Abstract
Here, we used reverse transcription-PCR (RT-PCR) and western blot to detect protease-activated receptor (PAR) 1, PAR 2 and PAR 4 expression in cancer tissues and cell lines of esophageal squamous cell carcinoma, and investigated the co-relationship between PAR expression and clinic-pathological data for esophageal cancer. The methylation of PAR4 gene promoter involved in esophageal carcinoma was also analyzed. By comparing the mRNA expressions of normal esophageal tissue and human esophageal epithelial cells (HEEpiC), we found that among the 28 cases of esophageal squamous cell carcinoma, PAR1 (60%) and PAR2 (71%) were elevated in 17 and 20 cases, respectively, and PAR4 (68%) expression was lowered in 19 cases. Whereas, in human esophageal squamous cells (TE-1 and TE-10), PAR1 and PAR2 expression was increased but PAR4 was decreased. Combined with clinical data, the expression of PAR1 in poorly differentiated (P=0.016) and middle and lower parts of the esophagus (P=0.016) was higher; expression of PAR4 in poorly differentiated carcinoma was lower (P=0.049). Regarding TE-1 and TE-10 protein expression, we found that in randomized esophageal carcinoma, PAR1 (P=0.027) and PAR2 (P=0.039) expressions were increased, but lowered for PAR4 (P=0.0001). In HEEpiC, TE-1, TE-10, esophageal and normal esophagus tissue samples (case No. 7), the frequency of methylation at the 19 CpG loci of PAR4 was 35.4%, 95.2%, 83.8%, 62.6% and 48.2%, respectively. Our results indicate that the expression of PAR1 and PAR2 in esophageal squamous cell carcinoma is increased but PAR4 is decreased. Hypermethylation of the promoter of the PAR4 gene may contribute to reduced expression of PAR4 in esophageal squamous cell carcinoma.
Keywords: PAR1, PAR2, PAR4, Esophageal squamous cell carcinoma, PCR, Western Blot, Methylation
The development of esophageal squamous cell carcinoma (ESCC) is a complicated process with multiple pathological stages. Among various regulatory factors, proteases play critical roles in activating signal transduction pathways and regulating gene expression (Ikeda et al, 1999). Protease-activated receptors (PARs) are a subfamily of the single seven-transmembrane G-protein-coupled receptors and include PAR1, PAR2, PAR3 and PAR4 (Macfarlane et al, 2001). PAR1, PAR3 and PAR4 are thrombin receptors, and PAR2 is trypsinase/tryptase receptor (Xu et al, 1998). Ribeiro et al (2009) found high expression of PAR1 but low expression of PAR2 in the tissues of ESCC; however, Wang et al (2010) found high expression of PAR2 in ESCC. PAR4 is a recently discovered novel subtype expressed in high amounts in colon cancer and hepatocarcinoma and induces the proliferation and migration of cancer cells (Gratio et al, 2009; Kaufmann et al, 2007). PAR4 expression in stomach cancer tissues (Zhang et al, 2011) and adenocarcinoma of the lung are low (Jiang et al, 2013), but its expression in ESCC remains unclear.
Here, we determined the expression of PAR1, PAR2 and PAR4 in tissues of ESCC and analyzed correlations between PAR4 promoter hypermethylation and the development of ESCC. Our aim was to provide theoretical evidence for clinical diagnosis, treatment and prognosis of ESCC and provide a basis for drug research.
MATERIALS AND METHODS
Experimental materials
Human esophageal epithelial cells (HEEpiC) and human esophageal squamous cells (TE-1, TE-10) were obtained from the Cell Bank of Kunming Institute of Zoology, Chinese Academy of Sciences. Tissues of 28 cases (male=21, female=5; 51-81 years old) of diagnosed ESCC were from hospitals affiliated with the Kunming Medical University, Yunnan, China. All patients were clean of any chemotherapy or radiation treatment prior to surgery. The carcinoma tissues and corresponding normal control tissues (at least 5 cm away from carcinoma tissue) obtained during surgery were quick frozen in liquid nitrogen and stored at -80 °C. All the experimental protocols were approved by the Ethics Committee of Kunming Medical University.
Experimental procedures Cell culture
TE-1 and TE-10 cells were cultured with RPMI1640 (Takara, Beijing, China) completed culture medium containing 10% FBS and incubated at 37 °C with 5% CO2 (relative humidity=95%). Culture medium was changed 1-2 days later. Cells were subcultured 3-5 days later and digested with 0.25% trypsinase: 0.03% EDTA (1:1). HEEpiC were cultured with HEEpiC-specific (Takara, Beijing, China) culture medium and the culture procedure was the same as that for TE-1 cells.
Reverse transcription-PCR (RT-PCR)
Total RNA of ESCC tissues, TE-1, TE-10 and HEEpiC were extracted using a RNA extraction kit (Tiangen Biotech, Beijing, China). The purity and integrity of total RNA were tested. cDNAs were reverse transcribed from total RNAs of tissues and cells (2 ng-2 µg) using a reverse transcription kit (Takara, Beijing, China) and then stored at -20 °C. Gene amplifications were performed by taking cDNAs as templates and GAPDH as internal reference. Primers for GAPDH, PAR1, PAR2 and PAR4 are shown in Table 1. PCR products were run by 2% agarose gel electrophoresis and observed under ultraviolet light and photographed.
Table 1.
Primers and reaction conditions used in RT-PCR
| Gene | Primer sequences | NCBI Accession Number | Annealing temperature (°C) | Product (bp) |
|---|---|---|---|---|
| GAPDH | F: 5′-ATGGGGAAGGTGAAGGTCG-3′ | NM_001101.3 | 60 | 308 |
| R: 5′-GGGGTCATTGATGGCAACAATA-3′ | ||||
| PAR1 | F: 5′-GCCGCCTGCTTCAGTCTGTGC-3′ | NM_001992.3 | 67 | 648 |
| R: 5′-GGCCAGACAAGTGAAGGAAGC-3′ | ||||
| PAR2 | F: 5′-CCATCCAAGGAACCAATAGATC-3′ | NM_005242.3 | 60 | 643 |
| R: 5′-ATGTCTCCCACCAAGAGCTGCTCA-3′ | ||||
| PAR4 | F: 5′-GGCAACCTCTATGGTGCCTA-3′ | NM_003950.2 | 58 | 244 |
| R: 5′-TTCGACCCAGTACAGCCTTC-3′ |
Western blot
Carcinoma tissues and corresponding normal control tissues were randomly selected from three cases. Cultured cells in logarithmic growth were rinsed twice with pre-chilled PBS (4 °C) and then lysed with 6× SDS (0.6 mL) and water-bathed (95 °C) for 15 min. After the polymerization of resolving gel (12%) and staking gel (5%), samples were loaded. Tris-glycine electrophoresis buffer (×l) was poured into the running chamber and loaded samples were run for 1 h at a constant current of 160 V. Then the gel was transferred onto the PVDF membrane for 2 h at a constant current of 250 mA and 4 °C. The blots were removed from the transfer unit and blocked by placing in 3% BSA-TBST blotting solution for 2 h with shaking at room temperature. After washing, the blot was incubated with the primary antibodies for PAR1, PAR2, PAR4 and β-actin over night at 4 °C. After washing, the blot was incubated with secondary antibodies. After another three washes, the blot was revealed via HRP-ECL chemiluminescence detection and scanned using the SynGene scanning system. The illumination densities of the detected protein bands were transferred into optical density values (OD values). The developing strength of each target protein band and its internal reference (β-actin) were determined by OD values. The ratio of the OD values of each target protein band to its internal reference (β-actin) was taken as the result of RT-PCR.
Bisulfite genomic sequencing PCR
Genomic DNAs of TE-1, TE-10, HEEpiC and the No. 7 tissue sample were extracted using a genome DNA extraction kit (Takara, Beijing, China) and their concentrations were determined. The CpG sites in PAR4 were amplified with the MethylCode Bisulfite Conversion Kit (Invitrogen, US). PCR products were eluted and purified with the AxyPrep DNA elution kit (Axygen). Primer sequences were: F-5′-TTTAAGGGTGATTTTAGGAAA GGTTTAGAG-3′and R-5′-ACTATAACCTCAAACTTC CTACCTC-3′. The products of ligation were transformed with DH5α component cells. Transformation products were spread on LB plates (with Ampicillin) and grown overnight. Clones were selected and sent for sequencing.
Statistical analyses
Data were analyzed using SPSS l7.0 (SPSS Inc., Chicago, USA). Correlations between PAR expression and clinical pathologies were tested using Fisher’s exact tests. The illumination strength of protein expression is expressed as mean±SD and comparisons between expressions were conducted using t-tests. Statistical significance was set at P<0.05.
RESULTS
mRNA expression of PAR1, PAR2 and PAR4 and correlation with clinical manifestation
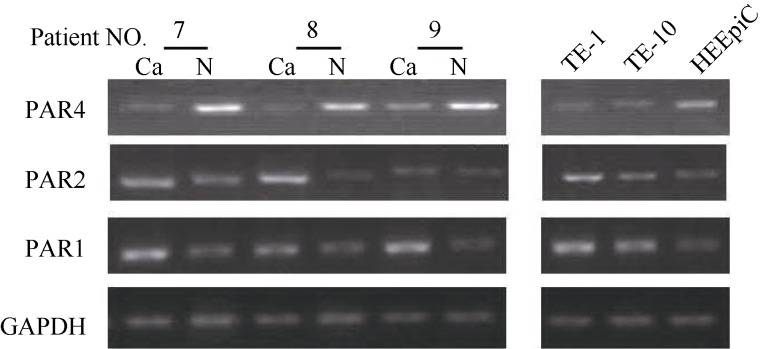
When the expression of GAPDH of each sample was at a similar level, the brighter the mRNA band was, the higher the expression level of PAR was, and vice versa. Among the 28 cases of ESCC, 17 (60%) and 20 (71%) cases were found with increased expression of PAR1 and PAR2, respectively. Totally, 19 (68%) cases were found with decreased PAR4. In TE-1 and TE-10, the expression of PAR1and PAR2 was high. The expression of PAR4 mRNA in TE-1 and TE-10 was lower than in HEEpiC. The expression level of PAR1, PAR2 and PAR4 mRNA in the tissues and cells from cases 7, 8 and 9 are shown in Figure 1. Combined with clinical pathological data, the expression of PAR1 in central and low located ESCC was significantly higher than in upper ESCC (P=0.007 and P=0.008, respectively). The expression of PAR2 in phase III+IV ESCC was significantly higher than in phase I+II (P=0.004). Significantly lower expression of PAR4 was found in lower ESCC, compared with central and upper ESCC (P=0.036). No significant correlation for the expression of PAR1, PAR2 and PAR4 and patients’ gender, sex and distant or lymph node metastases were found (Table 2).
Figure 1.
PAR1, PAR2 and PAR4 mRNA expression in esophageal squamous cell carcinoma and normal control tissues
Table 2.
Correlation between PAR1, PAR2 and PAR4 mRNA levels and clinical manifestation of esophageal squamous cell carcinoma
| Clinical data | Case numbers (n) | PAR1 | PAR2 | PAR4 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Decreased | Increased | P-value | Decreased | Increased | P-value | Decreased | Increased | P-value | ||
| Age (year) | ||||||||||
| ≤65 | 13 | 5 | 8 | 1.000 | 3 | 10 | 0.686 | 8 | 5 | 0.689 |
| >65 | 15 | 6 | 9 | 5 | 10 | 11 | 4 | |||
| Gender | ||||||||||
| Male | 21 | 9 | 12 | 0.668 | 4 | 17 | 0.142 | 15 | 6 | 0.646 |
| Female | 7 | 2 | 5 | 4 | 3 | 4 | 3 | |||
| Clinical stage | ||||||||||
| Ⅰ+Ⅱ | 16 | 8 | 8 | 0.253 | 7 | 9 | 0.088 | 11 | 5 | 1.000 |
| Ⅲ+Ⅳ | 12 | 3 | 9 | 1 | 11 | 8 | 4 | |||
| Location of the tumor | ||||||||||
| Upper | 4 | 4 | 0 | 0.016* | 0 | 4 | 0.549 | 2 | 2 | 0.574 |
| Central and lower | 24 | 7 | 17 | 5 | 16 | 17 | 7 | |||
| Differentiation | ||||||||||
| Well and moderated | 17 | 10 | 7 | 0.016* | 7 | 10 | 0.099 | 9 | 8 | 0.049* |
| Poor | 11 | 1 | 10 | 1 | 10 | 10 | 1 | |||
| Distant metastasis | ||||||||||
| Positive | 1 | 0 | 1 | 1.000 | 0 | 1 | 1.000 | 1 | 0 | 1.000 |
| Negative | 27 | 11 | 16 | 8 | 19 | 18 | 9 | |||
| Lymph node metastasis | ||||||||||
| Positive | 11 | 4 | 7 | 1.000 | 2 | 9 | 0.419 | 8 | 3 | 0.704 |
| Negative | 17 | 7 | 10 | 6 | 11 | 11 | 6 | |||
*: P<0.05.
Protein expression of PAR1, PAR2 and PAR4 in tissues and cells of ESCC
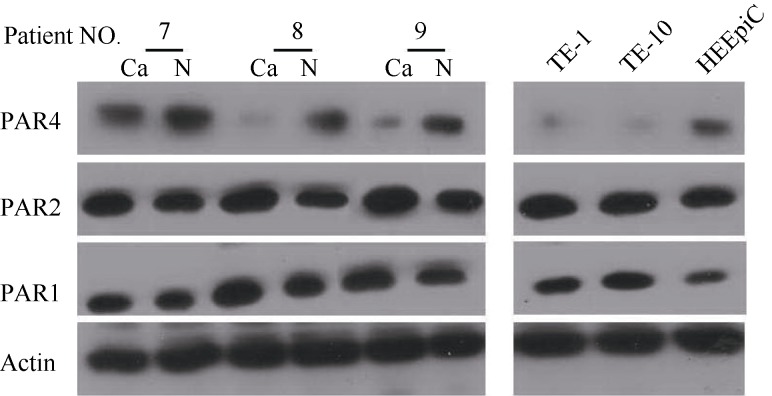
Protein expression was determined by taking β-actin as the internal reference (Figure 2). The expression of PAR1 and PAR2 proteins were increased (P<0.05), whereas, those of PAR4 were decreased (P<0.05). The light degrees of protein expression of PAR1, PAR2 and PAR4 in sample tissues are shown in Table 3. The expression of PAR1 and PAR2 were high and that of PAR4 was low in TE-1 and TE-10 cells.
Figure 2.
Protein expression of PAR1, PAR2 and PAR4 in tissues and cells of esophageal squamous cell carcinoma
Table 3.
Gray values of PAR expression in sample tissues
| Gene | Cancer tissue | Normal tissue | t | P |
|---|---|---|---|---|
| PAR4 | 0.18±0.05 | 0.42±0.12 | 9.77 | 0.0001** |
| PAR2 | 0.84±0.28 | 0.69±0.25 | 7.97 | 0.039* |
| PAR1 | 0.74±0.31 | 0.58±0.21 | 2.26 | 0.027* |
*: P<0.05; **: P<0.01.
PAR4 promoter hypermethylation in tissues and cells of ESCC
The BSP results of the PCR products of CpG sites in PAR4 are shown in Table 4. In HEEpiC, TE-1 and TE-10, esophageal and normal esophagus tissue from case No. 7, the methylation frequency of the 19 CpG sites in PAR4 was 35.4%, 95.2%, 83.8%, 62.6% and 48.2%, respectively.
Table 4.
Methylation frequency of the 19 CpG sites in PAR4 of cells and tissue of esophageal squamous cell carcinoma
| CpG position | Gene | 3 | 12 | 41 | 68 | 95 | 124 | 195 | 215 | 227 | 259 | 275 | 292 | 327 | 330 | 332 | 341 | 345 | 350 | 377 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Me-CpG (%) |
HEEPIC | 18 | 73 | 0 | 64 | 27 | 55 | 73 | 9 | 46 | 0 | 46 | 55 | 27 | 0 | 0 | 0 | 73 | 55 | 55 | 35.4 |
| TE-1 | 100 | 100 | 90.9 | 100 | 100 | 81.8 | 100 | 30 | 100 | 100 | 82 | 91 | 100 | 100 | 91 | 91 | 100 | 100 | 100 | 95.2 | |
| TE-10 | 82 | 100 | 100 | 100 | 100 | 100 | 100 | 91 | 100 | 100 | 46 | 73 | 55 | 55 | 100 | 100 | 100 | 55 | 30 | 83.8 | |
| Cancer tissue | 64 | 46 | 30 | 46 | 64 | 9 | 64 | 100 | 91 | 100 | 100 | 100 | 91 | 91 | 9 | 46 | 46 | 30 | 64 | 62.6 | |
| Normal tissue | 91 | 64 | 9 | 55 | 64 | 30 | 55 | 9 | 55 | 100 | 100 | 9 | 0 | 55 | 55 | 30 | 64 | 30 | 46 | 48.2 |
DISCUSSION
PAR2 over-expression is commonly found in malignant tumors. In this study, among 28 cases of ESCC, 20 (71%) cases were found with high PAR2 mRNA expression. The expression of PAR2 mRNA was also increased in TE-1 and TE-10 cells. The gray value indicates that PAR2 protein expression is increased in ESCC (P=0.039). PAR2 can be activated by trypsinase, human mast cell tryptase (MCT), coagulation factor VIIa and tissue factor complex (Caruso et al, 2006; Uusitalo-Jarvinen et al, 2007). The in vitro synthesized SLIGKV by the degradation of PAR2 can also activate PAR2 (Déry et al, 1998). Activated PAR2 may release vascular endothelial growth factor (VEGF), Interleukin-6 (IL-6) and IL-8 of tumor cells and thereafter promote the generation and invasion of novel vessels by malignant tumors (Knecht et al, 2007; Matej et al, 2007). Activated PAR2 may also increase the reverse activity of epidermal growth factor (EGF) and the release of transformation growth factor α (TGFα), and thus promote the proliferation of gastrointestinal cancer cells in stomach cancer, colon cancer, pancreatic cancer and ESCC (Darmoul et al, 2004; Fujimoto et al, 2006; Yada et al, 2005).
PAR1 and PAR4 are both thrombin receptors. Among the 28 cases of ESCC examined in the present study, 17 (60%) cases were found with high expression of PAR1 mRNA; however, 19 (68%) cases were found with low expression of PAR4 mRNA. The expression of PAR1 mRNA was increased in TE-1 and TE-10 cells, whereas, that of PAR4 was decreased. The gray value indicates that PAR1 protein expression was increased in ESCC (P=0.027) and that of PAR4 was significantly decreased (P=0.0001). High PAR1 expression was found in ESCC, particularly in central and lower (P=0.016) and poorly differentiated ESCC (P=0.049), whereas, the low expression of PAR4 was found in ESCC, particularly in poorly differentiated ESCC.
To investigate the underlying mechanisms of PAR4 and ESCC, methylation of the PAR4 promoter in tissues and cells of ESCC was evaluated. The results show that the methylation frequencies of PAR4 promoters in sample tissues of ESCC (62.6%) and TE-1 and TE-10 (83.8% and 95.2%, respectively) were both high. Together with the fact that the expression of PAR4 mRNA and protein is lower in tissues and cells of ESCC, these findings indicate that the methylation frequency of PAR4 promoters may play a role in its expression in ESCC.
Kawabata et al (1999) found the relaxation induced by PAR4 aggravates duodenal-gastric-esophageal reflux, and trypsinase within the reflux liquid may activate PAR4, thus promoting the incidence of ESCC. Han et al (2011) found that the promoting effects of PAR4 on anti-angiogenesis factors, including endostatin, thrombostondin 1, α2 macroglobulin, plasminogen activator, angiostatin and enzyme inhibitors may remarkably inhibit the generation of novel vessels and tumors. Human blood platelets only express PAR1 and PAR4. Activated PAR4 inhabits the release of VEGF but promotes the expression of endostatin, whereas, the effects of activated PAR1 are the opposite (Ma et al, 2005). Longitudinal gastrointestinal smooth muscles are intensified by PAR1 but relaxed by PAR4. PAR4 also prevents over intension in smooth muscles induced by PAR1 and thrombin (Lan et al, 2000). Cunningham et al (2012) found that similar interactions between PAR2 and PAR4 may play a role in locating receptors and cellular signal transduction.
In sum, here we investigated the expression characteristics of PAR1, PAR2 and PAR4 in ESCC tissues to broaden our understanding of PARs in ESCC. As G-protein-coupled receptors, the complicated biological distribution and function of PAR1, PAR2 and PAR4 remain unclear and their activity and interactions require further attention.
Funding Statement
This study was supported by the National Natural Foundation of China (81160302),the Major Research Project of Yunnan Province (2011FZ109),and Research project of Yunnan Education Bureau (2014Y153)
Footnotes
The authors have declared that no competing interests exist.
References
- [1].Caruso R, Pallone F, Fina D, Gioia V, Peluso I, Caprioli F, Stolfi C, Perfetti A, Spagnoli LG, Palmieri G, Macdonald TT, Monteleone G. 2006. Protease-activated receptor-2 activation in gastric cancer cells promotes epidermal growth factor receptor trans-activation and proliferation. The American Journal of Pathology, 169(1): 268-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cunningham MR, McIntosh KA, Pediani JD, Robben J, Cooke AE, Nilsson M, Gould GW, Mundell S, Milligan G, Plevin R. 2012. Novel role for proteinase-activated receptor 2 (PAR2) in membrane trafficking of proteinase-activated receptor 4 (PAR4). Journal of Biological Chemistry, 287(20): 16656-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Darmoul D, Gratio V, Devaud H, Laburthe M. 2004. Protease-activated receptor 2 in colon cancer: trypsin-induced MAPK phosphorylation and cell proliferation are mediated by epidermal growth factor receptor transactivation. Journal of Biological Chemistry, 279(20): 20927-34. [DOI] [PubMed] [Google Scholar]
- [4].Déry O, Corvera CU, Steinhoff M, Bunnett NW. 1998. Proteinase-activated receptors: novel mechanisms of signaling by serine proteases. The American Journal of Physiology, 274(6): C1429-52. [DOI] [PubMed] [Google Scholar]
- [5].Fujimoto D, Hirono Y, Goi T, Katayama K, Hirose K, Yamaguchi A. 2006. Expression of protease activated receptor-2 (PAR-2) in gastric cancer. Journal of Surgical Oncology, 93(2): 139-44. [DOI] [PubMed] [Google Scholar]
- [6].Gratio V, Walker F, Lehy T, Laburthe M, Darmoul D. 2009. Aberrant expression of proteinase-activated receptor 4 promotes colon cancer cell proliferation through a persistent signaling that involves Src and ErbB-2 kinase. International Journal of Cancer, 124(7): 1517-25. [DOI] [PubMed] [Google Scholar]
- [7].Han N, Jin K, He KF, Cao J, Teng LS. 2011. Protease-activated receptors in cancer: A systematic review. Oncology Letters, 2(4): 599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ikeda G, Isaji S, Chandra B, Watanabe M, Kawarada Y. 1999. Prognostic significance of biologic factors in squamous cell carcinoma of the esophagus. Cancer, 86(8): 1396-405. [DOI] [PubMed] [Google Scholar]
- [9].Jiang P, Yu GY, Zhang Y, Xiang Y, Hua HR, Bian L, Wang CY, Lee WH, Zhang Y. 2013. Down-regulation of protease-activated receptor 4 in lung adenocarcinoma is associated with a more aggressive phenotype. Asian Pacific Journal of Cancer Prevention, 14(6): 3793-8. [DOI] [PubMed] [Google Scholar]
- [10].Kaufmann R, Rahn S, Pollrich K, Hertel J, Dittmar Y, Hommann M, Henklein P, Biskup C, Westermann M, Hollenberg MD, Settmacher U. 2007. Thrombin-mediated hepatocellular carcinoma cell migration: cooperative action via proteinase-activated receptors 1 and 4. Journal of Cellular Physiology, 211(3): 699-707. [DOI] [PubMed] [Google Scholar]
- [11].Kawabata A, Kuroda R, Nishikawa H, Kawai K. 1999. Modulation by protease-activated receptors of the rat duodenal motility in vitro: possible mechanisms underlying the evoked contraction and relaxation. British Journal of Pharmacology, 128(4): 865-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Knecht W, Cottrell GS, Amadesi S, Mohlin J, Skåregärde A, Gedda K, Peterson A, Chapman K, Hollenberg MD, Vergnolle N, Bunnett NW. 2007. Trypsin IV or mesotrypsin and p23 cleave protease-activated receptors 1 and 2 to induce inflammation and hyperalgesia. Journal of Biological Chemistry, 282(36): 26089-100. [DOI] [PubMed] [Google Scholar]
- [13].Lan RS, Stewart GA, Henry PJ. 2000. Modulation of airway smooth muscle tone by protease activated receptor-1, -2, -3 and -4 in trachea isolated from influenza A virus-infected mice. British Journal of Pharmacology, 129(1): 63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ma L, Perini R, McKnight W, Dicay M, Klein A, Hollenberg MD, Wallace JL. 2005. Proteinase-activated receptors 1 and 4 counter-regulate endostatin and VEGF release from human platelets. Proceedings of the National Academy of Sciences of the United States of America, 102(1): 216-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Macfarlane SR, Seatter MJ, Kanke T, Hunter GD, Plevin R. 2001. Proteinase-activated receptors. Pharmacological Reviews, 53(2): 245-82. [PubMed] [Google Scholar]
- [16].Matej R, Mandáková P, Netíková I, Poucková P, Olejár T. 2007. Proteinase-activated receptor-2 expression in breast cancer and the role of trypsin on growth and metabolism of breast cancer cell line MDA MB-231. Physiological Research, 56(4): 475-84. [DOI] [PubMed] [Google Scholar]
- [17].Ribeiro FS, Simão TA, Amoêdo ND, Andreollo NA, Lopes LR, Acatauassu R, Rumjanek FD, Albano RM, Pinto LF, Monteiro RQ. 2009. Evidence for increased expression of tissue factor and protease-activated receptor-1 in human esophageal cancer. Oncology Reports, 21(6): 1599-604. [DOI] [PubMed] [Google Scholar]
- [18].Uusitalo-Jarvinen H, Kurokawa T, Mueller BM, Andrade-Gordon P, Friedlander M, Ruf W. 2007. Role of protease activated receptor 1 and 2 signaling in hypoxia-induced angiogenesis. Arteriosclerosis, Thrombosis, and Vascular Biology, 27(6): 1456-62. [DOI] [PubMed] [Google Scholar]
- [19].Wang X, Liu HT, Li S, Li K, Lin N, Fan QX, Zheng YL. 2010. Prognostic value of protease-activated receptor 2 expression in oesophageal squamous cell carcinoma. The Journal of International Medical Research, 38(4): 1381-8. [DOI] [PubMed] [Google Scholar]
- [20].Xu WF, Andersen H, Whitmore TE, Presnell SR, Yee DP, Ching A, Gilbert T, Davie EW, Foster DC. 1998. Cloning and characterization of human protease-activated receptor 4. Proceedings of the National Academy of Sciences of the United States of America, 95(12): 6642-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yada K, Shibata K, Matsumoto T, Ohta M, Yokoyama S, Kitano S. 2005. Protease-activated receptor-2 regulates cell proliferation and enhances cyclooxygenase-2 mRNA expression in human pancreatic cancer cells. Journal of Surgical Oncology, 89(2): 79-85. [DOI] [PubMed] [Google Scholar]
- [22].Zhang Y, Yu GY, Jiang P, Xiang Y, Li WL, Lee W, Zhang Y. 2011. Decreased expression of protease-activated receptor 4 in human gastric cancer. The International Journal of Biochemistry & Cell Biology, 43(9): 1277-83. [DOI] [PubMed] [Google Scholar]