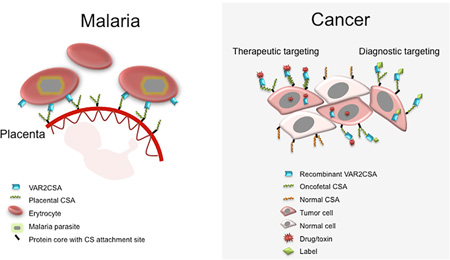
SUMMARY
Plasmodium falciparum engineer infected erythrocytes to present the malarial protein, VAR2CSA, which binds a distinct type chondroitin sulfate (CS) exclusively expressed in the placenta. Here, we show that the same CS modification is present on a high proportion of malignant cells and that it can be specifically targeted by recombinant VAR2CSA (rVAR2). In tumors, placental-like CS chains are linked to a limited repertoire of cancer-associated proteoglycans including CD44 and CSPG4. The rVAR2 protein localizes to tumors in vivo and rVAR2 fused to diphtheria toxin or conjugated to hemiasterlin compounds strongly inhibits in vivo tumor cell growth and metastasis. Our data demonstrate how an evolutionarily refined parasite-derived protein can be exploited to target a common, but complex, malignancy-associated glycosaminoglycan modification.
Graphical Abstract
INTRODUCTION
When the malaria parasite, Plasmodium falciparum, replicates within infected erythrocytes, the latter become susceptible to clearance through the spleen. To avoid host clearance, the parasite expresses adhesion proteins on the surface of infected erythrocytes, which effectively anchor these cells to specific receptors in the host vasculature (Baruch et al., 1995). The anchor protein, VAR2CSA, mediates binding of infected erythrocytes to placental syncytiotrophoblasts (Salanti et al., 2003, 2004). This is the key event underlying placental malaria pathogenesis. In the placenta, VAR2CSA binds a distinct type of chondroitin sulfate (CS) glycosaminoglycan (GAG) chain called CS A (CSA) (Fried and Duffy, 1996). The minimal CS binding region of VAR2CSA consists of the Duffy Binding Ligand-like (DBL) 2X domain with flanking interdomain (ID) regions. This domain binds CS with remarkably high specificity and affinity (KD ~15 nM) (Clausen et al., 2012; Dahlbäck et al., 2011).
CS is comprised of long linear polymers of repeated N-acetyl-D-galactosamine (GalNAc) and glucuronic acid (GluA) residues. These are present as modifications to proteoglycans (CSPGs) in the extracellular matrix (ECM) and in the cell plasma membrane. CSA chains are characterized by the presence of 4-O-sulfations (C4S) on the majority of the GalNAc residues of a given CS chain. Individual CSA chains show considerable variability with respect to the number of sulfated GalNAc residues, the density of sulfation modifications along the chain, as well as the chain length (Gama et al., 2006; Igarashi et al., 2013). P. falciparum-infected erythrocytes have been reported to bind CS oligosaccharides with 4-O-sulfated character (Alkhalil et al., 2000; Beeson et al., 2007). VAR2CSA expressing parasites only adhere in the placenta and do not bind to CS expressed elsewhere in the body (Fried and Duffy, 1996; Salanti et al., 2004). This suggests that placental CS, although incompletely resolved, is a distinct CS subtype expressed exclusively in the placenta. The function of placental-like CS (pl-CS) chains is not fully understood, but they are associated with the ability of trophoblasts to invade the uterine tissue and promote rapid cell proliferation as part of the normal placental implantation process (Baston-Büst et al., 2010). As proliferation and invasion are features shared with tumor cells, we hypothesized that placenta and cancer might express a similar type of CS. We explored the feasibility of using recombinant produced VAR2CSA proteins to target human cancer cells.
RESULTS
Recombinant Malarial VAR2CSA Detects a Distinct Placental-type CS
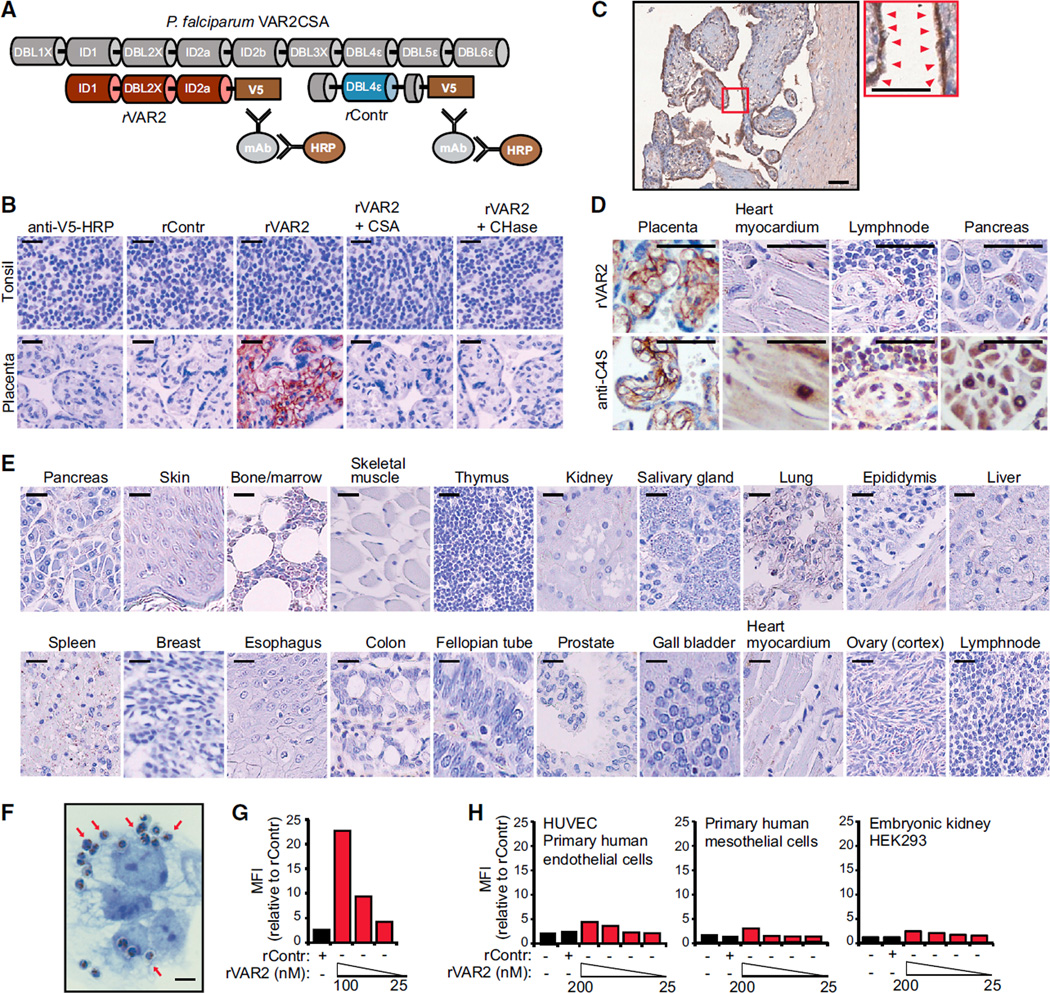
To examine the interaction between P. falciparum-infected erythrocytes and CS in the placenta, we developed an immunohistochemistry assay relying on the interaction between V5-tagged recombinant VAR2CSA (rVAR2) or control protein (rContr) (Figure 1A) and human tissue specimens. The rVAR2 protein efficiently bound both human and murine placental tissue, producing a grid-like staining pattern characteristic of plasma membrane binding, with no staining detected in normal tonsil control tissue (Figures 1B and S1A). Binding to placental tissues was completely inhibited by competition with purified CSA or by enzymatic removal of CS chains using chondroitinase AC, indicating that rVAR2 detects only pl-CS (Figure 1B). The majority of pl-CS was detected in the syncytiotrophoblast layer (Figure 1C, red arrows), with some staining in the underlying stromal cell compartment (Figures 1B and 1C). Staining with anti-C4S (2B6) revealed that CSA is abundant in most tissues despite the lack of rVAR2 staining. This confirms the tissue specificity of pl-CS (Figures 1D, S1B, and S1C). To evaluate the specific expression of pl-CS in other human tissues, we analyzed 20 different normal tissue types for their ability to bind rVAR2. While all tissues analyzed (other than placenta) displayed minimal to absent rVAR2 staining (Figure 1E), some tissues exhibited weak focal stromal staining not associated with cellular plasma membranes (Figure S1D). The choriocarcinoma cell line BeWo has been extensively used as a model for villous trophoblast function (Orendi et al., 2011). As expected, P. falciparum-infected human erythrocytes expressing VAR2CSA efficiently adhered to BeWo cells (Figure 1F, red arrows). Moreover, rVAR2 displayed strong binding to BeWo cells as measured by flow cytometry (Figure 1G), while primary endothelial, mesothelial, and embryonic kidney cells displayed low to absent binding of rVAR2 even at high concentrations (200 nM) (Figure 1H). Thus, in line with the fact that P. falciparum-infected VAR2CSA-expressing erythrocytes only sequester to the placenta, these data demonstrate that rVAR2 detects a distinct form of pl-CS expressed exclusively in the placenta and not on other normal cells or tissues.
Figure 1. rVAR2 Detects a Distinct CS Modification in Human Placenta.
(A) Schematic illustration of full-length P. falciparum VAR2CSA (gray), recombinant minimal CS-binding region (red), and recombinant non-CS binding region rContr (blue). The antibodies toward the C-terminal V5 tag are used to detect rVAR2.
(B) Representative images of indicated tissue specimens incubated with anti-V5 + anti-mouse-HRP alone (anti-V5-HRP) or in combination with recombinant rContr or rVAR2 with or without chondroitinase ABC (CHase ABC) or purified CSA. The scale bar represents 20 µm.
(C) Human placenta tissue stained as in (B), red arrows indicate pl-CS on the syncytium. The scale bars represent 10 µm.
(D) The denoted tissues stained for total CSA using enzymatic GAG end-processing and anti-C4S (2B6) antibody or for CS detected by rVAR2 as in (B). The scale bar represents 10 µm.
(E) A selection of 20 normal tissues stained as in (B). The scale bar represents 20 µm.
(F) Representative image of P. falciparum-infected, VAR2CSA-expressing erythrocytes bound to the plasma membrane (red arrows) of trophoblastic BeWo cells. The scale bar represents 1 µm.
(G) Relative mean fluorescence intensity (MFI) of trophoblastic BeWo cells incubated with recombinant rContr or rVAR2 as indicated and detected by flow cytometry using anti-V5-FITC.
(H) HUVECs, human primary mesothelial cells, and human embryonic kidney cells (HEK293) analyzed as in (G).
See also Figure S1.
pl-CS Is Expressed on Most Human Cancer Cells
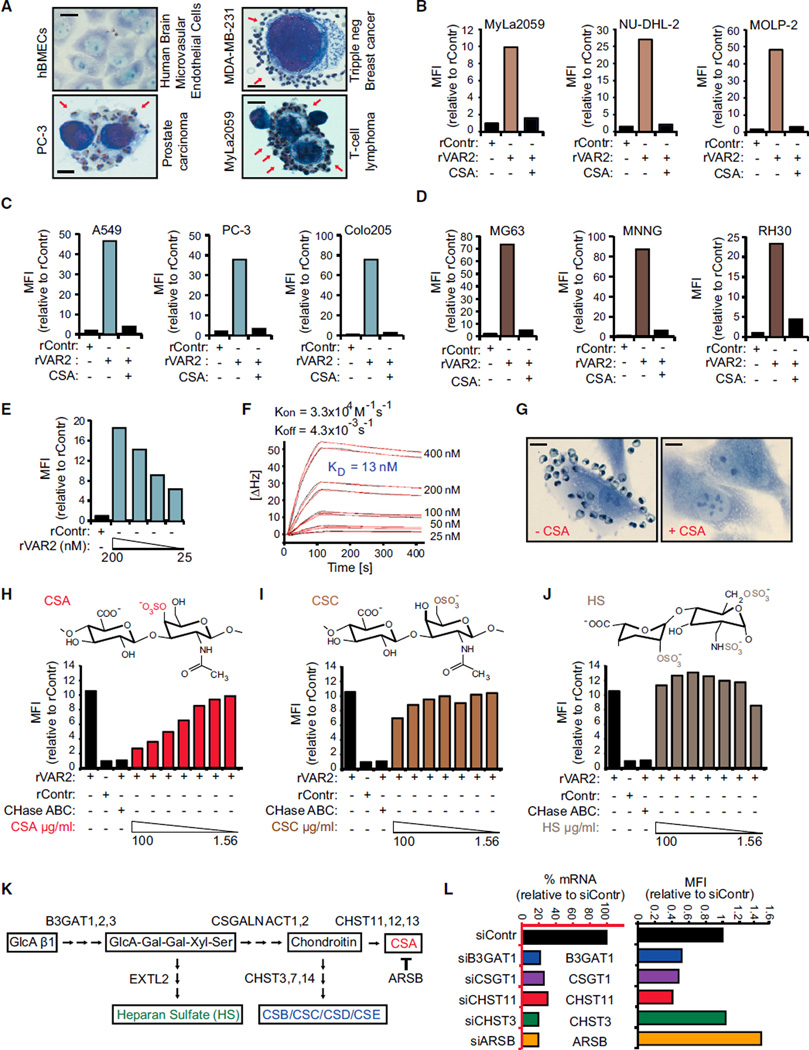
The CSPG component of the placenta has been associated with the ability of the placental cells to maintain high proliferation rates and the capacity of the villous trophoblasts to invade into uterine tissue during implantation (Baston-Büst et al., 2010; Van Sinderen et al., 2013). Invasion and enhanced proliferation are phenotypes shared with cancer cells. We therefore hypothesized that the placental- and malignant compartments display a common CS signature that binds malarial VAR2CSA. Accordingly, we first showed that VAR2CSA expressing P. falciparum-infected erythrocytes displayed binding to human cancer cell lines in vitro, while no binding was observed to normal primary cells (Figure 2A). We next tested binding of rVAR2 to cancer cell lines and found that rVAR2 reacted with 95% (106/111) of patient-derived human cancer cell lines of hematopoietic, epithelial, and mesenchymal origin (Figures 2B–2E; Table S1). The interaction was rVAR2-concentration dependent and could be blocked by competition with soluble CSA (Figures 2B–2E and S2A). Furthermore, the affinity of rVAR2 binding to C32 melanoma cells was high and occurred with a KD-value of 13 nM (Figure 2F). Also, VAR2CSA expressing P. falciparum-infected erythrocytes adhered to C32 cells and this interaction could be completely blocked by purified CSA (Figure 2G).
Figure 2. rVAR2 Detects pl-CS on Cancer Cells.
(A) Representative images of indicated normal primary cells (hBMECs) and tumor cells (PC-3, MDA-MB-231, and MyLa2059) with adherence P. falciparum-infected, VAR2CSA-expressing human erythrocytes (red arrows). The scale bar represents 1 µm.
(B–D) Relative mean fluorescence intensity (MFI) of representative hematological (B), epithelial (C), or mesenchymal (D) lineage human cancer cell lines incubated with recombinant rContr or rVAR2 in combination with soluble CSA as indicated and detected by flow cytometry using anti-V5-FITC.
(E) C32 human melanoma cells incubated with recombinant rContr or indicated concentrations of recombinant rVAR2 analyzed as in (B).
(F) Sensorgram showing binding between recombinant VAR2CSA and immobilized C32 cells measured in delta Hertz [ΔHz] as a function of time (s) using the indicated concentrations of recombinant protein. The black lines represent data and the red lines represent fitted curves attained by a 1:1 binding model. The affinity is given as KD values calculated from Kon and Koff.
(G) Representative images of C32 melanoma cells flushed over with P. falciparum-infected human erythrocytes in absence (−CSA) or presence (+CSA) of soluble CSA. The scale bar represents 1 µm.
(H–J) Relative mean fluorescence intensity (MFI) of C32 melanoma cells incubated with recombinant rContr or rVAR2 in combination with chondroitinase ABC (CHase ABC), soluble CSA, (H), CSC (I), or HS (J) as indicated and detected by flow cytometry using anti-V5-FITC.
(K) Schematic illustration of key enzymatic events in the CS synthesis pathway.
(L) rVAR2 binding (black bars, right) and RT-PCR readout of mRNA levels (blue bars, left) of U2OS cells transfected with control (C), B3GAT1 (B3), CSGALNACT1 (CSG), CHST11 (C11), CHST3 (C3), or ARSB (AB) siRNAs.
See also Figure S2 and Table S1.
We next determined the specificity of the rVAR2-CSA interaction using ELISA as well as a flow cytometry-based competition assay on C32 cells. While all batches of rVAR2 (rVAR2-1, 2, and 3) efficiently bound immobilized CSPG molecules, no binding was observed to heparin sulfate PG (HSPG) or BSA as measured by ELISA (Figure S2B). Furthermore, pre-treating C32 cells with chondroitinase ABC reduced the rVAR2 binding to background levels (rContr) (Figures 2H–2J). The rVAR2-C32 interaction was also inhibited by purified free CSA in a dose-dependent manner (Figure 2H), whereas competition with the structurally similar 6-O-sulfated CS C (CSC) (Figure 2I) or highly charged heparan sulfate (HS) (Figure 2J) had limited effect on rVAR2 binding. Furthermore, while rVAR2 bound to the HS deficient CHO-pgsD-677, no binding was observed to CHO-S745A cells that have been selected for low xylosyltransferase activity and therefore low overall GAG content (Esko et al., 1985) (Figures S2C–S2E). Thus, human cancer and placental trophoblast cells express a common and distinct form of CS, which can be specifically recognized by recombinant malarial VAR2CSA.
De novo CSA synthesis involves several enzymes (Sugahara et al., 2003). While B3GAT1 is required for synthesis of the basal GlcA-Gal-Gal-Xyl-Ser linker tetrasaccharide common to all GAGs, CSGALNACT1 commits the GAG to the CS pathway. Within the CS maturation pathway, CHST11 mediates CSA-specific 4-O-sulfation of the GalNAc residues of the CS chain (Figure 2K). Furthermore, the sulfation level of CSA is balanced by the 4-O-sulfatase ARSB, which removes C4S from the CSA chains (Litjens et al., 1989). RNAi-mediated knock down of B3GAT1, CSGALNACT1, and CHST11 expression inhibited the binding of rVAR2 to U2OS cells, while knock down of CHST3, an enzyme involved in 6-O-GalNAc sulfation, had no effect on binding (Figure 2L). Knock down of ARSB increased the binding of rVAR2 (Figure 2L), indicating that 4-O-sulfated CS residues constitute an important component of the pl-CS GAG chain.
To confirm the clinical relevance of CS expression, we analyzed available expression data from lung cancer patients linked to therapeutic outcome for expression of the key enzymes involved in CSA synthesis (B3GAT1, CSGALNACT1, and CHST11). While the expression pattern of enzymes required for synthesis of the CS GAG backbone (B3GAT1 and CSGALNACT1) had no predictive value in lung cancer, high expression of the enzyme specifically required for CSA 4-O-sulfation (CHST11) significantly correlated with poor relapse-free survival in three independent lung cancer cohorts (Figure S2F). The same pattern was observed for breast and colon cancer (Figure S2F). Together, these data indicate that major human cancers induce expression of a CS chain, which likely consists predominantly of GalNAc residues.
pl-CS Is Widely Expressed in Primary Human Cancer
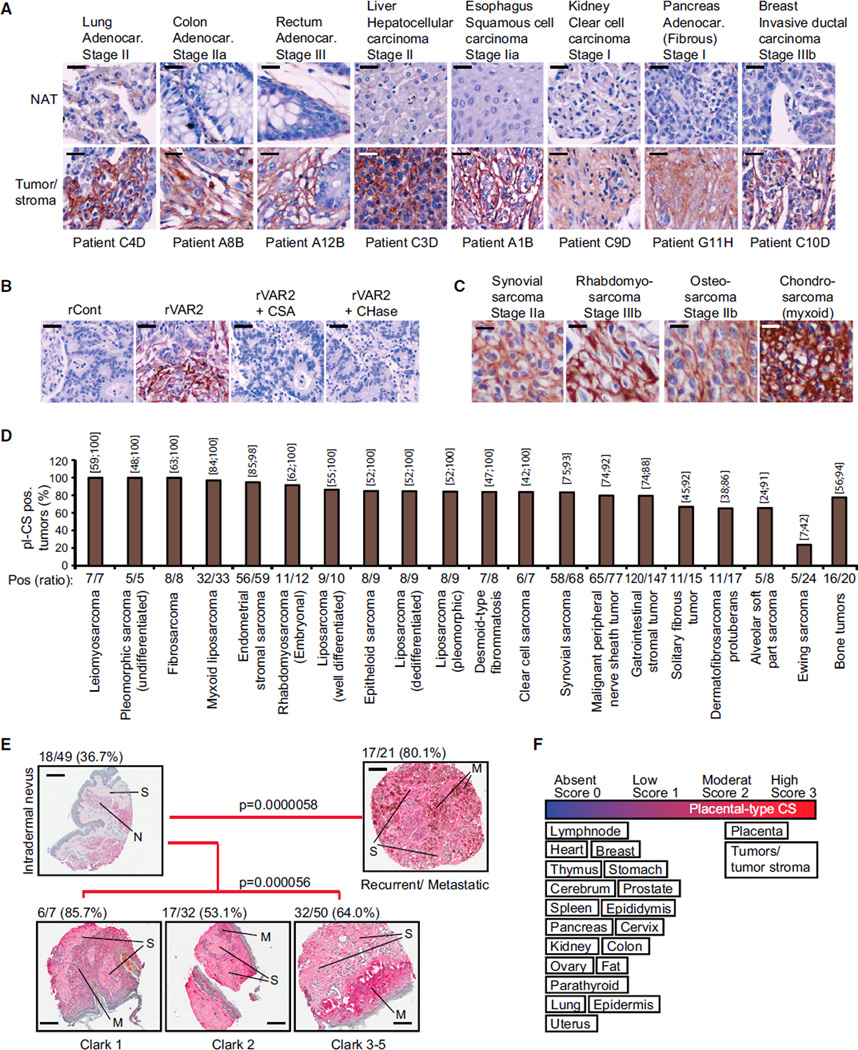
To extend these findings to human tumor specimens, we examined a panel of human epithelial tumors for rVAR2 binding as compared to adjacent normal tissue from the same patients, using the same methods as in Figures 1B–1E. Although differences in pl-CS staining intensity were observed among the different subtypes and tumor stages, all tumors displayed strong rVAR2 binding and only weak staining in matched normal tissue (Figure 3A). The staining pattern indicates that pl-CS may be expressed on both epithelial tumor cells and tumor-associated stromal cells. Binding of rVAR2 to primary human tumors could be completely inhibited by competing with soluble CSA chains or by enzymatic removal of CS from the tissue (Figure 3B). Moreover, pl-CS was not restricted to epithelial tumors, as rVAR2 also strongly reacted with mesenchymal soft-tissue and bone sarcomas (Figure 3C).
Figure 3. Diverse Types of Human Cancer Express pl-CS.
(A) Representative images of indicated patient-matched primary tumor and normal adjacent tissue (NAT) specimens stained with rVAR2-V5 and detected by anti-V5-HRP. The scale bar represents 10 µm.
(B) Representative images of stage IIa breast tumors stained with rContr or rVAR2 as in (A) with or without soluble CSA or chondroitinase ABC (CHase ABC). The scale bar represents 10 µm.
(C) Representative images of indicated soft-tissue and bone sarcomas stained with rContr or rVAR2 as in (A). The scale bar represents 5 µm.
(D) Column graph representation of pl-CS staining intensity in the indicated soft-tissue and bone sarcoma subtypes (n = 552) processed as in (A) and scored (0–3) for binding to recombinant VAR2CSA. The columns show the percentage and exact binomial 95% confidence interval of pl-CS positive (score 2–3) tumors.
(E) Representative images of a melanoma progression TMA stained with anti-V5-HRP in combination with recombinant rVAR2 and scored (0–3). The fraction of score 2–3 (moderate/high) pl-CS (pl-CS) positive tumors was identified and p values were generated by the Goodman-Kruskal-Gamma test (Nevus, N; Stroma, S; and Melanoma cells, M). The scale bar represents 40 µm.
(F) Schematic representation of pl-CS expression in the indicated tissue categories.
See also Figure S3 and Table S2.
We then analyzed 676 malignant tumors from patients with localized stage I–III invasive ductal breast carcinoma (n = 124), stage IIb bone sarcomas (n = 20), and stage I–III soft-tissue sarcomas (n = 532). Ranking the staining-intensity on a 0–3 scale (where 2 equals the staining intensity of the placenta) revealed that ~90% of the breast tumors, ~80% of the bone sarcomas, and ~85% of soft-tissue sarcoma specimens studied showed positive staining for pl-CS on the plasma membrane or in the tumor stroma (Figures 3D, S3A, and S3B). Of all the sarcoma subtypes investigated, only Ewing sarcomas showed weak or absent binding of rVAR2 in the majority of cases (Figure 3D). To investigate whether the weak binding of rVAR2 to Ewing sarcomas was due to a general lack of CS chains or a specific lack of pl-CS, we analyzed a tissue microarray (TMA) of 47 Ewing sarcoma specimens in triplicates from adult and pediatric patients. The TMA was subjected to staining with three different CS reagents, rVAR2, 2B6 (binding C4S following Chondroitinase ABC treatment), and CS56 (binding C4S and C6S) (Figures S3C–S3E). This analysis showed that 80.6% of all Ewing sarcoma cases were positive for at least one of the three reagents, while rVAR2 stained ~39% of the Ewing specimens (Figure S3F). These data suggest that Ewing sarcomas display a broad selection of CS chains, but only a fraction of these are placental-type CS that can be bound by rVAR2. Moreover, while many low-risk mesenchymal neoplasms (including lipoma, fibromatosis, neurofibroma, Schwannoma, and pigmented villonodular synovitis) displayed absent or low-to-moderate staining for pl-CS, the majority of malignant lesions showed strong staining (Figure S3G).
With a tool to broadly target pl-CS chains in primary human cancer, we next analyzed whether this malignancy-associated CS alteration was linked to progression or outcome of malignant disease. We analyzed an available melanoma progression TMA (n = 159) for expression of pl-CS. While the majority of benign nevi displayed an absent or low pl-CS expression, the presence of pl-CS in the tumor microenvironment increased significantly in Clark 2–5 staged melanoma (p = 0.000056) and in metastatic/recurrent disease (p = 0.000058) (Figure 3E; Table S2). These data suggest that pl-CS is a candidate marker for disease progression in melanoma. We subsequently investigated whether pl-CS detected by rVAR2 was associated with outcome in human non-small cell lung cancer. Analysis of a TMA comprised of 165 primary tumors linked to outcome demonstrated that high expression of pl-CS correlated with poor relapse-free survival of the patients (p = 0.016) (Figures S3H and S3I). Collectively, these data demonstrate that highly diverse human malignant tumor types originating from distinct germ layers display a common distinct pl-CS signature that predicts disease progression and outcome (summarized in Figure 3F).
Molecular Characterization and Expression of Placenta-like CS
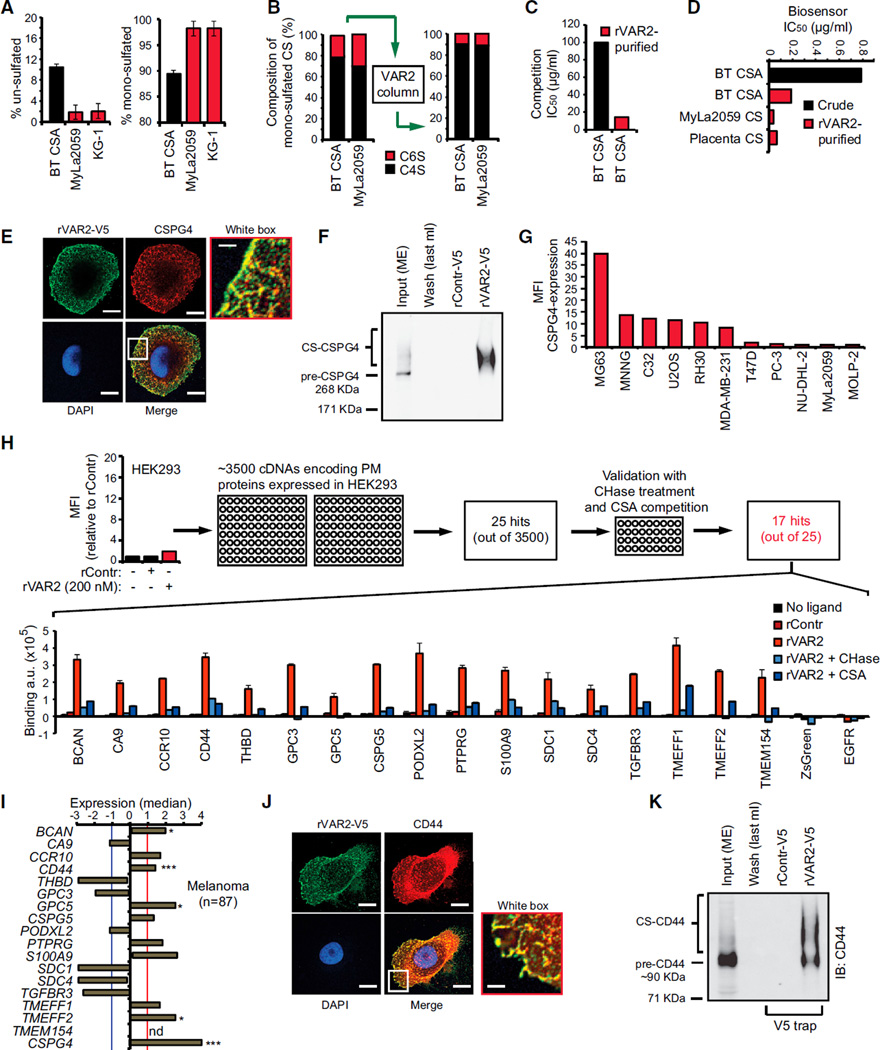
To better define the rVAR2 binding of CS from cancer cells, we extracted GAGs from different cancer cell types followed by rVAR2 affinity purification, structural analysis using tandem mass spectrometry (tandem-MS), and affinity analysis using Attana’s biosensor technology and flow cytometry. MS of GAG extracts showed that bovine trachea CSA (BT-CSA) was 10% un-sulfated and 90% mono-sulfated (no di-sulfation detected), whereas cancer-associated CS was 98% mono-sulfated (Figure 4; Table S3). Furthermore, tandem-MS revealed that BT-CSA mono-sulfated population contained 79.6% C4S and 20.4% of C6S, while CS from Myla2059 lymphoma cells contained 69.8% C4S and 30.2% C6S (Figure 4B, left). Although rVAR2 binding to cancer cells and tissue was outcompeted by BT-CSA, high amounts (100–400 µg/ml) were required to fully disrupt the rVAR2-cancer cell interaction (Figures 2B–2D). This indicated that rVAR2 only bound a fraction of the CSA chains present in the crude BT-CSA preparation. To investigate this, we affinity purified BT-CSA and MyLa2059 CS extract on an rVAR2 column (Figure 4B, right) and characterized it by MS. BT-CSA affinity purification on rVAR2 resulted in mono-sulfated CS that was enriched (90%) for C4S (Figure 4B; Table S3). Compared to bulk BT-CSA, the BT-CSA eluted from the column was considerably more effective in competitively inhibiting the binding between rVAR2 and C32 melanoma cells as measured by flow cytometry (Figure 4C), as well as the binding of rVAR2 to immobilized CSPG measured by the biosensor (Figure 4D). CS from MyLa2059 cells affinity purified on rVAR2 columns inhibited binding of rVAR2 to immobilized CSPG with an IC50 of 0.033 µg/ml compared to 0.063 µg/ml for placenta CS and 0.79 µg/ml for BT-CSA (Figures 4D and S4A–S4C; Table S4).
Figure 4. Defining the rVAR2 CS Epitope.
(A) Level of un-sulfated (left) and mono-sulfated (right) disaccharides of extracted CS from Sigma BT-CSA, T cell lymphoma (MyLa2059), and myeloid leukemia (KG-1) determined by liquid chromatography MS analysis. The values are given as a percentage of the total CS in the sample.
(B) BT-CSA and MyLa2059 CSA were affinity purified on a custom made rVAR2 column and eluted in a NaCl gradient. The composition of the mono-sulfated CS was analyzed by tandem MS before (left) and after (right) affinity purification.
(C) Binding inhibition capacity of BT-CSA before and after rVAR2 affinity purification is shown as the concentration needed to block 50% of the binding (IC50 values) between rVAR2 and the cells as measured by flow cytometry.
(D) Biosensor analyses of the capacity of BT-CSA (before and after rVAR2 affinity purification) and rVAR2 affinity-purified Myla2059 and placental CS, to inhibit rVAR2 binding to immobilized CSPG. The binding inhibition is shown as the concentration needed to block 50% of the binding (IC50 values) between rVAR2 and the cells.
(E) Representative picture of a C32 human melanoma cell co-stained for CS using rVAR2-V5 (green) and CSPG4 (red). The co-localization was analyzed by confocal microscopy. The scale bar represents 0.5 µm.
(F) Extracted membrane proteins (Input ME) from C32 melanoma cells were subjected to anon-column pulldown on a HiT rap NHS column coupled with rVAR2-V5 or rControl-V5. The figure shows Input (ME), last 1 ml of wash of the rVAR2-V5 column, and the 0.5 M NaCl elution following concentration. The samples were analyzed for precipitation of precursor (pre-CSPG4) and CSA-conjugated CSPG4 by immunoblotting (IB:CSPG4) as indicated.
(G) Relative mean fluorescence intensity (MFI) of indicated cell lines incubated with anti-CSPG4 antibody and detected by flow cytometry.
(H) Relative mean fluorescence intensity (MFI) of HEK293 cells incubated with recombinant rContr or rVAR2 as indicated and detected by flow cytometry using anti-V5-FITC. The HEK293 cells were transfected with 3,500 cDNAs encoding known tumor-associated plasma membrane proteins inserted in a ZsGreen expression system and analyzed for their ability to facilitate binding to recombinant rVAR2 detected by anti-V5-Alexa647. The column graph displays quantified anti-V5-Alexa647 detection (arbitrary units, a.u.) in HEK293 cells transfected with the indicated plasma membrane proteins and left un-treated (no ligand), or treated with recombinant rContr, rVAR2 alone, or in combination with chondroitinase ABC (rVAR2 + CHase) or purified CSA (rVAR2 + CSA).
(I) Median expression compared to overall average of the genes encoding the 17 proteins from (H) plus CSPG4 in primary melanoma (n = 87) patient specimens extracted from the Oncomine Riken melanoma array (*p < 0.05 and ***p < 0.001) (not determined: nd) (missing probe). The red and blue cross-lines designate the threshold for up and downregulated.
(J) Representative picture of a C32 human melanoma cell co-stained for CS using rVAR2-V5 (green) and CD44 (red). The co-localization was observed by confocal microscopy. The scale bar represents 0.5 µm.
(K) Extracted membrane proteins (Input ME) from C32 melanoma cells were subjected to an on-column pulldown on a HiTrap NHS column coupled with rVAR2-V5 or rControl-V5. The figure shows Input (ME), last 1 ml of wash of the rVAR2-V5 column, and the 0.5 M NaCl elution following up-concentration. The samples were analyzed for precipitation of precursor (pre-CD44) and CS-conjugated CD44 by immunoblotting (IB:CD44) as indicated. The error bars indicate ± SD.
See also Figure S4 and Tables S3–S5.
Placental cells, human melanoma cells, and some breast cancer and glioma cells have been found to express high levels of the CSA-modified proteoglycan, CSPG4 (Van Sinderen et al., 2013; Wang et al., 2010). To test whether CSPG4 carries pl-CS that can be bound by rVAR2, we analyzed co-localization and direct interaction between rVAR2 and CSPG4 on C32 melanoma cells. The rVAR2 stain co-localized with CSPG4 (Figure 4E) and specifically pulled down CS-conjugated CSPG4 from C32 melanoma cells (Figure 4F). However, rVAR2 could also bind tumor cells negative for CSPG4 expression (compare Figures 4G and 2B–2E), suggesting that pl-CS can be displayed on other CPSG protein cores. To identify these, we transfected non-malignant HEK293 cells (which do not bind rVAR2) with 3,500 different human plasma membrane proteins (Figure 4H). Following validation with CSA competition and enzymatic removal of CS chains by chondroitinase treatment, it was evident that 17 of the 3,500 proteins facilitated binding to rVAR2 (Figure 4H). Thus, in addition to CSPG4 (not included in the screen), at least 17 proteins including CD44, carbonic anhydrase IX (CA9), syndecan 1 (SDC1), or tomoregulins (TMEFF1 and −2) can carry pl-CS when overexpressed in HEK cells. Further studies are required to investigate if all of the proteoglycans can be modified with CS in vivo. Interestingly, ten of these proteins have previously been described as being conditional CS-conjugated and 15 have been directly associated with human malignant disease (Table S5). To investigate the inter-tumor diversity in expression of proteoglycans able to display pl-CS, we analyzed 1,555 primary human tumor specimens representing 17 major types of human cancers using the Bittner array (Rhodes et al., 2004) and the Riker melanoma array (Riker et al., 2008). This analysis showed that the proteoglycans associated with pl-CS were differentially, but complementarily, expressed in each of the 17 major cancer groups tested (Figures 4I and S4D). Accordingly, we validated the interaction of rVAR2 with the CS-modified form of CD44 in C32 melanoma cells (Figure 4J). Moreover, rVAR2 efficiently pulled down glycosylated CD44 from C32 melanoma protein lysates (Figure 4K). Together, these data suggest that rVAR2 can be utilized to broadly target pl-CS chains in human malignancies with different proteoglycan expression profiles.
Internalization and In Vivo Localization of rVAR2
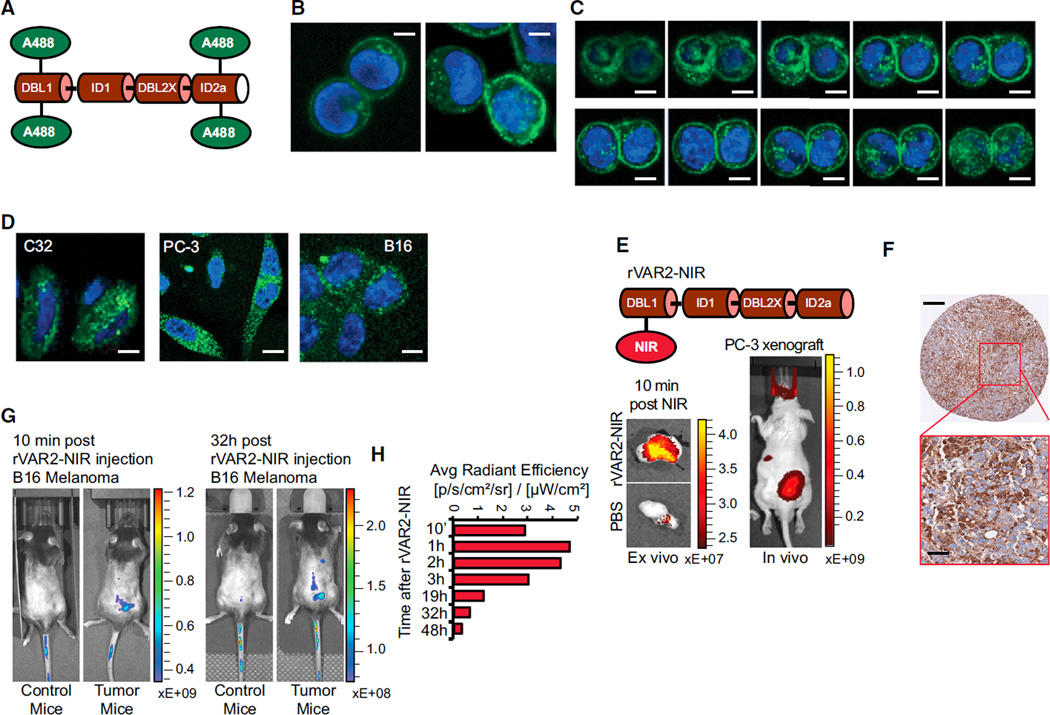
The specific presence of a pl-CS modification in malignant tumors suggests that VAR2CSA may be utilized to deliver cytotoxic payloads directly to the tumor microenvironment. We first investigated if rVAR2 is internalized upon binding to tumor cells. Alexa488-labeled rVAR2 (rVAR2-A488) (Figure 5A) rapidly bound to human colon cancer cells (Colo205) and was efficiently internalized within 30 min (Figures 5B and 5C). Similar internalization dynamics were observed with three other cancer cell lines (Figure 5D). We then investigated if rVAR2 was able to sequester to tumors in vivo. To visualize rVAR2 tumor sequestration, we conjugated an Alexa750 near-infra red (NIR) fluorescent probe to rVAR2 (rVAR2-NIR) (Figure 5E, top). The rVAR2-NIR was administrated IV into PC-3 tumor bearing mice and a NIR signal from the tumor region was observed after 10 min in vivo and ex vivo (Figure 5E, bottom). Moreover, ex vivo analysis of fixed PC-3 xenografts confirmed strong rVAR2 reactivity in the tumors as measured by immunohistochemistry (Figure 5F). Similarly, we injected the rVAR2-NIR IV into B16-tumor bearing mice and followed the rVAR2-NIR signal for 48 hr. As in the PC3 xenograft, rVAR2 rapidly located to the tumor and the signal was detectable 48 hr after injection (Figures 5G and 5H). Together, these data suggest that rVAR2 can be utilized to facilitate intracellular delivery of cytotoxic compounds to pl-CS expressing cells in vivo.
Figure 5. rVAR2 Internalization and In Vivo Tumor Localization.
(A) Schematic illustration of rVAR2 conjugated with Alexa-488 (rVAR2-A488).
(B) Colo205 colon carcinoma cells analyzed by confocal microscopy 5 (left) and 30 (right) min after addition of rVAR2-FITC (green) and DAPI (blue). The scale bar represents 0.5 µm.
(C) Confocal microscopy analysis of Colo205 cells as in (B) displayed as vertical depth at 30 min after addition of rVAR2-FITC. The scale bar represents 0.5 µm.
(D) C32 melanoma, PC-3 prostate adenocarcinoma, and B16 murine melanoma cells analyzed as in (B) 30 min after addition of rVAR2-FITC. The scale bar represents 0.5 µm.
(E) Schematic illustration of rVAR2 conjugated with NIR Alexa-750 (rVAR2-NIR) probe (upper) and in vivo (right) and ex vivo (left) detection of NIR signal in PC-3 tumor xenografts 10 min post-rVAR2-NIR injection in tail vein.
(F) IHC of PC-3 tumor xenografts stained with rVAR2-V5 and detected by anti-V5-HRP. The scale bar represents 40 µm.
(G) C57BL/6 mice with no tumors (Control mice) or carrying established B16 murine melanoma tumors (Tumor mice) were injected with rVAR2-NIR in the tail vein at day 10 and scanned in an IVIS Spectrum CT scanner.
(H) Quantification of IVIS signal from a subcutaneous B16 tumor (right flank) after rVAR2-NIR injection at different time intervals from 10 min to 48 hr. The right flank signal is considered as background and is subtracted from the initial signal.
Targeting Tumors through the rVAR2 pl-CS Signature
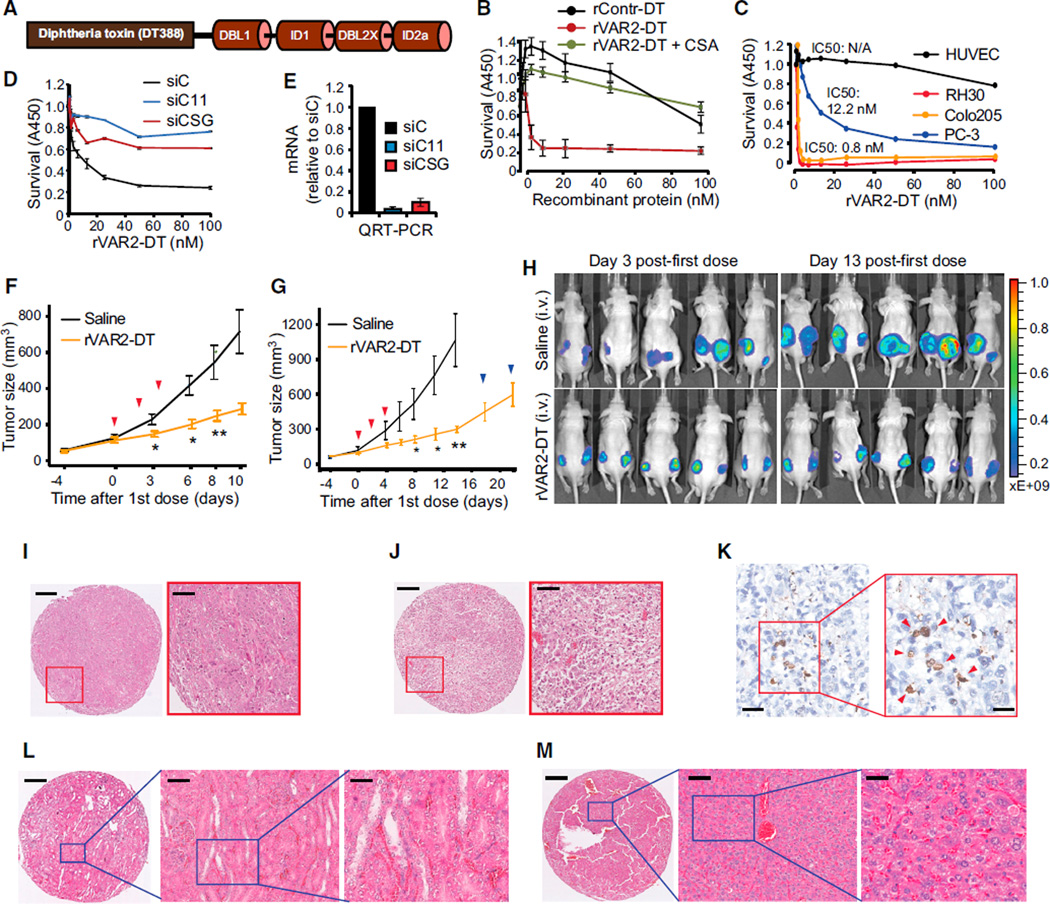
To test whether rVAR2 could be used as a pl-CS-specific tumor targeting system, we genetically fused the cytotoxic domain of Diphtheria toxin (DT388) to rVAR2, creating a recombinant rDT388-VAR2 (rVAR2-DT) fusion protein (Figure 6A). In vitro, the rVAR2-DT protein efficiently killed tumor cell lines of both epithelial and mesenchymal origin (Figures 6B and 6C) with low IC50 values (from 0.8 nM to 12.2 nM). The rVAR2-DT toxin had no effect on normal primary human endothelial cells (HUVEC) (Figure 6C), and competition with CSA abolished the cytotoxic effect of rVAR2-DT on cancer cells (Figure 6B). Moreover, small interfering (si)RNAs targeting C4S–sulfotransferase CHST11 or the CS backbone-conjugation enzyme CSGALNACT1 prevented the cytotoxic effects of rVAR2-DT in prostate cancer PC-3 cells (Figures 6D and 6E). These data demonstrate that the C4S conjugation machinery is required for rVAR2 to deliver a cytotoxic payload to tumor cells.
Figure 6. In Vivo Cancer Targeting Using rVAR2 Fusion Construct rVAR2-DT388.
(A) Schematic figure showing the architecture of the rVAR2-DT388 fusion protein.
(B) Survival of B16 melanoma cells treated with increasing concentrations (0–100 nM) of a DT-fused (rContr-DT) or rVAR2-DT with or without CSA competition as indicated and analyzed for WST1 staining 96 hr post-treatment. The error bars indicate ± SD.
(C) Survival of indicated cell lines treated with increasing concentrations (0–100 nM) of rVAR2-DT.
(D) PC-3 cells were transfected with control siRNA (siC) or siRNAs targeting CHST11 (siC11) or CSGALNACT1 (siCSG) and treated with the indicated concentrations (0–100 nM) of rVAR2-DT for 96 hr before analyzed for survival using methylene blue staining assay. The error bars indicate ± SD.
(E) Quantitative RT-PCR of indicated mRNA levels in PC-3 cells after 72 hr post-transfection with control siRNA (siC), CSGALNACT1 siRNA (siCSG), and CHST11 siRNA (siC11). The error bars indicate ± SD.
(F and G) Quantification of tumor volume over 10 days (Experiment 1, F) and 20 days (Experiment 2, G) in Foxn1nu mice xenotransplated with PC-3 cells and treated on day 0, 2, and 4 (red arrows) with either saline (black line) or 0.6 mg/kg rVAR2-DT (yellow line) (*p < 0.05 and **p < 0.01). The error bars indicate ± SEM.
(H) IVIS analysis of PC-3 tumor growth in mice (from G) on day 3 and 13 post-first dose treatment with saline or rVAR2-DT as indicated.
(I and J) Representative hematoxylin and eosin (H&E) images as indicated of PC-3 xenograft tumor after treatment with saline (I) or rVAR2-DT (J). The scale bar represents 40 µm.
(K) Representative image of PC-3 xenograft tumor stained with TUNEL reagent (in 20× and 40× magnification as indicated). The scale bar represents 10 µm (left) and 5 µm (right).
(L and M) Representative H&E images as indicated of kidney (L) and liver (M) extracted from rVAR2-DT treated mice in (G). The scale bar represents 40 µm.
Prostate cancer is a leading cause of morbidity and death among men in the Western world, and the prognosis for advanced castration resistant prostate cancer (CRPC) is poor (Kirby et al., 2011). We therefore tested the efficacy of rVAR2-DT in a mouse tumor xenograft model based on the metastatic CRPC cell line, PC-3. As few as three doses of rVAR2-DT were able to significantly inhibit growth of PC-3 CRPC tumors in vivo (Figure 6F). We next performed independent experiments to assess the longer-term effect of three doses of rVAR2-DT on CRPC growth in vivo. The rVAR2-DT treated group was monitored for tumor growth for up to 20 days after the first dose. Similar to the first setup (Figure 6F), we observed a strong inhibition of tumor growth in the rVAR2-DT treated group (Figure 6G). Interestingly, the inhibitory effect of rVAR2-DT on tumor growth persisted 14 days after first dose (10 days after last dose), before slow re-growth of the tumor was observed (day 14, blue arrows) (Figure 6G). The PC-3 cells used in the study expressed luciferase, allowing us to monitor the impact of rVAR2-DT on PC-3 xenografts by IVIS. Scanning of the mice at day 3 and day 13 after the initial dose fully corroborated the results from Figures 6F and 6G and showed marked inhibition of chemiluminiscence within the treated group (Figure 6H). Histopathology analysis revealed dramatic differences between rVAR2-DT treated and un-treated tumors. While the vehicle control displayed intact and dense tumor architecture with little necrosis (Figure 6I), rVAR2-DT treated tumors showed massive necrosis (Figure 6J), similar to what is seen in the liver after wild-type DT delivery (Saito et al., 2001). Furthermore, rVAR2-DT treated xenografts were also positive for TUNEL staining indicative of apoptosis (Figure 6K). Examination of kidney and liver tissues from the rVAR2-DT treated animals showed normal tissue architecture and no morphologic signs of toxicity (Figures 6L and 6M). These data demonstrate that rVAR2 can facilitate efficacious CS-dependent delivery of a cytotoxic compound to CRPC tumors in vivo with no morphologic evidence of adverse effects on normal tissues.
Development and Efficacy of an rVAR2-Hemiasterlin Drug Conjugate
From the human clinical trials with DT fusions, it is apparent that high drug concentrations are not well tolerated. We therefore chemically conjugated a hemiasterlin analog (KT886) to rVAR2 via a protease cleavable linker (Figure S5), utilizing free cysteines in the recombinant protein (Figures 7A and 7B). The rVAR2-KT886 drug conjugate (VDC886) carried an average of three toxins per rVAR2 molecule (Figure 7B). ELISA, biosensor, and flow cytometry confirmed the affinity and specificity of VDC886 to pl-CS. A total of 33 cancer cell lines of different origin were tested for sensitivity to VDC886. All lines were effectively killed in vitro by the VDC886, with IC50 values ranging from 0.2 pM to 30 nM (Figure 7C). We performed a dose escalation study of VDC886 in healthy female CD-1 mice, increasing the dose up to 15 mg/kg. The maximum tested dose of 15 mg/kg was well tolerated, and the animals did not display any signs of morbidity or physical distress. Histopathology examination of different organs did not show any evidence of adverse cytotoxic effects (Figure 7D). Importantly, the pl-CS modification was present in the murine placenta (Figure S1), as well as on murine tumor cells (Figures 5D, 5G, 7H, and 7K), indicating that the absence of adverse effects was not due to lack of pl-CS expression capability in the murine system. We subsequently tested the efficacy of the VDC886 in vivo using two different human xenograft mouse models of non-Hodgkin’s lymphoma (Karpas299) and prostate cancer (PC-3). VDC886 treatment significantly inhibited growth of both Karpas299 (Figure 7E) and PC-3 (Figure 7F) tumors as compared to the control groups. Remarkably, two out of six mice in the VDC886-treated PC-3 tumor group showed complete remission 32 days after the first treatment (Figure 7G). These data demonstrate that VDC886 can target diverse human tumor types in vivo. To further analyze the anti-tumor effects of pl-CS targeting in vivo, we made use of a highly aggressive syngeneic (immunocompetent) mouse model of metastatic murine breast cancer cells. 4T1 breast cancer cells were efficiently bound by rVAR2 in a concentration and CSA-dependent manner (Figure 7H). Moreover, VDC886 demonstrated strong cytotoxicity in 4T1 cells in vitro, which could be completely rescued by competition with soluble CSA (Figure 7I). Injection of luciferase-4T1 cells in the left ventricle of the heart of C57BL/6 mice resulted in aggressive bone metastasis with an overall penetrance of 50%–60% (Figure 7J). The bone metastases invaded into adjacent muscle and showed strong pl-CS expression as analyzed by rVAR2-based immunohistochemistry (IHC) (Figure 7K). Notably, of the mice with 4T1 bone metastases, five out of six mice in the VDC886-treated group were still alive at the end of the study (day 54) with no detectable metastases, while all control-treated mice died with metastatic disease (p = 0.0196; Figures 7L and 7M). Indeed, VDC886-treatment significantly increased survival of mice with 4T1 bone metastasis as compared to control-treated mice. Collectively, these data provide compelling evidence that diverse human and murine tumor types can be effectively targeted in vivo using an rVAR2 drug conjugate.
Figure 7. In Vivo Cancer Targeting Using VDC886.
(A) Structure of the hemiasterlin analog KT886.
(B) MS readout confirming KT886 loading on rVAR2 through a toxin-linker functionalized with a maleimide group to enable conjugation to protein-thiols. The D0 (blue) represents un-conjugated rVAR2 and D1-7 (red) designates the number of KT886 loaded. The VDC886 carries 3.5 KT886 toxins on average.
(C) Indicated human cancer cell lines were seeded in 96-well plates and treated with VDC886 in concentrations ranging from 0.01 pM to 1 µM. The column graph displays IC50 kill-values of VDC886 performance.
(D) Representative H&E images as indicated of kidney, spleen, prostate, bladder, and liver extracted from mice subjected to three doses of VDC886 on alternating days (15 mg/kg). The scale bar represents 10 µm.
(E) Female C.B-17/IcrHsd-Prkdc scid mice engrafted with Karpas299 non-Hodgkin’s lymphoma cells on the back were treated with 1× PBS (vehicle), naked rVAR2 (rVAR2), KT886 alone (KT886), or VDC886. The treatments were administered intravenously on day 0, 2, and 5 (red arrows) as indicated.
(F) Male Foxn1nu mice were implanted subcutaneously on the back with the PC3 prostate cancer cell line in 100 µl of Matrigel (in both right and left flanks). The test articles, as in (E), were administered intravenously on day 0, 2, and 5 (red arrows) as indicated. The animals remained on study until their combined tumor burden reached 1,000 mm3 in size or they otherwise required euthanasia due to distress (humane endpoint).
(G) Kaplan-Meier curve of Vehicle and VDC886 treated mice from (F). There were two of the six mice on study in the VDC886 treated group that had complete re-mission of disease (***p < 0.001).
(H) 4T1 murine breast cancer cells were analyzed by flow cytometry for binding to the indicated concentrations of rVAR2 or 100 nM rContr in the absence or presence of soluble CSA.
(I) The indicated concentration range of VDC886 was tested on 4T1 murine mammary cancer cells in the absence or presence of soluble CSA as indicated.
(J) Detection of bone metastasis detectable by IVIS (orange arrows) in C57BL/6 mice 2–3 weeks after injected with luciferase-expressing 4T1 cells from (H) and (I) in the left ventricle of the heart.
(K) Extracted bone metastasis from (J) subjected to matched H&E staining and immunohistochemical pl-CS staining using rVAR2-V5 +anti-V5-HRP as indicated. The lower image displays the sections within red boxes (from upper). The scale bar represents 1 mm (upper) and 50 µm (lower).
(L) Mice as in (J) with bone metastasis visible in the same IVIS detection range (n = 18) were randomized into three groups with six mice per group (n = 6) and subjected to four doses of VDC886 (15 mg/kg), rVAR2 alone (rVAR2), or KT886 alone (KT886) in equivalent molar ratios to VDC886 on day 21, 24, 27, and 30. All groups were monitored on day 15, 22, 29, 36, 45, and 54 as indicated for number of mice (n) and number of visible metastasis (M).
(M) Kaplan-Meier survival plot of (L), comparing the two control groups (rVAR2 and KT886) combined with VDC886 treated mice. The mice were sacrificed when reaching their humane endpoint. The red arrows designate dosing days. The p value was calculated with Chi2 log-rank test. The error bars represent ± SEM.
See also Figure S5.
DISCUSSION
The placenta is a fast-growing organ in which cells display high mitotic rates, the ability to invade the uterine tissue, and the capacity to establish an elaborate vasculature. These are features shared with cancer, and hence researchers have for decades attempted to identify molecules shared between the placental and malignant compartments (Holtan et al., 2009). We have demonstrated that recombinant versions of the evolutionarily refined malaria protein VAR2CSA can broadly detect pl-CS on trophoblastic cells as well as in human tumors.
The broad targeting potential of rVAR2 is likely facilitated by the redundant presentation of pl-CS chains on several different cancer-associated PGs such as CSPG4 and CD44. Several of the PGs that carry an rVAR2-reactive CS chain are currently being, or have been, tested as targets in clinical trials (Casucci et al., 2013; Wang et al., 2011). We propose that targeting the common CS chain present on different cancer-associated PGs may potentially offer a broad cancer targeting strategy.
Since rVAR2 is efficiently internalized into pl-CS expressing cells, rVAR2 could potentially facilitate the delivery of anti-cancer compounds directly into the tumor environment. This notion was supported by our in vivo and ex vivo imaging experiments. We subsequently demonstrated the therapeutic potential of targeting pl-CS on human tumors by two different approaches. First, rVAR2 genetically fused to a part of the diphtheria toxin (rVAR2-DT), efficiently killed tumor cells in vitro and in vivo in a CS-dependent manner. Second, chemical conjugation of a hemiasterlin toxin to rVAR2 created a highly potent VDC886 that specifically targeted pl-CS on diverse tumor cells in vitro and in vivo. Notably, non-pregnant mice injected with rVAR2-DT or VDC886 showed no adverse treatment effects, suggesting that pl-CS is expressed below rVAR2–based detection levels in non-malignant tissue in mammals. This is supported by the observation that P. falciparum-infected erythrocytes cannot bind anywhere in vascularized tissue compartments, except in the placenta.
Our data promote pl-CS as a candidate target for broad rVAR2-based anti-cancer therapies, as well as a progression marker for selected human tumor types such as melanoma. rVAR2 provides an example of how evolutionarily refined host-pathogen anchor molecules can be conveniently exploited to access and target cancer associated glycans.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture
The recombinant proteins were expressed in SHuffle T7 Express Competent E. coli (NEB) and purified using HisTrap from GE Healthcare, followed by size exclusion chromatography. Purified CSA, HS, and chondroitinase ABC were obtained from Sigma. CSC was obtained from Seikagaku, and Monoclonal anti-V5 and anti-V5-FITC antibodies were obtained from Invitrogen. Cells were transfected with siRNAs (QIAGEN) (10 nM final) against B3GAT1, CSGALNACT1, CHST11, CHST3, or ARSN using RNAiMAX (Invitrogen) and analyzed for rVAR2 binding by flow cytometry and for mRNA expression by RT-PCR.
IHC
Using the Ventana Discovery platform, sectioned paraffin-embedded tissue samples were stained with 500 picomolar V5-tagged rVAR2 without antigen retrieval, followed by 1:700 monoclonal anti-V5 step and a anti-mouse-HRP detection step. For a detailed description, please see Supplemental Information.
Flow Cytometry
Cells were grown to 70%–80% confluency in appropriate growth media and harvested in an EDTA detachment solution (Cellstripper). Cells were incubated with protein (200–25 nM) in PBS containing 2% fetal bovine serum (FBS) for 30 min at 4°C and binding was analyzed in a FACSCalibur (BD Biosciences) after a secondary incubation with an anti-V5-FITC antibody. For inhibition studies, protein was co-incubated with indicated concentration of GAGs (CSA, CSC, and HS).
Binding Kinetics Analysis
A quartz crystal microbalance biosensor (Attana Cell A200, Attana AB) was used for the kinetic analyses. Cells were seeded onto cell compatible sensor chips and incubated 24 hr at 37°C. Cells were then fixed in 3.7% formaldehyde and visualized using DAPI. The data, including kon, koff, and the calculated KD, were presented as sensorgrams showing VAR2CSA fragments binding to cells as response (in Hertz) as a function of time (s). Curve fitting was performed in the Attache evaluation software (Attana AB).
Immunocytochemistry
Internalization Assay
rVAR2 protein was conjugated with the Alexa488 fluorophore according to the manufacturer’s instructions. C32 cells were seeded to coverslips and grown to 60% confluency. There were 200 nM rVAR2–488 that were incubated with the cells for 1 hr at 4°C and then washed once to allow internalization of only surface-bound protein at either 10 min or 60 min at 37°C. Cells were subsequently washed with PBS prior to fixation with 4% paraformaldehylde for 15 min at room temperature. Coverslips were mounted in mounting media containing DAPI and analyzed by laser-scanning confocal microscopy.
Co-localization
C32 melanoma cells were grown on glass cover slides. Cells were fixed in 4% paraformaldehyde (PFA), blocked in 1% BSA/5% FBS, and stained for CS using rVAR2-V5 and CSPG4 (LHM2, Abcam) or CD44 (2C5, RnD Systems) overnight at 4°C. rVAR2 was detected by anti-V5 (Rabbit) and anti-Rabbit-Alexa488. CSPG4 and CD44 antibodies were detected with anti-mouse-Alexa568. Nuclei were stained with DAPI. Slides were analyzed by laser-scanning confocal microscopy.
CS Extraction from Cancer Cells
Myla-2059 T cell lymphoma and KG-1 leukemia cells were grown to 1 × 106 cells/ml in supplemented RPMI-1620. A total of 500 × 106 cells (Myla-2059 and KG-1) were pelleted and the pellet was treated with Trypsin-EDTA (Lonza), containing 1 mM NaSO4, for 30 min at 37°C. The supernatant was cleared of cells and the GAGs were extracted by ion exchange chromatography. Briefly, the supernatant was loaded onto a Q FF sepharose column (GE), the column was washed in 200 mM NaCl, and finally the GAGs were eluted in 1.5 M NaCl. The GAG’s were precipitated overnight (O/N) at 4°C in two volumes of ethanol and the precipitate was collected by centrifugation.
Purification of CS on a VAR2 Column
There were 1.5 mg rVAR2 (DBL1-ID2a) that were immobilized onto a Hitrap NHS HP Column (GE). The column was inactivated with ethanolamine and washed in PBS. Sigma CSA or Myla-2059 GAG extract was adjusted to 1 × PBS and loaded onto the column in five passages. The column was washed in PBS and the bound GAG eluted at 0.25 M NaCl, 0.5 M NaCl, 1 M NaCl, and 2 M NaCl in succession. The eluted GAGs were precipitated in ethanol O/N at 4°C in two volumes of ethanol and the precipitate was collected by centrifugation. The eluted fractions were tested for their ability to inhibit rVAR2 binding to CSPG (Attana) and C32 cancer cells (Flow Cytometry, see above).
Disaccharide Analysis of CS
The disaccharide compositions of CS samples were analyzed using chondroitinase ABC and size exclusion chromatography-MS (SEC-MS) as previously described (Shi and Zaia, 2009). Each type of CS disaccharides was extracted from the total ion chromatogram (TIC), integrated, and quantified by comparing with an external standard containing known amount of CS disaccharides. The sulfation position for the monoly-sulfated disaccharides was determined by tandem-MS experiments using previously established methods (Shao et al., 2013). The diagnostic fragment ions for the 4-O-sulfated disaccharide (Y1, m/z 300.0484) and for 6-O-sulfated disaccharide (Z1, m/z 282.0362) were extracted from the TIC, counted, and compared with a standard curve generated using commercial standards.
Biosensor Affinity Analysis
The analysis of the purified GAG species was performed on a quartz crystal microbalance biosensor (Attana A100, Attana AB). CSPG (Decorin, Sigma) was coupled to a LNB carboxyl chip using EDC and Sulfo-NHS. The chip was inactivated with ethanolamine. The sensor chips were inserted into the machine and allowed to stabilize in PBS running buffer, at 25°C using a flow rate of 25 µl per min. rVAR2 (30 nM) was mixed with a titration of inhibitor and injected onto the surface. Control rVAR2 was run repeatedly during analysis to account for changes in the binding surface. The binding surface was re-generated after each test injection with injections of 0.25% SDS in PBS. Peak response levels were recorded using the Attester Evaluation software (Attana AB) and presented as a ratio to the nearest rVAR2 injection. IC50 values were calculated in Excel.
IVIS In Vivo Imaging
rVAR2 was NIR labeled through available amines with an Alexa750 Succinimidyl ester (Invitrogen). This was done with an excess of NIR probe (10× molar) according to the manufacturer’s instructions. The coupled protein was injected (4 mg/kg) IV in the tail vein of healthy and tumor bearing mice 10 days post-establishment of a subcutaneous B16 melanoma tumor in the right flank. The mice were scanned using an IVIS spectrum CT scanner (Perkin Elmer). Scanning was done at time intervals ranging from 10 min to 48 hr. In vivo tumor signal quantification is presented as an absolute signal in reference to the signal of the flank of the healthy control mouse. Data analysis was performed using the Living Image Software (Caliper Life Sciences).
Patient Material
All human specimens were collected under full consent and according to the guidelines set forth and approved by the University of British Columbia (UBC) human ethics committee.
In Vivo Studies
The methodologies described were re-viewed and approved by the Institutional Animal Care Committee (IACC) at the University of British Columbia and the animal experiments inspectorate at the University of Copenhagen prior to conducting the study. During the study the care, housing, and use of animals was performed in accordance with the Canadian Council on Animal Care Guidelines and the Danish animal experiments inspectorate guidelines. For a detailed description of the in vivo studies and tolerability studies please see the Supplemental Information section.
Supplementary Material
Highlights.
The placenta and cancer express a similar type of oncofetal chondroitin sulfate
Oncofetal chondroitin sulfate is displayed on proteoglycans in cancer
Recombinant VAR2CSA proteins detect oncofetal chondroitin modifications
Human cancer can be broadly targeted by malarial VAR2CSA drug conjugates in vivo
Significance.
For decades researchers have sought to identify characteristics shared between placental or fetal development and cancer. This is based on the hypothesis that cancer cells, as part of their return to a less differentiated state, re-express genes involved in rapid growth and invasion during tissue development to facilitate cellular transformation and tumor progression. Using a specific placental glycan-binding malaria protein, we demonstrate that placental-like glycans are widely expressed in human tumors, with absent-to-low expression in normal tissues other than placenta. Furthermore, by conjugation of cytotoxic compounds to this protein, we demonstrate its capacity to specifically target cancer cells and block tumor growth in vivo.
Acknowledgments
The authors are thankful to Dr. Chao Sima at TGen, Phoenix, AZ, for assistance with TMA-related bioinformatics; Dr. Jeffrey Allen formerly of University of Tennessee Health Science Center, Memphis, TN, currently at Humboldt Medical Specialists, Eureka, CA for lung cancer tissue access and assistance with clinical annotation; Birita Fritleifsdottir for help with B16 xenograft model; and Monika Stints and Thomas Lavstsen for hBMEC cells. We would like to acknowledge the contributions of Geoffery Winters, Alex Mandel, Tom Hseih, Peter Bergqvist, Lawrence Amankwa, Siobhan O’Brien, Faisal Pany, Eric Cruz, and Natalia Neverova for their work on the rVAR2 drug conjugate at The Centre for Drug Research and Development.
The work was supported by funds from the Novo Nordisk Foundation; the Danish Cancer Society; and Stand Up To Cancer – St. Baldrick’s Pediatric Dream Team Translational Research Grant (SU2C-AACR-DT1113). Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research; Augustinus Foundation; U.S. Department of Defense (DoD); the Harboe Foundation; the European Research Councils (ERC) through the MalOnco program; The Danish Innovation Foundation; VAR2 Pharmaceuticals; Kairos Therapeutics; the Spar Nord Foundation; Team Finn and other riders in the Ride to Conquer Cancer; Safeway; and the Prostate Cancer Canada (PCC) Foundation. A.S., M.D., T.G.T., and P.H.S. are shareholders of VAR2 Pharmaceuticals ApS and members of the board of directors. J.R., B.H., and J.B. are employees of Kairos Therapeutics, Inc. J.F. and J.S. are employees of Retrogenix Ltd.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ccell.2015.09.003.
AUTHOR CONTRIBUTIONS
M.D., A.S., T.G.T., T.M.C., J.S.B., and P.H.B.S. designed the research; T.M.C., M.Dahlbäck, M.Ø.A., T.G., N.A.N., C.K.W., S.L., H.Z.O., L.F., M.E., J.S., L.B., F.F., M.A.N., J.F., Y.M., L.M., J.L., R.D., S.T., M.A.P., J.R.R., and B.J.H. performed the experiments; T.O.N., N.L.T., J.T., G.J.W., J.Z., P.J.H., and A.F.S. provided useful reagents and helpful discussions; and A.S., P.H.B.S., T.G.T., T.M.C., and M.D. wrote the manuscript.
REFERENCES
- Alkhalil A, Achur RN, Valiyaveettil M, Ockenhouse CF, Gowda DC. Structural requirements for the adherence of Plasmodium falciparum-infected erythrocytes to chondroitin sulfate proteoglycans of human placenta. J. Biol. Chem. 2000;275:40357–40364. doi: 10.1074/jbc.M006399200. [DOI] [PubMed] [Google Scholar]
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- Baston-Büst DM, Götte M, Janni W, Krüssel JS, Hess AP. Syndecan-1 knock-down in decidualized human endometrial stromal cells leads to significant changes in cytokine and angiogenic factor expression patterns. Reprod. Biol. Endocrinol. 2010;8:133. doi: 10.1186/1477-7827-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeson JG, Andrews KT, Boyle M, Duffy MF, Choong EK, Byrne TJ, Chesson JM, Lawson AM, Chai W. Structural basis for binding of Plasmodium falciparum erythrocyte membrane protein 1 to chondroitin sulfate and placental tissue and the influence of protein polymorphisms on binding specificity. J. Biol. Chem. 2007;282:22426–22436. doi: 10.1074/jbc.M700231200. [DOI] [PubMed] [Google Scholar]
- Casucci M, Nicolis di Robilant B, Falcone L, Camisa B, Norelli M, Genovese P, Gentner B, Gullotta F, Ponzoni M, Bernardi M, et al. CD44v6-targeted T cells mediate potent antitumor effects against acute myeloid leukemia and multiple myeloma. Blood. 2013;122:3461–3472. doi: 10.1182/blood-2013-04-493361. [DOI] [PubMed] [Google Scholar]
- Clausen TM, Christoffersen S, Dahlbäck M, Langkilde AE, Jensen KE, Resende M, Agerbæk MO, Andersen D, Berisha B, Ditlev SB, et al. Structural and functional insight into how the Plasmodium falciparum VAR2CSA protein mediates binding to chondroitin sulfate A in placental malaria. J. Biol. Chem. 2012;287:23332–23345. doi: 10.1074/jbc.M112.348839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlbäck M, Jørgensen LM, Nielsen MA, Clausen TM, Ditlev SB, Resende M, Pinto VV, Arnot DE, Theander TG, Salanti A. The chondroitin sulfate A-binding site of the VAR2CSA protein involves multiple N-terminal domains. J. Biol. Chem. 2011;286:15908–15917. doi: 10.1074/jbc.M110.191510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko JD, Stewart TE, Taylor WH. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. USA. 1985;82:3197–3201. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M, Duffy PE. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science. 1996;272:1502–1504. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- Gama CI, Tully SE, Sotogaku N, Clark PM, Rawat M, Vaidehi N, Goddard WA, Nishi A, 3rd, Hsieh-Wilson LC. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat. Chem. Biol. 2006;2:467–473. doi: 10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- Holtan SG, Creedon DJ, Haluska P, Markovic SN. Cancer and pregnancy: parallels in growth, invasion, and immune modulation and implications for cancer therapeutic agents. Mayo Clin. Proc. 2009;84:985–1000. doi: 10.1016/S0025-6196(11)60669-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi N, Takeguchi A, Sakai S, Akiyama H, Higashi K, Toida T. Effect of molecular sizes of chondroitin sulfate on interaction with L-selectin. Int. J. Carbohydr. Chem. 2013;2013:9. [Google Scholar]
- Kirby M, Hirst C, Crawford ED. Characterising the castration-resistant prostate cancer population: a systematic review. Int. J. Clin. Pract. 2011;65:1180–1192. doi: 10.1111/j.1742-1241.2011.02799.x. [DOI] [PubMed] [Google Scholar]
- Litjens T, Baker EG, Beckmann KR, Morris CP, Hopwood JJ, Callen DF. Chromosomal localization of ARSB, the gene for human N-acetylgalactosamine-4-sulphatase. Hum. Genet. 1989;82:67–68. doi: 10.1007/BF00288275. [DOI] [PubMed] [Google Scholar]
- Orendi K, Kivity V, Sammar M, Grimpel Y, Gonen R, Meiri H, Lubzens E, Huppertz B. Placental and trophoblastic in vitro models to study preventive and therapeutic agents for preeclampsia. Placenta. 2011;32(Suppl):S49–S54. doi: 10.1016/j.placenta.2010.11.023. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med. Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Iwawaki T, Taya C, Yonekawa H, Noda M, Inui Y, Mekada E, Kimata Y, Tsuru A, Kohno K. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat. Biotechnol. 2001;19:746–750. doi: 10.1038/90795. [DOI] [PubMed] [Google Scholar]
- Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, Arnot DE, Hviid L, Theander TG. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- Salanti A, Dahlbäck M, Turner L, Nielsen MA, Barfod L, Magistrado P, Jensen AT, Lavstsen T, Ofori MF, Marsh K, et al. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J. Exp. Med. 2004;200:1197–1203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Shi X, White M, Huang Y, Hartshorn K, Zaia J. Comparative glycomics of leukocyte glycosaminoglycans. FEBS J. 2013;280:2447–2461. doi: 10.1111/febs.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Zaia J. Organ-specific heparan sulfate structural phenotypes. J. Biol. Chem. 2009;284:11806–11814. doi: 10.1074/jbc.M809637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara K, Mikami T, Uyama T, Mizuguchi S, Nomura K, Kitagawa H. Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol. 2003;13:612–620. doi: 10.1016/j.sbi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Van Sinderen M, Cuman C, Winship A, Menkhorst E, Dimitriadis E. The chrondroitin sulfate proteoglycan (CSPG4) regulates human trophoblast function. Placenta. 2013;34:907–912. doi: 10.1016/j.placenta.2013.07.065. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang Y, Yu L, Sakakura K, Visus C, Schwab JH, Ferrone CR, Favoino E, Koya Y, Campoli MR, et al. CSPG4 in cancer: multiple roles. Curr. Mol. Med. 2010;10:419–429. doi: 10.2174/156652410791316977. [DOI] [PubMed] [Google Scholar]
- Wang X, Katayama A, Wang Y, Yu L, Favoino E, Sakakura K, Favole A, Tsuchikawa T, Silver S, Watkins SC, et al. Functional characterization of an scFv-Fc antibody that immunotherapeutically targets the common cancer cell surface proteoglycan CSPG4. Cancer Res. 2011;71:7410–7422. doi: 10.1158/0008-5472.CAN-10-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.