Abstract
OBJECTIVE
To determine whether the molecular epidemiological characteristics of methicillin-resistant Staphylococcus aureus (MRSA) had changed in a level III neonatal intensive care unit (NICU).
DESIGN
Retrospective review of medical records.
SETTING
Level III NICU of a university-affiliated children’s hospital in New York, New York.
PATIENTS
Case patients were neonates hospitalized in the NICU who were colonized or infected with MRSA.
METHODS
Rates of colonization and infection with MRSA during the period from 2000 through 2008 were assessed. Staphylococcal chromosomal cassette (SCC) mecA analysis and genotyping for S. aureus encoding protein A (spa) were performed on representative MRSA isolates from each clonal pulsed-field gel electrophoresis pattern.
RESULTS
Endemic MRSA infection and colonization occurred throughout the study period, which was punctuated by 4 epidemiologic investigations during outbreak periods. During the study period, 93 neonates were infected and 167 were colonized with MRSA. Surveillance cultures were performed for 1,336 neonates during outbreak investigations, and 115 (8.6%) neonates had MRSA-positive culture results. During 2001–2004, healthcare-associated MRSA clones, carrying SCC mec type II, predominated. From 2005 on, most MRSA clones were community-associated MRSA with SCC mec type IV, and in 2007, USA300 emerged as the principal clone.
CONCLUSIONS
Molecular analysis demonstrated a shift from healthcare-associated MRSA (2001–2004) to community-associated MRSA (2005–2008).
Rates of healthcare-associated infection among newborns hospitalized in the neonatal intensive care unit (NICU) are in the range 15%–20%.1,2 Staphylococcus aureus causes approximately 7.8% of episodes of late-onset sepsis in very low birth weight neonates (birth weight less than 1,500 g).3 However, despite the increasing prevalence of serious infections due to methicillin-resistant S. aureus (MRSA) in neonates,4 there are few data regarding endemic rates of MRSA in the NICU. Most published reports have focused on MRSA outbreak investigations in this setting,5–7 although we have recently described endemic and epidemic rates of methicillin-susceptible S. aureus and MRSA in our NICU.8
Community-associated MRSA (CA-MRSA) genotypes and phenotypes are now emerging as important causes of infection in people without traditional risk factors for MRSA infections.9,10 Recent reports of vaginal carriage of CA-MRSA in pregnant women11,12 and patient-to-patient transmission of CA-MRSA resulting in postpartum infections13 led us to hypothesize that community-associated clones, rather than healthcare-associated clones, would increasingly cause MRSA infections in the NICU. Thus, we evaluated the molecular epidemiological characteristics of the MRSA strains isolated from neonates colonized or infected with MRSA during the period from 2000 through 2008.
METHODS
Study Design and Study Site
A retrospective review was performed of medical records of patients discharged during the period from 2000 through 2008 to determine the epidemiological characteristics of infection and colonization with MRSA among newborns hospitalized in the level III NICU of a university-affiliated children’s hospital in New York, New York. In November 2003, construction was completed on a new NICU that expanded the number of beds from 45 to 62 and provided more space per newborn. Approximately 25%–30% of newborns were transferred from other institutions; approximately 40% of newborns were full term, many requiring surgical procedures for congenital anomalies; and by the end of the study period, approximately 1,100 newborns were discharged annually. The complexity of the patients’ conditions also increased during the study period, as documented by the increase in case mix index. The percentage of newborns who were transferred from other institutions decreased slightly from 30% to 25%, likely as a result of the development of a more efficient maternal transport system. The proportion of full-term newborns remained stable during the study period. The study was approved by the institutional review board of Columbia University, which granted a waiver of documentation of informed consent.
Molecular Studies
Isolates underwent pulsed-field gel electrophoresis (PFGE) analysis with the restriction enzyme SmaI, using a contour-clamped homogeneous electric field system (CHEF Mapper XA System; Bio-Rad) as described elsewhere.14 The PFGE patterns were digitized and analyzed using the BioNumerics software (Applied Maths), and strain relatedness was interpreted in accordance with established criteria.15 PFGE clonal types were designated with letter codes according to nomenclature established by the Clinical Microbiology Laboratory’s Molecular Epidemiology Unit. PFGE patterns were compared with those of major circulating MRSA clonal types (eg, USA300) by using representative clinical and laboratory strains run in parallel.
Isolates representative of each PFGE pattern were characterized by additional genotypic methods. DNA sequencing of the variable number of tandem repeat regions in the gene of S. aureus encoding protein A (spa)16 was performed. Sequences were entered into a spa search engine to determine the spa type of each test strain. The presence of Panton-Valentine leukocidin (PVL)17 was determined with the use of real-time polymerase chain reaction (PCR) technology.18 Multiplex PCR analysis to distinguish the 5 major staphylococcal chromosomal cassette (SCC) mec types, including some prevalent subtypes, was performed using 2 complementary methods.19,20
Case Definitions for MRSA Infection and Colonization
The hospital’s computerized information system was used to generate a list of neonates hospitalized in the NICU with MRSA-positive culture results from January 1, 2000, through December 31, 2008. Positive results of surveillance cultures were considered to represent colonization. MRSA-positive results of culture samples obtained for clinical purposes by the NICU staff were designated as either infection or colonization on the basis of review of the electronic medical record. Cases were considered to involve infection if the record contained documentation of MRSA-positive culture results for samples from sterile body sites, treatment of patients who had MRSA-positive skin and soft-tissue culture results with parenteral or topical antibiotics, or treatment of patients who had MRSA-positive conjunctival culture results with antibiotic ophthalmic drops together with documentation of clinical findings consistent with conjunctivitis, as described elsewhere.21 MRSA isolated from respiratory specimens was considered to indicate an infection if there was documentation of tracheitis or pneumonia in the medical record, treatment with parenteral antibiotics, and no concomitant infection at another body site (n = 2). Patients who had MRSA-positive culture results but were not treated with antibiotics were considered to have MRSA colonization.
Microbiologic Processing
Procedures used for specimen collection and for isolation of MRSA from clinical and surveillance cultures were performed as described elsewhere.7,22 The following assays were used for species identification and determination of the antimicrobial susceptibility profiles of staphylococcal strains: the Staphaurex latex agglutination test (Remel), the MicroScan Walk-Away SI microtiter system (Siemens Healthcare Diagnostics), CHROMagar MRSA-selective differential chromogenic medium (Becton Dickinson Diagnostics), and the MRSA-Screen latex agglutination test (Denka Seiken) for penicillin-binding protein 2′. Isolates were characterized as susceptible, intermediate, or resistant to oxacillin as defined in accordance with established criteria.23 The D-test to detect clindamycin resistance was introduced in April 2004.
Infection Control Strategies
Neonates born in our facility are not routinely screened for MRSA. However, neonates transferred from other facilities to our NICU do undergo surveillance culture sampling to detect MRSA colonization, as described elsewhere,7 and remain under contact precautions until the results are known. During the period from 2000 through 2006, surveillance culture samples were obtained only from the anterior nares. During an outbreak investigation in 2007, there was ongoing transmission and acquisition of MRSA by neonates who previously had negative results on surveillance cultures of samples of the anterior nares. Therefore, to optimize recovery of MRSA, surveillance culturing was expanded to include samples obtained from the periumbilical area, axilla, and groin, as well as the anterior nares.
When an MRSA infection was discovered in a neonate in the NICU, surveillance cultures of other neonates were performed in a stepwise fashion. First, culture samples were obtained from neonates in close proximity to the index patient who shared caregivers (ie, the same 2–3-bed “pod”). If any of these neonates were colonized with MRSA, then surveillance cultures were performed for neonates in an expanded area around the index patient (ie, the 10–20-bed “wing”). If additional MRSA-colonized neonates were identified, a unitwide surveillance effort was undertaken. Such an event was considered to be an outbreak, which was defined as 3 or more cases of temporally related infection or colonization found to be caused by the same clone. Neonates infected or colonized with MRSA remained under contact precautions for the duration of hospitalization. Measures to decolonize neonates included applying a 1-cm ribbon of mupirocin ointment to the anterior nares 3 times per day for 7 days and bathing neonates who weighed more than 1,500 g with an antiseptic soap, most recently chlorhexidine, 3 times during the mupirocin treatment period.
If ongoing transmission of a dominant clone persisted despite these interventions, targeted surveillance cultures of samples from the nares of staff who cared for new incident cases were performed,7 and the hands of staff were examined for skin lesions. Colonized staff were furloughed, treated with mupirocin applied to the nares and chlorhexidine showers, and permitted to return to work when 2 negative results of cultures of samples of the anterior nares were documented. In addition to healthcare worker surveillance efforts, the following infection control strategies were implemented: intensified environmental cleaning twice daily of high-touch and horizontal surfaces in colonized and infected neonates’ bed space, use of dedicated equipment, cohorting of MRSA-infected and colonized neonates, and monitoring of hand hygiene compliance. To document the success of these interventions, surveillance cultures were performed for all neonates weekly until no new MRSA-colonized neonates were identified during 3 consecutive unitwide surveillance efforts.
RESULTS
Molecular Epidemiology
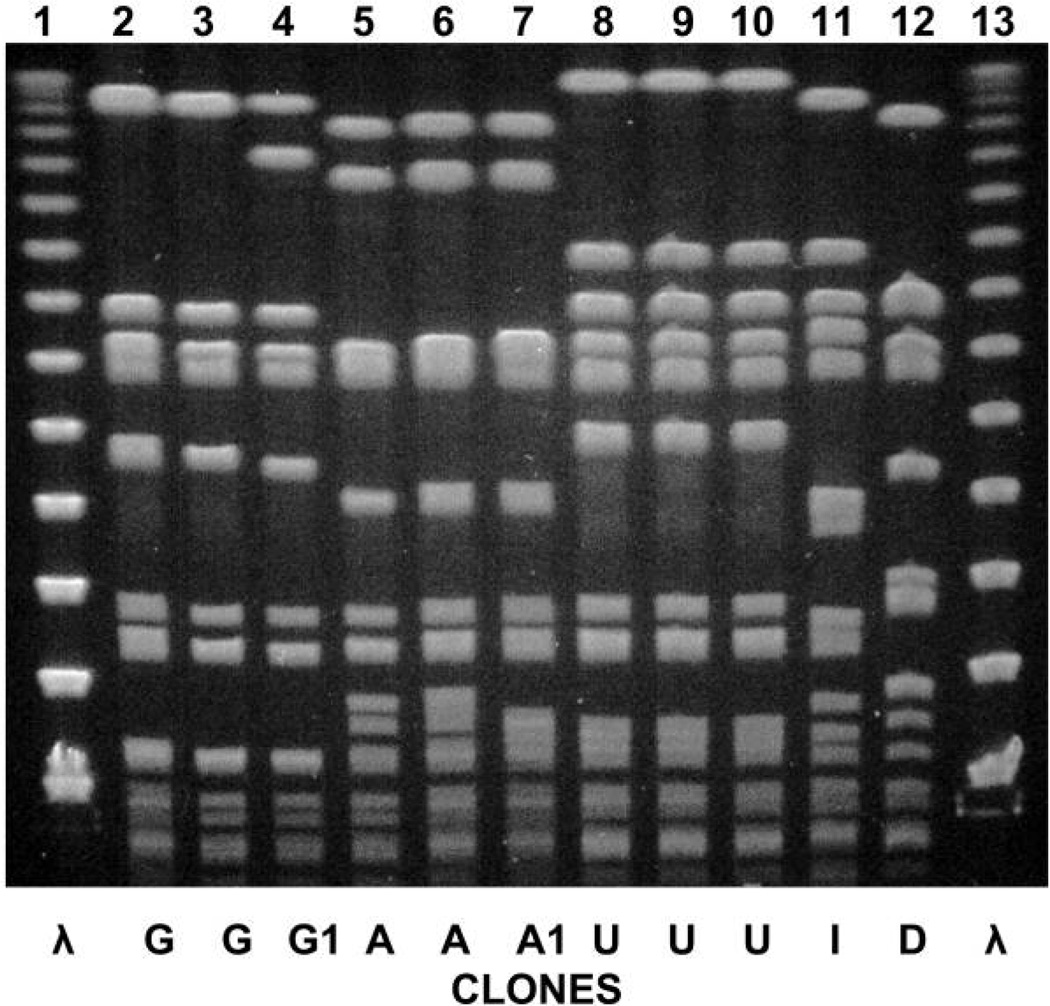
Overall, 149 (74%) of 201 MRSA-colonized or infected patients had an isolate available for molecular analysis (Table 1). During the period from 2001 through 2004, the healthcare-associated MRSA USA100 strain with SCC mec type II (PFGE-designated clone G) predominated, and clone G remained endemic in the NICU throughout the study period. In 2005, the predominant clone changed to CA-MRSA, SCC mec type IV (PFGE-designated clone A), which persisted in the NICU and caused endemic infections. In 2007, a new dominant CA-MRSA clone, USA300 with SCC mec type IV (PFGE-designated clone U) and PVL, appeared (Figure 1). Six additional sporadic MRSA clonal types, with various SCC mec elements and spa types, were detected in 14 patients, including the PVL-positive MW2/USA400 strain.
TABLE 1.
Molecular Epidemiological Characteristics of Methicillin-Resistant Staphylococcus aureus (MRSA) Strains in a Neonatal Intensive Care Unit, 2000–2008
| No. of MRSA isolates | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CA-MRSA | ||||||||||||
| No. of patients | HA-MRSA USA100 (clone G)a |
Clone Ab | USA300 (clone U)c | Sporadic clonesd | ||||||||
| Year | I | C | Total | Typede | I | C | I | C | I | C | I | C |
| 2000 | 5 | 1 | 6 | 0 | … | … | … | … | … | … | … | … |
| 2001 | 12 | 3 | 15 | 6 | 5 | 1 | … | … | … | … | … | … |
| 2002 | 21 | 11 | 32 | 26 | 17 | 7 | … | … | … | … | 2 | … |
| 2003 | 15 | 22 | 37 | 29 | 11 | 15 | … | … | … | … | … | 3 |
| 2004 | 7 | 3 | 10 | 7 | 5 | 1 | … | … | … | … | 1 | … |
| 2005 | 11 | 26 | 37 | 34 | … | 2 | 9 | 18 | … | 1 | 2 | 2 |
| 2006 | 6 | 11 | 17 | 13 | 3 | 5 | 1 | 1 | 1 | 1 | … | 1 |
| 2007 | 13 | 22 | 35 | 29 | 2 | … | … | 2 | 10 | 14 | … | 1 |
| 2008 | 3 | 9 | 12 | 5 | … | … | 1 | … | … | 1 | 1 | 2 |
| Total | 93 | 108 | 201 | 149 | 43 | 31 | 11 | 21 | 11 | 17 | 6 | 9 |
NOTE. C, colonization (without infection); CA-MRSA, community-associated MRSA; HA-MRSA, healthcare-associated MRSA; I, infection (with or without colonization); SCC, staphylococcal chromosomal cassette; PVL, Panton-Valentine leukocidin.
SCC mec type II, PVL negative, spa type 2.
SCC mec type IV, PVL negative, spa type 2.
SCC mec type IV, PVL positive, spa type 1.
Analysis for SCC mec type and presence of PVL was not completed for most of the sporadic clones.
Of the 149 neonates colonized or infected with MRSA who had an isolate available for molecular analysis, 134 neonates had 1 of the 3 dominant clones and 15 had strains of a sporadic MRSA clone.
FIGURE 1.
Pulsed-field gel electrophoresis patterns of each dominant clonal methicillin-resistant Staphylococcus aureus type isolated during the study period. Lane 1, molecular size ladder; lane 2, infected neonate 1; lane 3, colonized neonate 2; lane 4, colonized HCW 1; lane 5, infected neonate 3; lane 6, colonized neonate 4; lane 7, colonized HCW 2; lane 8, infected neonate 5; lane 9, colonized neonate 6; lane 10, colonized HCW 3; lane 11, infected neonate 7; lane 12, colonized neonate 8; lane 13, molecular size ladder.
Despite the different mec types, the 3 dominant clones had similar antimicrobial susceptibility patterns. All were susceptible to rifampin, clindamycin, tetracycline, and trimethoprim-sulbactam and resistant to erythromycin and levofloxacin. The USA300 clone was also resistant to gentamicin.
Infection and Colonization Rates
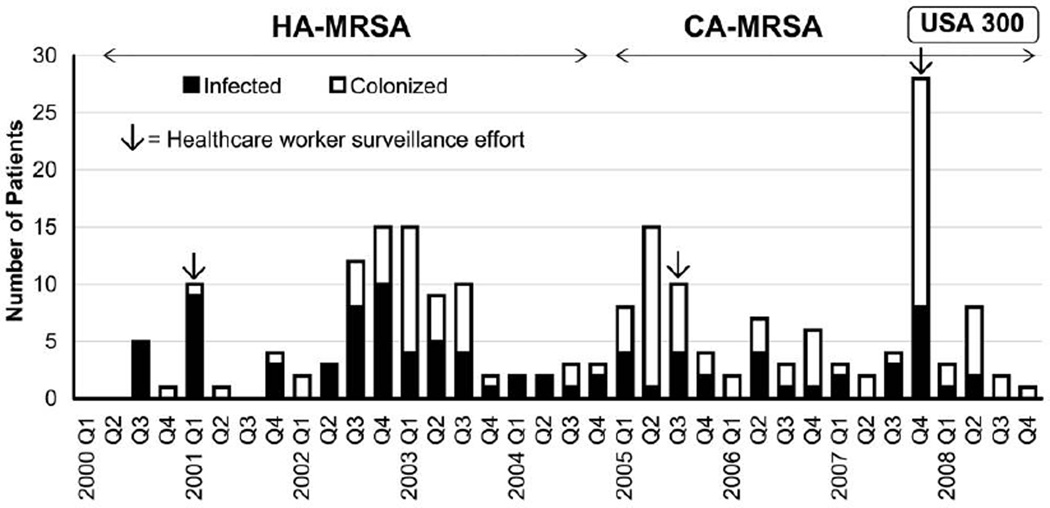
Endemic MRSA infection and colonization occurred throughout the study period (Figure 2), which was punctuated by 4 epidemiologic investigations during what were considered to be outbreak periods. These occurred during the first quarter of 2001, from the third quarter of 2002 through the third quarter of 2003, from the first quarter through the third quarter of 2005, and during the fourth quarter of 2007.
FIGURE 2.
Bar graph of number of neonatal intensive care unit patients infected or colonized with methicillin-resistant Staphylococcus aureus (MRSA) during each quarter of the study period. Three healthcare worker surveillance efforts are indicated by arrows. The periods when predominant MRSA clones were detected are shown. CA-MRSA, community-associated MRSA; HA-MRSA, healthcare-associated MRSA.
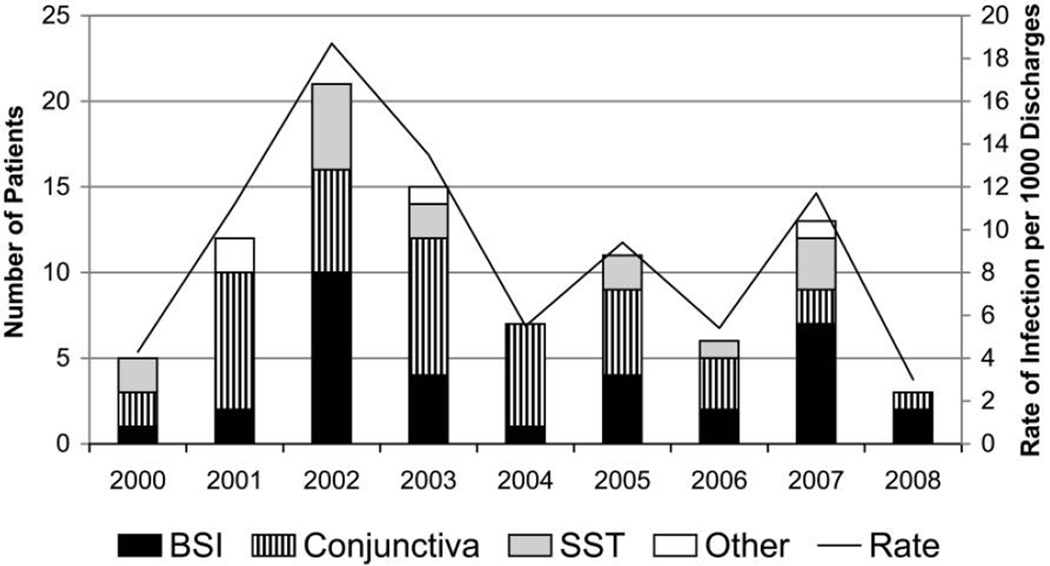
During 2000–2008, MRSA infections were diagnosed in 93 neonates (Figure 3). The most common sites of infection included the conjunctiva (41 [44%] patients), the bloodstream (33 [35%] patients), and skin and soft tissue (15 [16%] patients). The overall incidence of infection varied widely from year to year; the number of infections per 1,000 discharges ranged from 18 in 2002 to 2 in 2008. However, as a result of periodic outbreaks, there was no consistent trend of a change in incidence during the study period.
FIGURE 3.
Bar graph of number of neonates infected with methicillin-resistant Staphylococcus aureus (MRSA) (skin and soft-tissue [SST] infection, bloodstream infection [BSI], or conjunctivitis) in the neonatal intensive care unit from 2000 through 2008.
During the study period, surveillance cultures were performed for 1,336 neonates, of whom 115 (8.6%) had 1 or more MRSA-positive surveillance culture results. The proportion of colonized neonates detected during outbreak investigations ranged from 5.9% (6 of 102 infants tested in 2001) to 11.6% (22 of 189 infants tested in 2003); the proportion of positive surveillance culture results was highest during the 2002–2003 and 2007 outbreaks, in parallel with the highest rates of infection (approximately 15–20 colonized neonates per 1,000 discharges).
Of the 167 NICU patients colonized with MRSA, 31 had been transferred from other NICUs, 12 were pod neighbors of an infected or colonized index case patient, 101 were identified during outbreak investigations, and 23 were identified by means of a culture performed for clinical purposes. Fiftynine (35.3%) of these 167 colonized neonates developed an MRSA infection during their NICU hospitalization; 23 of these 59 infected neonates were documented to be colonized prior to infection, and 36 were found to be colonized during surveillance testing performed after their infections were diagnosed. For 34 of the 93 infected neonates, colonization was never documented during the hospitalization.
Healthcare Worker Surveillance
Three healthcare worker (HCW) surveillance efforts were conducted: in 2001, in 2005, and in 2007 (Figure 2). During these efforts, 0.4%–2.3% of HCWs were found to be colonized with the relevant outbreak clones, and overall, 8 (1.2%) of 695 HCWs were colonized with nonoutbreak clones. In all, 14 (2%) of 695 HCWs were colonized with MRSA.
DISCUSSION
This is one of the most extensive recent reports describing the molecular epidemiology of MRSA strains isolated from infected and colonized neonates in a single NICU. During the study period, we noted a change in the predominant clones from healthcare-associated MRSA to CA-MRSA. Previous reports have described infections in the NICU population due to CA-MRSA, including USA300 strains.5,24,25 Healy et al24 demonstrated that 4 of 8 neonates with MRSA bacteremia were infected with USA300 during a 12-month study period (C. Healy, written communication, 2009). David et al25 described the first documented transmission of CA-MRSA in a NICU located in the United Kingdom. However, these previous reports did not assess colonization or the potential source(s) of CA-MRSA clones.
Numerous potential reservoirs for MRSA have been described for neonates hospitalized in both the newborn nursery and the NICU. We previously described transmission of USA300 MRSA among postpartum women who presented with skin and soft-tissue infections, including mastitis and surgical wound infections.13 Notably, this outbreak occurred in 2002, prior to detection of USA300 in our NICU. We also investigated the rates of vaginal colonization among pregnant women in 2005 and found that 0.4% were colonized with CA-MRSA,11 but we did not assess vertical transmission in this study. In addition, cases of transmission of CA-MRSA through contaminated breast milk26 and from a father to his hospitalized newborn27 have been described. Skin infections with CA-MRSA (USA300) among otherwise healthy, full-term newborns occurring within 30 days of delivery have been reported in hospitals in Illinois,28 California,28 and Texas,29 as well as transmission of USA300 MRSA between a newborn nursery and the respective postpartum unit.30 Similarly, an outbreak of skin and soft-tissue infections (ie, pustulosis, cellulitis, and omphalitis) with USA400 occurred among neonates discharged from a newborn nursery in Brooklyn, New York.31 These reports confirm that multiple sources of MRSA can exist for the newborn population and that transmission of MRSA can occur quickly, because outbreaks have originated in newborn nurseries, where neonates typically have short hospital stays.
To prevent transmission of MRSA in our NICU, we have adopted a targeted surveillance strategy rather than universal surveillance. Our methods include strategies for eradication of MRSA in colonized neonates. A recent consensus document advocated that the following measures be performed during an outbreak investigation in the NICU: monitoring of hand hygiene compliance, surveillance cultures, molecular analysis of MRSA strains, and decolonization protocols “if deemed necessary by the affected institution.”32 However, data supporting the efficacy and cost-effectiveness of specific infection control strategies for MRSA in the NICU population are limited, because reports have described multiple strategies implemented simultaneously during outbreaks.7,33 Thus, further studies of specific eradication strategies are warranted.
We initiated surveillance cultures of samples from NICU staff when ongoing transmission of MRSA was documented despite implementation of the infection control strategies described above. Transmission seemed to be halted when colonized HCWs were detected and furloughed (Figure 2), although we cannot delineate the precise effect of any given intervention, because many interventions were implemented simultaneously. Others have similarly reported that HCWs can serve as a potential reservoir for both outbreak and nonoutbreak clones.6,7,34 However, limited guidance exists in the literature describing when to initiate HCW screening during outbreak investigations. Grant et al35 have suggested “clinical triggers” to initiate healthcare worker surveillance in the NICU, which could include the detection of at least 5 neonates colonized with MRSA during routine screening or at least 2 neonates infected with MRSA within 7 days, plus molecular typing evidence of clonal transmission. Both the Healthcare Infection Control Practices Advisory Committee and a consensus document coauthored by the Society for Healthcare Epidemiology of America and the Infectious Diseases Society of America endorsed obtaining surveillance cultures from HCWs if there was “epidemiologic support” to do so.36,37 Thus, additional studies to provide more guidance regarding the implementation of HCW surveillance for MRSA are needed.
We demonstrated that 35.3% of colonized neonates developed an MRSA infection during their hospitalization. Huang et al38 demonstrated a 28% attack rate in a Taiwanese NICU, and similar attack rates have been described in adults.39 In addition, Schultz et al40 compared the length of stay and hospital costs for MRSA-colonized neonates with those of MRSA-uncolonized neonates; no difference in the mean hospital costs per day were found, but MRSA-colonized neonates had a significantly greater length of stay, which was attributed to the inability to transfer these neonates to lower acuity facilities, which lacked the staff, bed space, and educational resources to accommodate MRSA-colonized neonates. To our knowledge, no treatment trials have been performed to assess the efficacy of eradication of MRSA colonization in the NICU. However, the high attack rate after colonization and the potential to discontinue transmission precautions after documentation of successful eradication support this strategy.
This study has limitations. This was an observational study performed in a single NICU in a university-affiliated children’s hospital, and thus the results may not be generalizable. We do not perform routine surveillance cultures for MRSA during nonoutbreak periods; therefore, both the colonization rates and the attack rates after colonization may be inaccurate. The success of eradication strategies for MRSA colonization in individual neonates was not assessed. Finally, we do not have longitudinal data on the rate of recolonization among HCWs.
In conclusion, MRSA caused both endemic and epidemic infections in our NICU. Rigorous, targeted infection control strategies led to successful control of outbreaks as documented by eradication of dominant clones. However, MRSA infection and colonization continued to occur, as a result of the introduction of new clones of MRSA into the NICU. Further investigations are warranted to assess the most successful strategies to prevent transmission of MRSA and to detect the relative contribution of different reservoirs of MRSA.
Acknowledgments
We appreciate the efforts of the NICU nurses in obtaining surveillance cultures.
Financial support. None.
Footnotes
Potential conflicts of interest. All authors report no conflicts of interest relevant to this article.
Presented in part: Annual Meeting of the Pediatric Academic Society, May 2006, San Francisco, CA and May 2009, Baltimore, MD.
REFERENCES
- 1.Banerjee SN, Grohskopf LA, Sinkowitz-Cochran RL, Jarvis WR. Incidence of pediatric and neonatal intensive care unit–acquired infections. Infect Control Hosp Epidemiol. 2006;27:561–570. doi: 10.1086/503337. [DOI] [PubMed] [Google Scholar]
- 2.Baltimore RS. Neonatal nosocomial infections. Semin Perinatol. 1998;22:25–32. doi: 10.1016/s0146-0005(98)80005-0. [DOI] [PubMed] [Google Scholar]
- 3.Stoll BJ, Hansen N, Fanaroff AA, et al. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics. 2002;110:285–291. doi: 10.1542/peds.110.2.285. [DOI] [PubMed] [Google Scholar]
- 4.Seybold U, Halvosa JS, White N, Voris V, Ray SM, Blumberg HM. Emergence of and risk factors for methicillin-resistant Staphylococcus aureus of community origin in intensive care nurseries. Pediatrics. 2008;122:1039–1046. doi: 10.1542/peds.2007-3161. [DOI] [PubMed] [Google Scholar]
- 5.McAdams RM, Ellis MW, Trevino S, Rajnik M. Spread of methicillin-resistant Staphylococcus aureus USA300 in a neonatal intensive care unit. Pediatr Int. 2008;50:810–815. doi: 10.1111/j.1442-200X.2008.02646.x. [DOI] [PubMed] [Google Scholar]
- 6.Bertin ML, Vinski J, Schmitt S, et al. Outbreak of methicillin-resistant Staphylococcus aureus colonization and infection in a neonatal intensive care unit epidemiologically linked to a healthcare worker with chronic otitis. Infect Control Hosp Epidemiol. 2006;27:581–585. doi: 10.1086/504933. [DOI] [PubMed] [Google Scholar]
- 7.Saiman L, Cronquist A, Wu F, et al. An outbreak of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2003;24:317–321. doi: 10.1086/502217. [DOI] [PubMed] [Google Scholar]
- 8.Carey AJ, Duchon J, Della-Latta P, Saiman L. The epidemiology of methicillin-susceptible and methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit, 2000–2007. J Perinatol. 2010;30:135–139. doi: 10.1038/jp.2009.119. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez BE, Martinez-Aguilar G, Hulten KG, et al. Severe staphylococcal sepsis in adolescents in the era of community-acquired methicillin-resistant Staphylococcus aureus. Pediatrics. 2005;115:642–648. doi: 10.1542/peds.2004-2300. [DOI] [PubMed] [Google Scholar]
- 10.Miller LG, Perdreau-Remington F, Rieg G, et al. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N Engl J Med. 2005;352:1445–1453. doi: 10.1056/NEJMoa042683. [DOI] [PubMed] [Google Scholar]
- 11.Chen KT, Huard RC, Della-Latta P, Saiman L. Prevalence of methicillin-sensitive and methicillin-resistant Staphylococcus aureus in pregnant women. Obstet Gynecol. 2006;108:482–487. doi: 10.1097/01.AOG.0000227964.22439.e3. [DOI] [PubMed] [Google Scholar]
- 12.Andrews WW, Schelonka R, Waites K, Stamm A, Cliver SP, Moser S. Genital tract methicillin-resistant Staphylococcus aureus: risk of vertical transmission in pregnant women. Obstet Gynecol. 2008;111:113–118. doi: 10.1097/01.AOG.0000298344.04916.11. [DOI] [PubMed] [Google Scholar]
- 13.Saiman L, O’Keefe M, Graham PL, III, et al. Hospital transmission of community-acquired methicillin-resistant Staphylococcus aureus among postpartum women. Clin Infect Dis. 2003;37:1313–1319. doi: 10.1086/379022. [DOI] [PubMed] [Google Scholar]
- 14.Roberts RB, de Lencastre A, Eisner W, et al. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J Infect Dis. 1998;178:164–171. doi: 10.1086/515610. [DOI] [PubMed] [Google Scholar]
- 15.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004;42:792–799. doi: 10.1128/JCM.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lina G, Piemont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 18.McDonald RR, Antonishyn NA, Hansen T, et al. Development of a triplex real-time PCR assay for detection of Panton-Valentine leukocidin toxin genes in clinical isolates of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:6147–6149. doi: 10.1128/JCM.43.12.6147-6149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas J, Larson E, Ross B, See B, Saiman L. Epidemiology and diagnosis of hospital-acquired conjunctivitis among neonatal intensive care unit patients. Pediatr Infect Dis J. 2005;24:586–589. doi: 10.1097/01.inf.0000168742.98617.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial disk susceptibility tests—approved standard. 10th. Wayne, PA: Clinical and Laboratory Standards Institute; 2009. CLSI document M2-A10. [Google Scholar]
- 23.Clinical and Laboratory Standards Institute (CLSI) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically—approved standard. 8th. Wayne, PA: Clinical and Laboratory Standards Institute; 2009. CLSI document M7-A8. [Google Scholar]
- 24.Healy CM, Hulten KG, Palazzi DL, Campbell JR, Baker CJ. Emergence of new strains of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Clin Infect Dis. 2004;39:1460–1466. doi: 10.1086/425321. [DOI] [PubMed] [Google Scholar]
- 25.David MD, Kearns AM, Gossain S, Ganner M, Holmes A. Community-associated methicillin-resistant Staphylococcus aureus: nosocomial transmission in a neonatal unit. J Hosp Infect. 2006;64:244–250. doi: 10.1016/j.jhin.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 26.Gastelum DT, Dassey D, Mascola L, Yasuda LM. Transmission of community-associated methicillin-resistant Staphylococcus aureus from breast milk in the neonatal intensive care unit. Pediatr Infect Dis J. 2005;24:1122–1124. doi: 10.1097/01.inf.0000189983.71585.30. [DOI] [PubMed] [Google Scholar]
- 27.Al-Tawfiq JA. Father-to-infant transmission of community-acquired methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2006;27:636–637. doi: 10.1086/505097. [DOI] [PubMed] [Google Scholar]
- 28.Watson J, Jones RC, Cortes C, et al. Community-associated methicillin-resistant Staphylococcus aureus infection among healthy newborns—Chicago and Los Angeles County, 2004. MMWR Morb Mortal Wkly Rep. 2006;55:329–332. [PubMed] [Google Scholar]
- 29.Fortunov RM, Hulten KG, Hammerman WA, Mason EO, Jr, Kaplan SL. Community-acquired Staphylococcus aureus infections in term and near-term previously healthy neonates. Pediatrics. 2006;118:874–881. doi: 10.1542/peds.2006-0884. [DOI] [PubMed] [Google Scholar]
- 30.Cannon JGA, Perlman J, DeLaMora P, et al. Abstracts of the 18th Annual Meeting of the Society for Healthcare Epidemiology of America. Arlington, VA: Society for Healthcare Epidemiology of America; 2008. Transmission of USA-300 methicillin-resistant Staphylococcus aureus in a newborn nursery also affecting post-partum women. Abstract 178. [Google Scholar]
- 31.Bratu S, Eramo A, Kopec R, et al. Community-associated methicillin-resistant Staphylococcus aureus in hospital nursery and maternity units. Emerg Infect Dis. 2005;11:808–813. doi: 10.3201/eid1106.040885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerber SI, Jones RC, Scott MV, et al. Management of outbreaks of methicillin-resistant Staphylococcus aureus infection in the neonatal intensive care unit: a consensus statement. Infect Control Hosp Epidemiol. 2006;27:139–145. doi: 10.1086/501216. [DOI] [PubMed] [Google Scholar]
- 33.Lepelletier D, Corvec S, Caillon J, Reynaud A, Roze JC, Gras-Leguen C. Eradication of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit: which measures for which success? AmJ Infect Control. 2009;37:195–200. doi: 10.1016/j.ajic.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 34.Stein M, Navon-Venezia S, Chmelnitsky I, et al. An outbreak of new, nonmultidrug-resistant, methicillin-resistant Staphylococcus aureus strain (SCCmec type IIIA variant-1) in the neonatal intensive care unit transmitted by a staff member. Pediatr Infect Dis J. 2006;25:557–559. doi: 10.1097/01.inf.0000219407.31195.44. [DOI] [PubMed] [Google Scholar]
- 35.Grant PS, Charns LG, Rawot BW, Benedetti SG. Consideration to culture health care workers related to increased methicillin-resistant Staphylococcus aureus activity in a neonatal intensive care unit. Am J Infect Control. 2008;36:638–643. doi: 10.1016/j.ajic.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Calfee DP, Salgado CD, Classen D, et al. Strategies to prevent transmission of methicillin-resistant Staphylococcus aureus in acute care hospitals. Infect Control Hosp Epidemiol. 2008;29(suppl 1):S62–S80. doi: 10.1086/591061. [DOI] [PubMed] [Google Scholar]
- 37.Siegel JD, Rhinehart E, Jackson M, Chiarello L. Management of multidrug-resistant organisms in health care settings, 2006. Am J Infect Control. 2007;35(10 suppl 2):S165–S193. doi: 10.1016/j.ajic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Huang YC, Chou YH, Su LH, Lien RI, Lin TY. Methicillin-resistant Staphylococcus aureus colonization and its association with infection among infants hospitalized in neonatal intensive care units. Pediatrics. 2006;118:469–474. doi: 10.1542/peds.2006-0254. [DOI] [PubMed] [Google Scholar]
- 39.Huang SS, Platt R. Risk of methicillin-resistant Staphylococcus aureus infection after previous infection or colonization. Clin Infect Dis. 2003;36:281–285. doi: 10.1086/345955. [DOI] [PubMed] [Google Scholar]
- 40.Schultz ED, Tanaka DT, Goldberg RN, Benjamin DK, Jr, Smith PB. Effect of methicillin-resistant Staphylococcus aureus colonization in the neonatal intensive care unit on total hospital cost. Infect Control Hosp Epidemiol. 2009;30:383–385. doi: 10.1086/596610. [DOI] [PMC free article] [PubMed] [Google Scholar]