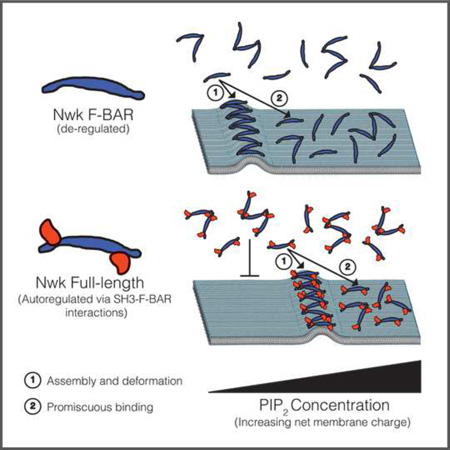
Summary
F-BAR domain proteins regulate and sense membrane curvature by interacting with negatively charged phospholipids and assembling into higher-order scaffolds. However, regulatory mechanisms controlling these interactions are poorly understood. Here, we show that Drosophila Nervous Wreck (Nwk) is autoregulated by a C-terminal SH3 domain module that interacts directly with its F-BAR domain. Surprisingly, this autoregulation does not mediate a simple “on-off” switch for membrane remodeling. Instead, the isolated Nwk F-BAR domain efficiently assembles into higher-order structures and deforms membranes only within a limited range of negative membrane charge, and autoregulation elevates this range. Thus, autoregulation could either reduce membrane binding or promote higher order assembly, depending on local cellular membrane composition. Our findings uncover an unexpected mechanism by which lipid composition directs membrane remodeling.
Keywords: Drosophila; Nwk; F-BAR domain; SH3 domain; membrane; PI(4,5)P2
Graphical abstract
Introduction
Membrane remodeling is critical for cellular processes such as cargo trafficking, signaling, cell motility, and organelle biogenesis, and requires the concerted action of scores of proteins that bind and actively shape cellular membranes. However, we still do not understand the underlying mechanisms that spatially and temporally target the activities of these proteins within the cell. BAR (Bin/Amphiphysin/Rvs) domain family proteins are important mediators of cellular membrane remodeling, and form banana-shaped α-helical dimers that use a positively charged binding surface to interact with negatively charged membrane phospholipids (McMahon and Boucrot, 2015). Further, they assemble into stable higher-order scaffolds that induce or stabilize membrane curvature over hundreds of nanometers (Frost et al., 2008; Mim et al., 2012; Becalska et al., 2013; Zhao et al., 2013). While it remains uncertain whether BAR domains actively generate or stabilize membrane curvature in vivo, in either case these activities require tight cellular regulation. However, the molecular mechanisms that regulate and target these proteins have only begun to be explored (Roberts-Galbraith and Gould, 2010).
Many BAR-domain proteins possess Src-homology 3 (SH3) domains that mediate interactions with regulators of the actin cytoskeleton, and other membrane-remodeling proteins such as dynamin (Owen et al., 1998; Takei et al., 1999; Itoh et al., 2005; Neumann and Schmid, 2013). Recently, SH3 domains have been implicated in directly regulating BAR domains via intramolecular interactions (Wang et al., 2009; Guerrier et al., 2009; Rao et al., 2010; Vazquez et al., 2013; Meinecke et al., 2013; Kast et al., 2014; Chen et al., 2014; Wu and Baumgart, 2014). In general, SH3 domain interactions inhibit the ability of BAR domains to induce membrane remodeling when overexpressed in cells and in vitro; however, only limited evidence exists for the role of SH3-mediated autoregulation of BAR domains in vivo (Kumar et al., 2009; Guerrier et al., 2009). Further, little is known about how SH3 domain autoregulation affects different steps of BAR domain membrane binding, higher order assembly, and deformation.
Nervous wreck (nwk) encodes a protein containing an N-terminal Fes/Cip4 homology-BAR (F-BAR) domain and two SH3 domains. Nwk regulates the traffic and signaling output of synaptic growth receptors at the Drosophila neuromuscular junction (NMJ) through its interactions with the membrane, actin nucleation machinery, and other endocytic proteins including dynamin, Dap160/Intersectin, and Sorting Nexin 16 (Rodal et al., 2008; O’Connor-Giles et al., 2008; Rodal et al., 2011). These activities and interactions define a recycling route by which activated receptors can be removed from signal-permissive early endosomes, downregulating their activities and controlling synaptic growth (Rodal et al., 2011). Mammalian Nwk homologs have been implicated in membrane remodeling and receptor traffic in stereocilia and in cerebellar granule neurons (Cao et al., 2013; Sun et al., 2014). Unraveling these trafficking pathways requires a deeper understanding of the membrane-deforming activities of Nwk. We previously reported that the F-BAR domain of Nwk binds to negatively charged phospholipids, similar to canonical F-BAR domains. The Nwk F-BAR self-assembles into zig-zags, distinct from canonical F-BAR proteins, and thus induces membrane scallops and ridges rather than membrane tubules (Becalska et al., 2013). Here, we describe regulatory mechanisms that direct the membrane remodeling activity of the Nwk F-BAR domain in the context of the full-length protein.
Results
The F-BAR domain and C-terminus of Nwk are required for in vivo localization and function
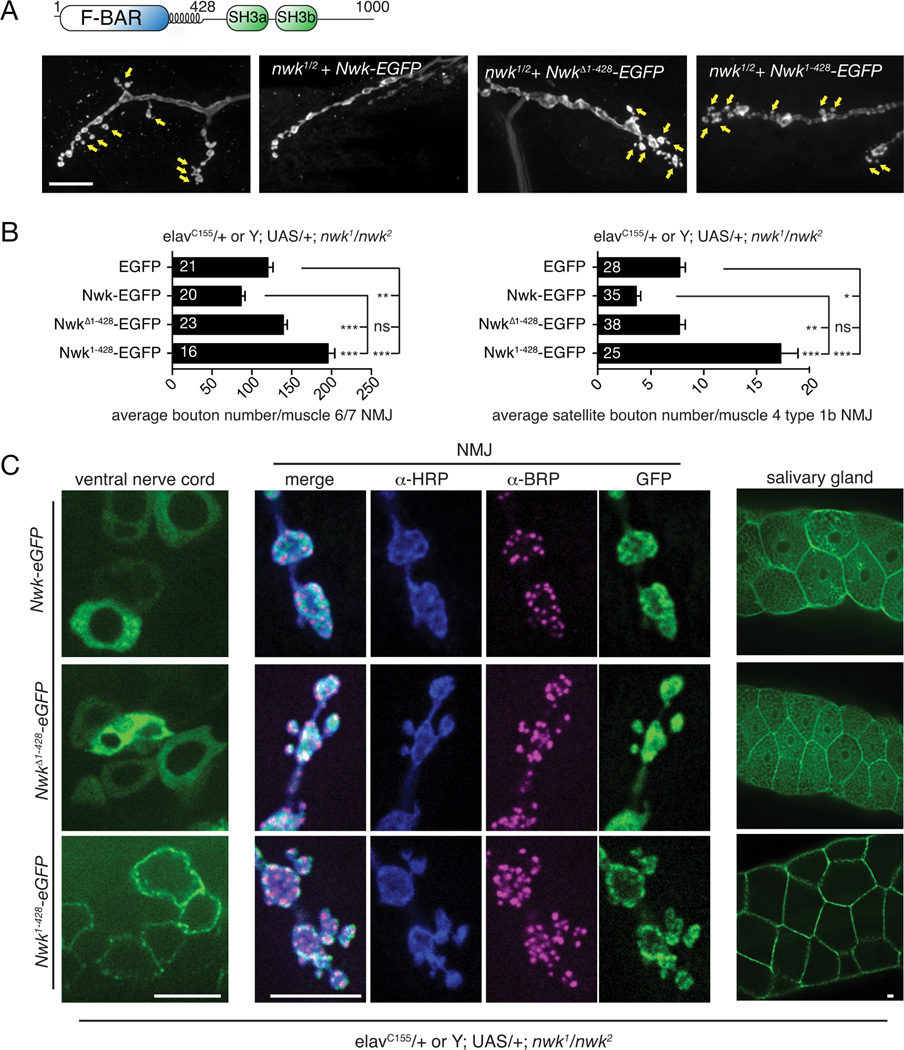
To investigate the importance of Nwk membrane-remodeling activity in vivo, we tested the role of the F-BAR domain in Nwk localization and synaptic growth at the Drosophila NMJ. We expressed EGFP-tagged Nwk variants (full-length Nwk (Nwk-EGFP), Nwk lacking its F-BAR domain (NwkΔ1–428-EGFP), and the Nwk F-BAR domain alone (Nwk1–428-EGFP)) in the nwk null mutant background (Fig. 1A, B, S1A, B). As has been previously reported for the nwk null and other endocytic traffic mutants (Coyle et al., 2004; Dickman et al., 2006), nwk mutants expressing GFP alone exhibit an increase in total synaptic bouton number at the NMJ, as well as an increase in “satellite” boutons that bud off the main axis of the axon terminal (arrows, Fig. 1A). This phenotype is a result of excess growth factor signaling in the absence of normal membrane recycling pathways (Rodal et al., 2008; O’Connor-Giles et al., 2008). Nwk-EGFP expression rescued the synaptic overgrowth and satellite bouton phenotypes of nwk mutants, while NwkΔ1–428-EGFP exhibited no rescuing activity (Fig. 1B). In contrast, Nwk1–428-EGFP exhibited a more severe synaptic overgrowth and satellite bouton phenotype than the nwk null mutant (Fig. 1B). These data indicate that the C-terminus of Nwk is required to properly regulate its function.
Figure 1. The Nwk F-BAR domain is required for its in vivo function.
(A) Schematic of Nwk domain structure and representative confocal images of α-Cpx-stained third instar larval NMJ morphology on muscle 4 for the indicated genotypes. Arrows indicate satellite boutons. (B) Quantification of synaptic growth on muscle 6/7 and satellite bouton number on muscle 4 by Nwk variants in a nwk1/nwk2 null background. Graphs show mean +/− s.e.m. Numbers in bar graphs represent the number of NMJs. (C) Localization of Nwk variants. GFP-tagged Nwk variants were expressed in the nwk1/nwk2 null background, under control of the GAL4 driver elavC155 (pan-neuronal and salivary glands). Third instar larvae were fixed and stained with α-BRP and α-HRP antibodies. Scale bars are 10 µm. Associated with Fig. S1.
To investigate the basis of the phenotypes of Nwk1–428-EGFP at the NMJ, we examined the localization of Nwk variants in fixed tissue in the nervous system, where endogenous Nwk is expressed (Coyle et al., 2004), At NMJs and in cell bodies in the ventral ganglion, Nwk-EGFP localized in a similar pattern to that previously reported for endogenous Nwk, in occasional puncta and in the “periactive zone” region surrounding Bruchpilot (BRP)-labeled active zones (Coyle et al., 2004). In contrast, NwkΔ1–428-EGFP exhibited a diffuse cytoplasmic localization, and Nwk1–428-EGFP was tightly localized to the plasma membrane (Fig. 1C). Similar results were obtained upon ectopic expression in larval salivary glands (Fig. 1C), where Nwk-EGFP and NwkΔ1–428-EGFP exhibited a very similar localization to the cytoplasm and to occasional puncta, while Nwk1–428-eGFP was strongly targeted to the plasma membrane. Taken together, these data indicate that selective membrane targeting of Nwk in vivo requires both the F-BAR domain and the C-terminal sequence including SH3 domains, and is essential for proper NMJ development.
Nwk C-terminal sequences regulate its membrane deforming activity
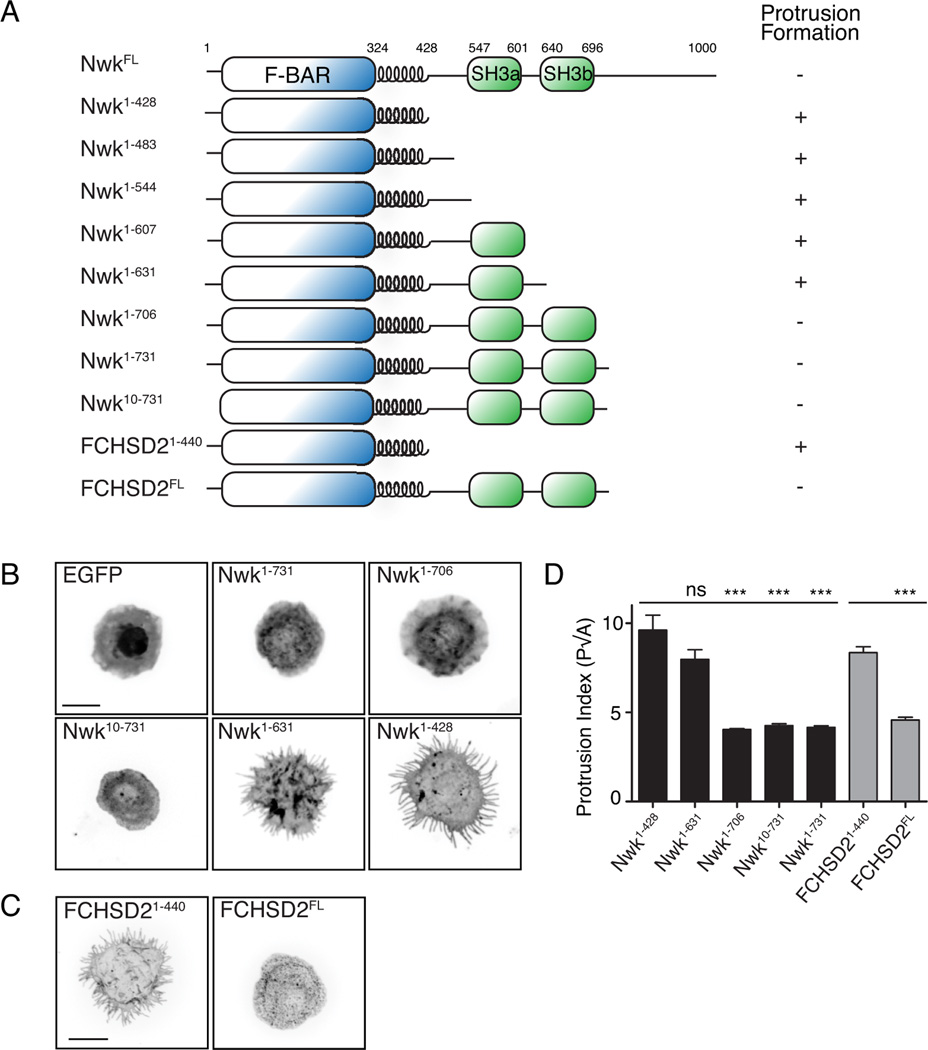
We next investigated the regulation of the Nwk F-BAR domain by C-terminal sequences, using a heterologous expression assay in Drosophila S2 cells, which do not express endogenous Nwk. We previously showed using this assay that the F-BAR domain of Nwk (Nwk1–428) localized to the S2 cell plasma membrane, and generated membrane buds that were extended into protrusions by the actin cytoskeleton (Becalska et al., 2013; Kelley et al., 2015). We tested the activities of GFP-tagged full-length Nwk or FCHSD2, a murine Nwk ortholog, and found that unlike their respective isolated F-BAR domains, they did not generate protrusions, and exhibited a predominantly cytoplasmic localization with occasional puncta (Fig. 2A–D), suggesting that the C-terminus of Nwk contains a conserved inhibitory activity for its F-BAR domain. We generated a series of C-terminal truncations of Nwk and quantified protrusion formation in S2 cells, and found that the inhibitory activity mapped to the second SH3 domain (SH3b; Fig. 2A, B, D). Given our previous finding that purified Nwk1–731 (which includes both SH3 domains) exhibits reduced membrane-binding compared to the isolated F-BAR domain (Becalska et al., 2013), SH3b domain-mediated inhibition is likely autoinhibitory, rather than due to a trans-acting factor. Further, inhibition did not require a short proline-rich stretch at the N-terminus of Nwk (Nwk10–731, Fig. 2A, B, D), suggesting that this sequence does not mediate autoinhibition via canonical SH3 domain-polyproline interactions.
Figure 2. The activity of the Nwk F-BAR domain in S2 cells is negatively regulated by its SH3b domain.
(A) Schematic of Nwk constructs and summary of protrusion formation in cells. (B,C) Activity of GFP-tagged Nwk truncations in S2 cells. Images show GFP fluorescence (in inverted contrast) of a maximum intensity Z-projection. Scale bars are 10 µm. (D) Quantification of cellular morphology (perimeter (P)/square root of area (A)). Data are represented as mean +/− s.e.m. from at least 9 cells per condition, ***p<0.001.
Nwk SH3 domains bind to the F-BAR domain tips via electrostatic interactions
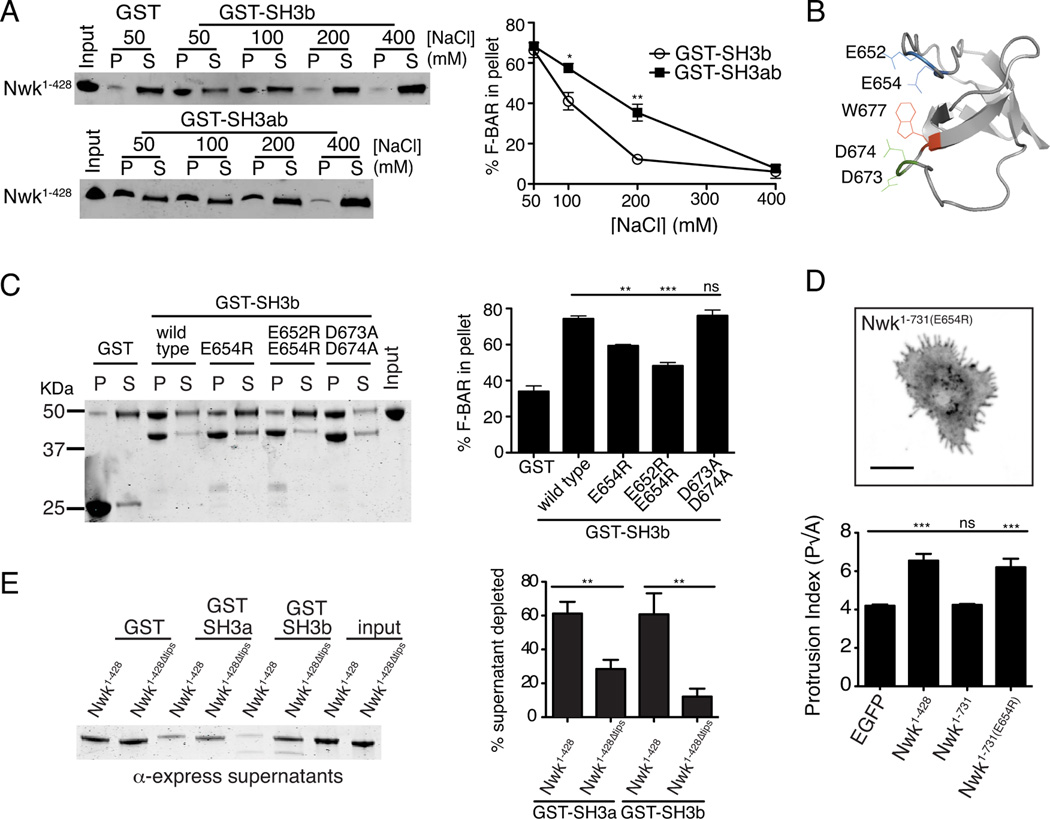
We next tested the prediction that autoinhibition is mediated by direct interactions between the Nwk F-BAR and SH3b domain using GST pull-down assays with purified Nwk fragments, and found that the F-BAR domain could co-precipitate with the SH3b domain (Fig. 3A). We then tested the salt sensitivity of Nwk F-BAR-SH3 interactions, to differentiate between hydrophobic interactions (required for canonical proline-rich ligands) or electrostatic interactions. Co-precipitation of the Nwk F-BAR domain with the GST-SH3b domain was sensitive to increasing ionic strength (Fig. 3A). Further, interaction of the F-BAR domain with a tandem GST-SH3ab domain was less sensitive than the GST-SH3b domain alone, suggesting that both SH3 domains contribute to the F-BAR C-terminus interaction (Fig. 3A).
Figure 3. Direct interactions between the Nwk SH3 and F-BAR domains.
GST fusion proteins were immobilized on glutathione agarose and incubated with the indicated purified proteins. Pellets and supernatants were fractionated by SDS-PAGE, immunoblotted or Coomassie stained, and quantified by densitometry: all graphs show the average +/− s.e.m. of three independent reactions. *p<0.05, **p<0.01, ***p<0.001. (A) Salt sensitivity of 6xHis-Xpress-Nwk1–428 (1.5 µM) co-sedimentation with GST-Nwk-SH3 domains (2–3 µM). Image shows representative anti-Xpress tag immunoblot of co-sedimentation assay. (B) Phyre model of Nwk SH3 domain (based on BIN1/AMPH2 SH3 (PDB C1mv3A)). (C) Co-sedimentation of Nwk1–428 (1.5 µM) with GST or GST-Nwk-SH3b variants (3 µM). Image shows representative Coomassie-stained gel. (D) Activity of Nwk1–731(E654R) in S2 cells Image shows GFP fluorescence (in inverted contrast) for a maximum intensity Z-projection. Scale bar is 10 µm. Quantification of cellular morphology (perimeter(P)/square root of area(A)) is shown below. (E) Co-sedimentation of Nwk1–428 (1.5 µM) with GST-Nwk-SH3b and GST-Nwk-SH3a (3–3.5 µM) depends on the charged dimer tips. Image shows a representative immunoblot. Associated with Fig. S2.
We then used the protein homology/analogy recognition engine program Phyre (Kelley and Sternberg, 2009) to model the Nwk SH3b domain against a fold library of all existing PDB crystal structures. The highest-scoring Phyre model was based on the structure of the BIN1/AMPH2 SH3 domain (PDB structure c1mv3A). Using this model (Fig. 3B, Fig. S2A), we mutated conserved charged residues in the RT-loop of the Nwk SH3b domain (E652 and E654), which are predicted to be in a similar orientation to the residues mediating electrostatic interactions of the Syndapin 1 SH3 domain with its F-BAR domain (Rao et al., 2010). In addition, we mutated conserved charged residues in the N-src loop of the Nwk SH3b domain (D673 and D674) (Fig. 3C, S2A). Nwk-SH3bE654R showed reduced F-BAR binding, while E652, D673 and D674 did not appear to play critical roles (Fig. 3C). Interestingly, Nwk-SH3bE654R did not disrupt binding to a previously identified ligand, Dap160 (Rodal et al., 2008), suggesting that the function of E654 in binding to the Nwk F-BAR is a specific rather than general feature of the SH3 domain (Fig. S2B). Finally, to test whether disrupting the interaction between the SH3b and F-BAR domains activates Nwk membrane-remodeling activity in cells, we transfected S2 cells with NwkE654R and examined protrusion formation. Cells expressing NwkE654R exhibited membrane-remodeling activity similar to Nwk1–428 (Fig. 3D). Thus, SH3-domain dependent inhibition of Nwk F-BAR activity is likely mediated primarily by electrostatic interactions and not through canonical SH3-proline motif interactions.
We next examined which site on the F-BAR domain is required for electrostatic interactions with the SH3 domain module. The tips of the Nwk F-BAR domain feature strong positive charge, which is important for high-affinity membrane association in vitro and membrane-deforming activity in S2 cells (Becalska et al., 2013). Nwk1–428Δtips lacks the positively charged tips, but exhibits a similar urea denaturation profile to Nwk1–428 and is likely to be well-folded in solution (Becalska et al., 2013). Nwk SH3a and SH3b exhibited significantly reduced binding to Nwk1–428Δtips compared to wild-type Nwk1–428 (Fig. 3E). These results indicate that the F-BAR tips are specifically required for Nwk SH3-F-BAR interactions in solution.
Nwk SH3 domains decrease F-BAR membrane binding
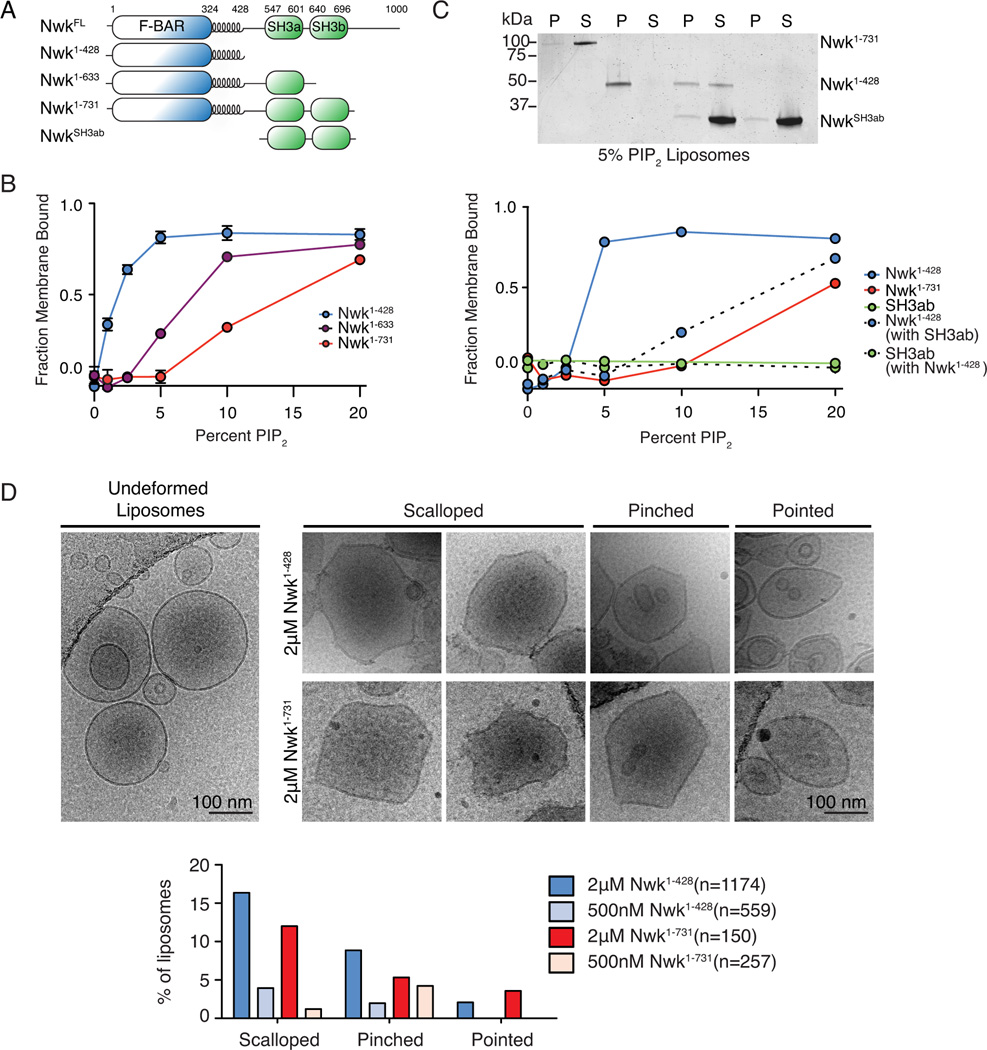
To test the role of the Nwk SH3 domains in regulation of F-BAR domain activity, we evaluated how the interaction between these domains affects membrane binding by purified Nwk. We previously showed that when compared to Nwk1–428 in bulk liposome co-sedimentation assays, Nwk1–731 requires more PI(4,5)P2 to bind to membranes (Becalska et al., 2013). To determine if Nwk SH3 domains were required and sufficient for inhibition of membrane binding, we compared liposome co-sedimentation of Nwk1–428, Nwk1–731, and Nwk1–633 (SH3b domain truncated; Fig. 4A). Compared to Nwk1–428 and Nwk1–731, Nwk1–633 required an intermediate PI(4,5)P2 concentration for binding (Fig. 4B), suggesting that the SH3b domain is involved in, but not sufficient for, inhibiting membrane binding by purified Nwk1–731, and that intervening sequences (including the SH3a domain) contribute to the remaining autoinhibitory effect. In contrast, in S2 cells, relief of autoinhibition depends more strictly on the electrostatic surface of the SH3b domain (Fig. 2A, B, D), suggesting that in a cellular context, a binding partner may modulate the contribution of SH3a to autoinhibition.
Figure 4. Nwk SH3b inhibits membrane deformation by the Nwk F-BAR domain.
(A) Nwk constructs used for in vitro assays. (B, C) Purified proteins (10 µM NwkSH3ab (Nwk residues 536–731); 1 µM all other proteins) were incubated with liposomes of the following composition: 80-X% PC, 15% PE, 5% PS and X% PI(4,5)P2 (where X is the concentration indicated in the graph) and subjected to liposome cosedimentation assays. Graphs show mean densitometry from one (B) or three (mean +/− s.e.m.) (C) independent experiments. (C) Image shows representative Coomassie staining of supernatant (S) and pellet (P) fractions at 10% PI(4,5)P2. (D) Cryo-EM of control and Nwk deformed liposomes. Both purified Nwk1–428 and Nwk1–731 induce membrane scalloping, pointing, and pinching of 10% PI(4,5)P2 liposomes. Scale bar is 100 nm. Bar graph summarizes vesicle morphology after 30-minute incubation of Nwk1–428 or Nwk1–731 (2µM and 500nM) with 0.3 mM [DOPC:DOPE:DOPS:PI(4,5)P2] = 70:15:5:10 liposomes. n represents the number of liposomes examined; Associated with Fig. S3.
We then tested the effects of a purified SH3ab fragment in trans on Nwk1–428 membrane binding. We found that pre-incubation of the F-BAR domain with SH3ab dramatically shifted the PI(4,5)P2-dependent binding profile of Nwk1–428 to more closely resemble Nwk1–731 (Fig. 4C). Furthermore the SH3ab fragment did not co-sediment with liposomes, either on its own or with low or high concentrations of Nwk1–428 (Fig. 4C, Fig. S3A). These results suggest that the interaction with the Nwk F-BAR domain directly inhibits membrane binding in solution, and that the SH3 domains do not bind to the F-BAR domain when it is membrane-associated.
To test if autoregulation alters the properties of Nwk once bound to the membrane, we examined liposomes incubated with Nwk1–428 and Nwk1–731 by cryo-electron microscopy (cryo-EM). Liposomes incubated with Nwk1–731 exhibited the same types of protein-dependent deformations as we previously observed for Nwk1–428 (pointing, pinching and scalloping, Fig. 4D). These types of deformations did not change with longer protein-liposome incubation times (Fig S3B), and were observed more frequently with increasing protein concentration, suggesting that they are actively generated by Nwk (Fig. 4D, Fig. S3E, F). Finally, we did not observe any new properties such as vesiculation or tubulation for Nwk1–731 compared to Nwk1–428 under these conditions (Fig. 4D, Fig. S3C–E). Thus, SH3 domain-mediated autoregulation via F-BAR domain tips inhibits membrane binding in solution, without changing the membrane-deforming properties of Nwk.
Nwk-mediated membrane deformation is associated with assembly of F-BAR dimers into stable scaffolds
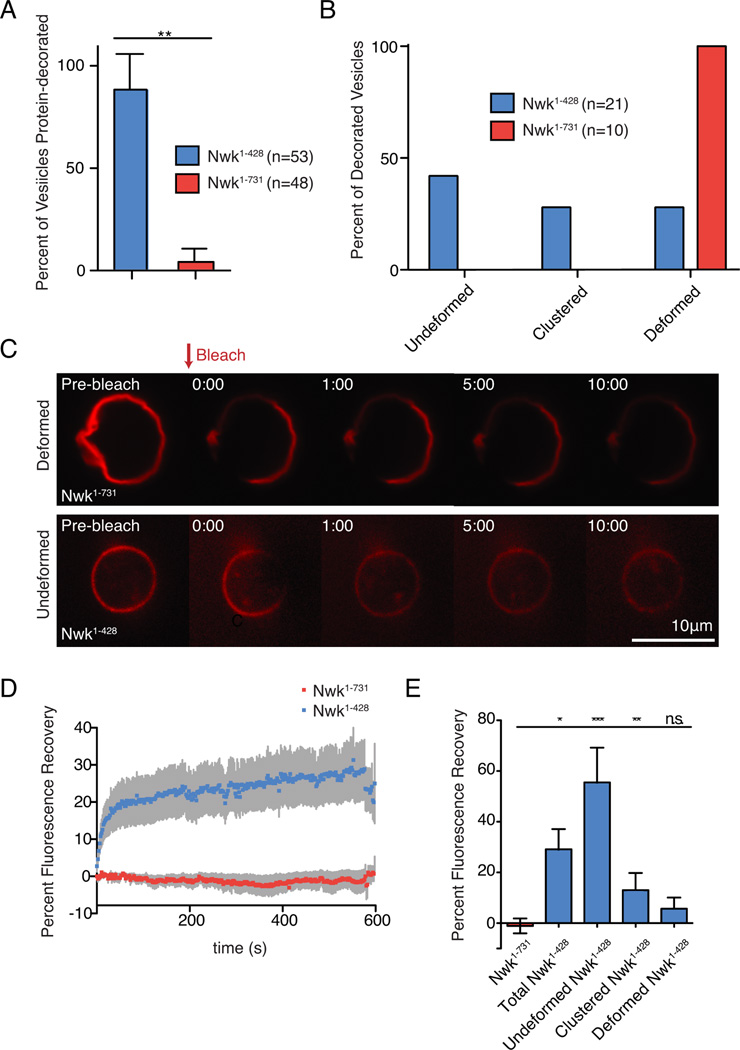
We next considered two possible explanations for reduced membrane binding for Nwk1–731 compared to Nwk1–428 in bulk assays: either Nwk1–731 could be uniformly reduced compared to Nwk1–428 on all liposomes (suggesting a general reduction in binding), or alternatively the fraction of liposomes bound by Nwk1–731 could be decreased (suggesting a more complex or cooperative model for binding). To distinguish between these possibilities, we imaged Alexa-549-SNAP-tagged Nwk1–428 or Nwk1–731 on NBD-phosphatidylethanolamine (PE)-labeled GUVs by confocal microscopy. Nwk1–731 strongly decorated a dramatically smaller fraction of GUVs compared to Nwk1–428 (Fig. 5A), supporting the second model. We previously showed that Nwk induces deformations including membrane flattening, pinching, and clustering on GUVs (Becalska et al., 2013). Strikingly, all Nwk1–731-bound GUVs were highly deformed, compared to a much smaller fraction of Nwk1–428-bound GUVs (Fig. 5B). Thus, though SH3 domain-mediated inhibition limits F-BAR interaction with the membrane, it unexpectedly enhances F-BAR membrane remodeling activity on the few vesicles that are decorated.
Figure 5. Nwk SH3 domains limit promiscuous membrane binding, and promote stable F-BAR protein scaffolds associated with membrane remodeling.
NBD-PE-labeled GUVs incubated with SNAP-549-tagged Nwk variants (500 nM) were imaged by spinning disk confocal microscopy. Lipid composition was [DOPC:POPE:DOPS:PI(4,5)P2:NBD-PE] = 75:14.5:5:5:.5. n represents the number of vesicles examined. (A) Percent of protein-decorated GUVs after 30-minute incubation with Nwk1–428 (blue) or Nwk1–731 (red). Graph represents mean +/− s.e.m. from 3 independent experiments. Data are identical to 5% PI(4,5)P2 data point in Figs. 6B, C. (B) Quantification of the morphology of protein-decorated GUVs after 30-minute incubation with purified Nwk1–428 or Nwk1–731. GUVs were imaged from n independent reactions. (C) Single spinning disc confocal slices of GUVs showing Nwk1–731 displaying limited recovery on deformed membranes, while partial recovery is observed on spherical, undeformed Nwk1–428-coated GUVs. Scale bar is 10 µm. Associated with Movies S1–S3. (D) Quantification of recovery of protein fluorescence for Nwk1–428 and Nwk1–731. The data are a mean of at least 10 independent experiments and the error bars indicate ± s.e.m. (E) Quantification of protein fluorescence recovery according to vesicle morphology. One-way ANOVA *p<0.05,**p<.005,***p<.001.
Higher-order assemblies are a conserved feature of BAR domains, and likely central to their membrane-remodeling properties. To determine if Nwk autoregulation affects its propensity to assemble on (and by extension, deform) membranes, we next examined higher-order assembly of SNAP-tagged Nwk using fluorescence recovery after photobleaching (FRAP) experiments. BAR domains can form stable scaffolds on membrane, limiting lipid fluidity and lateral diffusion of proteins (Zhao et al., 2013). Conversely, individually bound or incompletely oligomerized BAR domain dimers are expected to be mobile, either through exchange between the membrane and solution or rapid lipid diffusion. Therefore, assembled BAR domains should recover very slowly after photobleaching, while individually bound or disordered BAR domains should recover rapidly.
We photobleached a region corresponding to less than ∼10% of the GUV surface, and examined the recovery of fluorescent Nwk into this region (Fig. 5C and Movies S1–S3). For the entire population of GUVs, recovery of Nwk1–731 was significantly lower than Nwk1–428 ten minutes post-bleach (Fig. 5D,E), and the distribution of total recovery for Nwk1–428 was much wider, whereas Nwk1–731 recovery never exceeded 11.5%. We assessed recovery based on GUV morphology, and found that spherical vesicles had significantly higher total recovery (up to 75%) compared to deformed vesicles (less than 10%, Fig.5E). Notably, protein mobility on Nwk1–428 and Nwk1–731 GUVs that were deformed was not significantly different, indicating similar behavior of assembled F-BAR proteins at sites of deformation for both Nwk1–428 and Nwk1–731. These results suggest that under these conditions, the isolated F-BAR domain is capable of interacting with the membrane in a range of different assembly states with distinct FRAP profiles, from individually bound and disordered with rapid turnover and protein fluorescence recovery, to assembled stable protein scaffolds with slower turnover and protein fluorescence recovery. In contrast, Nwk1–731 protein coats do not recover after photobleaching, indicating that under these conditions all membrane-bound Nwk1–731 is assembled into stable higher-order scaffolds.
Nwk autoregulation increases the phosphoinositide requirement for membrane deformation without altering the mechanism of membrane binding
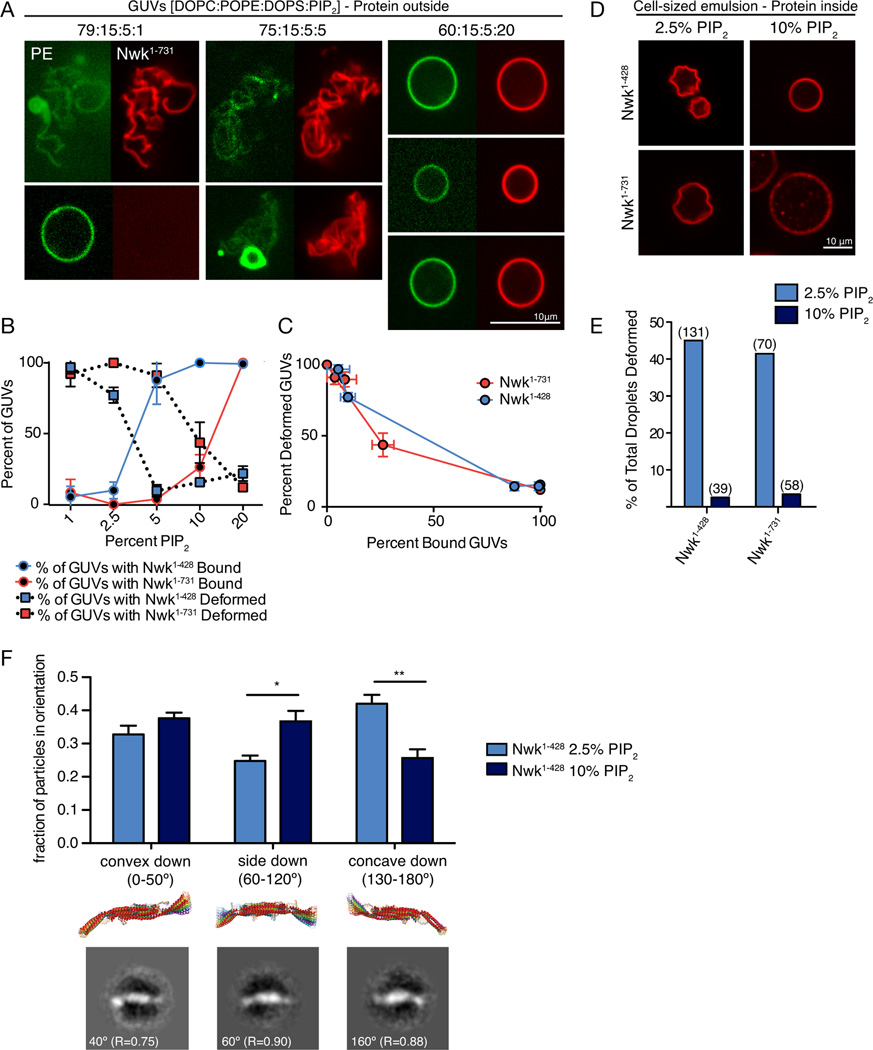
We next tested whether SH3 domains specifically enhance assembly of Nwk F-BAR domains, or if the differences in the assembly and deformation properties of Nwk1–428 and Nwk1–731 on GUVs of a fixed lipid composition (Fig. 5) reflect inherent differences in Nwk F-BAR behavior at different points on the PI(4,5)P2 binding curve (Fig. 4B). We first tested the ability of Nwk1–731 and Nwk1–428 to bind and deform GUVs with different PI(4,5)P2 compositions. Remarkably, at 1% PI(4,5)P2, a concentration at which Nwk1–428 is only partially membrane-associated in co-sedimentation assays (Fig. 4B), we found that it decorated only a small fraction of highly deformed GUVs, much like Nwk1–731 at 5% PI(4,5)P2 (Fig. 6A, B). Further, at 20% PI(4,5)P2, a concentration at which all of Nwk1–731 is bound to liposomes in co-sedimentation assays (Fig. 4B), all GUVs were bound but poorly deformed, much like Nwk1–428 at 5% PI(4,5)P2 (Fig. 6A, B). To rule out any quantitative differences in the behavior of Nwk1–428 and Nwk1–731, reflecting different mechanisms of assembly or deformation, we normalized the fraction of membrane-bound Nwk to the extent of membrane deformation at the full range of PI(4,5)P2 concentrations. Nwk1–428 and Nwk1–731 exhibited quantitatively indistinguishable behavior in this analysis (Fig. 6C). These results suggest an inherent bias of the F-BAR domain towards high-order scaffolds and membrane deformation when binding conditions are more stringent.
Figure 6. PI(4,5)P2 levels regulate F-BAR membrane binding and deformation.
Scale bars are 10 µM. (A) Representative images of NBD-PE-labeled GUVs of varying PI(4,5)P2 concentrations incubated with 500nM SNAP-549-Nwk1–731 for 30 minutes and imaged by spinning disk confocal microscopy. (B) Percent of vesicles bound by Nwk1–428 and Nwk1–731 at various PI(4,5)P2 concentrations (solid line) and percent of bound vesicles that are deformed (dotted line). Graph represents mean +/− s.e.m. from 2–3 independent experiments. 5% PI(4,5)P2 data is identical to Fig. 5A. (C) A plot of percent bound vesicles vs. percent-deformed (from data in (B)) shows an inverse relationship between the number bound and frequency of deformation (D) Representative images of cell-sized water droplets with 1 µM SNAP549-labeled BAR proteins. Emulsions were made with 23 mM lipid mixes of DPHPC +/− PI(4,5)P2 in decane and incubated for 1 hour before imaging. Associated with Movies S4–S5. (E) Quantification of the percentage of total vesicles with protein-induced deformation at 2.5% and 10% PI(4,5)P2 for Nwk1–428 and Nwk1–731. Number above bars represents the number of droplets examined. Associated with Fig. S4. (F) Comparison of the orientation of Nwk1–428 dimers on lipid monolayers with 2.5% or 10% PI(4,5)P2. Error bars represent fraction of particles +/− s.e.m. from 3–4 independent EM grids. Shown below are superimposed 10° rotations of the predicted Nwk1–313 structure (Becalska et al., 2013), and representative class averages from 10% PI(4,5)P2, grid A, Table S1. R represents correlation coefficients to the 20 Å-filtered predicted structure. Associated with Table S1.
Several different parameters could together account for the observation that a small number of GUVs are highly decorated and deformed under stringent binding conditions. F-BAR proteins may exhibit nucleation behavior, resulting in recruitment of protein molecules to a small fraction of vesicles, until the concentration of protein in solution is depleted below a critical assembly threshold (nucleation mechanism). However, our observations also raise the possibility that F-BAR proteins have inherently different activities on membranes of low versus high PI(4,5)P2 content (lipid-directed mechanism). To examine this possibility, we tested F-BAR domain-mediated membrane deformation in cell-sized water droplets encapsulated within a phospholipid membrane. In this system, protein is emulsified in a mix of lipid and oil, resulting in the formation of droplets of protein surrounded by an interface of lipid, with head groups facing the protein in the aqueous phase (Fig. S4A, (Hase and Yoshikawa, 2006; Miyazaki et al., 2015)). Compared to GUVs, droplets are isolated from each other and therefore not subject to nucleation effects. Both SNAP-Nwk1–428 and SNAP-Nwk1–731 induced deformation and crumpling of PI(4,5)P2-encapsulated droplets, but not droplets without PI(4,5)P2 (Fig. 6D, E, S4B and C, Movies S4 and S5). Further, the SNAP tag alone did not deform PI(4,5)P2-encapsulated droplets, indicating that crumpling is a specific activity of Nwk (Fig. S4B). Remarkably, deformation was prevalent at 2.5% PI(4,5)P2 for both Nwk1–428 and Nwk1–731, but rare at 10% PI(4,5)P2, supporting a lipid-directed mechanism that favors deformation at lower negative membrane charge (Fig. 6D, E).
One possible mechanism to account for differential activities of the Nwk F-BAR domains with changing negative charge could be the orientation of the F-BAR on the membrane. F-BAR domains interact with membranes through positively charged concave surfaces when assembled on highly curved membranes, but can also interact with membranes in a number of other orientations, including a deformation-inactive side-lying state (Frost et al., 2008; Yu and Schulten, 2013; Becalska et al., 2013). To test whether different modes of protein-membrane interaction may be favored depending on membrane charge, we used single particle electron microscopy on lipid monolayers to compare the orientations of the Nwk F-BAR domain at 2.5% and 10% PI(4,5)P2 (Fig. 6F). The Nwk F-BAR forms an S-shaped dimer whose chirality allows single particle averages to be assigned to concave-surface down, side-lying, or convex-surface down orientations (Fig. 6F and (Becalska et al., 2013)). We found that at 2.5% PI(4,5)P2, the concave surface-down orientation predominates, whereas at 10% PI(4,5)P2 Nwk does not show a strong proclivity for any one orientation. These data support a model by which limited membrane charge results in an orientation-selective binding mode, favoring assembly and membrane deformation, while high membrane charge favors promiscuous binding and diminishes the likelihood of assembly.
Nwk deforms cellular membranes within a sweet spot of PI(4,5)P2 concentration
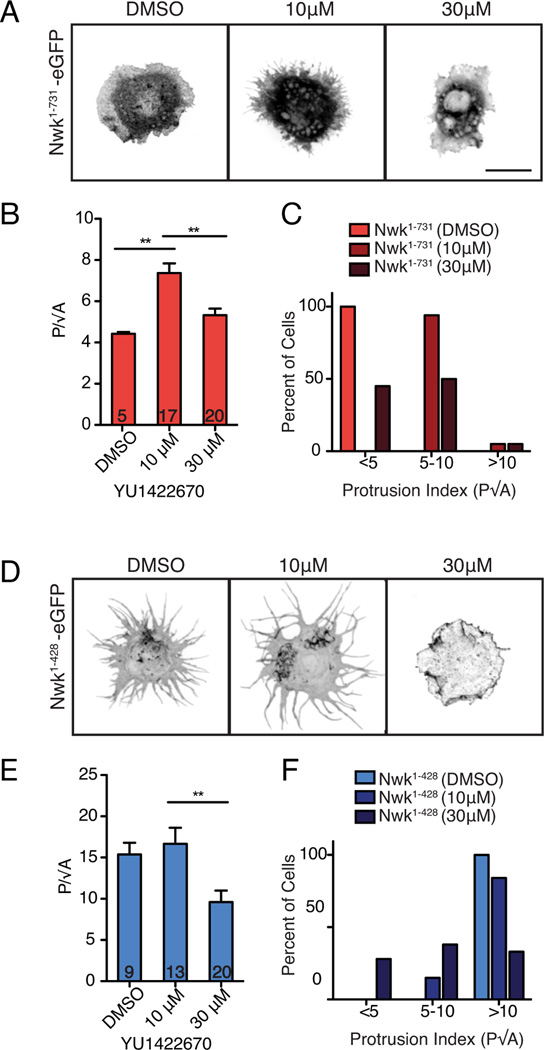
In order to test the role of PI(4,5)P2 levels in regulating Nwk F-BAR activity in vivo, we used our S2 cell assay. We acutely increased PI(4,5)P2 using YU1422670, a small molecule inhibitor of OCRL (Pirruccello et al., 2014), which is an important phosphoinositide-5-phosphatase in S2 cells (Ben El Kadhi et al., 2011). This compound is active in Drosophila, as it recapitulates the reported OCRL mutant phenotype of increased F-actin levels in S2 cells (Ben El Kadhi et al., 2011, Fig. S5). Treatment with low concentrations of the drug (10 µM) for 15 min increased cellular deformation in Nwk1–731-EGFP expressing S2 cells, supporting our in vitro finding that PI(4,5)P2 promotes membrane binding by Nwk1–731 (Fig. 7A–C). In contrast, high levels of drug (30 µM) decreased deformation of Nwk1–428-EGFP expressing cells, though Nwk was still localized to the membrane (Fig. 7D–F), supporting our finding that high levels of PI(4,5)P2 decouple membrane binding and deformation. YU1422670-treated cells that were not expressing Nwk had normal cellular morphology and actin localization (despite modestly increased levels of F-actin) (Fig. S5), indicating that the differences observed in Nwk-expressing cells after drug treatment are due to differential F-BAR activity. These results provide strong in vivo support for the model that PI(4,5)P2 levels can shift the F-BAR domain between unbound, membrane-deforming, and promiscuous binding states, and that autoregulation increases the PI(4,5)P2 concentration at which these states occur.
Figure 7. Cellular PI(4,5)P2 levels regulate Nwk-induced cellular deformation.
(A,D) Increasing cellular PI(4,5)P2 levels with the OCRL inhibitor YU1422670 alters Nwk-induced deformation. Images show GFP fluorescence (in inverted contrast) of maximum intensity z-projections. Scale bar is 10 µm. (B) Nwk1–731-EGFP expressing cells exhibit a significantly higher protrusion index (P/√A; mean +/− s.e.m.; number in bar graph represents n cells) at low concentrations of YU1422670 compared to DMSO control-treated cells, (C) with a higher percentage of cells with a protrusion index of >5. (E) Nwk1–428-EGFP expressing cells have a significantly lower protrusion index (P/√A; mean +/− s.e.m.; number in bar graph represents n cells) at high concentration (30µM) of YU1422670 compared to DMSO and 10µM YU1422670, (F) and a lower percentage of cells with a protrusion index of 10 or higher after drug treatment. Associated with Fig. S5
Discussion
Mechanisms of Nwk regulation by intramolecular interactions and membrane composition
Here, we report that the F-BAR domain of Nwk is autoregulated by its C-terminal SH3 domains, and that the Nwk C-terminus is critical for regulation of its F-BAR localization and membrane remodeling function in vivo. Surprisingly, membrane composition is a key factor in the efficiency of remodeling, and autoregulation alters the optimal membrane charge requirement for maximum F-BAR activity.
Nwk is one of several F-BAR proteins with two SH3 domains (Roberts-Galbraith and Gould, 2010), and we found that the SH3b domain mediates the majority of the inhibitory effect in a cellular context. This inhibition occurs via non-canonical electrostatic interactions between the SH3b and F-BAR domains, similar to Syndapin (Rao et al., 2010). Further, we show that the autoregulatory SH3 domains are displaced from the F-BAR domain upon lipid binding. Our results indicate that regulation may directly reduce the probability of membrane binding by occluding residues important for interactions with the membrane. Our finding that binding and deformation scale similarly for Nwk1–428 and Nwk1–731 (Fig. 6C) argues that they do not bind, oligomerize, or deform membranes by distinct mechanisms, and that SH3 domains do not actively promote higher-order assembly via inter-dimer interactions. Instead, the SH3 domains shift the threshold for binding so that binding and deformation occur at higher negative charge. Importantly, we show that both the isolated F-BAR domain and the full-length protein actively generate similar types of membrane deformations (Fig. 4D, Fig. 6D and Movies S4 and S5), arguing against the model that these proteins merely sense curvature, even under stringent binding conditions.
To date, autoregulation of BAR domain proteins has been thought of as an “on-off switch” for membrane binding, released by binding partners (Roberts-Galbraith and Gould, 2010; Meinecke et al., 2013) or by increased membrane charge (Wu and Baumgart, 2014). However, our data support a more nuanced explanation of these inhibitory interactions that depends critically on local membrane composition. While the net effect of autoinhibition is to shift the membrane-binding and membrane-deformation curves to higher negative charge, we found that the membrane-binding and membrane-deforming curves do not overlap, as would be predicted by the simple “on-off switch” model. Rather, our FRAP, GUV, and droplet deformation data indicate that higher-order assembly and deformation occur efficiently only within a range or “sweet spot” of negative charge, and that high levels of PI(4,5)P2 promote promiscuous membrane binding of the F-BAR domain.
A combination of protein-directed and lipid-directed mechanisms may account for this unexpected property of the Nwk F-BAR domain. Under stringent binding conditions and when the protein to lipid ratio is limiting (e.g. on GUVs), F-BAR domains exhibit nucleation behavior, resulting in favored binding and assembly on a small population of vesicles. Importantly, this is a physiologically relevant regime, since cellular membranes and proteins are present in limiting amounts, lipid composition is constantly changing, and F-BARs are acutely released from autoinhibition by regulatory binding partners. Nucleation of F-BAR domains may occur by several potential, nonexclusive mechanisms. F-BAR domains can induce clustering of PI(4,5)P2 (Zhao et al., 2013), which could drive further protein binding and deformation in a feed-forward mechanism. Conversely, PI(4,5)P2 clusters may form in areas of spontaneous local membrane curvature (Koldso et al., 2014), promoting curvature-sensitive or properly-oriented binding of F-BAR domains. Finally, membrane-bound F-BAR-F-BAR oligomers may increase the propensity for further protein assembly through the effects of avidity. In addition to nucleation effects, our cell-sized water droplet experiments (which eliminate protein sequestration due to nucleation) suggest that the Nwk F-BAR domain preferentially deforms membranes with lower negative charge. One mechanism for this behavior, supported by our single particle electron microscopy data, is that the F-BAR domain is biased towards a deformation-promoting (concave surface-down) conformation at low PI(4,5)P2. Another nonexclusive possibility is that at high PI(4,5)P2, the organization or rigidity of membrane itself may inhibit assembly, either by forming lipid clusters of a size that disfavor deformation, or by restricting dynamic rearrangements of individual proteins into higher assemblies (Ruiz-Herrero and Hagan, 2015). Assessing the relative contributions of these forces will require developing a theoretical framework that describes the nucleation, oligomerization, and lipid-binding behaviors of Nwk.
Other proteins in the BAR domain family are sensitive to negative membrane charge and may be similarly regulated by nucleation behavior and membrane composition. Indeed, elevated PI(4,5)P2 levels suppress the membrane deforming activity of the F-BAR protein FBP17 in vivo (Tsujita et al., 2015), suggesting that the “sweet spot” may be conserved feature of BAR domain activity. Further investigation will be required to determine whether BAR domains have different ranges of optimal membrane charge, which could contribute to the specificity of these proteins in the context of their cellular roles.
Contributions of membrane composition and autoregulation to the role of Nwk in vivo
Despite advances in our knowledge of the molecular mechanisms regulating BAR domain membrane curvature-sensing and sculpting properties, as well as the effects of their binding partners, we still lack a thorough understanding of how these activities are regulated and targeted in their cellular contexts. Our data show that Nwk requires both F-BAR activity and the C-terminal regulatory SH3 domains for its in vivo role in growth signal regulation at the Drosophila NMJ. Removing the Nwk C-terminus produces a more severe phenotype than the null mutant, suggesting that constraining and regulating Nwk F-BAR activity and targeting to the membrane via autoregulation is required for proper synapse formation. A critical next step is to consider the role of binding partners in modulating the state of the Nwk F-BAR on the membrane. Nwk interacts with dynamin and the endocytic scaffold Dap160/intersectin via the SH3a and SH3b domains respectively (Rodal et al., 2008; O’Connor-Giles et al., 2008). These interactions, in addition to localizing BAR domains, may modulate Nwk autoinhibition to actively fine-tune membrane binding and deformation at specific levels of negative membrane charge.
Our data also raise an important issue: the physiological significance of autoregulatory mechanisms depend not only on the effects of Nwk-binding partners on its activity, but also critically on the local PI(4,5)P2 concentration at which Nwk operates in vivo. A primary defining characteristic of biological membrane compartments is their lipid composition. BAR domains selectively associate with intracellular membranes rich in charged phosphoinositides (Becalska et al., 2013; Di Paolo and De Camilli, 2006; Saarikangas et al., 2010). PI(4,5)P2 concentrations have been estimated to be about 0.5–1% of total cellular membranes but are tightly regulated, and may transiently accumulate to much higher concentrations (Di Paolo and De Camilli, 2006). Submicrometer clusters of PI(4,5)P2 have been detected composed of as much as 80% PI(4,5)P2 (Milosevic et al., 2005; James et al., 2008; van den Bogaart et al., 2011). This suggests that PI(4,5)P2 exists in cells at a diverse range of concentrations that are dynamically altered by phosphoinositide kinases and phosphatases. Local, regulated changes in lipid composition could shift membrane-bound Nwk between a disassembled state and an assembled state that is capable of deforming membranes. Using a pharmacological approach to acutely increase PI(4,5)P2 levels in S2 cells, we found that similar to our in vitro results, membrane association and deformation by Nwk occur at a sensitive range of cellular PI(4,5)P2 that is elevated by autoregulation. Further tests of this lipid-directed mechanism of membrane remodeling in the nervous system will require the development of new methods to visualize endogenous Nwk-mediated membrane deformation and to acutely manipulate PI(4,5)P2 levels at the NMJ.
In summary, SH3-domain mediated autoinhibition of the Nwk F-BAR domain shifts the PI(4,5)P2-dependence of higher-order assembly and promiscuous binding of individual dimers to the membrane. We predict that through this mechanism, activating or inhibitory binding partners work in concert with temporal and spatial regulation of negative membrane charge to control membrane remodeling by BAR domains.
Materials and Methods
Fly stocks and NMJ morphology analysis
UAS-Nwk constructs were generated as described previously (Becalska et al., 2013), and injected into flies at Genetic Services Inc. Cambridge, MA, using Φc381 recombinase at the Attp40 locus. NMJs on muscle 6/7, segment A3 and muscle 4, segments A2-A3 were selected for analysis of morphology, in fixed 3rd instar larval fillets immunostained with α-Cpx (Huntwork and Littleton, 2007) and α-Dlg (Developmental Studies Hybridoma Bank) antibodies. Both type 1b and type 1s boutons were quantified on muscle 6/7. Only type Ib innervation, delineated by extensive postsynaptic α-Dlg staining, was quantified on muscle 4. Satellite boutons were defined as strings of five or fewer boutons extending from the main axis of the NMJ.
Cell culture
S2 cells were cultured, transfected with Effectene, spread for 1 h on Concanavalin-A-coated coverslips, and imaged on a spinning disk confocal microscope (see Supplemental methods) as described previously (Becalska et al., 2013). For S2 cell protrusion quantification, perimeter and area were calculated from maximum intensity projection images of confocal stacks of at least 9 cells per sample. For the OCRL inhibitor experiment, cells were incubated with YU1422670 or 1% DMSO control for 15 minutes before spreading for 1 hour on Concanavalin-A-coated coverslips and fixing.
Cell-sized Water Droplet Assay
Lipids (DPHPC (1,2-diphytanoyl-sn-glycero-3-phosphocholine, Avanti Polar Lipids Inc.) or DPHPC:PI(4,5)P2) were mixed in chloroform, dried down, and rehydrated to 23 mM (20 mg/mL) in decane. SNAP-tagged proteins were added to the lipid mix at a 1:50 volume ratio, and pipetted vigorously until cloudy, before imaging by spinning disk confocal microscopy.
GUV Assays
GUVs were generated by electroswelling on indium titanium oxide (ITO)-coated slides. Approximately 450 µM (350ug/mL) GUVs were mixed with 500nM SNAP-tagged F-BAR proteins in 5 mM HEPES and 150 mM KCl, pH 7.5, incubated for 30 min and imaged. For analysis of fraction of GUVs bound, fields of GUVs imaged were identified using only the NBD-PE signal. For analysis of GUV morphology, the SNAP-549 signal was used to identify bound vesicles. All non-spherical vesicles were classified as deformed. For FRAP experiments, GUVs were imaged for 10 min at 2 sec intervals, with a pause for bleaching after timepoint 10 (20 sec). Prebleach fluorescence was normalized to 1.0 in order to calculate the fraction of fluorescence recovery. Signal intensity over time from a non-bleached region of the GUV was used to correct for photobleaching.
Statistical Analyses
All error bars shown are mean +/− s.e.m. Statistical significance was calculated with GraphPad Prism 6 software using ANOVA followed by pairwise Tukey’s tests, or using Student’s t-tests where only two groups were compared, with * denoting p < 0.05, ** p < 0.01, and *** p < 0.005. χ-squared tests for independence were used to evaluate the liposome morphology from EM images.
Supplementary Material
Acknowledgements
Monoclonal antibodies were provided by the Developmental Studies Hybridoma Bank. We thank Bruce Goode, Brian Graziano, Mike Vahey, Shae Padrick, Tony Dinsmore, Jeff Gelles and Jerome Fung for helpful discussions. This work was supported by NIH/NIGMS genetics training grant (T32 GM007122 C.F.K.), a Basil O’Connor Scholar Award from the March of Dimes and a Pew Scholar Award (A.A.R.), NSF (NSF MRI DBI-1228757 to A.A.R.)., A Brandeis-Leir Foundation award (A.A.R. and D.D.), NIGMS (R01GM108021 to M.F.H.), and the Swedish Research Council (13473), Parkinsonfonden and Hjärnfonden to O. S. D.D. acknowledges the support of the Russell-Berrie Nanotechnology Institute (RBNI), and the assistance of Inbal Abutbul, Ludmila Abezgauz, and Ellina Kesselman with cryoEM imaging. Negative stain TEM experiments were performed at the User Facility Center for EM at Moscow State University. Single particle analysis was supported by a grant from Russian scientific foundation (#14-14-00234) to O.S.S. We particularly acknowledge support from the Brandeis Center for Bioinspired Soft Materials, an NSF MRSEC (DMR-1420382).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
C.F.K., E.M.M. and A.A.R. conceived of the project and designed the experiments. C.F.K. and E.M.M performed and analyzed most of the experiments, with the help of A.N.B., A.A.R., D.D., M.F.H., K.S., O.S.S., K.V. and S.W. C.F.K., E.M.M., O.S., and A.A.R. wrote the manuscript.
References
- Becalska AN, Kelley CF, Berciu C, Stanishneva-Konovalova TB, Fu X, Wang S, Sokolova OS, Nicastro D, Rodal AA. Formation of membrane ridges and scallops by the F-BAR protein Nervous Wreck. Mol Biol Cell. 2013;24:2406–2418. doi: 10.1091/mbc.E13-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben El, Kadhi K, Roubinet C, Solinet S, Emery G, Carreno S. The inositol 5-phosphatase dOCRL controls PI(4,5)P2 homeostasis and is necessary for cytokinesis. Curr Biol. 2011;21:1074–1079. doi: 10.1016/j.cub.2011.05.030. [DOI] [PubMed] [Google Scholar]
- Cao H, Yin X, Cao Y, Jin Y, Wang S, Kong Y, Chen Y, Gao J, Heller S, Xu Z. FCHSD1 and FCHSD2 are expressed in hair cell stereocilia and cuticular plate and regulate actin polymerization in vitro. PLoS One. 2013;8:56516. doi: 10.1371/journal.pone.0056516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Chang K, Capraro BR, Zhu C, Hsu CJ, Baumgart T. Intradimer/Intermolecular interactions suggest autoinhibition mechanism in endophilin A1. J Am Chem Soc. 2014;136:4557–4564. doi: 10.1021/ja411607b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle IP, Koh YH, Lee WC, Slind J, Fergestad T, Littleton JT, Ganetzky B. Nervous wreck, an SH3 adaptor protein that interacts with Wsp, regulates synaptic growth in Drosophila. Neuron. 2004;41:521–534. doi: 10.1016/s0896-6273(04)00016-9. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Dickman DK, Lu Z, Meinertzhagen IA, Schwarz TL. Altered synaptic development and active zone spacing in endocytosis mutants. Curr Biol. 2006;16:591–598. doi: 10.1016/j.cub.2006.02.058. [DOI] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase M, Yoshikawa K. Structural transition of actin filament in a cell-sized water droplet with a phospholipid membrane. J Chem Phys. 2006;124:104903. doi: 10.1063/1.2174004. [DOI] [PubMed] [Google Scholar]
- Huntwork S, Littleton JT. A complexin fusion clamp regulates spontaneous neurotransmitter release and synaptic growth. Nat Neurosci. 2007;10:1235–1237. doi: 10.1038/nn1980. [DOI] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- James DJ, Khodthong C, Kowalchyk JA, Martin TF. Phosphatidylinositol 4,5-bisphosphate regulates SNARE-dependent membrane fusion. J Cell Biol. 2008;182:355–366. doi: 10.1083/jcb.200801056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast DJ, Yang C, Disanza A, Boczkowska M, Madasu Y, Scita G, Svitkina T, Dominguez R. Mechanism of IRSp53 inhibition and combinatorial activation by Cdc42 and downstream effectors. Nat Struct Mol Biol. 2014;21:413–422. doi: 10.1038/nsmb.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley CF, Becalska AN, Berciu C, Nicastro D, Rodal AA. Assembly of actin filaments and microtubules in Nwk F-BAR-induced membrane deformations. Commun Integr Biol. 2015;8:1000703. doi: 10.1080/19420889.2014.1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Koldso H, Shorthouse D, Helie J, Sansom MS. Lipid clustering correlates with membrane curvature as revealed by molecular simulations of complex lipid bilayers. PLoS Comput Biol. 2014;10:1003911. doi: 10.1371/journal.pcbi.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Fricke R, Bhar D, Reddy-Alla S, Krishnan KS, Bogdan S, Ramaswami M. Syndapin promotes formation of a postsynaptic membrane system in Drosophila. Mol Biol Cell. 2009;20:2254–2264. doi: 10.1091/mbc.E08-10-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. Membrane curvature at a glance. J Cell Sci. 2015;128:1065–1070. doi: 10.1242/jcs.114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinecke M, Boucrot E, Camdere G, Hon WC, Mittal R, McMahon HT. Cooperative recruitment of dynamin and BIN/amphiphysin/Rvs (BAR) domain-containing proteins leads to GTP-dependent membrane scission. J Biol Chem. 2013;288:6651–6661. doi: 10.1074/jbc.M112.444869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milosevic I, Sorensen JB, Lang T, Krauss M, Nagy G, Haucke V, Jahn R, Neher E. Plasmalemmal phosphatidylinositol-4,5-bisphosphate level regulates the releasable vesicle pool size in chromaffin cells. J Neurosci. 2005;25:2557–2565. doi: 10.1523/JNEUROSCI.3761-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C, Cui H, Gawronski-Salerno JA, Frost A, Lyman E, Voth GA, Unger VM. Structural basis of membrane bending by the N-BAR protein endophilin. Cell. 2012;149:137–145. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Chiba M, Eguchi H, Ohki T, Ishiwata S. Cell-sized spherical confinement induces the spontaneous formation of contractile actomyosin rings in vitro. Nat Cell Biol. 2015;17:480–489. doi: 10.1038/ncb3142. [DOI] [PubMed] [Google Scholar]
- Neumann S, Schmid SL. Dual role of BAR domain-containing proteins in regulating vesicle release catalyzed by the GTPase, dynamin-2. J Biol Chem. 2013;288:25119–25128. doi: 10.1074/jbc.M113.490474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor-Giles KM, Ho LL, Ganetzky B. Nervous wreck interacts with thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron. 2008;58:507–518. doi: 10.1016/j.neuron.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Wigge P, Vallis Y, Moore JD, Evans PR, McMahon HT. Crystal structure of the amphiphysin-2 SH3 domain and its role in the prevention of dynamin ring formation. EMBO J. 1998;17:5273–5285. doi: 10.1093/emboj/17.18.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirruccello M, Nandez R, Idevall-Hagren O, Alcazar-Roman A, Abriola L, Berwick SA, Lucast L, Morel D, De Camilli P. Identification of inhibitors of inositol 5-phosphatases through multiple screening strategies. ACS Chem Biol. 2014;9:1359–1368. doi: 10.1021/cb500161z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y, Ma Q, Vahedi-Faridi A, Sundborger A, Pechstein A, Puchkov D, Luo L, Shupliakov O, Saenger W, Haucke V. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc Natl Acad Sci U S A. 2010;107:8213–8218. doi: 10.1073/pnas.1003478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Gould KL. Setting the F-BAR: functions and regulation of the F-BAR protein family. Cell Cycle. 2010;9:4091–4097. doi: 10.4161/cc.9.20.13587. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Blunk AD, Akbergenova Y, Jorquera RA, Buhl LK, Littleton JT. A presynaptic endosomal membrane trafficking pathway controls synaptic growth signaling. J Cell Biol. 2011;193:201–217. doi: 10.1083/jcb.201009052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Motola-Barnes RM, Littleton JT. Nervous Wreck and Cdc42 cooperate to regulate endocytic actin assembly during synaptic growth. J Neurosci. 2008;28:8316–8325. doi: 10.1523/JNEUROSCI.2304-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrero T, Hagan MF. Simulations show that virus assembly and budding are facilitated by membrane microdomains. Biophys J. 2015;108:585–595. doi: 10.1016/j.bpj.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev. 2010;90:259–289. doi: 10.1152/physrev.00036.2009. [DOI] [PubMed] [Google Scholar]
- Sun X, Pinacho R, Saia G, Punko D, Meana JJ, Ramos B, Gill G. Transcription factor Sp4 regulates expression of nervous wreck 2 to control NMDAR1 levels and dendrite patterning. Dev Neurobiol. 2014 doi: 10.1002/dneu.22212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- Tsujita K, Takenawa T, Itoh T. Feedback regulation between plasma membrane tension and membrane-bending proteins organizes cell polarity during leading edge formation. Nat Cell Biol. 2015;17:749–758. doi: 10.1038/ncb3162. [DOI] [PubMed] [Google Scholar]
- van den Bogaart G, Meyenberg K, Risselada HJ, Amin H, Willig KI, Hubrich BE, Dier M, Hell SW, Grubmuller H, Diederichsen U, et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479:552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez FX, Unger VM, Voth GA. Autoinhibition of endophilin in solution via interdomain interactions. Biophys J. 2013;104:396–403. doi: 10.1016/j.bpj.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Navarro MV, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci U S A. 2009;106:12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Baumgart T. BIN1 Membrane Curvature Sensing and Generation Show Autoinhibition Regulated by Downstream Ligands and PI(4,5)P2. Biochemistry. 2014;53:7297–7309. doi: 10.1021/bi501082r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Schulten K. Membrane sculpting by F-BAR domains studied by molecular dynamics simulations. PLoS Comput Biol. 2013;9:1002892. doi: 10.1371/journal.pcbi.1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Michelot A, Koskela EV, Tkach V, Stamou D, Drubin DG, Lappalainen P. Membrane-sculpting BAR domains generate stable lipid microdomains. Cell Rep. 2013;4:1213–1223. doi: 10.1016/j.celrep.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.