Abstract
Pearl millet [Pennisetum glaucum (L.) R. Br.] a widely used grain and forage crop, is grown in areas frequented with one or more abiotic stresses, has superior drought and heat tolerance and considered a model crop for stress tolerance studies. Selection of suitable reference genes for quantification of target stress-responsive gene expression through quantitative real-time (qRT)-PCR is important for elucidating the molecular mechanisms of improved stress tolerance. For precise normalization of gene expression data in pearl millet, ten candidate reference genes were examined in various developmental tissues as well as under different individual abiotic stresses and their combinations at 1 h (early) and 24 h (late) of stress using geNorm, NormFinder and RefFinder algorithms. Our results revealed EF-1α and UBC-E2 as the best reference genes across all samples, the specificity of which was confirmed by assessing the relative expression of a PgAP2 like-ERF gene that suggested use of these two reference genes is sufficient for accurate transcript normalization under different stress conditions. To our knowledge this is the first report on validation of reference genes under different individual and multiple abiotic stresses in pearl millet. The study can further facilitate fastidious discovery of stress-tolerance genes in this important stress-tolerant crop.
Plants being sessile in nature are forced to thrive in adverse environmental conditions. Drought, salinity and temperature extremes (heat and cold) are major abiotic stresses that challenge production and productivity of crop plants1,2. The adverse effects of these stresses on crop plants are further compounded due to changing climate worldwide3. Further it has also been reported that levels of abscisic acid (ABA), a plant growth regulator, also increases under abiotic stresses4,5. Though individual stress conditions such as drought, salinity or heat have been the focus of intense research, not much study has been carried out on the combination of different abiotic stresses such as drought and heat, drought and salinity, salinity and heat etc. which frequently affect growth and yield potential of crops and other plants in field conditions across the globe6,7. There have been quite a few reports suggesting that response of plants to two or more different abiotic stresses is unique and quite distinct from the response of those exposed to individual stress8. Generally, more than one stress factors occurring simultaneously, interact with each other in additive manner and trigger more damage than when present individually9,10. Therefore it is necessary to focus on development of crops varieties that can endure multiple environmental stress factors. Since crop plants are subjected to one or more abiotic stresses concurrently under field conditions, it is thus necessary to make conscious efforts towards mimicking these conditions in laboratory as well7,9,11. Further an overlap in the expression patterns of stress responsive genes after drought, salt, heat, cold, or ABA treatments or after combinations of stresses has also been reported8,12. Subsequently several studies have been taken up recently to monitor the effects of abiotic stress combinations on crop plants as well as to elucidate the molecular mechanism(s) of stress tolerance8,12,13,14,15.
Pearl millet [Pennisetum glaucum (L.) R. Br.] is an important small-grained C4 panicoid crop grown for forage, grain and stover in the arid and semi-arid regions of Asia and Africa16. It is the sixth most important cereal crops after rice, wheat, maize, barley and sorghum with excellent nutrient composition17 and is also considered a potential biofuel grain feedstock2,18; http://ag.fvsu.edu/index.php/research/bioenergy/). It usually thrive in areas with scanty rainfall and is also well adapted to various abiotic stresses such as drought, high temperature, salinity etc. whether occurring individually or in combination making it an ideal crop for functional genomic studies to understand the molecular basis of abiotic stress tolerance and adaptation2,16,19. Only a limited amount of genome sequence information is available in pearl millet that greatly hinders gene discovery, functional validation, expression profiling, and ultimately crop improvement programs. Quantification of variable transcriptomes and analysing differential expression of stress responsive genes in diverse biological samples and experimental conditions are important functional genomic approaches to investigate the molecular basis of stress tolerance involving complex regulatory gene networks20. In this regard, qRT-PCR is a widely used technique to quantify transcriptional abundance of numerous individual genes owing to its high sensitivity, specificity and synchronized quantification of gene expression in diverse samples with a broad quantification range of up to several orders of magnitude in comparison to conventional techniques such as reverse transcription (RT)-PCR or northern hybridization20. However relative quantification of gene expression using qRT-PCR is highly influenced by the expression stability of internal control or reference genes used for transcript normalization of target genes. The use of inappropriate or unstable reference gene(s) can seriously impact the transcript quantification results leading to false inferences or misinterpretations21,22. Accurate normalization is thus necessary for obtaining biologically meaningful expression data and hence qRT-PCR analysis greatly depends upon careful selection of reliable reference gene(s) which should be stably expressed across various tissue samples, developmental stages and experimental conditions23,24. Numerous studies have been carried out until now in various economically important cereal crops for determining the stability of reference genes in various developmental stages and abiotic stress conditions25,26,27. However to our knowledge no systematic study has been carried out in any cereal crop for validation of reference genes under different individual abiotic stresses and their various combinations at different durations of stress which are a must for the normalization of gene transcripts that are expressed and regulated under multiple stress conditions. Hence considering the inevitability of crop- as well as stress-specific internal control genes for accurate normalization of transcripts in qRT-PCR, the present study was carried out to evaluate the expression stability of 10 candidate reference genes in pearl millet subjected to individual or combination of abiotic stresses and or abscisic acid (ABA), a stress hormone at early (1h) and late (24h) stress durations as well as in different developmental tissues of two contrasting genotypes.
Results
Selection of candidate reference genes, sample size, primer specificity and amplification efficiency
A total of ten potential candidate reference genes including both traditional housekeeping as well as new reference genes namely, actin (ACT), elongation factor 1α (EF-1α), eukaryotic initiation factor 4A (eIF4A), glutaredoxin (GlutR), heat shock protein 90 (HSP90), malate dehydrogenase (MDH), protein phosphatase 2A (PP2A), ribosomal protein L20 (RPL20), tonoplast intrinsic protein 41 (TIP41) and ubiquitin-conjugating enzyme E2 (UBC-E2) were identified for gene expression studies using qRT-PCR in pearl millet. Out of 10 reference genes sequences of 8 were retrieved from pearl millet and those of PP2A and TIP41were obtained from foxtail millet.
The quantification of cycling threshold (Ct) or quantitative cycle (Cq) values through qRT-PCR and calculation of coefficient of variation (CV) for 10 candidate reference genes was carried out in an experimental set of a total of 204 samples (n = 204) from two contrasting pearl millet genotypes namely PRLT2/89-33 and H77/833-2, which are also parents of a mapping population, subjected to different individual abiotic stresses or ABA or combinations of individual stresses and ABA or in developmental tissues (Table 1). The individual abiotic stress experimental set comprised a total of 60 samples from 4 stresses namely, drought, salt, heat and cold, and from a hormone stimulus ABA at 1 h and 24 h of stress, whereas multiple stresses set included 96 samples from 8 treatments i.e. drought + salt, drought + heat, drought + ABA, salt + ABA, heat + ABA, cold + ABA, drought + salt + ABA, and drought + salt + heat + ABA from both genotypes. Both individual and multiple stresses included one control sample of each genotype without stress (n = 6). The experimental set of developmental tissues consisted of 42 samples including leaf, root, stem, stem sheath, panicle at booting stage, panicle at grain filling stage and seeds of both genotypes.
Table 1. Details of Ct and CV values of each of the selected candidate reference genes tested in pearl millet.
| Gene name | Description | Accession no. | Ct ± SD | CV ± SD |
|---|---|---|---|---|
| ACT | Actin | HM243500 | 26.4 ± 2.3 | 5.8 ± 0.3 |
| EF-1α | Elongation factor-1 alpha | EF694165 | 23.4 ± 2.1 | 4.5 ± 0.2 |
| eIF4A | Eukaryotic initiation factor 4A | EU856535 | 23.8 ± 3.1 | 6.6 ± 0.4 |
| GlutR | Glutaredoxin | GD180652 | 21.9 ± 2.5 | 5.3 ± 0.2 |
| HSP 90 | Heat shock protein 90 | ADP89125 | 22.8 ± 2.9 | 9.6 ± 0.9 |
| MDH | Malate dehydrogenase | CD724779 | 22.2 ± 2.3 | 15.7 ± 2.6 |
| PP2A | Protein phosphatase 2A | Si017892m | 25.6 ± 2.3 | 5.1 ± 0.2 |
| RPL20 | Ribosomal protein L20 | KJ490012 | 21.7 ± 3.4 | 6.5 ± 0.4 |
| TIP41 | Tonoplast intrinsic protein | Si036884m | 28.9 ± 2.5 | 6.7 ± 0.6 |
| UBC-E2 | Ubiquitin-conjugating enzyme E2 | CD724586 | 23.2 ± 2.5 | 5.7 ± 0.3 |
The specificity and accuracy of each qRT-PCR primer pair was determined through agarose gel electrophoresis and melt curve analysis for each gene (Supplementary Fig. S1) where single amplicon or single peak, respectively suggested the absence of primer dimmers or non-specific PCR products. The PCR amplification efficiency (E) is dependent on the assay, master mix performance, and sample quality. The amplification efficiencies and correlation coefficients (R2) of the ten candidate reference genes were determined using the slopes of the standard curves obtained by serial dilutions which fall in the acceptable range of 90–100% for E, and 0.80–1.00 for the R2. The description of 10 candidate reference genes, accession numbers, primer sequences, amplicon lengths, and amplification efficiency are enlisted in Supplementary Table S1.
Expression profiling of candidate reference genes
Expression levels of all 10 candidate reference genes were determined by qRT-PCR and the Ct values showed differential transcript levels in the samples examined with lower Ct values suggesting higher transcript abundance and vice versa. The Ct values were monitored under four groups including all samples, individual abiotic stresses, multiple abiotic stresses, and developmental tissue samples. The mean Ct values of 10 potential reference genes ranged from 15.5 to 34.7 (Table 2; Fig. 1). The distribution of Ct values is listed in Supplementary Tables S2, S3 and S4. In the all samples set, the mean Ct values showed a minimum of 21.7 ± 3.4 and maximum of 28.9 ± 2.5 for highest and lowest expression levels for RPL20 and TIP41, respectively. RPL20 and TIP41 also showed minimum and maximum average Ct values, respectively in individual stress sample set as well as in multiple stress samples set. On the other hand in the developmental tissue set, UBC-E2 has minimum average Ct value of 21.1 ± 4.8 while TIP41 has maximum average Ct value of 29.4 ± 3.7. Further CV of the Ct values was also calculated to evaluate the expression levels of candidate reference genes under all four experimental sets, where lower values represent lower variability or maximum stability. The CV of 10 reference genes among all samples ranged between 4 to 15%. EF-1α was the least variable reference gene with a CV of 4.5% among 10 candidate reference genes studied and MDH was the most variable with a CV of 15.7%. The stability ranking of all candidate reference genes on the basis of CV values is as follows: EF-1α < PP2A < GlutR < UBC-E2 < ACT < RPL20 < eIF4A < TIP41 < HSP 90 < MDH (Table 1).
Table 2. Expression levels of different housekeeping genes under study in all four experimental sets.
| Gene name | All samples Ct ± SD | Individual stresses Ct ± SD | Multiple stresses Ct ± SD | Developmental tissues Ct ± SD |
|---|---|---|---|---|
| ACT | 26.4 ± 2.3 | 26.8 ± 2.7 | 26.4 ± 1.8 | 24.8 ± 3.7 |
| EF-1α | 23.4 ± 2.1 | 23.7 ± 2.0 | 23.2 ± 1.6 | 23.8 ± 4.0 |
| eIF4A | 23.8 ± 3.1 | 23.6 ± 2.5 | 24.1 ± 2.2 | 21.2 ± 4.5 |
| GlutR | 21.9 ± 2.5 | 21.5 ± 2.5 | 21.8 ± 1.9 | 23.6 ± 4.3 |
| HSP 90 | 22.8 ± 2.9 | 23.3 ± 2.7 | 22.6 ± 2.5 | 21.8 ± 4.2 |
| MDH | 22.2 ± 2.3 | 21.5 ± 2.0 | 22.3 ± 1.6 | 22.9 ± 3.2 |
| PP2A | 25.6 ± 2.3 | 26.0 ± 2.3 | 25.7 ± 1.7 | 23.5 ± 3.3 |
| RPL20 | 21.7 ± 3.4 | 20.1 ± 2.4 | 21.5 ± 2.0 | 26.7 ± 4.3 |
| TIP41 | 28.9 ± 2.5 | 28.2 ± 1.8 | 29.0 ± 2.0 | 29.4 ± 3.7 |
| UBC-E2 | 23.2 ± 2.5 | 23.3 ± 1.9 | 23.3 ± 1.7 | 21.1 ± 4.8 |
Figure 1. Ct values of candidate reference genes across all samples.
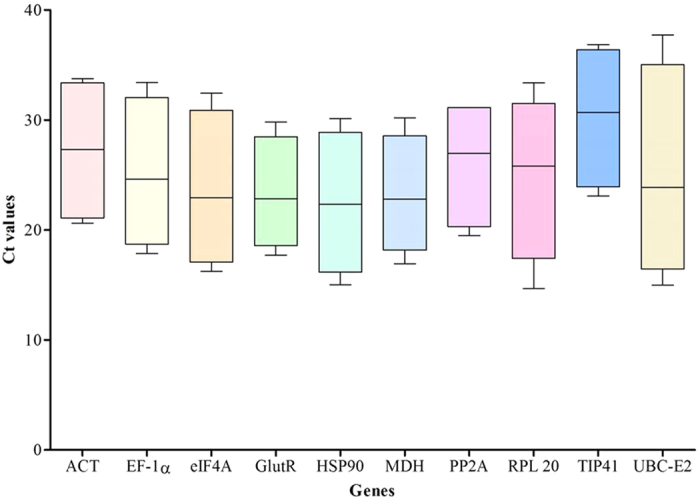
Line across the box depict the median value and inside the box show the Ct values. The top and bottom whiskers are determine by the 5th and 95th percentiles, respectively.
Stability ranking of the candidate reference genes
Expression stability of the candidate reference genes were determined by geNorm, NormFinder and RefFinder which evaluated the stability ranking of each reference gene using the Ct values across all the experimental sets and tissue samples (Tables 3, 4, 5, 6). The stability ranking analyses done by each of the three softwares is detailed in the following sections:
Table 3. Stability of candidate reference genes under individual stress conditions in pearl millet.
| Rank | geNorm |
NormFinder |
RefFinder |
|||
|---|---|---|---|---|---|---|
| Genes | Normalization Value (MV) | Genes | Stability Value (SV) | Genes | Geomean Of Ranking Value | |
| 1 | EF-1α│UBC-E2 | 0.286 | UBC-E2 | 0.048 | EF-1α | 1.19 |
| 2 | PP2A | 0.103 | UBC-E2 | 2.28 | ||
| 3 | PP2A | 0.317 | ACT | 0.104 | PP2A | 3.03 |
| 4 | ACT | 0.363 | EF-1α | 0.112 | TIP41 | 4.60 |
| 5 | RPL20 | 0.455 | RPL20 | 0.140 | MDH | 4.73 |
| 6 | HSP90 | 0.482 | HSP90 | 0.140 | ACT | 5.42 |
| 7 | eIF4A | 0.492 | GlutR | 0.154 | GlutR | 5.6 |
| 8 | TIP41 | 0.561 | eIF4A | 0.167 | RPL20 | 6.47 |
| 9 | GlutR | 0.593 | MDH | 0.177 | eIF4A | 9.24 |
| 10 | MDH | 0.603 | TIP41 | 0.197 | HSP90 | 9.46 |
Table 4. Stability of candidate reference genes under multiple stress conditions in pearl millet.
| Rank | geNorm |
NormFinder |
RefFinder |
|||
|---|---|---|---|---|---|---|
| Genes | Normalization Value (MV) | Genes | Stability Value (SV) | Genes | Geomean Of Ranking Value | |
| 1 | EF-1α│PP2A | 0.335 | PP2A | 0.097 | EF-1α | 1.19 |
| 2 | EF-1α | 0.159 | PP2A | 3.72 | ||
| 3 | UBC-E2 | 0.419 | UBC-E2 | 0.161 | MDH | 5.00 |
| 4 | MDH | 0.438 | MDH | 0.168 | TIP41 | 6.00 |
| 5 | TIP41 | 0.445 | HSP90 | 0.183 | GlutR | 7.45 |
| 6 | GlutR | 0.517 | eIF4A | 0.194 | ACT | 7.74 |
| 7 | eIF4A | 0.579 | TIP41 | 0.201 | GlutR | 5.6 |
| 8 | ACT | 0.597 | GlutR | 0.215 | RPL20 | 8.74 |
| 9 | RPL20 | 0.606 | ACT | 0.242 | HSP90 | 10.00 |
| 10 | HSP90 | 0.677 | RPL20 | 0.244 | eIF4A | 11.24 |
Table 5. Stability of candidate reference genes in the developmental tissues of pearl millet.
| Rank | geNorm |
NormFinder |
RefFinder |
|||
|---|---|---|---|---|---|---|
| Genes | Normalization Value (MV) | Genes | Stability Value (SV) | Genes | Geomean Of Ranking Value | |
| 1 | EF-1α│MDH | 0.094 | MDH | 0.032 | MDH | 1.57 |
| 2 | HSP90 | 0.034 | HSP90 | 2.45 | ||
| 3 | HSP90 | 0.151 | EF-1α | 0.053 | EF-1α | 2.82 |
| 4 | eIF4A | 0.187 | UBC-E2 | 0.059 | ACT | 4.16 |
| 5 | PP2A | 0.261 | eIF4A | 0.060 | PP2A | 4.30 |
| 6 | UBC-E2 | 0.271 | ACT | 0.074 | eIF4A | 4.76 |
| 7 | ACT | 0.294 | RPL20 | 0.080 | GlutR | 5.18 |
| 8 | GlutR | 0.376 | PP2A | 0.082 | TIP41 | 7.77 |
| 9 | RPL20 | 0.427 | TIP41 | 0.102 | RPL20 | 8.24 |
| 10 | TIP41 | 0.476 | GlutR | 0.117 | UBC-E2 | 10.01 |
Table 6. Stability of candidate reference genes across all samples tested in pearl millet.
| Rank | geNorm |
NormFinder |
RefFinder |
|||
|---|---|---|---|---|---|---|
| Genes | Normalization Value (MV) | Genes | Stability Value (SV) | Genes | Geomean Of Ranking Value | |
| 1 | PP2A│UBC-E2 | 0.218 | EF-1α | 0.061 | EF-1α | 1.00 |
| 2 | PP2A | 0.064 | UBC-E2 | 2.38 | ||
| 3 | EF-1α | 0.292 | UBC-E2 | 0.089 | PP2A | 3.00 |
| 4 | eIF4A | 0.293 | eIF4A | 0.089 | MDH | 3.13 |
| 5 | ACT | 0.355 | ACT | 0.110 | GlutR | 5.00 |
| 6 | HSP90 | 0.374 | HSP90 | 0.114 | TIP41 | 6.09 |
| 7 | MDH | 0.462 | MDH | 0.124 | ACT | 6.24 |
| 8 | TIP41 | 0.485 | TIP41 | 0.132 | eIF-4A | 8.24 |
| 9 | RPL20 | 0.498 | GlutR | 0.146 | HSP90 | 8.74 |
| 10 | GlutR | 0.516 | RPL20 | 0.148 | RPL20 | 10.00 |
geNorm analysis
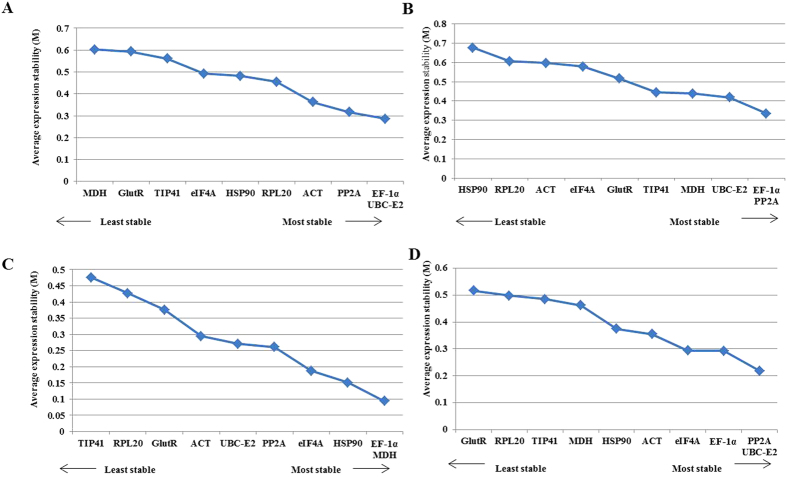
The gene expression stability of all the 10 candidate reference genes was evaluated using geNorm statistical algorithm. This software determines the normalization value based on the geometric mean of various candidate reference genes and mean pair wise variation of each gene from all the reference genes in a given set of samples. According to geNorm analysis the cut-off range of stability value (M) is <1.5, so the gene with lowest M value is considered to be the most stable reference gene in terms of gene expression and vice versa. We analyzed our data for all four experimental sets and found that all the 10 candidate reference genes exhibited high expression stability with low (<0.7) M values which were much below the default limit of 1.5. Among all samples, PP2A and UBC-E2 have the least M value of 0.22 followed by EF-1α and eIF4A with M value of 0.29, however GlutR exhibited highest M value of 0.52 indicating that PP2A and UBC-E2 were most stable in expression and GlutR the least. On the other hand in the individual and multiple stress sample sets, EF-1α/UBC-E2 and Ef-1α/PP2A had the lowest M values of 0.28 and 0.33, respectively among all 10 reference genes studied suggesting their higher expression stability, whereas MDH (0.60) and RPL20 (0.68), respectively were referred as least stable genes for normalization in both these sample sets. In developmental tissues experimental set, EF-1α and MDH with M value 0.094 were ranked as most stable reference genes while TIP41 (0.48) was ranked least stable. Our results thus suggested EF-1α to be the most stable candidate reference gene in three out of four experimental sets (Supplementary Table S5, Fig. 2).
Figure 2.
Expression stability and ranking of 10 candidate reference genes as calculated by geNorm in the individual stress samples set (A), multiple stress samples set (B), developmental tissue samples set (C) and all samples set (D). A lower value of average expression stability (M) indicates most stable expression.
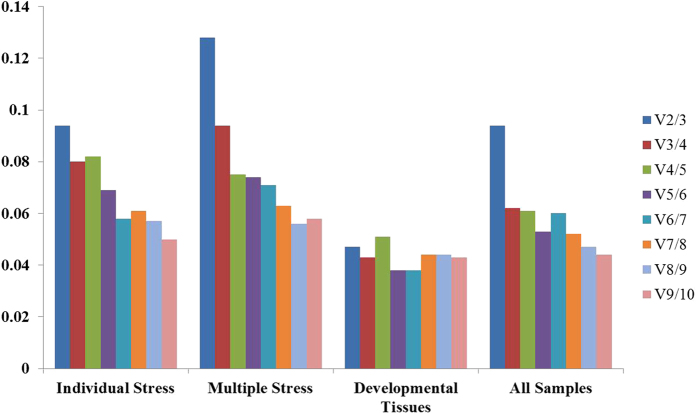
geNorm algorithm was also used to determine the optimal number of reference genes as inclusion of two or more internal control genes for transcript normalization in qRT-PCR experiments is considered better for obtaining precise and steady results. geNorm determines the pairwise variation (Vn/Vn+1) between sequential normalization factors NF (NFn and NFn+1) in each sample set28. geNorm first calculates the NF for two highly expressed reference genes and then for remaining genes by adding each gene one by one in order of their decreasing expression stability20,25,28,29. For the entire four sets, all pairwise variations were below the cut-off value of 0.15 (Fig. 3).
Figure 3. Pairwise variation (V) analysis to determine the optimal number of control genes for accurate transcript normalization in all the four experimental sets.
geNorm calculates pairwise variation (Vn/Vn+1) for the normalization factors NFn and NF+1 to determine (V<0.15) the optimal number of reference genes.
NormFinder analysis
Expression level of 10 candidate reference genes were also evaluated using NormFinder which is an excel based mathematical tool that analyzes each sample set individually and also estimates intra- and inter-group variation in expression across different sample sets. NormFinder ranks the control genes on the basis of their stability value (SV) where lower stability value represents higher gene expression stability and vice versa. For the all samples set EF-1α, PP2A and UBC-E2 were identified as the best three optimal internal control genes with SV of 0.061, 0.064 and 0.089 while RPL20 with SV 0.148 exhibited utmost variation. UBC-E2 (0.048) and PP2A (0.103) were the top two reference genes in the individual stress sample set, and PP2A (0.097) and EF-1α (0.159) topped the multiple stress samples set. While MDH (0.032) and HSP90 (0.034) were ranked the most stable genes in the developmental tissue samples set (Supplementary Table S6).
RefFinder analysis
Stability ranking of 10 candidate reference genes across four experimental sets was further confirmed using comprehensive ranking method by RefFinder. It is an online available web tool that compares data generated from different software’s (geNorm, NormFinder, SI, ΔCt and BestKeeper) and gives comprehensive ranking to confirm the stability ranking via different programs. In this study the data generated by geNorm and NormFinder across all four sets were compared by RefFinder. EF-1α with Geomean ranking values of 1.00 and 1.19 for all and individual as well as multiple stress samples set, was ranked the most stable, and hence best reference gene for accurate transcript normalization except the developmental tissue set where MDH was given the highest Geomean ranking value of 1.57 (Supplementary Table S7).
Expression analysis of an AP2-like ERF gene from pearl millet under individual and multiple abiotic stresses for reference genes validation
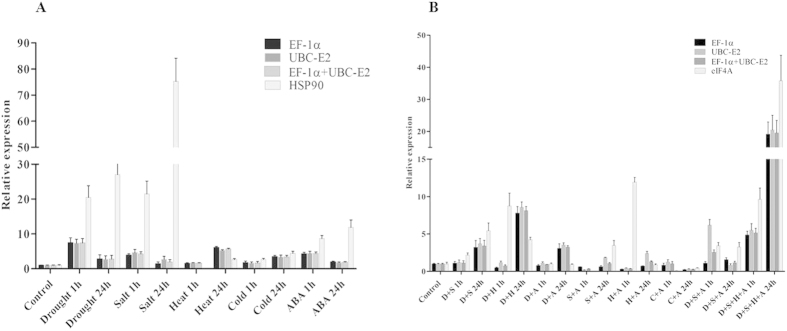
In order to validate the selection of most and least stable reference genes from this study, the relative expression patterns of an objective gene namely PgAP2-like ERF, a transcription factor found to be up-regulated (>2.0 fold) during transcriptome analysis of PRLT2/89-33 and H77/833-2 subjected to drought stress (data from another study; not published), was analyzed under individual and combinations of abiotic stresses at 1 h and 24 h in the tolerant genotype PRLT2/89-33 (Fig. 4). For this overall ranked two most stable (EF-1α and UBC-E2) reference genes for both single and multiple abiotic stresses, and two least stable reference genes namely, HSP90 for individual abiotic stresses, and eIF4A for multiple abiotic stresses as deduced by RefFinder, were used for the validation (Tables 3 and 4). Relative transcript accumulation of PgAP2-like ERF under individual or multiple abiotic stresses was found to be unbiased when EF-1α, UBC-E2 or combination of EF-1α and UBC-E2 were used confirming high level of expression stability of these two genes. Using EF-1α, UBC-E2 or EF-1α + UBC-E2, the expression of PgAP2-like ERF in leaves of the tolerant pearl millet genotype increased under drought, salinity and ABA at 1 h and 24 h with strong expression at 1 h of stress, while the gene was found to be up-regulated under heat and cold stress at 24 h (Fig. 4A). However PgAP2-like ERF gene showed relative up-regulation at late time point (24 h) for all stress combinations when EF-1α, UBC-E2 or EF-1α + UBC-E2 were used for transcript normalization (Fig. 4B). The relative expression pattern of AP2-like ERF showed strong fluctuations and failed to achieve consistency when HSP90 and eIF4A was used for transcript normalization under individual and multiple abiotic stresses, respectively.
Figure 4.
Relative expression of PgAP2 like-ERF using selected reference genes including the most and the least stable reference genes for transcript normalization following (A) individual, and (B) multiple stress treatments after 0, 1 and 24 h. Bar indicate the standard error (±SE) evaluated from three biological replicates.
Discussion
Abiotic stress conditions such as drought, salinity, heat and cold individually cause extensive losses to agricultural production globally, which further aggravates under field conditions due to the simultaneous exposure of plants to more than one abiotic stress factors9. It has also been revealed that simultaneous occurrence of more than one stress leads to huge complexity in plant responses, as the responses and adaptation to such combination of stresses are mostly influenced by diverse and at times by antagonistic signaling pathways that may interact and impede each other6,7. Therefore lately focus of several stress biology research has been directed towards understanding the molecular mechanisms of plant responses to abiotic stresses and their combinations through transcriptomics or functional genomics approaches. qRT-PCR has emerged as a useful technique for validation of transcriptomics data owing to its precision, accuracy, convenience, speed and sensitivity20. However accurate normalization of gene expression remains a major criterion for precise qRT-PCR analysis as normalization helps in adjusting variations introduced at various steps of qRT-PCR arising from sample-to-sample variations, variations in RNA integrity, PCR efficiency and cDNA sample loading22,30. An ideal reference gene thus has constant expression despite of experimental conditions, developmental stages, tissues and cell types20,31. Actin, tubulin,18S, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and elongation factor 1a (EF-1a) have been commonly used as reference genes and their expression is considered to be stable since they are present in all nucleated cell types and are involved in primary metabolism or other cellular processes necessary for cell survival32. However there are a number of reports on variable expression of these housekeeping genes in different plant species under different treatments as they could also go through substantial molecular regulations in different environmental conditions and in specific cellular functions5,28,30,33,34,35. Erroneous results and inaccurate gene expression data could be obtained if suitable reference genes are not used36. As for example, in tall fescue the expression of FaWRKY was found to be increased when SAND and TUB or GAPDH and TUB were used for normalization in salt-treated roots and polyethylene glycol-treated leaves, respectively but fluctuations in expression pattern was observed when EF-1α was used for gene quantification34. Therefore it is necessary to identify and utilize suitable internal control gene(s) with stable expression in different crop species, biological samples and experimental conditions for reliable transcript measurements through qRT-PCR20,37. In fact more than one study has also been carried out in a single crop to evaluate different control genes for optimal normalization in different experimental conditions eg. rice25,38,39, cucumber32,40, coffee41,42, soybean36,43,44, sugarcane45,46 and pearl millet47,48.
Pearl millet is one of the most important small-grained annual cereal crops grown in arid and semi-arid regions of Africa and Asia which are usually characterized by scanty rainfall, poor soil fertility and high temperature16. It is thus enforced to thrive in such harsh environments, where co- occurrence of one or more than one stress factors is a common phenomenon. Despite these it is well adapted to such adverse environmental conditions and known for its excellent nutritional benefits, and hence is attracting plant researchers to examine its cellular and molecular mechanisms of stress tolerance through transcriptomics or functional genomics approaches. A good understanding of the molecular mechanisms of abiotic stress responses in crop plants will not only help in improving yield but also aid to breeding and transgenic approaches2. This study thus investigates 10 potential housekeeping genes for the gene expression analysis in a total of 204 pearl millet samples out of which four are traditional (ACT, EF-1α, eIF4A, and UBC-E2) and six new (GlutR, HSP90, MDH, PP2A, RPL20, and TIP41) reference genes under different abiotic stresses and their combinations at early (1 h) and late (24 h) stress durations in parents of a mapping population differing for drought tolerance as compared to previous studies in pearl millet that were performed under individual stress conditions at one time point only where no contrasting genotypes were used for analysis47,48. Further the best and worst reference gene sets were validated through expression profiling of a PgAP2-like ERF gene from pearl millet.
Several statistical algorithms such as geNorm28, NormFinder49, BestKeeper50, Stability Index37, ΔCt51 and RefFinder52 have been developed to assess the expression stability of candidate reference genes for accurate normalization in gene expression studies and it is also assumed that a comparison of different algorithms allows reliable evaluation32. geNorm analysis revealed MV < 1.5 for all 10 reference genes under different experimental conditions in this study indicating their potential expression stability28,29. The comprehensive results obtained on the basis of statistical analysis by geNorm, NormFinder and RefFinder showed consistency in determining the most stable candidate internal control genes in this study. All the three algorithms recommended EF-1α and UBC-E2 as the top two most stable reference genes in the all samples set across all treatments with some variability in their ranking order indicating the accuracy of our experimental findings. EF-1α also constituted the most stable reference gene pair with either UBC-E2 or PP2A according to geNorm in the individual abiotic stress and combinations of abiotic stresses samples set, respectively. Our findings are in confirmation of several previous studies where EF-1α was also established as the most stable reference gene as for example, wheat under different abiotic and biotic stresses26, cucumber under osmotic and salt stress32, soybean under drought and salinity stress36, sugarcane under drought and salinity stress45, poplar under cold stress53, and bermuda grass under drought stress52. Interestingly out of six new reference genes used in this study, only PP2A could make to the top three stable genes. There are several reports where both old and new reference genes are found to be suitable for normalization26,36,54. In fact PP2A was also ranked among best five reference gene in pearl millet in an earlier study47. Rest five new genes have low stability with RPL20 ranked as the least stable reference gene by NormFinder and RefFinder in all samples set. geNorm also ranked RPL20 as the second least stable gene after GlutR. However in the developmental tissues MDH, HSP90 and EF-1α were the top three most stably expressed reference genes. The ubiquitous expression of HSP90 and EF-1α is not surprising because they were also reported as valid reference genes across various developmental stages in chickpea29. EF-1α was also reported to be the most stable reference gene in different tissues of pearl millet however MDH showed constant expression in different abiotic stresses only according to a previous study48. HSP90 was not included for analysis in earlier two studies on pearl millet but our study showed it to be the second most stable internal control gene after MDH across different developmental stages. However HSP90 was one of the most variably expressed reference genes in individual and multiple stress conditions, thus limiting its use as reference gene under abiotic stress conditions.
Several studies in various plants have shown that inclusion of more than one reference gene would help in more precise normalization of gene transcript for qRT-PCR analysis28,55. Pairwise variation analysis by the geNorm applet suggests inclusion of one or more gene for accurate normalization when the value of pairwise variation is above the cut off range of 0.15. Both the earlier studies in pearl millet suggested the inclusion of one or more genes for normalization of gene transcripts47,48. However in this study, across all the four experimental sets the value of pairwise variation was found to be lower than the geNorm pairwise cut off range indicating that inclusion of additional gene for optimal normalization in pearl millet is not necessary against the suggested gene pair similar to some previous reports28,30,34,56.
The suitability of the identified two most stable genes namely EF-1α and UBC-E2 and their combination was assessed by quantifying the expression pattern of an AP2-like ERF gene, PgAP2-like ERF under both individual and multiple abiotic stresses. PgAP2-like ERF showed clear amplification profiles when these two genes were used as internal controls for both individual and multiple abiotic stresses. While severe disparities in the expression pattern of PgAP2-like ERF were observed when the least stable reference genes HSP90 and eIF4A were used for normalization in qRT-PCR under individual and combination of abiotic stresses, respectively. Our results indicated that EF-1α and UBC-E2 either singly or in combination are suitable for transcript normalization in pearl millet subjected to different abiotic stresses. Similar kind of study was also carried out in tall fescue34. Therefore we recommend the use of EF-1α and UBC-E2 as reference genes for diverse experimental conditions including individual or multiple abiotic stresses, hormone stimulus and developmental tissues in different pearl millet genotypes. It is unlikely that the reference genes other than those identified in this study, may act as better candidates for transcript normalization under various experimental conditions in pearl millet. To the best of our knowledge this is the first report on identification and validation of suitable candidate reference genes for accurate transcript normalization for gene expression studies using qRT-PCR in any crop under various individual abiotic stresses, hormone stimulus and their combinations at early and late durations of stress in contrasting genotypes. The identified reference genes would thus facilitate accurate and consistent qRT-PCR gene expression data analysis over a broad range of developmental tissue samples and multiple abiotic stress conditions in pearl millet and related bioenergy crops for functional genomic studies.
Methods
Plant materials and stress treatments
Seeds of two pearl millet cultivars namely, PRLT2/89-33 (drought tolerant) and H77/833-2 (drought sensitive) were kindly provided by Dr. Rakesh K. Srivastava, International Crops Research Institute for Semi-Arid Tropics (ICRISAT), Patancheru, India. Seeds were sown in composite soil (peat : vermiculite : sand mixture, 2:2:1) in glass house at 32 ± 2 °C day/28 ± 2 °C night with relative humidity oscillating between 40–70% and natural sunlight at National Phytotron Facility (IARI), New Delhi, India. Plants were supplemented with water and 1/3rd strength Hoagland solution on alternate days. After two weeks of germination, pearl millet seedlings of each genotype were used for stress treatment. They were precultured for 24 h in one-third strength Hoagland’s solution4,57. The seedlings were then divided into 14 groups with each group containing 6 seedlings, out of which 13 groups were subjected to individual or multiple abiotic stresses or ABA treatment by transferring the seedlings in the following solutions: drought, solutions containing 20% PEG 6000; salinity, saline solutions containing 250 mM NaCl; heat, solutions containing 1/3rd strength Hoagland solution at 42 °C in a growth chamber; cold, solutions containing 1/3rd strength Hoagland solution at 4 °C in a growth chamber; ABA, solutions containing 100 μM ABA; and in 8 different combinations of stresses including drought + salt, solutions containing 20% PEG 6000 and 250 mM NaCl; drought + heat, solutions containing 20% PEG 6000 exposed to 42 °C in a growth chamber; drought + ABA, solutions containing 20% PEG 6000 and 100 μM ABA; salt + ABA, solutions containing 250 mM NaCl and 100 μM ABA; heat + ABA, solutions containing 100 μM ABA exposed to 42 °C in a growth chamber; cold + ABA, solutions containing 100 μM ABA exposed to 4 °C in a growth chamber; drought + salt + ABA, solutions containing 20% PEG 6000, 250 mM NaCl and 100 μM ABA; and drought + salt + heat + ABA, solutions containing 20% PEG 6000, 250 mM NaCl and 100 μM ABA exposed to 42 °C for 1 h (early) and 24 h (late) according to previous studies4,45,58. Unstressed seedlings were maintained as control and were cultured in the same way as those subjected to stress treatments but without the addition of PEG, NaCl, ABA, or temperature stress. Leaves from a total of 162 samples from 13 different stress treatments at two time points as mentioned above including one untreated control of two genotypes in three biological replicates were carefully harvested and immediately frozen in liquid nitrogen and stored at −80 °C until total RNA isolation.
Developmental tissue samples
For collection of developmental tissue samples, seeds of both cultivars were sown in separate 9′ pots filled with 5 kg autoclaved composite soil under standard green house conditions as mentioned above. A total of 42 tissue samples including leaf, stem, stem sheath, root, panicle at booting stage and grain filling stages, and seeds in three biological replicates from both genotypes were collected at different developmental stages. Tissue samples of leaf, stem, stem sheath and root were harvested after 30 days of sowing (DAS) from plants grown under normal conditions. Panicle at booting stage and grain filling stages were collected at 40DAS and 55DAS, respectively. Fully mature seeds were harvested at 75DAS.
Sequence retrieval and primer designing
Sequence information of 10 candidate internal control genes were obtained either from pearl millet expressed sequence tag (EST) database at National Centre for Biotechnology (NCBI) or by searching foxtail millet orthologous locus Ids from Phytozome v.9.0. Sequences of 8 reference genes namely, ACT, EF-1α, eIF4A, GlutR, HSP90, MDH, RPL20 and UBC-E2 were retrieved from respective pearl millet EST sequences available at NCBI, while sequences of PP2A and TIP41 were retrieved from foxtail millet Phytozome database (http://phytozome.jgi.doe.gov/pz/portal.html) for subsequent qRT-primer designing and amplification. Primer pairs were designed from the non-conserved regions of the gene or EST sequences using Primer Express 3.0 (Applied Biosystems, USA) with the following parameters: 20 ± 2 mers primer length; 75–150 bp amplicon size; 60 ± 1 °C melting temperature (Tm); 50 ± 5% guanine-cytosine (GC) content including no hairpin structures, self-dimers or self-complementarities at the 3′-end (Supplementary Table 1).
Total RNA isolation and cDNA synthesis
The total RNA from samples was isolated using RNAiso plus Reagent (TaKaRa, Japan) according to manufacturer’s instructions. RNA concentration of each sample was determined using NanoDrop- 1000 spectrophotometer (NanoDrop Technologies). The OD260/OD280 nm absorption ratio (1.98–2.01) and OD260/OD230 (≥2.0), was used to determine the quality and purity of RNA preparations. RNA integrity of each sample was also verified by electrophoresis in 1.2% formaldehyde-agarose gel in 1 × 3-(N-morpholino) propanesulfonic acid (MOPS) buffer prepared using diethyl pyrocarbonate (DEPC) treated water. Approximately 5 μg of total RNA was DNase treated to completely eliminate DNA contamination using RNase free DNase set (Qiagen, Germany). The first-strand cDNA synthesis was carried out using 1 μg of total Dnase free total RNA primed with oligodT primers in a final reaction volume of 20 μl using Maxima H Minus M-MuLV reverse transcriptase (Thermoscientific, USA) following manufacturer’s instructions and stored at −20 °C.
PCR and qRT-PCR
Specific amplification of all 10 primer pairs from cDNA was checked by PCR. The 20 μl PCR cocktail contained 10× PCR buffer (Sigma, USA), 10 mM dNTPs mix (Thermosceintific, USA), 10 μM each of forward and reverse primers, 1 μl of cDNA, and 0.2 μl of Taq polymerase. The PCR amplification program was as follows: an initial denaturation step of 3 min at 94 °C, and 30 cycles of 30 s at 94 °C, 45 sec at 60 °C and 45 sec at 72 °C followed by an extension step of 10 min at 72 °C. PCR products were separated on 2% agarose-Tris Borate EDTA (TBE) gels. Real-time PCR was performed utilizing SYBR Green detection chemistry on a 7500 fast Real- time PCR system v2.0.6 (Applied Biosystems). qRT-PCR reaction mixture contained 1 μl of 5-fold diluted cDNA (equal to 50 ng of initial amount of RNA), 10 μM of each gene-specific forward and reverse primer and 5 μl of 2× Power SYBR Green PCR Master Mix (Applied Biosystems, USA) in a total reaction volume of 10 μl. The qRT-PCR cycling conditions were as follows: an initial denaturation step of 20 s at 50 °C, 10 min at 95 °C, and 40 cycles of 15 s at 95 °C, and 1 min at 60 °C followed by melt curve analysis using default parameters in order to verify the PCR specificity by constant increase in temperature from 60 °C to 90 °C. The PCR efficiency of each primer pair was determined through slope of the amplification curve in the exponential phase, obtained by serial dilution using E = 10 (−1/slope)–1 (3.6 ≥ slope ≥ 3.1) by the software itself (Applied Biosystems). Three biological replicates of each sample were used for real-time PCR analysis and three technical replicates were analyzed for each biological sample.
Data analysis for expression stability of candidate reference genes
Expression stability of the 10 potential internal control genes across all treatments and samples were determined using three statistical algorithms namely geNorm28, NormFinder49 and RefFinder52 http://omictools.com/reffinder-s2857.html) following developer’s instructions. The Cq or Ct values of all reference genes used in the geNorm and NormFinder were converted into relative quantities using the formula 2−ΔCt where ΔCt is the value obtained after subtracting minimum Ct value from each corresponding Ct value (ΔCt = each corresponding Ct value - minimum Ct value). The geNorm program calculated the expression stability value (M) for each candidate reference gene on the basis of the average variations in the expression level of a particular gene against expression levels of all the other control genes. The geNorm software provided in qBasePlus (v2.4) was also used to calculate the pairwise variation (Vn/Vn+1) between the two sequential normalization factors (NFn and NFn+1) in order to determine the minimum number of reference genes for optimal normalization. NormFinder, another Microsoft Excel based software package, calculated expression stabilities of the candidate reference genes by combining the intra- and inter-group variations in a sample set containing any number of samples organized in any number of groups. The overall recommended inclusive geomean ranking values of the best reference gene(s) were obtained using the ranking results of geNorm and NormFinder algorithms in the RefFinder online tool.
Validation of reference genes
For the validation of selected reference gene for qRT-PCR analysis under different individual and abiotic stress combinations in pearl millet, the relative transcript accumulation of an objective gene, Pg AP2-like ERF was analyzed using the two most stable namely EF-1α and UBC-E2; and the two most varying reference genes namely HSP90 and eIF4A identified from this study. Sequence of the objective gene PgAP2-like ERF was obtained from a pearl millet transcriptome data from another study (data not published). Primer designing and qRT-PCR reactions were carried out as mentioned earlier. PgAP2-like ERF was amplified using PgAP2-like ERF_RT_ F: 5′GCAGAAGAGATTGCTGATGA 3′ and PgAP2-like ERF_RT_ R: 5′GAGGGCTTTGAAGAAGAGAG 3′ primer pair with an amplicon length of 101 bp. The amount of transcript accumulated for PgAP2-like ERF normalized to the internal control genes used was analyzed using 2−ΔΔCt method59. The relative mRNA expression data of PgAP2-like ERF were the means of at least three biological replicates with the results presented as the mean fold change values ± SE.
Additional Information
How to cite this article: Shivhare, R. and Lata, C. Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Sci. Rep. 6, 23036; doi: 10.1038/srep23036 (2016).
Supplementary Material
Acknowledgments
C.L. acknowledges the research grant from INSPIRE Faculty Award [IFA-11LSPA-01] by DST, GoI. Authors acknowledge Dr. Rakesh K. Srivastava, International Crops Research Institute for Semi-Arid Tropics (ICRISAT), Patancheru, India for providing pearl millet seed materials. Authors are also thankful to the Director, CSIR-National Botanical Research Institute, Lucknow, India and the Director, National Research Centre on Plant Biotechnology, New Delhi, India for providing facilities and support during the study.
Footnotes
Author Contributions C.L. conceived and designed the experiment, and provided new reagents. R.S. and C.L. carried out the experiments, analyzed data and wrote the manuscript. Both authors reviewed and approved the manuscript.
References
- Bray E. A., Bailey-Serres J. & Weretilnyk E. Responses to abiotic stresses. Biochemistry and molecular biology of plants [Gruissem W., Buchannan B., Jones R. (eds)], [1158–1249] (American Society of Plant Physiologists, Rockville, MD 2000). [Google Scholar]
- Lata C., Muthamilarasan M. & Prasad M. Drought stress responses and signal transduction in plants. Elucidation of abiotic stress signaling in plants [Pandey G. K. (ed.)] [195–225] (Springer, New York, 2015). [Google Scholar]
- Varshney R. K., Bansal K. C., Aggarwal P. K., Datta S. K. & Craufurd P. Q. Agricultural Biotechnology for crop improvement in a variable climate: hope or hype. Trends Plant Sci. 16, 363–371 (2011). [DOI] [PubMed] [Google Scholar]
- Lata C., Bhutty S., Bahadur R. P., Majee M. & Prasad M. Association of SNP in a novel DREB2-like gene SiDREB2 with stress tolerance in foxtail millet [Setaria italica (L.)]. J. Exp. Bot. 62, 3387–3401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata C. Advances in omics for enhancing abiotic stress tolerance in millets. Proc. Indian Natl. Sci. Acad. 81, 397–417 (2015). [Google Scholar]
- Mittler R. & Blumwald E. Genetic engineering for modern agriculture: challenges and perspectives. Annu. Rev. Plant Biol. 61, 443–62 (2010). [DOI] [PubMed] [Google Scholar]
- Suzuki N. et al. Enhanced tolerance to environmental stresses in transgenic plants expressing the transcriptional co-activator MBF1. Plant Physiol. 139, 1313–22 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S. et al. Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol. 161, 1783–1794 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11, 15–19 (2006). [DOI] [PubMed] [Google Scholar]
- Barnabas B., Jäger K. & Fehér A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 31, 11–38 (2008). [DOI] [PubMed] [Google Scholar]
- Atkinson, N., J. & Urwin P. E. The interaction of plant biotic & abiotic stresses: From gene to the field. J. Exp. Bot. 10, 3523–3543 (2012). [DOI] [PubMed] [Google Scholar]
- Iyer N. J., Tang Y. & Mahalingam R. Physiological, biochemical and molecular responses to a combination of drought and ozone in Medicago truncatula. Plant, Cell Environ. 36, 706–720 (2013). [DOI] [PubMed] [Google Scholar]
- Prasad P. V. V., Pisipati S. R., Mom_cilovi. C. I. & Ristic Z. Independent and combined effects of high temperature and drought stress during grain filling on plant yield and chloroplast EF-Tu expression in spring wheat. J. Agronomy Crop Sci. 197, 430–441 (2011). [Google Scholar]
- Prasch C. M. & Sonnewald U. Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol. 162, 1849–1866 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syvertsen J. P. & Garcia-Sanchez F. Multiple abiotic stresses occurring with salinity stress in citrus. Environ. Exp. Bot. 103, 128–137 (2014). [Google Scholar]
- Vadez V., Hash T., Bidinger F. R. & Kholova J. Phenotyping pearl millet for adaptation to drought. Front. Physiol. 3, 386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R. S., Sehgal D. & Vadez V. Using genetic mapping and genomics approaches in understanding and improving drought tolerance in pearl millet. J. Exp. Bot. 62, 397–408 (2011). [DOI] [PubMed] [Google Scholar]
- Lata C., Gupta S. & Prasad M. Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses. Crit. Rev. Biotechnol. 33, 328–343 (2013). [DOI] [PubMed] [Google Scholar]
- Rajaram V. et al. Pearl millet [Pennisetum glaucum (L.) R. Br.] consensus linkage map constructed using four RIL mapping populations and newly developed EST-SSRs BMC Genomics. 14, 159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K., Muthamilarasan M. & Prasad M. Reference genes for quantitative real-time PCR analysis in the model plant foxtail millet (Setaria italica L.) subjected to abiotic stress conditions. Plant Cell Tiss. Organ Cult. 115, 13–22 (2013). [Google Scholar]
- Gutierrez L. et al. The lack of a systemic validation of reference genes: serious pitfall undervalued in reverse transcription–polymerase chain reaction (RT–PCR) analysis in plants. Plant Biotechnol. J. 6, 609–618 (2008). [DOI] [PubMed] [Google Scholar]
- Gue´nin S. et al. Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions specific, validation of references. J. Exp. Bot. 60, 487–493 (2009). [DOI] [PubMed] [Google Scholar]
- Udvardi M. K., Czechowski T. & Scheible W. R. Eleven golden rules of quantitative RT-PCR. Plant Cell 20, 1736–1737 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A. et al. MIQE précis: practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol. 11, 74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Tyagi A. K. & Khurana J. P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345, 646–651 (2006). [DOI] [PubMed] [Google Scholar]
- Long X. Y. et al. Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol. Biol. 74, 307–311 (2010). [DOI] [PubMed] [Google Scholar]
- Manoli A., Sturaro A., Trevisan S., Quaggiotti S. & Nonis A. Evaluation of candidate reference genes for qPCR in maize. J. Plant Physiol. 169, 807–815 (2012). [DOI] [PubMed] [Google Scholar]
- Vandesompele J. et al. Accurate normalisation of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research, 00341–0034.11 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R., Sahoo A., Tyagi A. K. & Jain M. Validation of internal control gene expression studies in chickpea (Cicer arietinum L.). Biochem. Biophys.Res. Commun. 396, 283–288 (2010). [DOI] [PubMed] [Google Scholar]
- Huggett J., Dheda K., Bustin S. & Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6, 279–284 (2005). [DOI] [PubMed] [Google Scholar]
- Fan C. et al. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS One. 8, e56573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migocka M. & Papierniak A. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol. Breed. 28, 343–357 (2011). [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M. K. & Scheible W. R. Genome wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Chen Y., Hu B., Tan Z. & Huang B. Identification and validation of reference gene for quantification of target gene expression with quantitative real-time PCR for tall Fescue under four abiotic stresses. PLoS One. 10, e0119569 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. et al. Reference gene selection for quantitative real- time PCR normalization in Caragana intermedia under different abiotic stress conditions. PLoS One. 8, e53196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S. et al. Expression stabilities of candidate reference genes for RT-qPCR under different stress conditions in soybean. PLoS One. 8, e75271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner A. M., Yakovlev I. A. & Strauss S. H. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 4, 14 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo-Ra K. et al. Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol. Lett. 25, 1869–1872 (2003). [DOI] [PubMed] [Google Scholar]
- Li Q.-F. et al. Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Mol. Bio. 28, 49–57 (2010). [Google Scholar]
- Wan H. et al. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 399, 257–261 (2010). [DOI] [PubMed] [Google Scholar]
- Cruz F. et al. Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol. Breed. 23, 607–616 (2009). [Google Scholar]
- Barsalobres-Cavallari C. F., Severino F. E., Maluf M. P. & Maia I. G. Identification of suitable internal control genes for expression studies in Coffea arabica under different experimental conditions. BMC Mol. Biol. 10, 1 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R., Fan C., Li H., Zhang Q. & Fu Y. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 10, 93 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M. et al. Clough, Stacey G. Identification of four soybean reference genes for gene expression normalization. Plant Genome. 1, 1 (2008). [Google Scholar]
- Guo J., Ling H., Wu Q., Xu L. & Que Y. The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Sci. Rep. 4, 7042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H., Wu Q., Guo J., Xu L. & Que Y. Comprehensive selection of reference genes for gene expression normalization in sugarcane by real time quantitative RT-PCR. PLoS One. 9, e97469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P. & Blumwald E. Assessing reference genes for accurate transcript normalization using quantitative real-time PCR in pearl millet [Pennisetum glaucum (L.) R. Br]. PLoS One. 9, e106308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P. S., Reddy D. S., Sharma, Kiran K., Bhatnagar-Mathur P. & Vadez V. Cloning and validation of reference genes for normalization of gene expression studies in pearl millet [Pennisetum glaucum (L.) R. Br.] by quantitative real-time PCR. Plant Gene 1, 35–42 (2015). [Google Scholar]
- Andersen C. L., Jensen J. L. & Ørntoft T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250 (2004). [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C. & Neuvians T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515 (2004). [DOI] [PubMed] [Google Scholar]
- Silver N., Best S., Jiang J. & Thein S. L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Bio. 7, 33 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. et al. Selection of reference genes for quantitative real-time PCR normalization in creeping bentgrass involved in four abiotic stresses. Plant Cell Rep. 34, 1825–1834 (2014). [DOI] [PubMed] [Google Scholar]
- Wang Z. et al. Selection of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in Brassica napus under various stress conditions. Mol. Genet. Genomics 289, 1023–1035 (2014). [DOI] [PubMed] [Google Scholar]
- Tian C., Jiang Q., Wang F., Wang G., Xu Z. & Xiong A. Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PLoS One. 10, e0117569 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le D. T. et al. Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS One. 7, e46487 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condori J., Nopo-Olazabal C., Medrano G. & Medina-Bolivar F. Selection of reference genes for qPCR in hairy root cultures of peanut. BMC Res Notes. 4, 392 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. Construction and application of EST library from Setaria italica in response to dehydration stress, Genomics 90, 121–131 (2007). [DOI] [PubMed] [Google Scholar]
- Lata C. et al. Genome-wide investigation and expression profiling of AP2/ERF transcription Factor superfamily in foxtail millet (Setaria italica L.). PLoS One. 9, 11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.