SUMMARY
The regenerative capacity of the injured CNS in adult mammals is severely limited, yet axons in the peripheral nervous system (PNS) regrow, albeit to a limited extent, after injury. We reasoned that coordinate regulation of gene expression in injured neurons involving multiple pathways was central to PNS regenerative capacity. To provide a framework for revealing pathways involved in PNS axon regrowth after injury, we applied a comprehensive systems biology approach, starting with gene expression profiling of dorsal root ganglia (DRGs) combined with multi-level bioinformatic analyses and experimental validation of network predictions. We used this rubric to identify a drug that accelerates DRG neurite outgrowth in vitro and optic nerve outgrowth in vivo by inducing elements of the identified network. The work provides a functional genomics foundation for understanding neural repair and proof of the power of such approaches in tackling complex problems in nervous system biology.
INTRODUCTION
The regenerative capacity of the injured adult mammalian CNS is extremely limited, which leads to permanent neurological deficits following CNS injury. In contrast, injured axons in the adult mammalian peripheral nervous system (PNS) maintain the capacity to regenerate, providing potential for functional recovery after peripheral nerve injury (Abe and Cavalli, 2008; Ramón y Cajal et al., 1991). The failure of CNS axons to regenerate is due to many factors, primarily a lack of induction of a cell-intrinsic growth capacity after injury (Afshari et al., 2009; Giger et al., 2010) and the presence of extrinsic inhibitory effects (Filbin, 2003; Yiu and He, 2006), both mechanisms supported by many lines of experimental evidence (Hoffman, 2010; Neumann and Woolf, 1999; Sun et al., 2011; Yiu and He, 2006). The concept that specific intrinsic molecular differences contribute to the divergent neuronal growth states after PNS and CNS injuries is supported by the manipulation of individual candidate genes induced in neurons by PNS, but not CNS, injury, which can promote limited CNS regrowth after injury (Hoffman, 2010; Neumann and Woolf, 1999; Sun et al., 2011). The relative importance of intrinsic neuronal signals during injury in CNS regeneration failure (Sun and He, 2010) is highlighted by the very limited axon regeneration observed even after eliminating combinations of known extrinsic inhibitory signals (Yiu and He, 2006). Furthermore, a conditioning lesion of the peripheral axon of dorsal root ganglion (DRG) neurons in the PNS increases the intrinsic growth state of the neurons sufficiently to enable the regeneration of their central axons in the CNS (Neumann and Woolf, 1999).
One of the intrinsic molecular mechanisms contributing to the regenerative process is the retrograde transport of injury signals to the cell body of the neuron, leading to expression of regeneration-associated genes (RAGs; Abe and Cavalli, 2008). For example, injured PNS axons activate RAGs such as Atf3, Jun, Hsp27, Sprr1a, Gap43, and genes involved in the JAK-STAT3 pathway, whereas injury to CNS axons does not result in the activation of these RAGs (Afshari et al., 2009). Axonal injury also induces local activation and retrograde transport of several MAPKs, including ERK (Hanz et al., 2003; Perlson et al., 2005) and JNK (Cavalli et al., 2005; Lindwall and Kanje, 2005), while elevation of cAMP levels in the soma after axonal injury to the PNS neurons enhances axonal re-growth (Qiu et al., 2002). Elevated cAMP levels also increase regenerative responses of retinal ganglion cells (Chierzi et al., 2005) and the damaged spinal cord (Qiu et al., 2002). PNS nerve injury also triggers the expression of several cytokines, such as interleukin-6, leukemia inhibitory factor, and ciliary neurotrophic factor, at the lesion sites (Cao et al., 2006; Sendtner et al., 1992; Subang and Richardson, 2001). Subsequently, these cytokines activate the JAK-STAT pathway through gp130-containing receptor complexes (Taga and Kishimoto, 1997), resulting in the accumulation of phospho-STAT3 in the nucleus (Schweizer et al., 2002), which further activates the axon regeneration program (Bareyre et al., 2011).
Overexpression of a single RAG alone, including the transcription factors (TFs) ATF3 and STAT3 or the cytoskeletal protein GAP43, is usually not sufficient for effective successful regeneration (Bareyre et al., 2011; Bomze et al., 2001; Seijffers et al., 2007). For example, constitutive expression of a single TF (ATF3) increases axonal growth in the PNS after injury, but it does not overcome myelin inhibition in culture or enhance neuronal regeneration in the CNS (Seijffers et al., 2007). These findings demonstrate the necessity for the combined and cooperative intrinsic regulation of many RAGs involved in various pathways for the regeneration program to occur. This is consistent with the growing appreciation that genes do not act in isolation, but rather in coordinated networks in conjunction with other genes (Parikshak et al., 2015).
Recently, combinatorial knockout of two genes, Pten and Socs3, responsible for negative regulation of the mammalian target of rapamycin (mTOR) and JAK-STAT pathways, was shown to enhance regeneration in the CNS, providing important evidence that it is possible to reactivate central axonal growth in the injured adult (Sun et al., 2011). In many cases, modulation of expression of individual genes in these pathways has led to some increase in axonal outgrowth after injury, consistent with their functional role in neural repair. However, these findings also suggest that multiple pathways act in parallel to stimulate neuronal regeneration after injury.
We reasoned that differences in the regenerative potential between injured PNS neurons and injured CNS neurons likely reflect major differences in intrinsic transcriptional networks, rather than changes in expression of a few individual genes. We applied a multi-staged approach to characterize the transcriptional network associated with axon outgrowth in the PNS, and we experimentally validated several network predictions using bioinformatic and experimental approaches. By integration of protein-protein interactions (PPIs) with the RNA co-expression network, we identified a core RAG co-expression module highly enriched in a core set of hub genes, including TFs known to promote neurite outgrowth as well as novel TFs not previously associated with axonal outgrowth. Our analysis indicates that, rather than acting in isolation, these enriched TFs provide crucial cross-talk between the signaling pathways involved in neuronal regeneration after PNS injury. We show, moreover, that the expression of these TFs is coordinately upregulated following PNS injury, but not after CNS injury, consistent with the notion that coordinate regulation of this core network, rather than individual components, is necessary for axonal outgrowth. As a proof of principle, we show that top two hub TFs present in the core regeneration-associated gene network, ATF3 and JUN, when simultaneously over-expressed increased axonal outgrowth in mouse DRG neurons, when compared to the individual TF over-expression. Finally, we show that a compound that increases the expression of several components of the PNS coordinate transcriptional network, can promote CNS regeneration, validating this approach to screen for CNS growth-promoting treatments.
RESULTS
Construction of RAG Co-expression Networks
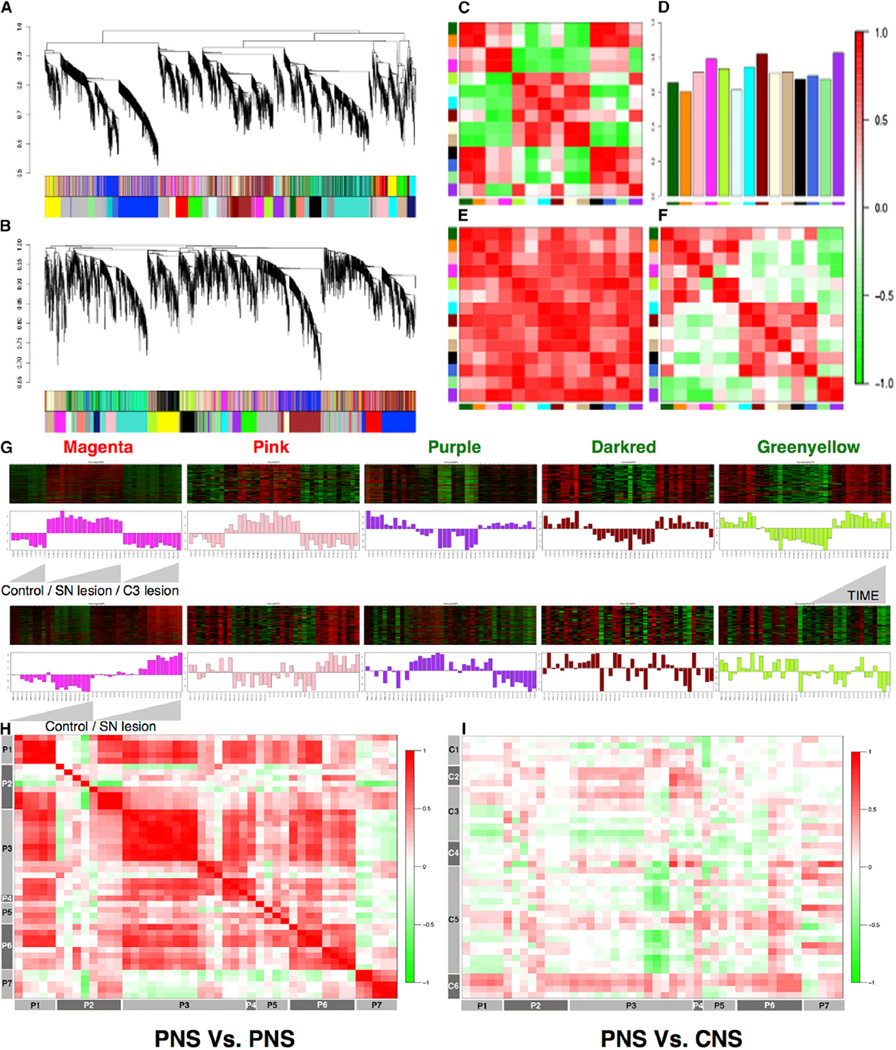
We first analyzed the genome-wide transcriptional changes that occur in DRG neurons during axon outgrowth following peripheral nerve injury at multiple time points, ranging from hours to days, after injury (see Table S1) in several experimental models of rodent peripheral nerve injury (Beggs and Salter, 2006) generated independently in collaborating laboratories as follows: sciatic nerve (SN) lesion (SN transection), spinal nerve ligation, spared nerve injury, and chronic nerve constriction, as well as a model where regeneration does not occur after a C3 lesion (cervical cord hemi-section at the C3 level cutting the central axons of DRG neurons in the dorsal columns). In total, 147 microarrays for 31 time points were analyzed in this initial study. We performed weighted gene co-expression network analysis (WGCNA) (Geschwind and Konopka, 2009; Langfelder and Horvath, 2008; Zhang and Horvath, 2005), a powerful method for understanding the modular network structure of the transcriptome, to identify the peripheral nerve regrowth-related network following SN lesions. WGCNA permits identification of modules of highly co-expressed genes whose grouping reflects shared biological functions and key functional pathways, as well as key hub genes within the modules (Langfelder and Horvath, 2008; Parikshak et al., 2015).
We first conducted WGCNA on two different microarray-based time series experiments on DRGs during the period of regrowth after nerve injury, performed independently in two different laboratories (Experimental Procedures). We applied consensus network analysis (Langfelder and Horvath, 2008) to define 14 robust and reproducible co-expression modules (Experimental Procedures; Figure 1) between the two SN lesion datasets generated 1, 3, 7, 14, and 49 days after injury and 1, 3, 8, 12, 16, 18, 24, and 28 hr post-injury (Figures 1A–1F). This analysis indicated that the gene co-expression relationships in the DRG following SN injury are highly preserved; these 14 modules represent common pathways associated with regeneration after nerve injury (Figures 1C–1F).
Figure 1. Network Analysis of Sensory Neuron Profile Changes after SN Lesions.
(A and B) Gene dendrograms for two SN lesion datasets are shown.
(C–F) Consensus module preservation across datasets. (C and F) Eigengene (first principal component of gene expression) adjacencies of two datasets are shown; rows and columns correspond to one eigengene consensus module (red, positive correlation; green, negative correlation). (D) Preservation measure for each consensus eigengene is shown. (E) Overall module preservation among SN lesion datasets is shown; rows and columns correspond to a consensus module; red saturation denotes module preservation.
(G) Heatmaps depicting expression of genes (rows) across samples (columns) for five modules (red corresponds to gene upregulation and green to down-regulation). First principal component of gene expression is shown as a bar-plot. (Top) SN and C3 lesion datasets 1, 3,7,14, and 49 days after injury (left to right, ascending order) are shown. (Bottom) Same genes in another SN lesion dataset 1, 3, 8, 12, 16, 18, 24, and 28 hr post-injury are shown.
(H and I) Plots comparing direction of correlation of top 50 hub genes in magenta module in 16 (eight PNS and eight CNS) neuronal injury datasets (H) PNS versus PNS and (I) CNS versus PNS; correlation scores encoded −1 (green, anti-correlated) to +1 (red, correlated).
On the basis of the module eigengene correlation with time-dependent changes after injury, we first classified modules as upregulated, downregulated, and early regulated following injury (Experimental Procedures; Figure S1A). Next, based on the significant module-trait relationships (Experimental Procedures; Figure S1B; Bonferroni corrected p value < 0.01), we identified five modules strongly associated with regeneration as follows: two modules (magenta and pink) whose genes are unregulated, and three (purple, dark red, and green-yellow) whose genes are downregulated after nerve injury (Figures 1A, 1B, and 1G; Table S2), all of which were conserved in a third, independent peripheral nerve injury dataset (Costigan et al., 2002; Table S1).
As a first step toward functional annotation, we examined each module’s association with neuronal regeneration from published literature by testing association with the key words neuronal regeneration, axonal regeneration, and nerve injury in the PubMed database for every gene (Experimental Procedures). This analysis identified the magenta module as significantly enriched for genes associated with neuronal regeneration. Nearly 24% of the genes (108 of 435 genes in the module; hyper-geometric p value 3.3 × 10−11; Experimental Procedures) were associated with neural regeneration and/or axon outgrowth (Table S2). To further annotate module function at a general level, we applied gene ontology (GO) enrichment analyses, which showed enrichment (Benjamini-corrected p values < 0.05) for several GO categories in the upregulated RAG co-expression modules (magenta and pink) that are functionally associated with neuronal regeneration. Significant clusters included regulation of transcription, neuron differentiation, inflammation, stimulus related, signaling related, and cell proliferation/growth/migration (Table S3). GO functional analysis for downregulated RAG co-expression modules (purple and dark red) revealed enrichment for various categories related to plasma membrane, ion/gated channel, ion binding, and synapse/cell junction (Table S3). Several previous studies have observed the differential regulation of many TFs (Abe and Cavalli, 2008; Michaelevski et al., 2010) and membrane ion channel regulation after nerve injury (Abe and Cavalli, 2008; Yang et al., 2004), consistent with these observations.
Validation of RAG Co-expression Modules
To validate these co-expression modules, we first compared the direction of differential expression of the top 50 hub genes, which represent the most central genes in the co-expression network in all five modules, in 16 independent datasets (eight PNS and eight CNS) containing 382 microarrays related to either PNS or CNS neuronal injury (Figures 1H and 1I; Figures S1C and S1D; Table S4). The co-expression relationships observed in our PNS data were also observed in these independent PNS datasets (r2 range, 0.5 to 1.0; Figures 1H and 1I; Figures S1C and S1D). In contrast, these relationships were not as well preserved in datasets after CNS injury, where we observed a higher degree (>1.5-fold versus PNS) of anti-correlation (≤−0.5; Figure S1C and S1D), especially significant for magenta and purple modules. This analysis demonstrates that the core PNS injury-related co-expression network in the DRG is highly reproducible and is not observed in injured CNS neurons following a CNS lesion (Figures 1H and 1I).
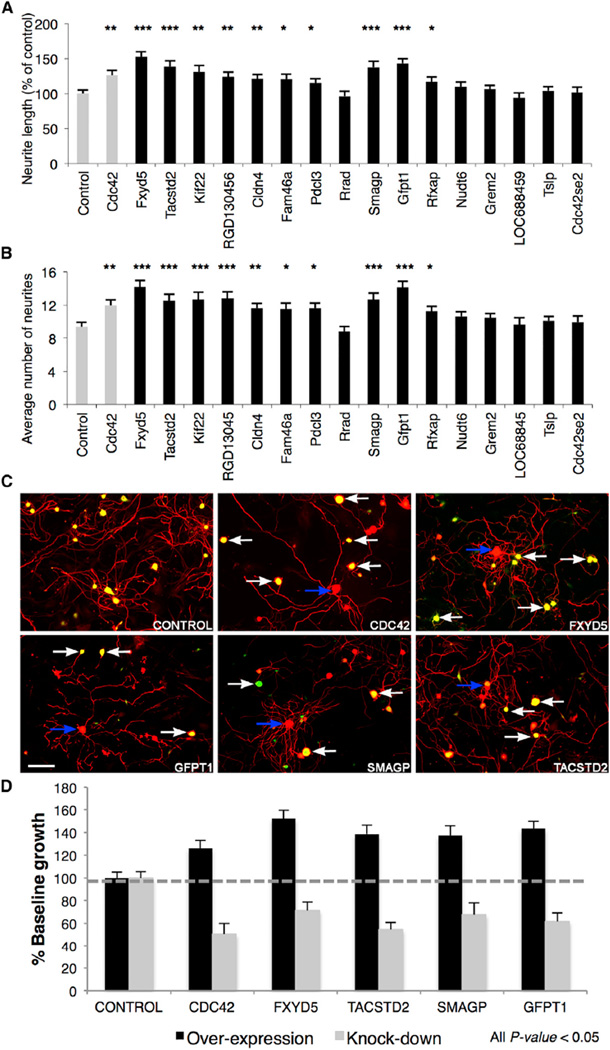
The association of nearly 24% of magenta module genes with terms related to neuronal regeneration in the literature predicts that the other genes in this module and the other upregulated module also would modulate regeneration. To validate these network-based predictions, we selected the following upregulated genes from the magenta and pink modules based on their high intramodular connectivity (hub status; Experimental Procedures), requiring that they had not been previously associated with neuronal regeneration in the literature (i.e., were putative novel RAGs): Smagp (small trans-membrane and glycosylated protein), Gfpt1 (glutamine fructose-6-phosphate transaminase 1), Tslp (thymic stromal lymphopoietin), Nudt6 (nucleoside diphosphate linked moiety X-type motif 6), Cdc42se2 (CDC42 small effector 2), Rfxap (regulatory factor X-associated protein), Grem2 (gremlin 2), and LOC688459. We augmented this validation set utilizing a knowledge-based semi-supervised approach (Experimental Procedures) to include the following additional genes with strong co-expression relationships in our datasets to neuronal regeneration: Fxyd5 (FXYD domain containing ion transport regulator 5), Tacstd2 (tumor-associated calcium signal transducer 2), Kif22 (kinesin family member 22), RGD1304563, Cldn4 (claudin 4), Fam46a (family with sequence similarity 46, member A), Pdcl3 (phosducin-like 3), and Rrad (Ras-related associated with diabetes).
To determine if these nominated genes were indeed RAGs, we first performed an in vitro assay by monitoring acute neurite outgrowth following overexpression of each candidate gene in adult mouse DRG neurons (Figures 2A and 2B; Figure S1E). Of the 16 putative RAGs tested, ten caused significant increases in both neurite length and the number of neurites after overexpression (ANOVA with Bonferroni-Holm post hoc test, p < 0.05; Figures 2A, 2B, and 2D; Figure S1E) (genes Fxyd5, Gfpt1, Smagp, Tacstd2, Kif22, RGD1304563, Cldn4, Fam46a, Rfxap, and Pdcl3). We selected the top four (Fxyd5, Gfpt1, Smagp, and Tacstd2) for further loss-of-function validation, and we performed an in vitro assay monitoring neurite outgrowth in replated adult mouse DRG neurons following knockdown of the candidate RAGs Fxyd5, Gfpt1, Smagp, Tacstd2, and Cdc42 by RNAi, compared with a small hairpin RNA (shRNA) control vector (containing a non-specific shRNA). In all cases, target knockdown significantly (ANOVA with Bonferroni-Holm post hoc test, p < 0.05) reduced neurite outgrowth (Figures 2C and 2D).
Figure 2. Experimental Validation of Novel Candidate RAGs.
(A and B) Differences in neurite outgrowth produced by overexpression of 16 cDNA clones in lentiviral expression vectors with an IRES eGFP expression tag in cultured adult C57BL/6 DRG neurons with Cdc42 as positive control. (A) Total neurite length and (B) number of neurites per neuron were quantified using ImageJ software (NeuronJ plugin), from 50 to 150 cells per view. Significant differences were determined by ANOVA with Bonferroni-Holm post hoc test; 10/16 candidates induce greater neurite growth.
(C) Knockdown of the top four selected genes using lentiviral delivery of shRNA with eGFP reporter in C57BL/6 DRGs. Transfected (white arrow) and non-transfected (blue arrow) individual DRG neurons are highlighted. Scale bar, 100 µm.
(D) Average total neurite length relative to control. All data shown is significant relative to control (p < 0.05, mean ± SEM).
TF-Binding Site Enrichment in RAG Co-expression Modules
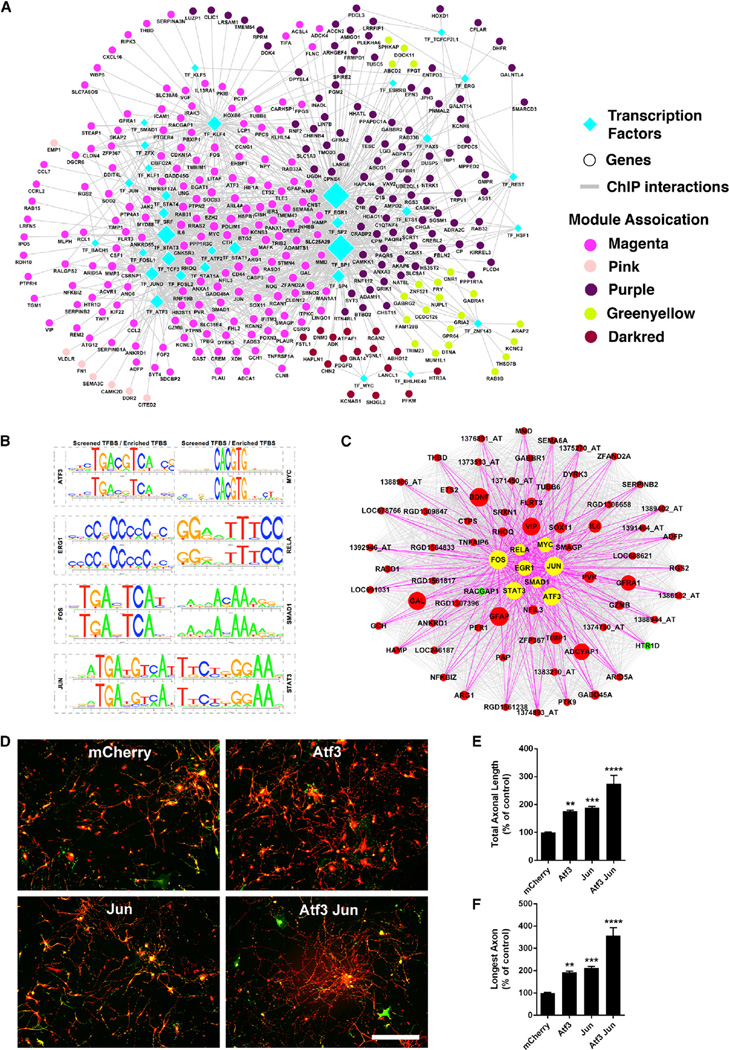
To uncover the potential regulatory network contributing to the consistent co-expression of multiple genes after nerve injury, we performed TF-binding site (TFBS) enrichment analysis for each of the RAG co-expression modules. To avoid confounders and identify the most statistically robust sites, we used three different background datasets (1,000-bp sequences upstream of all rat genes, rat CpG islands, and the rat chromosome 20 sequence). We identified 62 TFs (Experimental Procedures) significantly enriched in RAG co-expression modules (p value < 0.05 relative to all three background datasets; Table S5). Of these TFs, 39 had experimental data available, providing confirmation of predicted binding sites via chromatin immunoprecipitation (ChIP) experiments (hypergeometric p value 3.4 × 10−25; Figure 3A; Table S6; Experimental Procedures). Interestingly, up-(magenta and pink) and downregulated modules (purple, green-yellow and dark red) occupied relatively distinct network territories that share different core TF regulators, as defined by network co-expression and ChIP data, suggesting quite distinct control of the regulation of these gene sets after peripheral nerve injury (Figure 3A). The two upregulated modules showed enrichment for 15 TFs previously associated with the axonal growth and neuronal injury response (Figure 3A; Table S5), including ATF3, EGR1, EGR2, FOS, JUN, KLF4, REL, RELA, SMAD1, SP1, SP2, STAT1, and STAT3 (Abe and Cavalli, 2008; Figure 3A). Remarkably, eight of these over-represented TFs (ATF3, EGR1, FOS, JUN, MYC, RELA, SMAD1, and STAT3) are present in the magenta module, the major upregulated module after PNS injury (Figures 3B and 3C).
Figure 3. TF-Binding Site Enrichment in Injured Regenerating Neurons.
(A) Regulatory network of differentially expressed genes after nerve injury. Nodes correspond to genes or TFs and edges to ChIP interaction. Node color represents their corresponding module associations as denoted in the legends and over-represented TFs (diamond) are shown in cyan. Node size is based on its centrality.
(B) Sequence logo plots of reference (JASPAR/TRANSFAC) and identified position weight matrix for each TF significantly over-represented in magenta module are shown.
(C) Magenta module, showing eight over-represented TFs as hub genes. Nodes correspond to genes and edges to significant correlation. Larger nodes correspond to number of PubMed hits with co-occurrence of gene and neuronal regeneration, axonal regeneration, and nerve injury tags; upregulated (red), downregulated (green), and over-represented TFs (yellow) are shown. Only edges connected by over-represented TFs are highlighted.
(D–F) Combined TF overexpression. (D) Photomicrographs of dissociated DRG neurons transduced with the indicated viruses at 1 DIV, replated at 7 DIV, and cultured for an additional 20 hr on laminin. Red, bIII-tubulin; green, EGFP. (E and F) Quantification of total axonal length (E) and longest axon (F) reveal significant enhancement of axonal growth for ATF3 and JUN expression individually, compared to control (mCherry). Combined ATF3 and JUN overexpression enhances axonal growth significantly more than ATF3 or JUN individually. Data are mean ± SE. n = 30 wells. **p < 0.01, ***p < 0.001, ****p < 0.0001, one-way ANOVA with Bonferroni’s post hoc test. Scale bar, 200 microns.
Next, we cross-validated the TFBS enrichment analysis of the sequences upstream of the orthologous genes in mouse and human. We found that 51% of TFs that were over-represented in rats also were over-represented in the sequences of mouse or human homologs, demonstrating phylogenetic conservation of TFBS in the promoter regions of these co-expressed genes after nerve injury (Table S6). It is notable that the enriched TFs related to upregulated modules (ATF3, EGR2, FOS, JUN, KLF4, REL, RELA, SMAD1, SP1, SP2, and STAT3 [Abe and Cavalli, 2008]) are well studied for involvement in the nerve injury process compared to TFs enriched in the downregulated modules (candidate transcriptional repressors).
During regeneration, many genes are dynamically down-regulated (Shim and Ming, 2010). To specifically identify TFs that could regulate this process, we screened for experimentally validated TFBS over-represented in modules down-regulated after injury—purple, dark red, and green-yellow (candidate transcriptional repressors; Experimental Procedures), which are enriched in genes related to the GO terms plasma membrane, ion/gated channel, ion binding, and synapse/cell junction related (Table S3). By screening for highly phylogenetically conserved regulatory regions (rat, mouse, and human) in these down-regulated modules, we found that TFBS for SP1, 2, EGR1, 2, KLF4, 5, and MZF1 were enriched in the promoter regions of these orthologous genes (Table S6b). A recent study has shown that MZF1 expression increases after nerve injury in DRG neurons and regulates a long noncoding RNA that contributes to neuropathic pain (Zhao et al., 2013), but little is known about its regulatory targets. We validated the network-predicted co-expression relationships via over-expression of MZF1 in adult DRG neurons and examination of its putative target’s mRNA levels, validating two-thirds of the predicted targets (6 out of 9) including Ntrk1 (neurotrophic tyrosine kinase receptor type 1), Htr3a (5-hydroxytryptamine serotonin receptor 3A), and Gabbr2 (gamma-aminobutyric acid B receptor 2) (Figure S1F). Both Htr3a and Gabbr2 are present in the down-regulated modules whose members are enriched with MZF1 TFBS in their promoter regions, and both had been previously shown to be down regulated by nerve injury (Li et al., 2015).
Co-expression of Two Hub TFs Induces a Combinatorial Increase in Axonal Growth
Next, to provide an experimental test of our TFBS enrichment predictions, we assessed the action of two of the transcription factors by over-expression in DRG neurons in vitro. We selected two critical TFs (ATF3 and JUN) based on their presence in the core regeneration associated magenta module (Figure 3C) and their increased expression levels after nerve injury in vivo (Figure S2). We assessed neurite outgrowth in the context of over-expression of each TF individually, as well with their combined over-expression. We observed that ATF3 and JUN increase neurite outgrowth significantly when over-expressed individually (Figures 3E and 3F), consistent with previous findings (Dragunow et al., 2000; Seijffers et al., 2006). Their combinatorial over-expression synergistically increased neurite outgrowth; the total neurite outgrowth per neuron and the longest neurite per neuron were enhanced by 2.7 ± 0.3-fold, and 3.6 ± 0.4-fold, respectively (Figures 3D–3F) relative to the control. The magnitude of the increased growth on laminin induced by these two factors (2.7-fold increase in total neurite outgrowth per neuron) is significantly higher when compared to the over-expression of each TF individually, supporting the predictions of the network analysis.
Co-regulated Genes Represent Convergent Pathways
To extend this work to the level of specific proteins and identify potential mammalian conserved protein-signaling pathways represented by the RAG co-expression modules, we determined the PPI network represented by the genes in all the five modules. We reasoned that this would not only provide independent validation of the relationships inferred by RNA co-expression, but that the PPIs would provide important pathways for possible therapeutic intervention.
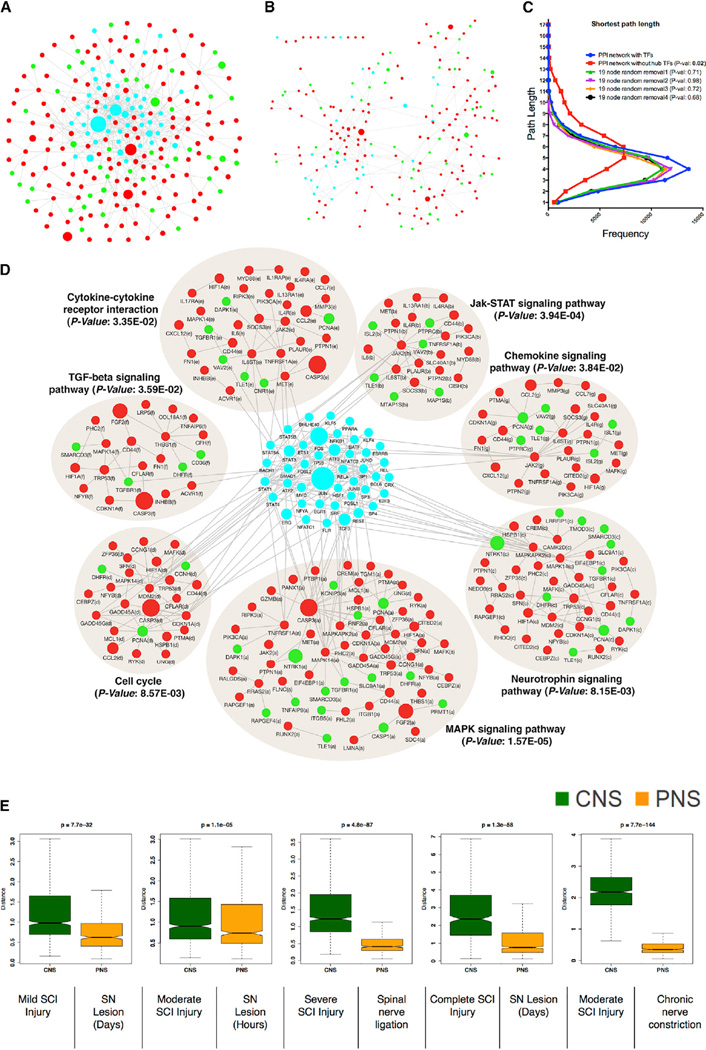
We screened experimentally validated PPIs among all possible combinations of gene pairs present in the co-expressed modules and over-represented TFs, obtaining a PPI network consisting of 280 nodes and 496 edges (Figure 4A; Experimental Procedures). Strikingly, despite the relatively small number of proteins in this network, we observed higher connectivity when compared to a random PPI network consisting of a similar number of nodes (clustering coefficient = 0.114, for random network = 0.016; average number of neighbors = 4.12, for random network = 1.83). We also observed enrichment of several important signaling pathways that contribute to the regulation of neuronal regeneration (Abe and Cavalli, 2008), including the Neurotrophin-, MAPK-, TGF-beta-, chemokine-, and JAK-STAT-signaling pathways (Abe and Cavalli, 2008; Figure 4D). Many of the TFs whose binding sites were over-represented in the RAG mRNA co-expression modules were also hubs in this protein network (Figures 4A and 4D; 19/62; hypergeometric p value 5.8 × 10−20); based on network centrality, 68% (19 TFs of the top 10%, n = 28) of the most highly connected nodes represented TFs. Most genes belonging to the enriched signaling pathways also were enriched for the TFBSs of hub TFs in the PPI network (for 12 TFs with experimental ChIP data, 42 of 280 genes had three or more TFBSs for hub TFs; hypergeometric p value 1.1 × 10−12).
Figure 4. Over-Represented TFs Are Involved in Transcriptional Cross-Talk between Regeneration-Associated Pathways.
(A) Protein-protein interaction (PPI) network of differentially expressed genes after nerve injury. Nodes correspond to genes and edges to PPI. Larger nodes correspond to number of PubMed hits with co-occurrence of gene and neuronal regeneration, axonal regeneration, and nerve injury tags. Node color represents upregulated (red), downregulated (green), and over-represented (cyan) TFs.
(B) PPI network dissociation after in silico removal of 19 hub TFs is shown.
(C) Distribution of the shortest path between pairs of nodes in the PPI network with or without in silico removal of 19 hub TFs. Random removal of a similar number of nodes is shown for comparison.
(D) Significantly enriched KEGG pathways (Benjamini-corrected p values < 0.05) in the PPI network.
(E) Boxplot representation of the variability in the expression levels of the over-represented TFs between CNS and PNS injuries (see Figures S1C and S1D). Time series data after CNS or PNS injury (see Figure S2) were used to create distance matrix using Euclidean distance measure to create the boxplot. Non-parametric Kruskal-Wallis test was used to compare differences between CNS and PNS injury datasets.
The presence of TFBS enrichment within genes representing these seven signaling pathways (Figure 4D) suggested that the coordinate regulation of core regeneration-associated hub TFs could provide the key regulatory cross-talk connecting these distinct pathways. To test this prediction, we removed these 19 TFs from the PPI network (Figure 4B), and we examined the resulting network mean path length, which is a measure of the connectivity of the remaining protein interactions in the absence of these TFs. We observed a drastic and significant reduction in protein connectivity, essentially causing nearly complete module disintegration (from 0.10 to 0.04 average clustering coefficient, p = 1.2 × 10−4; Figures 4B and 4C). However, random removal of a similar number of nodes from the PPI network did not cause module disintegration (Figure 4C). This illustrates that these TFs provide critical cross-talk that coordinately links the core signaling pathways responsible for neuronal regeneration (Figures 4A and 4D).
Based on these observations, we reasoned that, rather than each of these pathways acting independently to facilitate regeneration, coordinate regulation of these regeneration-associated pathways is necessary for effective regeneration to occur after injury and that the identified hub TFs contribute to connecting these signaling pathways. To test this, we examined the gene expression levels of these over-represented TFs in independent published PNS and CNS spinal cord injury profiling data (Di Giovanni et al., 2003; Ryge et al., 2010; Table S4). Regardless of the model, these TFs were significantly co-expressed and upregulated after PNS injury (Figure 4E; Figure S2). In contrast, in five independent CNS injury datasets (spinal cord injury: mild, moderate, severe, and complete transection), the levels of these TFs were significantly variable or downregulated (Figure 4E) (non-parameteric Kruskal-Wallis p < 1.1 × 10−5; Figure S2). These data provide strong independent evidence that the coordinate regulation of all these TFs is associated with or related to nerve outgrowth after injury, as their coordinate upregulation is observed only in the PNS, and not in the CNS. This is also supported by the data shown in Figure 3, that two hub TFs present in the core regeneration-associated gene network, ATF3 and JUN, show cooperative (partially additive) effects when simultaneously over-expressed (increasing axonal outgrowth in mouse DRG neurons), when compared to either over-expressed individually (Figures 3D–3F).
In many cases, TF regulation of gene expression is often activated in a cooperative way by direct physical contact between two or more TFs forming transcriptional complexes (Ravasi et al., 2010). We next asked whether the over-represented TFs are known to physically interact, identifying experimentally validated interactions between several TFs, including ATF3, JUN, STAT3, and SP1 (Kiryu-Seo et al., 2008; Experimental Procedures; Figure S3). The TF SP1, when bound to the promoter region, recruits ATF3, JUN, and STAT3 and physically interacts with them to regulate gene expression in a synergistic fashion (Kiryu-Seo et al., 2008). Minimal regenerative effect after individual overexpression of these TFs in isolation has been observed previously (ATF3, Seijffers et al., 2007; STAT3, Bareyre et al., 2011). Hence, targeting the neuronal growth state network, rather than a single gene, may be more effective to mobilize a more complete range of signaling pathways necessary for recovery after injury, consistent with the cooperative effects that we observe with concurrent ATF3 and JUN over-expression.
Regulation of Neurite Outgrowth in DRG Neurons Using Small Molecules
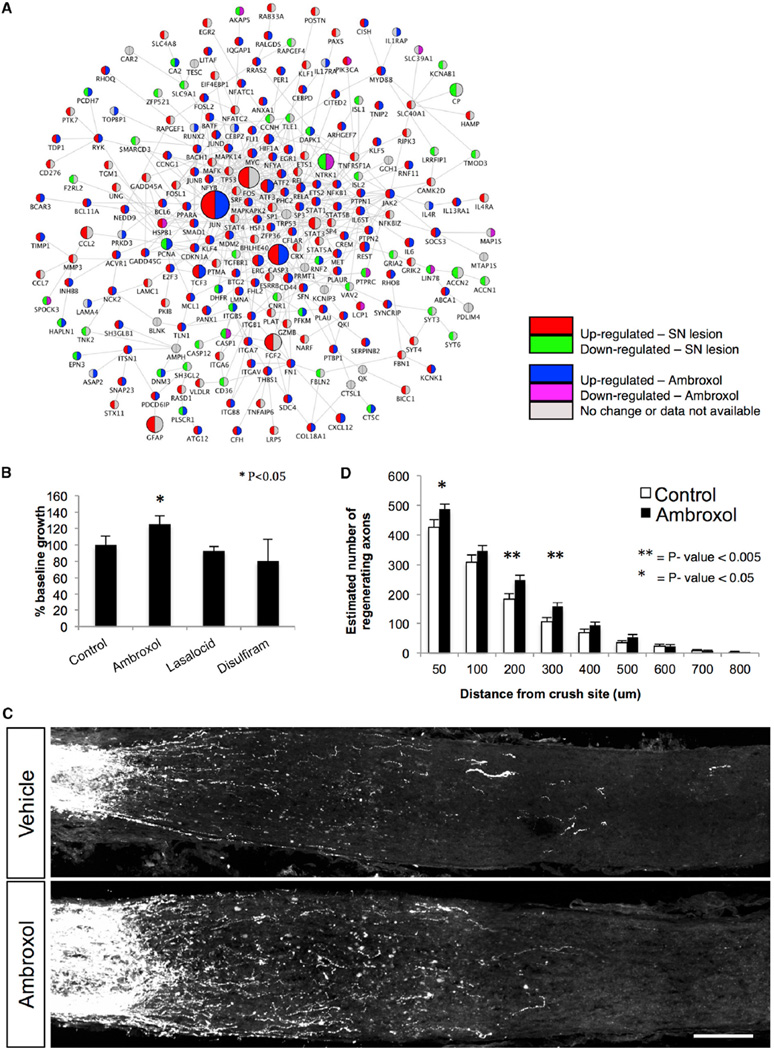
We reasoned that, if we could identify a small molecule whose effect on gene expression in injured neurons closely approximated the core signaling network associated with regeneration in the PNS, we could formally test the network biology prediction that such a compound should promote neurite outgrowth. In doing so, we extend this work to the level of initial screen for the discovery of therapeutics and provide further support for our strategy. We utilized the gene expression levels from the identified candidate up- and downregulated genes in the core 280-gene PPI network (M280; Figure 5A; Experimental Procedures) as a signature with which to query a publicly available database of drug-related expression profiles derived from non-neuronal cell lines known as the Connectivity Map (Lamb et al., 2006). We chose the top three matching expression patterns based on the connectivity and specificity score (Lamb et al., 2006), identifying the drugs ambroxol, lasalocid, and disulfiram (Experimental Procedures) for further analysis. Only ambroxol showed significant enhancement of axonal outgrowth in DRG neurons (Figure 5B).
Figure 5. Targeting Candidate RAG Regulatory Network Using Small Molecules.
Gene expression signatures after PNS injury were used to query drug-related expression profiles in the Connectivity Map. Using a pattern-matching algorithm, we selected three drugs (ambroxol, disulfiram, and lasalocid) based on enrichment and specificity scores.
(A) PPI (edges) network of co-expressed and differentially expressed genes (nodes) after PNS injury is shown. Upregulation (red) and downregulation (green) after SN lesion; upregulation (blue) and downregulation (purple) after ambroxol treatment (from Connectivity Map).
(B) Differences in DRG neurite outgrowth after treatment with drugs. Ambroxol elicited more neuronal growth than control (p < 0.05, t test).
(C and D) Ambroxol promotes retinal ganglion cell axonal regeneration. Ambroxol (Amb 25 mg/ml) or vehicle (Veh) was injected into the eye just before ON crush. Animals received daily 120 ml ambroxol (25 mg/ml) or vehicle by intraperitoneal (i.p.) injection from day 1 to day 14. At day 7 ambroxol (25 mg/ml) or vehicle was injected into the eye. Tracer CTB was injected into the eye on day 11 and animals were sacrificed on day 14. (C) Representative confocal images show ON sections from WT animals treated with vehicle (n = 10) and WT animals treated with ambroxol (25 mg/ml, n = 13). Axons are labeled with CTB. Scale bar, 100 mm. Measurements were made blinded to treatment. (D) Quantification of number of axons in (C) is shown (t test, **p < 0.001 and *p < 0.05).
Next, since the original pattern was derived from non-neuronal cell lines (Lamb et al., 2006),we tested if ambroxol regulates target marker genes in DRG neurons, observing that ambroxol-treated DRG neurons showed significant differential expression of eight genes (p value < 0.05) among 14 tested genes from the M280 network (Figure S4). Interestingly, this included five of the core hub TFs (ATF3, FOS, JUN, SMAD1, and SP1) in the M280 network (Figure 5A; Figure S4). Ambroxol is a potent blocker of neuronal Na+ and Ca2+ channels and is used clinically as a mucolytic (Weiser, 2008). In animal models, ambroxol effectively suppresses symptoms of peripheral and central neuropathic pain (Gaida et al., 2005); but, the relationship of ambroxol to peripheral or central regeneration has not been described or predicted previously.
Ambroxol Enhances CNS Regeneration In Vivo
Another prediction from our network analyses is that an appropriate co-regulation of the core regeneration-associated module, M280, which does not normally occur in CNS injury, might enable CNS regeneration. Since ambroxol recapitulates many of the core expression changes in the M280, we reasoned that it could potentially promote CNS regeneration. Optic nerve (ON) regeneration is a standard model for CNS regeneration (Sun et al., 2011), so we examined ON regeneration in C57BL/6 mice after a crush injury following treatment with ambroxol (Experimental Procedures). We observed a limited, but significant increase in axon regeneration beyond the site of the lesion (>1.5-fold increase between 200 and 500 µm; p < 0.04) after 2 weeks in animals treated with ambroxol compared with control animals (Figures 5C and 5D; Figure S5A), confirming the predictive properties of the approach.
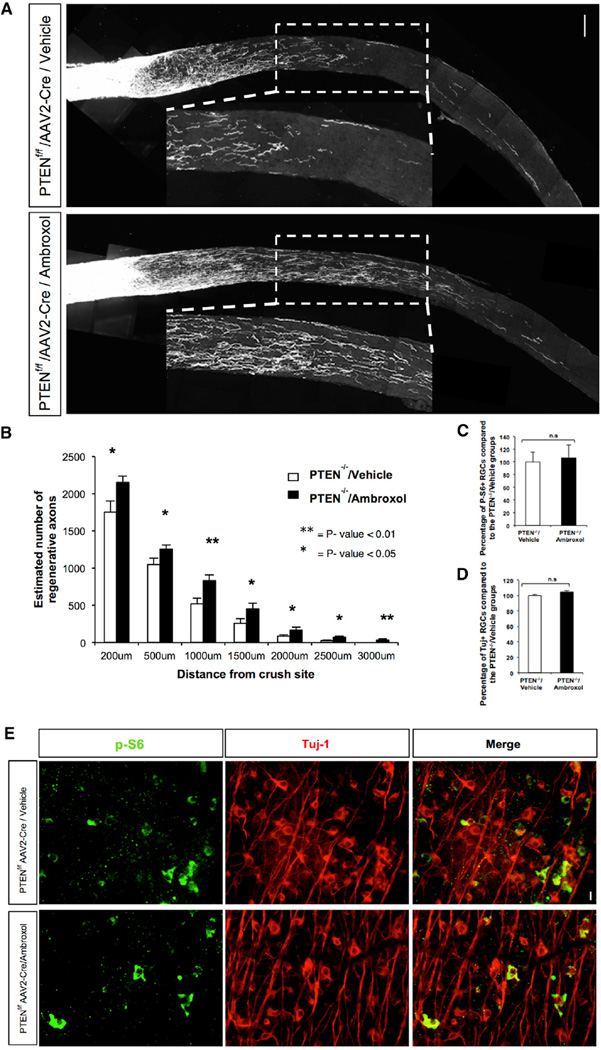
Next we compared ambroxol to one of the few proven genetic approaches to determine whether it would have a similar additive effect of enabling axonal regeneration after injury, on top of a genetic manipulation already shown to do so (Sun et al., 2011). For this, we examined the combinatorial activity of ambroxol treatment in PTEN homozygous knockout mice, which have significantly improved axonal regeneration after ON crush relative to wild-type (WT) animals (Park et al., 2008), which is further enhanced by double PTEN/SOCS knockout (Sun et al., 2011). We observed enhanced regeneration beyond the site of the lesion (>2.9-fold increase at 2,500 µm; p value 0.02) after 2 weeks in PTEN−/− (PTENf/f) animals treated with ambroxol compared with PTEN−/− animals treated with vehicle (Figure 6; Figure S5B), confirming that ambroxol administration in vivo enhances ON regeneration combined with a known regeneration-promoting genetic manipulation.
Figure 6. Ambroxol Promotes Retinal Ganglion Cell Axonal Regeneration in PTEN Knockout Mice.
Ambroxol (Amb 25 mg/ml) or vehicle was injected into the eye just before ON crush. Animals received daily 300 mg/kg ambroxol or vehicle by i.p. injection for the first 5 days after the crush and then they received 150 mg/kg until day 14. At day 7 ambroxol (25 mg/ml) or vehicle was injected into the eye. Tracer CTB was injected into the eye on day 11 and animals were sacrificed on day 14.
(A) Representative confocal images of ON sections from PTEN−/− animals treated with vehicle (n = 4) and PTEN−/− animals treated with ambroxol (n = 4). Axons are labeled with CTB. Scale bar, 100 µm. Measurements were made blinded to treatment.
(B) Quantification of number of axons in (A) is shown (t test, **p < 0.01 and *p < 0.05).
(C and D) Quantifications of retinal ganglion cell (RGC) survival measured by Tuj1 and P-S6 antibody staining are shown.
(E) Representative retina whole-mount images with Tuj1 and P-S6 antibody staining 2 weeks post injury are shown. Scale bar, 25 µm.
DISCUSSION
The limited regenerative potential of CNS neurons is due to both cell-extrinsic and cell-intrinsic factors (Afshari et al., 2009; Filbin, 2003; Giger et al., 2010; Yiu and He, 2006). Several decades of work devoted to identifying cell-intrinsic factors has resulted in dozens of individual RAGs, including cytoskeletal proteins, cell adhesion and axon guidance molecules, TFs, trophic factors, and their receptors (Costigan et al., 2002; Giger et al., 2010; Sun and He, 2010). Individual manipulation of these RAGs causes, though, only limited regeneration (Ferreira et al., 2012), while combinatorial reductions in cell-extrinsic factors Nogo, MAG, and OMgp have yielded inconclusive results (Cafferty et al., 2010; Lee et al., 2010).
We hypothesized that changing the growth state of differentiated adult CNS neurons by using drugs that activate relevant signaling pathways, TFs, and thereby regeneration effector genes would allow the neurons to re-grow without changing their molecular identity, making it possible to activate and enhance CNS regeneration. To achieve this, we applied a comprehensive systems genomics approach, starting with gene expression profiling combined with multi-level bioinformatic analyses and then experimental validation of network predictions, to provide a framework for understanding the precise molecular pathways involved in nerve regrowth after injury. We provide multiple lines of experimental support for the predictions of the regeneration network that we make, and we further apply this rubric for pathway discovery to identify a drug that accelerates nerve outgrowth after CNS injury, albeit modestly. This work provides a key proof of principle for the power of such systems genomic approaches in tackling complex problems in nervous system biology.
Our data describe that the injury-induced regulation of neuronal transcription in sensory neurons in the PNS includes a specific network of genes that are associated by coordinate action with the capacity for autonomous regeneration. The network includes many previously identified RAGs and related pathways, as well as many novel RAGs and the structure of the regulatory network in which they operate, building upon decades of research in this field. We speculate that failure of induction of this network after CNS injury at least partially explains why regeneration in CNS injury does not occur.
In support of this view, we show that ambroxol, a compound that increases expression of elements of the PNS coordinate transcriptional network, can promote CNS regeneration. We acknowledge that the effects of ambroxol are modest compared with combinatorial genetic manipulations (Sun et al., 2011), which is perhaps not unexpected given the paucity of drugs that enhance regeneration to any extent. We view this drug-screening component as a proof of principle that should be further refined in future experiments but that demonstrates the potential of coordinate regulation of the identified core network. Understanding the mechanism of ambroxol activity in this regard will also aid in the further development of mechanistic-based treatments. We also emphasize that the identified core network can be used as tool for screening additional drugs that may enhance neuronal regeneration. Moreover, the additive activity of ambroxol in combination with PTEN reduction provides support for the notion that combinatorial activation of RAGs and their pathways enhances CNS regeneration.
Combining approaches that activate the intrinsic regeneration network with suppression of inhibitory extrinsic cues will almost certainly further optimize recovery. An obvious conceptual parallel that was deemed improbable prior to its demonstration was the overexpression of four TFs to change the state of a cell from fully differentiated to pluripotent (Takahashi and Yamanaka, 2006). Technical challenges to coordinate overexpression of multiple TFs in vivo in the CNS exist. But, our network and bioinformatic analyses lead to the expectation that coordinate overexpression of the core set of TFs identified here will enhance both peripheral and central regeneration. We predict that changing the transcriptional state of differentiated adult CNS neurons, either by coordinate expression of sets of TFs or by using drugs that activate the relevant downstream functional signaling pathways, represents a worthwhile strategy, enhancing the possibility of creating CNS regenerative capacity after injury.
EXPERIMENTAL PROCEDURES
Nerve Injury Expression Data
In total, 382 gene expression data from microarray experiments related to nerve injury were utilized in this study; preprocessing and normalization procedures were calculated according to the R functions as described in the Supplemental Experimental Procedures (see Table S4 for data-set details).
Weighted Gene Co-expression Network Analysis
We used the R package WGCNA (Langfelder and Horvath, 2008) to construct co-expression networks, as previously done (Oldham et al., 2006; Zhang and Horvath, 2005) and as described in detail in the Supplemental Experimental Procedures. To identify hub genes and modules shared across independent nerve injury datasets, we applied consensus network analysis (Langfelder and Horvath, 2007). GO and pathway enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) platform (https://david.ncifcrf.gov/; Huang da et al., 2009). All network plots were constructed using the Cytoscape software (Saito et al., 2012).
Overexpression of Novel Candidate RAGs
The 16 lentiviral open reading frame (ORF) expression clones (candidate RAGs) and control vector (pReceiver-Lv122) were purchased from GeneCopoeia. Viral stocks were used to transduce DRGs for 24 hr. Then 1 week after infection, the cells were fixed with 4% paraformaldehyde 24 hr after replating. Fixed cells were immunostained for anti-beta-tubulin (1:800, Sigma, RRID:AB_1844090). Neurite initiation and the longest neurite length of the cells expressing GFP and immunostained for beta-tubulin were quantified using NeuroMath and ImageJ. Data were obtained from at least three separate experiments.
Knockdown of Novel Candidate RAGs
Mission control plasmid containing either shRNA sequences to Fxyd5, Gfpt1, Smagp, Tacstd2, and Cdc42 were purchased from Sigma-Aldrich and shRNA control vector, containing a non-specific shRNA, was purchased from Open-Biosystems. Viral particles were produced as previously described (Sena-Esteves et al., 2004). Then 1 hr after plating the DRG neurons, 100 MOI shRNA was added to the DRG cultures and incubated for 24 hr. Then 1 week after infection, the cells were fixed with 4% paraformaldehyde 17 hr after replating. Fixed cells were immunostained for anti-beta-tubulin (1:800, Sigma, RRID: AB_1844090). Neurite initiation and the longest neurite length of the cells expressing GFP and immunostained for beta-tubulin were quantified using NeuroMath. Data were obtained from at least three separate experiments repeated in quadruplicate. Measurements were made blinded to treatment (see the Supplemental Experimental Procedures for further details).
TFBS Enrichment
TFBS enrichment analysis was performed by scanning the canonical promoter region (1,000 bp upstream of the transcription start site) for the genes (kME > 0.5) present in the regeneration-associated co-expression modules. We utilized TFBS position weight matrices from JASPAR and TRANSFAC databases (Matys et al., 2003; Portales-Casamar et al., 2010) to examine the enrichment using Clover algorithm (Frith et al., 2004; see the Supplemental Experimental Procedures for further details). We also integrated the existing ChIP data for TFs from either the Encyclopedia of DNA Elements (ENCODE, Landt et al., 2012) or other compiled genome-wide ChIP data (Lachmann et al., 2010) to generate the transcriptional network (Figure 3A).
PPI Network Analyses
We constructed an experimentally validated PPI network using all the regeneration-associated co-expression gene network modules (two upregulated modules after nerve injury and three downregulated modules after injury). We created all possible combinations of gene pairs present in these co-expression networks and identified all experimentally verified interaction data (in human, mouse, or rat dataset) for their corresponding proteins in the STRING database (Franceschini et al., 2013), constructing the protein network by force-directed layout (Figure 4A).
In Silico Small Molecule Screening
The Connectivity Map (cmap) database was used for screening and the database details have been described previously (Lamb et al., 2006). In this method, the similarity between the query signature (RAG signature) and more than 7,000 expression profiles for 1,309 compounds (reference signatures) in the cmap database was evaluated (Lamb et al., 2006). Enrichments of both the up- and downregulated nerve injury-induced genes in the profiles of each treatment instance were estimated as described previously (Lamb et al., 2006) and as described in detail in the Supplemental Experimental Procedures. Permutated results were used to evaluate the significance of the scores and to rank the molecules in order to select the top molecules for experimental validation (see the Supplemental Experimental Procedures). This methodology was applied for two query signatures related to nerve injury as follows: (1) PPI network consisting of 280 genes (Figure 4A), and (2) regeneration-associated co-expression modules (Figure 1; Table S2). The top three enriched small molecules intersecting in both the signature lists were utilized for experimental validation.
Neurite Outgrowth Assay in Primary Adult DRG Neuron Culture
Adult C57BL/6J dissociated DRG neurons were plated in a tissue culture eight-well chamber slide dish (Nalge Nunc) coated with poly-D-lysine and laminin (Sigma) cultured in Neurobasal medium (Invitrogen) supplemented with B27 supplement, penicillin, streptomycin, 1 mM L-glutamine, 50 ng/ml NGF, 2 ng/ml GDNF, and 10 mM AraC at 37°C. For drug treatment, DRG neurons were cultured for 24 hr in the presence of 40 and 60 µM concentrations of drugs. The cells were fixed with 4% paraformaldehyde. Fixed cells were immunostained for b-III-tubulin (RRID: AB_2313773). Neurite initiation and the longest neurite length of the cells were quantified using NeuroMath. Data were obtained from at least three separate experiments repeated in quadruplicate.
Mice Surgery, Drug Administration, and ON Injury
All experimental procedures were performed in compliance with animal protocols approved by Institutional Animal Care and Use Committee (IACUC) at Boston Children’s Hospital. ONs from 6-week-old C57BL/6 mice were crushed as described previously (Park et al., 2008) just after intravitreal injection of 1 µl ambroxol (Sigma, 25 mg/ml in 5% Tween 80%-5% Polyethylen glycol 400 in water) or vehicle (5% Tween 80%-5% PEG 400 in water). A second intravitreal injection of ambroxol or vehicle was performed 7 days post-ON crush. Daily mice received 120 µl (25 mg/ml) ambroxol or vehicle intraperitoneally, i.e., from the day after the ON crush to the day prior to termination. For PTEN−/− animal experiments, P21 PTEN floxed mice received AAV2-Cre intraorbital injection. Then 2 weeks later we performed ON crush and the first ambroxol injection within the eye. We injected (intraperitoneally) ambroxol at 300 mg/kg for the first 5 days after the crush and then we injected 150 mg/kg. Mice were injected twice a day every day. Regenerating axons were traced by intravitreal injection of 1 µl CTB-Alexa-488 (1 µg/µl in PBS, Invitrogen) 2 days before termination.
Supplementary Material
Highlights.
We identify a transcriptional program observed after PNS, but not CNS injury
This program links known signaling pathways via a core set of transcription factors
We experimentally and bio-informatically validate several network predictions
We use the core transcriptional profile to identify a drug that promotes regeneration
Acknowledgments
We gratefully acknowledge support from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691) (G.C.), NIH R01 NS038253 (C.J.W.) and NS074430 (M.C.), and the Bertarelli Foundation (C.J.W.).
Footnotes
ACCESSION NUMBERS
The accession numbers for the data reported in this paper are GEO: GSE30691, GSE26350, GSE2884, GSE33175, GSE21007, GDS63, and GSE19701. The network datasets generated and analyzed in this study also are available at https://coppolalab.ucla.edu/gclabapps/nb/browser?id=Consensus_Tuszynski;ver=vijay and https://coppolalab.ucla.edu/gclabapps/nb/browser?id=Consensus_Fainzilber;ver=vijay.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.neuron.2016.01.034.
AUTHOR CONTRIBUTIONS
V.C., G.C., and D.H.G. designed the experiments and V.C., C.J.W., and D.H.G. wrote the manuscript. G.C. and V.C. performed network analyses on the two initial PNS datasets, and V.C. performed all subsequent bioinformatic analyses with input from D.H.G. V.C. conducted or coordinated the in vitro validation experiments via gene overexpression. T.O. performed the in vitro knockdown experiments. E.A.H. performed the transcription factor co-expression experiments. H.N. performed the ON crush experiments with supervision by Z.H. R.V., A.Y., and L. Barrett provided technical assistance with experiments. G.C., F.G., J.D.-T., P.L., S.H., and A.Z. also contributed to aspects of the data analyses. M.T., M.F., C.J.W., M.C., A.B., I.M., and L. Benowitz provided microarray data utilized in this study. All authors discussed the results and provided comments and revisions on the manuscript.
REFERENCES
- Abe N, Cavalli V. Nerve injury signaling. Curr. Opin. Neurobiol. 2008;18:276–283. doi: 10.1016/j.conb.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshari FT, Kappagantula S, Fawcett JW. Extrinsic and intrinsic factors controlling axonal regeneration after spinal cord injury. Expert Rev. Mol. Med. 2009;11:e37. doi: 10.1017/S1462399409001288. [DOI] [PubMed] [Google Scholar]
- Bareyre FM, Garzorz N, Lang C, Misgeld T, Büning H, Kerschensteiner M. In vivo imaging reveals a phase-specific role of STAT3 during central and peripheral nervous system axon regeneration. Proc. Natl. Acad. Sci. USA. 2011;108:6282–6287. doi: 10.1073/pnas.1015239108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs S, Salter MW. Neuropathic pain: symptoms, models, and mechanisms. Drug Dev. Res. 2006;67:289–301. [Google Scholar]
- Bomze HM, Bulsara KR, Iskandar BJ, Caroni P, Skene JH. Spinal axon regeneration evoked by replacing two growth cone proteins in adult neurons. Nat. Neurosci. 2001;4:38–43. doi: 10.1038/82881. [DOI] [PubMed] [Google Scholar]
- Cafferty WB, Duffy P, Huebner E, Strittmatter SM. MAG and OMgp synergize with Nogo-Atorestrict axonal growth and neurological recovery after spinal cord trauma. J. Neurosci. 2010;30:6825–6837. doi: 10.1523/JNEUROSCI.6239-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Gao Y, Bryson JB, Hou J, Chaudhry N, Siddiq M, Martinez J, Spencer T, Carmel J, Hart RB, Filbin MT. The cytokine inter-leukin-6 is sufficient but not necessary to mimic the peripheral conditioning lesion effect on axonal growth. J. Neurosci. 2006;26:5565–5573. doi: 10.1523/JNEUROSCI.0815-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli V, Kujala P, Klumperman J, Goldstein LS. Sunday Driver links axonal transport to damage signaling. J. Cell Biol. 2005;168:775–787. doi: 10.1083/jcb.200410136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chierzi S, Ratto GM, Verma P, Fawcett JW. The ability of axons to regenerate their growth cones depends on axonal type and age, and is regulated by calcium, cAMP and ERK. Eur. J. Neurosci. 2005;21:2051–2062. doi: 10.1111/j.1460-9568.2005.04066.x. [DOI] [PubMed] [Google Scholar]
- Costigan M, Befort K, Karchewski L, Griffin RS, D’Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giovanni S, Knoblach SM, Brandoli C, Aden SA, Hoffman EP, Faden AI. Gene profiling in spinal cord injury shows role of cell cycle in neuronal death. Ann. Neurol. 2003;53:454–468. doi: 10.1002/ana.10472. [DOI] [PubMed] [Google Scholar]
- Dragunow M, Xu R, Walton M, Woodgate A, Lawlor P, MacGibbon GA, Young D, Gibbons H, Lipski J, Muravlev A, et al. c-Jun promotes neurite outgrowth and survival in PC12 cells. Brain Res. Mol. Brain Res. 2000;83:20–33. doi: 10.1016/s0169-328x(00)00191-1. [DOI] [PubMed] [Google Scholar]
- Ferreira LM, Floriddia EM, Quadrato G, Di Giovanni S. Neural regeneration: lessons from regenerating and non-regenerating systems. Mol. Neurobiol. 2012;46:227–241. doi: 10.1007/s12035-012-8290-9. [DOI] [PubMed] [Google Scholar]
- Filbin MT. Myelin-associated inhibitors of axonal regeneration in the adult mammalian CNS. Nat. Rev. Neurosci. 2003;4:703–713. doi: 10.1038/nrn1195. [DOI] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaida W, Klinder K, Arndt K, Weiser T. Ambroxol, a Nav1.8-preferring Na(+) channel blocker, effectively suppresses pain symptoms in animal models of chronic, neuropathic and inflammatory pain. Neuropharmacology. 2005;49:1220–1227. doi: 10.1016/j.neuropharm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Konopka G. Neuroscience in the era of functional genomics and systems biology. Nature. 2009;461:908–915. doi: 10.1038/nature08537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giger RJ, Hollis ER, 2nd, Tuszynski MH. Guidance molecules in axon regeneration. Cold Spring Harb. Perspect. Biol. 2010;2:a001867. doi: 10.1101/cshperspect.a001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Hoffman PN. A conditioning lesion induces changes in gene expression and axonal transport that enhance regeneration by increasing the intrinsic growth state of axons. Exp. Neurol. 2010;223:11–18. doi: 10.1016/j.expneurol.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kiryu-Seo S, Kato R, Ogawa T, Nakagomi S, Nagata K, Kiyama H. Neuronal injury-inducible gene is synergistically regulated by ATF3, c-Jun, and STAT3 through the interaction with Sp1 in damaged neurons. J. Biol. Chem. 2008;283:6988–6996. doi: 10.1074/jbc.M707514200. [DOI] [PubMed] [Google Scholar]
- Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Geoffroy CG, Chan AF, Tolentino KE, Crawford MJ, Leal MA, Kang B, Zheng B. Assessing spinal axon regeneration and sprouting in Nogo-, MAG-, and OMgp-deficient mice. Neuron. 2010;66:663–670. doi: 10.1016/j.neuron.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xue C, Yuan Y, Zhang R, Wang Y, Wang Y, Yu B, Liu J, Ding F, Yang Y, Gu X. The transcriptional landscape of dorsal root ganglia after sciatic nerve transection. Sci. Rep. 2015;5:16888. doi: 10.1038/srep16888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindwall C, Kanje M. Retrograde axonal transport of JNK signaling molecules influence injury induced nuclear changes in p-c-Jun and ATF3 in adult rat sensory neurons. Mol. Cell. Neurosci. 2005;29:269–282. doi: 10.1016/j.mcn.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gössling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelevski I, Segal-Ruder Y, Rozenbaum M, Medzihradszky KF, Shalem O, Coppola G, Horn-Saban S, Ben-Yaakov K, Dagan SY, Rishal I, et al. Signaling to transcription networks in the neuronal retrograde injury response. Sci. Signal. 2010;3:ra53. doi: 10.1126/scisignal.2000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S, Woolf CJ. Regeneration of dorsal column fibers into and beyond the lesion site following adult spinal cord injury. Neuron. 1999;23:83–91. doi: 10.1016/s0896-6273(00)80755-2. [DOI] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc. Natl. Acad. Sci. USA. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Gandal MJ, Geschwind DH. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat. Rev. Genet. 2015;16:441–458. doi: 10.1038/nrg3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, He Z. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45:715–726. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cai D, Dai H, McAtee M, Hoffman PN, Bregman BS, Filbin MT. Spinal axon regeneration induced by elevation of cyclic AMP. Neuron. 2002;34:895–903. doi: 10.1016/s0896-6273(02)00730-4. [DOI] [PubMed] [Google Scholar]
- Ramnó y Cajal S, DeFelipe J, Jones EG. Cajal’s Degeneration and Regeneration of the Nervous System. Oxford University Press; 1991. [Google Scholar]
- Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryge J, Winther O, Wienecke J, Sandelin A, Westerdahl AC, Hultborn H, Kiehn O. Transcriptional regulation of gene expression clusters in motor neurons following spinal cord injury. BMC Genomics. 2010;11:365. doi: 10.1186/1471-2164-11-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat. Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer U, Gunnersen J, Karch C, Wiese S, Holtmann B, Takeda K, Akira S, Sendtner M. Conditional gene ablation of Stat3 reveals differential signaling requirements for survival of motoneurons during development and after nerve injury in the adult. J. Cell Biol. 2002;156:287–297. doi: 10.1083/jcb.200107009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seijffers R, Allchorne AJ, Woolf CJ. The transcription factor ATF-3 promotes neurite outgrowth. Mol. Cell Neurosci. 2006;32:143–154. doi: 10.1016/j.mcn.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Seijffers R, Mills CD, Woolf CJ. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J. Neurosci. 2007;27:7911–7920. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena-Esteves M, Tebbets JC, Steffens S, Crombleholme T, Flake AW. Optimized large-scale production of high titer lentivirus vector pseudotypes. J. Virol. Methods. 2004;122:131–139. doi: 10.1016/j.jviromet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Sendtner M, Stöckli KA, Thoenen H. Synthesis and localization of ciliary neurotrophic factor in the sciatic nerve of the adult rat after lesion and during regeneration. J. Cell Biol. 1992;118:139–148. doi: 10.1083/jcb.118.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Ming GL. Roles of channels and receptors in the growth cone during PNS axonal regeneration. Exp. Neurol. 2010;223:38–44. doi: 10.1016/j.expneurol.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subang MC, Richardson PM. Synthesis of leukemia inhibitory factor in injured peripheral nerves and their cells. Brain Res. 2001;900:329–331. doi: 10.1016/s0006-8993(01)02323-x. [DOI] [PubMed] [Google Scholar]
- Sun F, He Z. Neuronal intrinsic barriers for axon regeneration in the adult CNS. Curr. Opin. Neurobiol. 2010;20:510–518. doi: 10.1016/j.conb.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Park KK, Belin S, Wang D, Lu T, Chen G, Zhang K, Yeung C, Feng G, Yankner BA, He Z. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011;480:372–375. doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu. Rev. Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Weiser T. Ambroxol: a CNS drug? CNS Neurosci. Ther. 2008;14:17–24. doi: 10.1111/j.1527-3458.2007.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang EK, Takimoto K, Hayashi Y, de Groat WC, Yoshimura N. Altered expression of potassium channel subunit mRNA and alpha-dendrotoxin sensitivity of potassium currents in rat dorsal root ganglion neurons after axotomy. Neuroscience. 2004;123:867–874. doi: 10.1016/j.neuroscience.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Yiu G, He Z. Glial inhibition of CNS axon regeneration. Nat. Rev. Neurosci. 2006;7:617–627. doi: 10.1038/nrn1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article 17. [DOI] [PubMed] [Google Scholar]
- Zhao X, Tang Z, Zhang H, Atianjoh FE, Zhao JY, Liang L, Wang W, Guan X, Kao SC, Tiwari V, et al. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat. Neurosci. 2013;16:1024–1031. doi: 10.1038/nn.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.