly using synthetic biology strategies in microbial cell factories. Previous works have indicated that at least four different diterpenoid biosynthetic pathways exist in S. miltiorrhiza (Fig. 1)6. Among them, the tanshinone biosynthetic pathway is uniquely initiated by a sequential pair of cyclization reactions catalyzed by SmCPS1 and SmKSL1 to produce abietane miltiradiene, which is a precursor of at least cryptotanshinone7,8. Then, SmCYP76AH1 catalyzes the turnover of miltiradiene to form ferruginol, thereby providing a solid foundation to elucidate the tanshinone biosynthetic pathway.
Figure 1. Four different diterpenoid biosynthetic pathways in S. miltiorrhiza.
(A) Gibberellin biosynthetic pathway. (A1) Proposed mechanism for SmKO conversion of ent-kaurene into ent-kaurenoic acid35. (B) Tanshinone biosynthetic pathway. (C,D) Unknown diterpenoid biosynthetic pathways.
However, only two diterpene synthases (diTPSs) in the S. miltiorrhiza GA biosynthetic pathway have been reported to date, and the roles of GAs in S. miltiorrhiza root and rhizome development and the total yield of tanshinones per plant are less clear. GAs are formed from GGPP via a set of reactions catalyzed by different enzymes, including two consecutive diTPSs, cytochrome P450 (CYP) and 2-oxoglutarate-dependent dioxygenases (2ODDs) in plants9. As a group of plant-growth regulators, these GAs control different aspects of plant development, such as seed germination, stem elongation, flowering, fruit set and fruit development. Understanding GA biosynthesis will allow us to improve the tanshinone contents by regulating the expression of the genes involved in the S. miltiorrhiza GA biosynthetic pathway. Here, we cloned three genes (SmCPSent, SmKS and SmKO) from S. miltiorrhiza hairy roots and then identified their functions by co-expressing them in Saccharomyces cerevisiae. Biochemical studies suggested that CPS and KS might interact with one another10; therefore, we constructed a fused SmCPSent and SmKS protein and showed that the production of ent-kaurene was significantly improved.
Results
Cloning and sequence analysis of SmCPS ent , SmKS and SmKO from S. miltiorrhiza hairy roots
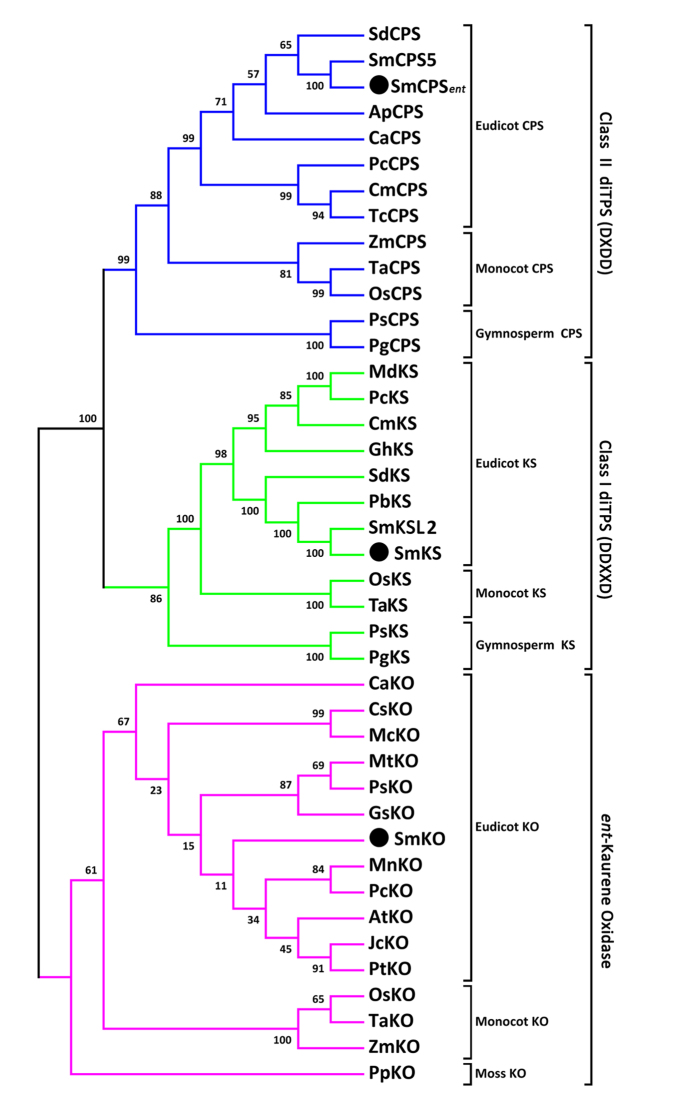
The full-length SmCPSent and SmKS cDNAs were determined by 5′ RACE and 3′ RACE, and the corresponding cDNA sequences were submitted to the National Center for Biotechnology Information (Supplementary Fig. S1). The full-length SmCPSent cDNA (GenBank accession number KT934789) is 2413 nt and encodes a polypeptide of 793 amino acids. SmCPSent clusters most closely to SmCPS5 of S. miltiorrhiza f. alba and to SdCPS from Scoparia dulcis (Fig. 2). The first 21 N-terminal amino acids are rich in serine and threonine (19%), which is a common characteristic of transit peptides that target the diTPS to plastids11,12. This information was supported by our analysis using ChloroP 1.1 13. The amino acid sequence also contains a conserved DIDD motif (Fig. 3), which strongly suggests that SmCPSent can catalyze GGPP to CPP as a class II diTPS. The SmKS cDNA (GenBank accession number KT934790) is 2636 nt in length and encodes a predicted protein of 806 amino acid residues. At the protein level, the KS sequence from the hairy roots of S. miltiorrhiza exhibits 99% identity with the SmKSL2 from S. miltiorrhiza f. alba (Fig. 2). The first 27 N-terminal amino acids are rich in serine and threonine (22%), suggesting that SmKS is also localized in plastids. Its amino acid sequence contains a DDFFD motif but lacks the DxDD motif (Fig. 3), indicating that SmKS is a plant KS protein with monofunctional class I diTPS activity. The SmKO cDNA (GenBank accession number KJ606394) is 1930 nt in length and has an open reading frame (ORF) encoding 519 amino acid residues, containing a cytochrome P450 conserved site (amino acids 451–460, Fig. 3). The deduced amino acid sequence shows 64% and 66% identity with AtKO (Arabidopsis thaliana, AAC39507) and PsKO (Pisum sativum, AAP69988) (Fig. 2). The gene was identified as a multifunctional kaurene oxidase catalyzing three sequential oxidations (ent-kaurene to ent-kaurenoic acid) in the GA biosynthetic pathway14,15.
Figure 2. Phylogenetic tree of CPS, KS and KO from different species.
The neighbor-joining phylogenetic trees were constructed using the bootstrap method in MEGA 5.1. The number of bootstrap replications was 1000. Descriptions of the three different types of synthases used in the phylogeny are listed in Supplementary Table 2.
Figure 3. Alignment of plant CPS, KS and KO.
The boxes represent the conserved regions. (A) AtCPS (Q38802), SmCPS5 (AHJ59324), SmCPSent (ALX18648), PpCPS/KS (BAF61135), AgAS (Q38710), GbLPS (Q947C4), AtKS (AAC39443), SmKSL2 (AHJ59325), SmKS (ALX18649). (B) AtKO (AAC39507), OsKO (AAT81230), SmKO (AJF93403).
Recombinant expression and functional characterization of SmCPS ent and SmKS
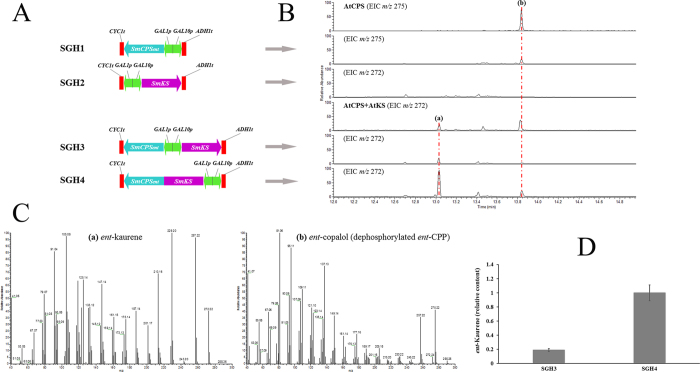
Previously reported evidence suggested that SmCPSent and SmKS might be involved in the S. miltiorrhiza GA biosynthetic pathway6. To confirm the biochemical functions of SmCPSent and SmKS in vivo, the SmCPSent ORF and SmKS ORFs were ligated individually or in combination in the yeast expression vector pESC-Trp (Fig. 4A) and expressed in the yeast strain BY-T20 (provided by Prof. Xueli Zhang’s lab, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, China). As expected, CPP was detected as the SmCPSent product in the products of the yeast strain SGH1 (BY-T20/pESC-Trp::SmCPSent) compared with the product of the A. thaliana AtCPS using GGPP as the substrate. No CPP was found in the yeast strain carrying the empty pESC-Trp vector. Using the product of the A. thaliana AtKS as the authentic standard, ent-kaurene was detected as the SmKS product in the products of the yeast strain SGH3 (BY-T20/pESC-Trp::SmCPSent/SmKS) but not SGH2 (BY-T20/pESC-Trp::SmKS), confirming that SmKS possessed monofunctional class I diTPS activity and catalyzed the formation of ent-kaurene using the SmCPSent product CPP as the substrate. Therefore, the absolute configuration of the SmCPSent product CPP was identified as an enantiomer (i.e., ent-CPP) (Fig. 4B).
Figure 4. Extracts from yeast strain BY-T20 cultures expressing the S. miltiorrhiza SmCPSent or/and SmKS using GC-MS.
(A) Construction of the recombinant plasmid in yeast strains. BY-T20: BY4742, ΔTrp1, Trp1::HIS3-PPGK1-BTS1/ERG20-TADH1-PTDH3-SaGGPS-TTPI1-PTEF1-tHMG1-TCYC1, SGH1: BY-T20/pESC-Trp::SmCPSent, SGH2: BY-T20/pESC-Trp::SmKS, SGH3: BY-T20/pESC-Trp::SmCPSent/SmKS, SGH4: BY-T20/pESC-Trp::SmKS-SmCPSent. (B) Extracted ion chromatograms showing the SmCPSent and SmKS products. AtCPS (ent-copalol, the dephosphorylated product of ent-CPP), AtCPS + AtKS (ent-kaurene). (C) The mass spectra of the SmCPSent and SmKS products. (D) Relative ent-kaurene contents in the yeast strains SGH3 and SGH4 based on their peak area ratio. The data represent the mean ± SD from three independent experiments.
Protein complexes have been reported to improve the efficiency of specific pathways by protecting substrates and intermediates from diffusion and degradation16. Zhou et al. reported that a recombinant strain containing the fused enzyme SmKSL1-SmCPS1 produced 2.8-fold more miltiradiene compared with another recombinant strain in which SmCPS1 and SmKSL1 were expressed separately17. Hence, we constructed the fused enzyme SmKS-SmCPSent in the yeast strain SGH4 (BY-T20/pESC-Trp::SmKS-SmCPSent) using the RF cloning method, and the results showed that SGH4 produced approximately 4.25-fold more ent-kaurene than SGH3 (Fig. 4D).
Recombinant expression and functional characterization of SmKO in vivo
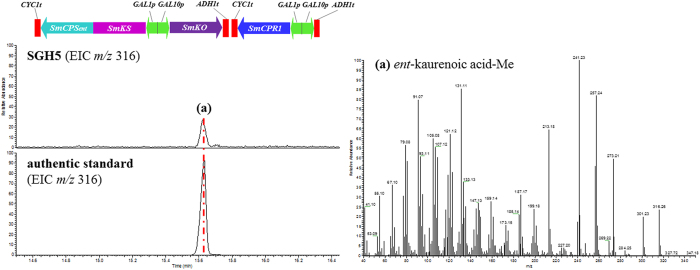
As a strategy to characterize the biochemical function of SmKO in vivo, first we constructed the fused enzyme SmKS-SmCPSent, which improved the ent-kaurene precursor supply as expected. Then, SmKO was coexpressed with the fused enzyme SmKS-SmCPSent and a NADPH-cytochrome P450 reductase (SmCPR1) in the yeast strain SGH5 (BY-T20/pESC-Trp::SmKS-SmCPSent/SmKO + pESC-Leu::SmCPR1). After extraction and methylation, the ent-kaurenoic acid methyl ester was detected by a comparison with the methylated authentic standard (Sigma, USA) (Fig. 5). This result confirmed that SmKO encoded a functional ent-kaurene oxidase that was involved in the three-stage oxidation of ent-kaurene to ent-kaurenoic acid in the S. miltiorrhiza GA biosynthetic pathway.
Figure 5. The product from BY-T20 yeast strains expressing the S. miltiorrhiza SmKO. SGH5: BY-T20/pESC-Trp::SmKS-SmCPSent/SmKO + pESC-Leu::SmCPR1.
Extracted ion chromatograms showing methyl ester derivatives of the SmKO product with its corresponding mass spectra.
Discussion
We identified three consecutive enzymes (SmCPSent, SmKS and SmKO) involved in the S. miltiorrhiza GA biosynthetic pathway. SmCPSent catalyzes the formation of ent-CPP from GGPP; then, SmKS converts ent-CPP to ent-kaurene. Subsequently, SmKO converts ent-kaurene to ent-kaurenoic acid via a three-stage oxidation reaction. ent-Kaurene biosynthesis was reported to be catalyzed by a one-to-one CPS/KS complex in which CPP could be channeled from CPS to the KS catalytic site10. Therefore, we fused SmCPSent and SmKS to obtain a close proximity between the active sites of the two consecutive enzymes. As expected, the fused enzyme SmKS-SmCPSent produced 4.25-fold more ent-kaurene than the separate expression of SmCPSent and SmKS in the yeast strain, suggesting that the protein fusion treatment was an efficient approach to improve the catalytic activity and enlarge the heterologous production of ent-kaurene. With an increased supply of the ent-kaurene precursor, SmKO catalyzed the formation of ent-kaurenoic acid. However, the intermediates ent-kaurenol and ent-kaurenal were not detected. One possible explanation is that the intermediates were unstable and were changed into other intermediates during the extraction process. The enzymes involved in the early steps of the GA biosynthetic pathway (i.e., CPS, KS, KO, and KAO) are primarily encoded by single genes, whereas those involved in the later steps (i.e., GA20ox, GA3ox, and GA2ox) are encoded by gene families18. The SmCPSent, SmKS and SmKO genes are likely single copy genes responsible for GA biosynthesis in S. miltiorrhiza.
In addition to the identification and characterization of SmCPSent, SmKS and SmKO, we provided insights into the genes encoding the enzymes involved in all steps of the GA biosynthetic pathway from GGPP to ent-kaurenoic acid. Our results provide a foundation for further characterization of the subsequent enzymes (i.e., SmKAO and the CYP88A subfamily) involved in the GA biosynthetic pathway using this yeast expression system. In plants, GA levels vary at different sites and during different development processes19. It is possible to control the GA levels by regulating the expression of these genes to acquire better growth of the S. miltiorrhiza roots and rhizomes, thereby improving the total yield of tanshinones per plant.
In conclusion, we functionally characterized three consecutive enzymes (SmCPSent, SmKS and SmKO) involved in the GA biosynthetic pathway from GGPP to ent-kaurenoic acid, thereby laying the foundation for further characterization of GA biosynthesis. Based on these results, we could regulate the expression of all genes involved in the GA biosynthetic pathway to acquire better growth and an increased accumulation of the bioactive tanshinones involved in the S. miltiorrhiza developmental processes. Protein fusion is an applicable and efficient approach that can be used to direct metabolic flux to the bioactive diterpenoid tanshinones pathway for the heterologous production of isoprenoids in microbial cell factories.
Methods
RNA isolation and cDNA cloning
Hairy roots were induced from the S. miltiorrhiza leaf explants under the mediation of A. rhizogenes strain ACCC10060 as described previously20 and maintained in 6,7-V liquid medium21 at 25 °C on a gyratory shaker (80 rpm) in the dark. Total RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The 5′ and 3′ ends of the targeted SmCPSent and SmKS genes were cloned by RACE (Invitrogen) according to the manufacturer’s directions using the corresponding S. miltiorrhiza genome sequences released by the National Center for Biotechnology Information (NCBI)22. The primer sequences are shown in Supplementary Table 1. An aliquot (1 μg) of the total RNA was used to synthesize the first strand cDNA according to the PrimeScript 1st Strand cDNA Synthesis Kit (Takara Bio, Dalian, China) manufacturer’s protocol. The full-length cDNA for each ORF was cloned using the PrimeSTAR DNA polymerase (Takara Bio). The PCR products were purified and cloned into the pEASY-T3 cloning vector (TransGen Biotech, Beijing, China), transformed into Escherichia coli Trans5α cells (TransGen Biotech), and then cultured in Luria-Bertani (LB) medium at 37 °C in the dark. Positive clones were sequenced. The full-length cDNA of SmKO was cloned previously23.
Bioinformatics analysis
The SmCPSent, SmKS and SmKO sequences were confirmed at NCBI (http://www.ncbi.nlm.nih.gov/). The open reading frames (ORFs) and deduced amino acid sequences were analyzed using the online tool ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and the ExPASy online tool (http://web.expasy.org/translate/), respectively. The ChloroP 1.1 Server (http://www.cbs.dtu.dk/services/ChloroP/) was used to predict chloroplast transit peptides. The sequences of the SmCPSent, SmKS and SmKO as well as other corresponding sequences downloaded from GenBank were aligned using the DNAMAN program, and the phylogenetic trees for SmCPSent, SmKS and SmKO were constructed using sequences from other plants (Supplementary Table 2) using the neighbor-joining method in MEGA5.1 24.
Recombinant expression and functional characterization of SmCPS ent and SmKS
The SmCPSent and SmKS ORFs (alone or in combination) were subcloned into the yeast epitope-tagging vector pESC-Trp under the control of the GAL1 or GAL10 inducible promoter (Agilent Technologies, USA) via digestion by the corresponding restriction endonucleases. The resulting constructs were verified by complete gene sequencing and then transformed into the yeast strain BY-T20 (BY4742, ΔTrp1, Trp1::HIS3-PPGK1-BTS1/ERG20-TADH1-PTDH3-SaGGPS-TTPI1-PTEF1-tHMG1-TCYC1, provided by Prof. Xueli Zhang’s lab, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, China)25,26,27. Then, the recombinant strains SGH1 (containing the plasmid pESC-Trp::SmCPSent), SGH2 (containing the plasmid pESC-Trp::SmKS), and SGH3 (containing the plasmid pESC-Trp::SmCPSent/SmKS) were selected on synthetic dropin medium -Trp-His (SD-Trp-His) containing 20 g/L glucose and grown at 30 °C for 2–3 d. Single transformed yeast colonies were grown in SD-Trp-His liquid medium supplemented with 20 g/L glucose at 30 °C for approximately 2 d. The yeast cells were pelleted and resuspended in 100 mL of SD-Trp-His liquid induction medium supplemented with 20 g/L galactose and grown at 30 °C for 3 d. Finally, the induced yeast cells were extracted three times with an equal volume of hexane. The organic fractions were pooled and dried using a Nitrogen Evaporator (Baojingkeji, Henan, China). The dried samples were dissolved in 100 μL of hexane for GC-MS analysis as described previously28. To confirm the products of these strains, the identified products ent-CPP and ent-kaurene of A. thaliana AtCPS and AtKS were used as the authentic standards29,30. The detailed protocols for the constructions of the recombinant plasmids and strains and the recombinant expression and enzymatic assay for AtCPS and AtKS are described in the Supplementary Methods.
Construction of the module producing the fused protein SmKS-SmCPS ent and the functional characterization of SmKO
To prepare the module producing the fused protein SmKS-SmCPSent, a restriction-free (RF) cloning method was used31. The genes encoding the fusion enzyme were constructed by inserting a widely used GGGS linker encoded by a “GGT GGT GGT TCT” sequence between the two corresponding genes32,33. The recombinant plasmid pESC-Trp::SmKS-SmCPSent was transformed into the yeast strain BY-T20 to generate SGH4 and induced with D-galactose as described above. Then, the products of SGH4 were analyzed by GC-MS. The detailed protocols for the RF cloning are described in the Supplementary Methods.
The ORF region of SmKO was ligated into the recombinant plasmid pESC-Trp::SmKS-SmCPSent as described above and then transformed into the yeast strain BY-T20 with another recombinant plasmid pESC-Leu::SmCPR1 (SmCPR1, S. miltiorrhiza cytochrome P450 reductase8). The recombinant strain SGH5 (containing the plasmids pESC-Trp::SmKS-SmCPSent/SmKO and pESC-Leu::SmCPR1) was induced with D-galactose and extracted once with an equal volume of hexane and twice with an equal volume of ethyl acetate. The organic fractions were pooled and dried and then dissolved in 50 μL of methanol and methylated with approximately 200 μL of (trimethylsilyl)diazomethane (Aladdin Industrial Inc., Shanghai, China). The methylated samples were redried and then dissolved in 100 μL of ethyl acetate for GC-MS using a Thermo TRACE 1310/TSQ 8000 gas chromatograph (splitless; injector temperature 250 °C) with a DB-5 ms (30 m × 0.25 mm × 0.25 μm) capillary column. The GC conditions were the same as those described previously34.
Additional Information
How to cite this article: Su, P. et al. Functional characterization of ent-copalyl diphosphate synthase, kaurene synthase and kaurene oxidase in the Salvia miltiorrhiza gibberellin biosynthetic pathway. Sci. Rep. 6, 23057; doi: 10.1038/srep23057 (2016).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81422053 and 81373906 to W.G and 81325023 to L.H.) and National High Technology Research and Development Program of China (863 Program: 2015AA0200908) to W.G. and the Author of National Excellent Doctoral Dissertation of China (201188) to W.G.
Footnotes
Author Contributions L.H. and W.G. conceived and designed the study. P.S. and Y.T. performed the experiments and wrote the manuscript. Q.C., Y.H., M.Z., J.Y. and Z.T. participated in the research and analyzed the data. All authors read and approved the final manuscript.
References
- Jiang W. Y. et al. PF2401-SF, standardized fraction of Salvia miltiorrhiza shows anti-inflammatory activity in macrophages and acute arthritis in vivo. Int Immunopharmacol 16, 160–164 (2013). [DOI] [PubMed] [Google Scholar]
- Kim S. J., Kwon D. Y., Kim Y. S. & Kim Y. C. Peroxyl radical scavenging capacity of extracts and isolated components from selected medicinal plants. Arch Pharm Res 33, 867–873 (2010). [DOI] [PubMed] [Google Scholar]
- Wang S. et al. Tanshinone I selectively suppresses pro-inflammatory genes expression in activated microglia and prevents nigrostriatal dopaminergic neurodegeneration in a mouse model of Parkinson’s disease. J Ethnopharmacol 164, 247–255 (2015). [DOI] [PubMed] [Google Scholar]
- Munagala R., Aqil F., Jeyabalan J. & Gupta R. C. Tanshinone IIA inhibits viral oncogene expression leading to apoptosis and inhibition of cervical cancer. Cancer Lett 356, 536–546 (2015). [DOI] [PubMed] [Google Scholar]
- Cong M. et al. Cryptotanshinone and dihydrotanshinone I exhibit strong inhibition towards human liver microsome (HLM)-catalyzed propofol glucuronidation. Fitoterapia 85, 109–113 (2013). [DOI] [PubMed] [Google Scholar]
- Cui G. H. et al. Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza Bunge. Plant Physiol 169, 1607–1618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W. et al. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett 11, 5170–5173 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc Natl Acad Sci USA 110, 12108–12113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. et al. Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J Plant Growth Regul 20, 319–331 (2001). [DOI] [PubMed] [Google Scholar]
- Duncan J. D. & West C. A. Properties of kaurene synthetase from Marah macrocarpus endosperm. Evidence for the participation of separate but interacting enzymes. Plant Physiol 68, 1128–1134 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E. et al. Molecular cloning and characterization of a cDNA encoding ent-cassa-12,15-diene synthase, a putative diterpenoid phytoalexin biosynthetic enzyme, from suspension-cultured rice cells treated with a chitin elicitor. Plant J 37, 1–8 (2004). [DOI] [PubMed] [Google Scholar]
- Günnewich. N. et al. A diterpene synthase from the clary sage Salvia sclarea catalyzes the cyclization of geranylgeranyl diphosphate to (8R)-hydroxy-copalyl diphosphate. Phytochemistry 91, 93–99 (2013). [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H. & von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8, 978–984 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C. A., Poole A., Peacock W. J. & Dennis E. S. Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol 119, 507–510 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S. E., Smith J. J., Helliwell C. A., Poole A. T. & Reid J. B. The pea gene LH encodes ent-kaurene oxidase. Plant Physiol 134, 1123–1134 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srere P. A. Complexes of sequential metabolic enzymes. Annu Rev Biochem 56, 89–124 (1987). [DOI] [PubMed] [Google Scholar]
- Zhou Y. J. et al. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J Am Chem Soc 134, 3234–3241 (2012). [DOI] [PubMed] [Google Scholar]
- Sakamoto T. et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134, 1642–1653 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebers M. et al. Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. Plant J 17, 241–250 (1999). [DOI] [PubMed] [Google Scholar]
- Cheng Q. Q. et al. RNA interference-mediated repression of SmCPS (copalyl diphosphate synthase) expression in hairy roots of Salvia miltiorrhiza causes a decrease of tanshinones and sheds light on the functional role of SmCPS. Biotechnol Lett 36, 363–369 (2014). [DOI] [PubMed] [Google Scholar]
- Veliky I. A. & Martin S. M. A fermenter for plant cell suspension cultures. Can J Microbiol 16, 223–226 (1970). [DOI] [PubMed] [Google Scholar]
- Ma Y. et al. Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J Exp Bot 63, 2809–2823 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. T. et al. Cloning and bioinformatics analysis of ent-kaurene oxidase synthase gene in Salvia miltiorrhiza. Zhongguo Zhong Yao Za Zhi 39, 4174–4179 (2014). [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z. B. et al. Producing aglycons of ginsenosides in bakers’ yeast. Sci Rep 4, 3698 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z. B. et al. Metabolic engineering of Saccharomyces cerevisiae for production of ginsenosides. Metab Eng 20, 146–156 (2013). [DOI] [PubMed] [Google Scholar]
- Shi M. Y. et al. Construction of Saccharomyces cerevisiae cell factories for lycopene production. Zhongguo Zhong Yao Za Zhi 39, 3978–3985 (2014). [PubMed] [Google Scholar]
- Zhang M. et al. Identification of geranylgeranyl diphosphate synthase genes from Tripterygium wilfordii. Plant Cell Rep 34, 2179–2188 (2015). [DOI] [PubMed] [Google Scholar]
- Sun T. P. & Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6, 1509–1518 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Sun T. P., Kawaide H. & Kamiya Y. The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol 116, 1271–1278 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F. & Lowe J. RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods 67, 67–74 (2006). [DOI] [PubMed] [Google Scholar]
- Ukibe K., Hashida K., Yoshida N. & Takagi H. Metabolic Engineering of Saccharomyces cerevisiae for Astaxanthin Production and Oxidative Stress Tolerance. Appl Environ Microbiol 75, 7205–7211 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan F. M., Yu Y., Mohr S. C. & Cantor C. R. Engineered single-chain dimeric streptavidins with an unexpected strong preference for biotin-4-fluo. Proc Natl Acad Sci USA 102, 8507–8512 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L. S., Faergestad E. M., Poole A. & Chandler P. M. Grain Development Mutants of Barley ([alpha]-Amylase Production during Grain Maturation and Its Relation to Endogenous Gibberellic Acid Content). Plant Physiol 114, 203–214 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone D., Chen X. M., Coates R. M. & Peters R. J. Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin biosynthesis. BiochemJ 431, 337–344 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.