Abstract
The grass carp (Ctenopharyngodon idella) is one of the most important cultivated fish species in China. Mounting evidences suggests that microRNAs (miRNAs) may be key regulators of skeletal muscle among the grass carp, but the knowledge of the identity of myogenic miRNAs and role of miRNAs during skeletal muscle anabolic state remains limited. In the present study, we choose 8 miRNAs previously reported to act as muscle growth-related miRNAs for fasting-refeeding research. We investigated postprandial changes in the expression of 8 miRNAs following a single satiating meal in grass carp juveniles who had been fasting for one week and found that 7 miRNAs were sharply up-regulated within 1 or 3 h after refeeding, suggesting that they may be promising candidate miRNAs involved in a fast-response signaling system that regulates fish skeletal muscle growth.
Keywords: MicroRNA, Grass carp, Fasting, Refeeding, Skeletal muscle
The grass carp (Ctenopharyngodon idella) is one of the most important cultivated species in Chinese freshwater aquaculture. Generally, fast skeletal muscle comprises the largest tissue of the fish body (Zhang et al, 2009), and forms the main edible part of this species. Given the complicated development process as well as the importance of these tissues, gaining a clearer understanding of the mechanism underlying muscle development may provide key data for both developmental biologists and researchers attempting to improve grass carp musculature for aquaculture.
Several lines of research have implicated growth factors, regulatory proteins, and transcription factors as key actors involved in the regulation and maintenance of skeletal muscle mass among fish (Nihei et al, 2006; Steinbacher et al, 2006; Chu et al, 2010). Recent studies found that alongside transcriptional factors involved in muscle proliferation and differentiation, a set of microRNAs (miRNAs) may also play important roles in skeletal muscle development among vertebrate animals (Ge & Chen, 2011; Güller & Russell, 2010; Chu et al, 2013). MicroRNAs (miRNAs) are approximately 22-nt noncoding RNAs that act as negative regulators of gene expression, either via inhibiting mRNA translation or promoting mRNA degradation through base pairing to the 3' untranslated region (UTR) of target mRNAs (Xie et al, 2005; Liu, 2008; Zhang & Wen, 2010). Furthermore, miRNAs regulate the expression of transcription factors and signaling mediators critical to both cardiac and skeletal muscle development and function (Callis & Wang, 2008; van Rooij et al, 2008). Studies using the mouse myogenic C2C12 cell line demonstrated miR-1 and miR-133 are involved in myoblast proliferation and differentiation via regulation of the expression of HDAC4 and SRF, respectively (Chen et al, 2006). Meanwhile, miR-1, miR-133a and miR-206 (muscle-specific miRNAs), were also noted to be differentially expressed in Japanese flounder (Paralichthys olivaceus) during metamorphosis, as well as in Nile tilapia (Oreochromis niloticus) among several developmental stages. Taken together, these findings imply that miRNAs likely play an important role in regulating muscle development (Fu et al, 2011; Yan et al, 2012a). Further evidences suggest that miRNAs act as key regulators of myogenesis, but unfortunately characterizing the identity of myogenic miRNAs and delineating the role of miRNAs during skeletal muscle anabolic state remains unclear.
The maintenance of skeletal muscle mass is a complex and controlled process, largely influenced by both nutritional and physiological states of different animals (Fuentes et al, 2012). Fasting-refeeding protocols have been commonly used as models to investigate the regulation of muscle growth in fish species that experience the transition from catabolic to anabolic states. In the present study, we analyzed the expression of 8 select miRNAs during skeletal muscle anabolic state using a fasting-refeeding experiment. The goal of our study was to better parse out the potential role of these miRNAs in skeletal muscle proliferation and differentiation. The 8 miRNAs (miR-1a, miR-133a-3p, miR-133b-3p, miR-146, miR-181a-5p, miR-206, miR-214 and miR-26a) were previously reported to act as muscle growth- related miRNAs (McCarthy & Esser, 2007; Flynt et al, 2007; Kuang et al, 2009; Yan et al, 2012b). In theory, these miRNAs and their target genes may comprise a coordinated regulating net to favor resumption of myogenesis as an early response to refeeding.
MATERIALS AND METHODS
Fasting-refeeding experiments and sampling
All grass carp individuals were reared under standard conditions at the Che Tian Jiang Reservoir in Loudi, Hunan, China. Two homogeneous groups of grass carp juveniles (average body weight 150 g, 90 days post-hatching (dph)) were reared respectively in two net cages (5 m×5 m×2 m) with fifty fish per tank. All juveniles were fed under standard conditions for 3 weeks, after which the juveniles underwent fasting for 1 week, and were then fed a single meal, which was distributed to all individuals until they appeared to be visually satiated. At each time point from 0 h (before the recovery meal), and at 1, 3, 6, 12, 24, 48 and 96 h (hours after the single meal), six fish were sampled, wherein fast muscles were dissected from the dorsal myotome of individuals. The resulting samples were then snap-frozen in liquid nitrogen and stored at -80 °C until further processing.
Quantitative real-time PCR for the miRNAs
Tissue samples were ground in liquid nitrogen, and total RNAs were extracted using TRIzol (Invitrogen, USA), and then treated with RNAse-free DNAse I (Promega, USA) in the presence of RNAse inhibitor (Sigma, China Branch) followed by ethanol precipitation. The obtained RNAs were polyadenylated by poly (A) polymerase, and then reverse transcribed with one step PrimeScript miRNA cDNA synthesis Kit (TaKaRa, Dalian, China) and a Universal Adaptor Primer (a poly (T) primer ligated with an adapter) for miRNA quantitative assays.
The miRNA expression levels were quantified using real-time PCR with grass carp β-actin gene (GenBank No. DQ211096.1) as an internal control. The cDNA samples were used as templates for quantitative RT-PCR assays with SYBR Premix Ex Taq II (TaKaRa, Japan) and its amplification reaction was carried out on a Bio-Rad CFX96 system (USA). Each 2 μL cDNA template was added to a total volume of 25 μL reaction mix containing 12.5 μL SYBR Green mix, 1 μL of each miRNA or gene specific forward primer (as shown in Table 1, 10 μmol/L) and 1μL of universal downstream primer (Uni-miR qPCR Primer, 10 μmol/L, TaKaRa) or gene specific reverse primer, 8.5 μL nuclease-free water. The protocols used are as follows: (i) pre-denaturation at 95 °C for 60 s; (ii) amplification and quantification, repeated 40 cycles of at 95 °C for 5 s and at 60 °C for 25 s; (iii) melting curve program (65-95 °C with heating rate of 0.1 °C/S and fluorescence measurement) (Zhou et al, 2010). The relative expression ratio (R) of target miRNA was calculated by R=2-ΔΔCt (Livak & Schmittgen, 2001; Bustin et al, 2009), where Ct is the cycle threshold. The basic equation employed was:
Table 1.
Primers used for miRNA detection
| Name | Primer Sequence (5′-3′) | Name | Primer Sequence (5′-3′) |
|---|---|---|---|
| miR-1a-F | TGGAATGTAAAGAAGTATGTAT | miR-146-F | CGTGAGAACTGAATTCCATAGATGG |
| miR-133a-3p-F | CGCGTTTGGTCCCCTTCA | miR-133b-3p-F | TTGGTCCCCTTCAACCAGCTA |
| miR-206-F | CGTGGAATGTAAGGAAGTGTGTGG | miR-214-F | ACAGCAGGCACAGACAGGCAG |
| miR-181a-5p-F | CGAACATTCAACGCTGTCGGT | miR-26a-F | CGTTCAAGTAATCCAGGATAGGCT |
| β-actin-F | GCCGTGACCTGACTGACTACCT | β-actin-R | CGCAAGACTCCATACCCAAGAAG |
Forward primers used for detection were detailed in the Methods; the reverse primer used for detection was universal downstream primer (Uni-miR qPCR Primer, 10 μmol/L, Takara).
ΔΔCt=(Cttarget gene -Cthousekeeping gene)experiment- (Cttarget gene -Cthousekeeping gene)control
The miRNA expression levels were then analyzed by one-way ANOVA procedures and regression analysis of SPSS 17.0 (SPSS inc., Chicago, USA). Duncan’s multiple range tests were used to compare the control (0 h before the recovery meal) and experimental (# hours after refeeding) groups. The differences were considered statistically significant when P<0.05. Data are shown as means±SE (n=6). Correlation of gene expression was analyzed by the Spearman rank order correlation test. Hierarchical clustering was performed using Cluster3.
RESULTS
Effect of fasting and refeeding on the expression of the miRNAs
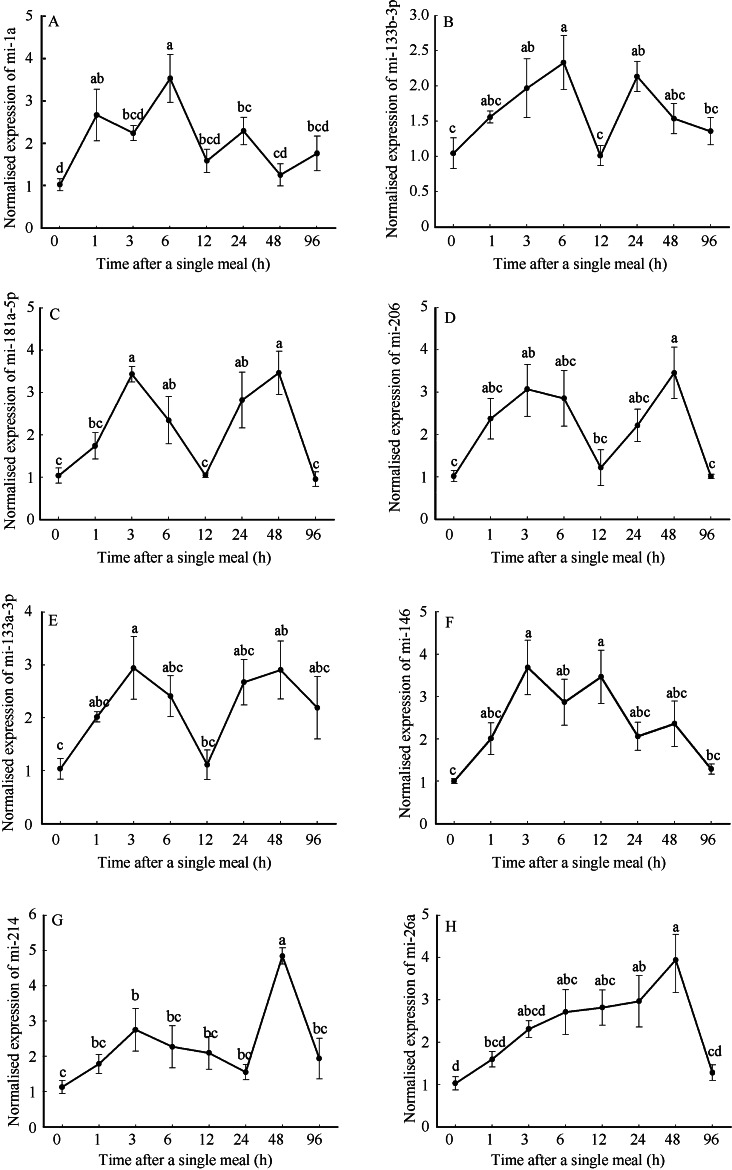
A significant up-regulation of each of the 8 miRNAs was observed between 1-6 hours after the single meal (P<0.05) (Figure 1). MiR-1a was sharply up-regulated within 1 h after refeeding and peaked at 6 h (Figure 1A). The expression of miR-133a-3p, miR-133b-3p, miR-146, miR-181a-5p, miR-206 and miR-214 were significantly increased at 3h (P<0.05), reached the maximal levels at 3, 6 or 48 h postprandial (Figure 1B-G). While miR-26a responded slowly to refeeding, it significantly increased at 6 h (P<0.05) and peaked at 48 h (Figure 1F). However, miR-1a, miR-133b-3p and miR-181a-5p significantly decreased at 12 h. All 8 miRNAs returned to the initial baseline values at 96 h.
Figure 1.
Relative expression of miRNAs with significant up-regulation following refeeding after 1 week fasting β-actin expression was detected as the internal control. All values are presented as mean±SE, n=6. Different letters indicate significant differences between columns (P<0.05).
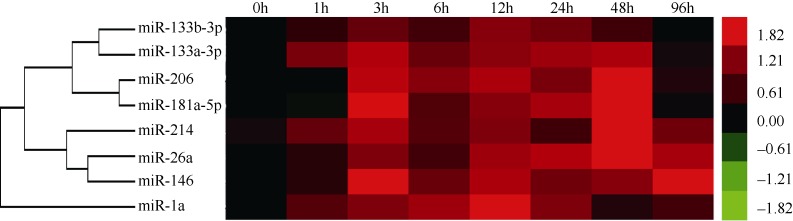
Heat map summary of hierarchical clustering of miRNAs in skeletal muscle during fasting-refeeding periods
Hierarchical clustering analysis of miRNAs in muscle was done according to the similarity in their expression across different postprandial times (0-96 h). Hierarchical clustering of the miRNAs throughout the trial showed three clades (Figure 2). The first clade including 2 pairs of closely linked miRNAs (miR-133b-3p and miR-133a-3p, miR-206 and miR-181a-5p) that clustered together. The second clade clustered miR-26a with miR-146 expression.
Figure 2.
Heat map summary of hierarchical clustering of miRNAs in skeletal muscle during fasting-refeeding periods
Blue and yellow respectively denote a decrease or increase. The absolute signal intensity ranged from -1.82 to +1.82, with corresponding color changes from green to red.
DISCUSSION
MicroRNAs (miRNAs) are noncoding RNA molecules that regulate the stability and/or the translational efficiency of target mRNAs, and several miRNAs have been found to be specifically expressed or highly enriched in skeletal muscle. The expression of muscle-specific miR-1, miR-133, miR-206, while miR-208 is regulated by muscle transcriptional networks involving SRF, MyoD and MEF2. (Flynt & Lai, 2008; Latronico et al, 2007; Thum et al, 2008; Van Rooij et al, 2008; Callis et al, 2008). Interestingly, non-muscle-specific miRNAs, e.g., miR-26a and miR-181, also regulate skeletal muscle differentiation (Wong & Tellam, 2008; Naguibneva et al, 2006). While a great deal of attention has been paid to miRNAs involved in control of muscle development, a recent study suggests that several other miRNAs, including miR-499, miR-208b and miR-23a, also play an important role in human muscle growth (Drummond et al, 2009). Among fish species, Huang et al (2012) previously detected differentially expressed miRNA between 2 strains of Nile tilapia and identified miR-140, miR-192, miR-204, miR-218a, miR-218b, miR-301c, miR-460, miR-133, miR-152, miR-15a, miR-193a, miR-30b and miR-34 as being associated with body growth in tilapia.
Our present study focused on exploring the potential role(s) of miRNAs as a new layer of control in the postprandial regulation of the muscle development among grass carp. Typically, nutrient availability is among the most important environmental variable altering muscle growth (Valente et al, 2012). As such, starvation and refeeding experiments have served as an effective model for studying the regulation of muscle growth in fish, including the Atlantic salmon (Salmo salar) (Bower et al, 2009), rainbow trout (Oncorhynchus mykiss) (Montserrat et al, 2007), and Atlantic halibut (Hippoglossus hippoglossus) (Hagen et al, 2009). Similarly, in humans MiRNAs turned over quite rapidly (i.e. hours) in skeletal muscle following amino acid ingestion. However, little information is available regarding the early transcriptional changes of miRNA during the postprandial period, especially among fish. We therefore focused on exploring the postprandial regulation of growth-related miRNAs shortly after feeding a single meal in grass carp. Our results showed that miR-1a, miR-133a-3p, miR-133b-3p, miR-146, miR-181a-5p, miR-206 and miR-214 were significantly elevated at 1 or 3 h after refeeding in the fast muscle of grass carp. Drummond et al (2009) previously found a rapid up-regulation of the miR-1, miR-208b, miR-23a and miR-499 following the amino acid ingestion in humans. These findings suggest that the identified miRNAs may be promising candidate miRNAs involved in a fast-response signaling system that regulates fish skeletal muscle growth. The other finding of significant decreased of miR-1a, miR-133b-3p and miR-181a-5p at 12 h after single meal suggests there may be other signaling pathways regulated by the miRNAs that limit excessive regulation of muscle growth.
A further finding of our study was that miR-206 and miR-181a-5p showed a dramatic and simultaneous up-regulation following feeding by a single meal. MiR-206 is known to be a muscle-specific miRNA, with its role in muscle development having been verified in some animal models, including mice, rats and zebrafish (Anderson et al, 2006; Kim et al, 2006; Mishima et al, 2009; Shan et al, 2009). MyoD acts as a transcriptional activator of the miR-206 pre-miRNA transcript, and subsequently the induced high levels of the mature miR-206 result in the down-regulation of specific target muscle growth-related genes (Rosenberg et al, 2006). MiR-181 is also thought to function partly through inhibition of Hox-A11 expression, a known repressor of MyoD, which is required for new muscle growth (Naguibneva et al, 2006). Taken together, these different lines of evidence suggest that miR-181 may indirectly promote miR-206 expression, though some further study is needed. Moreover, our finding that two miRNAs (miR-181a-5p and miR-206) clustered together suggest a close relation and coordination regulation of these miRNAs towards resumption of myogenesis following refeeding.
In conclusion, the present results show that several miRNAs likely involved in fast skeletal muscle in grass carp respond quickly to refeeding of a single meal following fasting. Results of our analysis indicate that refeeding induced a coordinated regulation of several miRNAs involved in a strong resumption of myogenesis, wherein the 8 tested miRNAs transcripts were sharply up-regulated in muscle tissues in response to refeeding. This finding suggests that these miRNAs may be promising candidate miRNAs involved in regulating fish fast muscle growth. Further study is needed to experimentally assess the targets of these miRNAs and elucidate how they contribute to the regulation of skeletal muscle growth during anabolic state.
Acknowledgements
We would like to thank Yu-Long LI, Dun-Xue CHEN, Kai-Zhuo WANG and Jun-Zhi ZHANG for their help and support in sample collection.
Funding Statement
This study was supported by the National Natural Science Foundation of China (31230076; 31340054), the Natural Science Foundation of Hunan province (14JJ2135) and the State Key Laboratory of Freshwater Ecology and Biotechnology (2012FB01)
Footnotes
The authors have declared that no competing interests exist.
References
- [1].Anderson C, Catoe H, Werner R. 2006. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Research, 34(20): 5863-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bower NI, Taylor RG, Johnston IA. 2009. Phasing of muscle gene expression with fasting-induced recovery growth in Atlantic salmon. Frontiers in Zoology, 6: 18-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, 55(4): 611-22. [DOI] [PubMed] [Google Scholar]
- [4].Callis TE, Deng Z, Chen JF, Wang DZ. 2008. Muscling through the microRNA world. Experimental Biology and Medicine, 233(2): 131-8. [DOI] [PubMed] [Google Scholar]
- [5].Callis TE, Wang DZ. 2008. Taking microRNAs to heart. Trends in Molecular Medicine, 14(6): 254-60. [DOI] [PubMed] [Google Scholar]
- [6].Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature Genetics, 38(2): 228-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chu WY, Liu LS, Li YL, Chen L, Wang K Z, Li HH, Du SJ, Zhang JS. 2013. Systematic identification and differential expression profiling of microRNAs from white and red muscles of Siniperca chuatsi. Current Molecular Medicine, 13(8): 1397-407. [DOI] [PubMed] [Google Scholar]
- [8].Chu WY, Xia XJ, Chen DG, Fu GH, Liu C, Chen J, Liu F, Lu SQ, Zhang JS. 2010. Gene expression profiles of the muscle tissues of the commercial important teleost, Siniperca chuatsi L. Aquaculture International, 18: 667-78. [Google Scholar]
- [9].Drummond MJ, Glynn EL, Fry CS, Dhanani S, Volpi E, Rasmussen BB. 2009. Essential amino acids increase microRNA-499, -208b, and -23a and downregulate myostatin and myocyte enhancer factor 2c mRNA expression in human skeletal muscle. The Journal of Nutrition, 139(12): 2279-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Flynt AS, Lai EC. 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nature Reviews Genetics, 9(11): 831-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. 2007. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nature Genetics, 39(2): 259-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fu Y, Shi Z, Wu M, Zhang J, Jia L, Chen X. 2011. Identification and differential expression of microRNAs during metamorphosis of the Japanese flounder (Paralichthys olivaceus). PLoS ONE, 6(7): e22957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fuentes EN, Ruiz P, Valdes JA, Molina A. 2012. Catabolic signaling pathways, atrogenes, and ubiquitinated proteins are regulated by the nutritional status in the muscle of the fine flounder. PLoS ONE, 7(9): e44256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ge Y, Chen J. 2011. MicroRNAs in skeletal myogenesis. Cell Cycle, 10(3): 441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Güller I, Russell AP. 2010. MicroRNAs in skeletal muscle: their role and regulation in development, disease and function. The Journal of Physiology, 588(pt21): 4075-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hagen O, Fernandes JMO, Solberg C, Johnston IA. 2009. Expression of growth-related genes in muscle during fasting and refeeding of juvenile Atlantic halibut, Hippoglossus hippoglossus L. Comparative Biochemistry and Physiology. Part B, Biochemistry & Molecular Biology, 152(1): 47-53. [DOI] [PubMed] [Google Scholar]
- [17].Huang CW, Li YH, Hu SY, Chi JR, Lin GH, Lin CC, Gong HY, Chen JY, Chen RH, Chang SJ, Liu FG, Wu JL. 2012. Differential expression patterns of growth-related microRNAs in the skeletal muscle of Nile tilapia (Oreochromis niloticus). Journal of Animal Science, 90(12): 4266-79. [DOI] [PubMed] [Google Scholar]
- [18].Kim, HK, Lee, YS, Sivaprasad U, Malhotra A, Dutta A. 2006. Muscle specific microRNA miR-206 promotes muscle differentiation. The Journal of Cell Biology, 174(5): 677-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kuang W, Tan J, Duan Y, Duan J, Wang W, Jin F, Jin Z, Yuan X, Liu Y. 2009. Cyclic stretch induced miR-146a upregulation delays C2C12 myogenic differentiation through inhibition of Numb. Biochemical and Biophysical Research Communications, 378(2): 259-63. [DOI] [PubMed] [Google Scholar]
- [20].Latronico MV, Catalucci D, Condorelli G. 2007. Emerging role of microRNAs in cardiovascular biology. Circulation Research, 101(12): 1225-36. [DOI] [PubMed] [Google Scholar]
- [21].Liu J. 2008. Control of protein synthesis and mRNA degradation by microRNAs. Current Opinion in Cell Biology, 20(2): 214-21. [DOI] [PubMed] [Google Scholar]
- [22].Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods, 25(4): 402-8. [DOI] [PubMed] [Google Scholar]
- [23].McCarthy JJ, Esser KA. 2007. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. Journal of Applied Physiology, 102(1): 306-13. [DOI] [PubMed] [Google Scholar]
- [24].Mishima Y, Abreu-Goodger C, Staton AA, Stahlhut C, Shou C, Cheng C, Gerstein M, Enright AJ, Giraldez AJ. 2009. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes & Development, 23(5): 619-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Montserrat N, Gabillard JC, Capilla E, Navarro MI, Gutiérrez J. 2007. Role of insulin, insulin-like growth factors, and muscle regulatory factors in the compensatory growth of the trout (Oncorhynchus mykiss). General and Comparative Endocrinology, 150(3): 462-72. [DOI] [PubMed] [Google Scholar]
- [26].Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. 2006. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nature Cell Biology, 8(3): 278-84. [DOI] [PubMed] [Google Scholar]
- [27].Nihei Y, Kobiyama A, Ikeda D, Ono Y, Ohara S, Cole NJ, Johnston IA, Watabe S. 2006. Molecular cloning and mRNA expression analysis of carp embryonic, slow and cardiac myosin heavy chain isoforms. The Journal of Experimental Biology, 209(1): 188-98. [DOI] [PubMed] [Google Scholar]
- [28].Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. 2006. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. The Journal of Cell Biology, 175(1): 77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shan ZX, Lin QX, Fu YH, Deng CY, Zhou ZL, Zhu JN, Liu XY, Zhang YY, Li Y, Lin SG, Yu XY. 2009. Upregulated expression of miR-1/miR- 206 in a rat model of myocardial infarction. Biochemical and Biophysical Research Communications, 381(4): 597-601. [DOI] [PubMed] [Google Scholar]
- [30].Steinbacher P, Haslett JR, Six M, Gollmann HP, Sänger AM, Stoiber W. 2006. Phases of myogenic cell activation and possible role of dermomyotome cells in teleost muscle formation. Developmental Dynamics: An Official Publication of the American Association of Anatomists, 235(11): 3132-43. [DOI] [PubMed] [Google Scholar]
- [31].Thum T, Catalucci D, Bauersachs J. 2008. MicroRNAs: novel regulators in cardiac development and disease. Cardiovascular Research, 79(4): 562-70. [DOI] [PubMed] [Google Scholar]
- [32].Valente LM, Bower NI, Johnston IA. 2012. Postprandial expression of growth related genes in Atlantic salmon (Salmo salar L.) juveniles fasted for 1 week and fed a single meal to satiation. The British Journal of Nutrition, 108(12): 2148-57. [DOI] [PubMed] [Google Scholar]
- [33].Van Rooij E, Liu N, Olson EN. 2008. MicroRNAs flex their muscles. Trends in Genetics: TIG, 24(4): 159-66. [DOI] [PubMed] [Google Scholar]
- [34].Wong CF, Tellam RL. 2008. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. Journal of Chemical Biology, 283(15): 9836-43. [DOI] [PubMed] [Google Scholar]
- [35].Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. 2005. Systematic discovery of regulatory motifs in human promoters and 3’UTRs by comparison of several mammals. Nature, 434(7031): 338-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yan B, Guo JT, Zhao LH, Zhao JL. 2012. a. microRNA expression signature in skeletal muscle of Nile tilapia. Aquaculture, 364-365: 240-6. [Google Scholar]
- [37].Yan X, Ding L, Li Y, Zhang X, Liang Y, Sun X, Teng CB. 2012. b. Identification and profiling of microRNAs from skeletal muscle of the common carp. PLoS ONE, 7(1): e30925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang JS, Fu GH, Chu WY, Chen J, Liu Z, Liu F, Lu SQ, Liang P. 2009. cDNA cloning and expression analysis of myosin heavy chain gene(MHC)of the Mandarin fish, Sniperca kneri. Aquaculture Research, 40(4): 412-8. [Google Scholar]
- [39].Zhang YQ, Wen JF. 2010. Mirna system in unicellular eukaryotes and its evolutionary implications. Zoological Research, 31(1): 39-42 (in Chinese) [DOI] [PubMed] [Google Scholar]
- [40].Zhou RX, Meng T, Meng HB, Cheng DX, Bin SY, Cheng J, Fu GH, Chu WY, Zhang JS. 2010. Selection of reference genes in transcription analysis of gene expression of the Mandarin fish, Siniperca chuasti. Zoological Research, 31(2): 141-6. [DOI] [PubMed] [Google Scholar]