Abstract
Protein- and peptide-based structural biopolymers are abundant building blocks of biological systems. Either in their natural forms, such as collagen, silk or fibronectin, or as related synthetic materials they can be used in various technologies. An emerging area is that of biomimetic materials inspired by protein-based biopolymers, which are made up of small molecules rather than macromolecules and can therefore be described as supramolecular polymers. These materials are very useful in biomedical applications because of their ability to imitate the extracellular matrix both in architecture and their capacity to signal cells. This article describes important features of the natural extracellular matrix and highlight how these features are being incorporated into biomaterials composed of biopolymers and supramolecular polymers. We particularly focus on the structures, properties, and functions of collagen, fibronectin, silk, and the supramolecular polymers inspired by them as biomaterials for regenerative medicine.
Keywords: self-assembly, biomedical, biomimetic (assembly), nanostructure
Introduction
The extracellular matrix (ECM) is the component present within all tissues and organs, and provides mechanical support as well as can guide and direct cell function.1 The ECM is composed of two main classes of macromolecules, proteoglycans and fibrous proteins with distinct hierarchical assemblies at various length scales.2,3 The hierarchical structure–function relationship of these biopolymers regulates the functions of cells, tissues, and organs. In the context of biomaterials for regenerative medicine, proteins that can be used to construct scaffolds that mimic the structure and function of the extracellular matrix are of particular interest.4 Specifically, fibrillar proteins that are abundant components of the native ECM, such as collagen and fibronectin, are natural choices for tissue engineering applications.5–7 Collagen is responsible for the structural support and elasticity of tissues.8 Fibronectin supports cell adhesion to the ECM, as well as cell migration, proliferation, and differentiation.9,10 In addition to collagen and fibronectin, silk proteins (originating from silkworms and spiders) have attracted considerable interest for tissue engineering applications because of their tailorable mechanical properties, good biocompatibility, and facile processability.11–13 Critical to the function of these and other protein biopolymers is their ability to spontaneously assemble from smaller subunits into long uniform structures stabilized by many noncovalent interactions. In some cases, the dynamic features of these interactions are coupled to the capacity of the resulting fibrillar structures to rapidly polymerize and depolymerize and guide specific function through biomechanical and biochemical signals.14–16 The intracellular fibers known collectively as the cell’s cytoskeleton are the best example of the central functional role of highly dynamic fibrous structures.
The hierarchical self-assembly of proteins into well-defined structures has inspired research on artificial supramolecular architectures intended to mimic the function of native proteins. In these supramolecular assemblies, the monomeric units are held together by multiple noncovalent intermolecular interactions such as hydrogen bonding,17–20 metal–ligand coordination,21–23 π−π stacking,24 hydrophobic interactions,25 and host-guest interactions.26,27 Unlike covalent polymerization, all of these processes are highly reversible, and dynamic, which endow the new polymer materials with many attractive properties. Such properties include a structurally responsive nature, easy synthesis and functionalization, and the possibility of incorporating an array of different ligands through co-assembly of the building blocks.28 Although a variety of building blocks have been employed to design bioinspired polymeric architectures,29–30 peptide-based supramolecular polymers are the most common. Peptide building blocks are ideal mainly because they are the primary signaling components in the ECM. Also, the peptide sequences are biocompatible, degradable and can be quickly and reliably synthesized.
In this article, we discuss the common structural features of the biopolymers collagen, fibronectin, and silk and how their characteristics have inspired the synthesis of peptide-based supramolecular polymers as bioactive materials with tailored properties and customizable functions. We describe the use of these two classes of polymers for regenerative medicine and biomedical applications and address the remaining challenges for designing future biomaterials for clinical applications.
Structure and organization of protein biopolymers
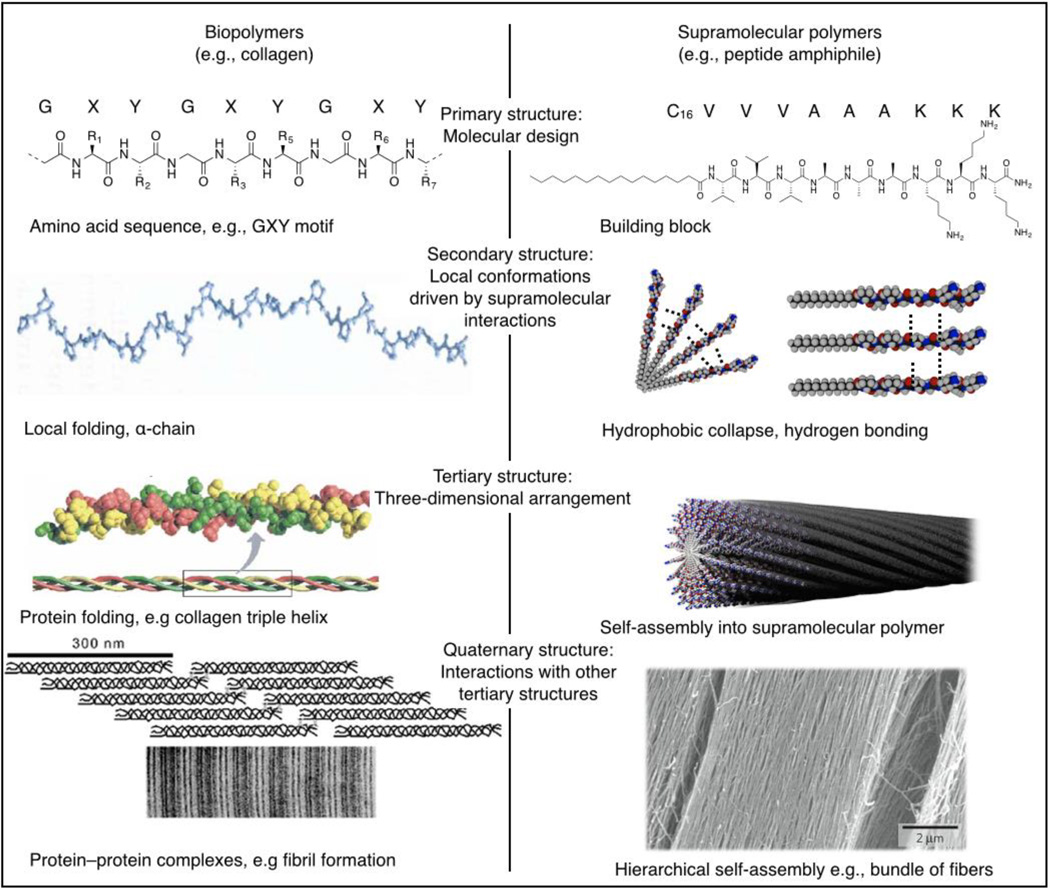
Nature provides many examples of structurally complex and functional architectures obtained by integrating multiple simple interactions that act in concert to produce the final macromolecular compound. Proteins are an example of this design strategy. The base level of protein structure is the primary structure, a sequence of amino acid residues covalently linked together to form the linear backbone of the biomolecule. Although the arrangement of amino acid residues might seem rather simple, the primary structure prescribes the folding and higher-order conformation of the protein. The secondary structure describes the local conformation of the protein backbone. The hydrogen bonding of the peptide backbone can result in regular folding patterns such as α helices and β sheets. The folding of these peptide chains into tertiary structures is largely determined by hydrophobic interactions and contributions from disulfide bonds. Some proteins are assembled from multiple polypeptide chains; the specific arrangement of these subunits into a multimeric molecule is defined as the quaternary structure of the protein (Figure 1, left). Taken separately, interactions such as hydrogen-bonding and hydrophobic interactions seem rather limited, yet when combined, they contribute to hierarchical self-assembly resulting in an impressive number of uniquely functional proteins.31 This hierarchical assembly process is directly responsible for the mechanical properties and bioactivity of protein biopolymers.
Figure 1.
Hierarchical self-assembly of (left) biopolymers and (right) supramolecular polymers. Both biopolymer folding and supramolecular polymer assembly start with a molecular design, coding for organization on several hierarchical levels. Second, the structures order based on local interactions with monomers, neighboring amino acids in the case of biopolymers or adjacent building blocks for supramolecular polymers. On a third level, the monomers act to form a three-dimensional structure such as a fibrillar protein or a supramolecular polymer. Finally, the fibrillar proteins can be organized with other proteins into a functional system. Similarly, self-assembled structures can be assembled hierarchically to form a superstructure.
Reproduced with permission from Reference 33 © 2008 Elsevier and 34 © 2007 Elsevier (left) and 87 © 2010 Nature Publishing Group (right).
One of the most researched families of protein biopolymers is that of collagen, the main component of the extracellular matrix (ECM), which provides strength and elasticity to tissues, blood vessels, ligaments, and bone.32 Collagen type I, the most abundant collagen in the human body, supports and stabilizes the ECM as a whole, provides attachments sites for adhesion proteins and glycosaminoglycans, and interacts with the cytoskeleton to maintain dynamic exchange between the cell and the matrix. Collagen type I consists of three polypeptides, so-called α chains with the repetitive sequence Gly-X-Y, where X and Y can be any amino acid including glycine (Gly), proline (Pro), or hydroxyproline (Hyp). The three alpha chains assemble into a triple helix structure, Figure 1, left. The triple helixes themselves assemble further into fibrils stabilized primarily by hydrogen bonding.8,33–34 On the next hierarchical level, these fibrils assemble into supramolecular complexes, where different tissues or organs contain fibrils of different diameters.35 In the cornea, for example, 20-nm-diameter collagen fibrils are arranged orthogonally to maintain its structure while retaining optical transparency.35 In mature tendons, larger-diameter (500-nm) fibrils align in parallel bundles to support the high tensile demands.17 Thus, the repetitive primary structure of collagen, its coordinated self-assembly, and the demands from its local microenvironment give rise to its ultimate biological function.
Another ECM protein, fibronectin (FN), is structurally and functionally different from collagen but exploits similar design principles to form fibrils. The FN primary structure contains three repeating β-sheet structures, designated I, II, and III.36–40 Two FN polypeptides associate into a dimer linked by disulfide bonds. This association creates a series of paired ligand-binding domains, spaced at strategic points, with a high affinity for cell membrane receptors, including integrins, and various other ECM components and factors (e.g., heparin sulfate, collagen,41 and fibrin). By binding collagen, FN stabilizes its overall scaffold structure, and by linking integrins, FN participates in the cell-matrix cross-talk that controls cell proliferation, migration, and differentiation. When FN recruits other globular molecules from the extracellular space, it initiates polymerization through noncovalent bonding at its N terminus.42 Fibrillar FN is not static. Once it is coupled to cells, it can be rearranged, remodeled, and recycled to meet the demands of its local microenvironment.43 Thus, a key feature of FN assembly into fibers is its ability to undergo reversible conformational changes that convert soluble FN from its initial inactivated, compact conformation into a surface-activated, extended conformation.
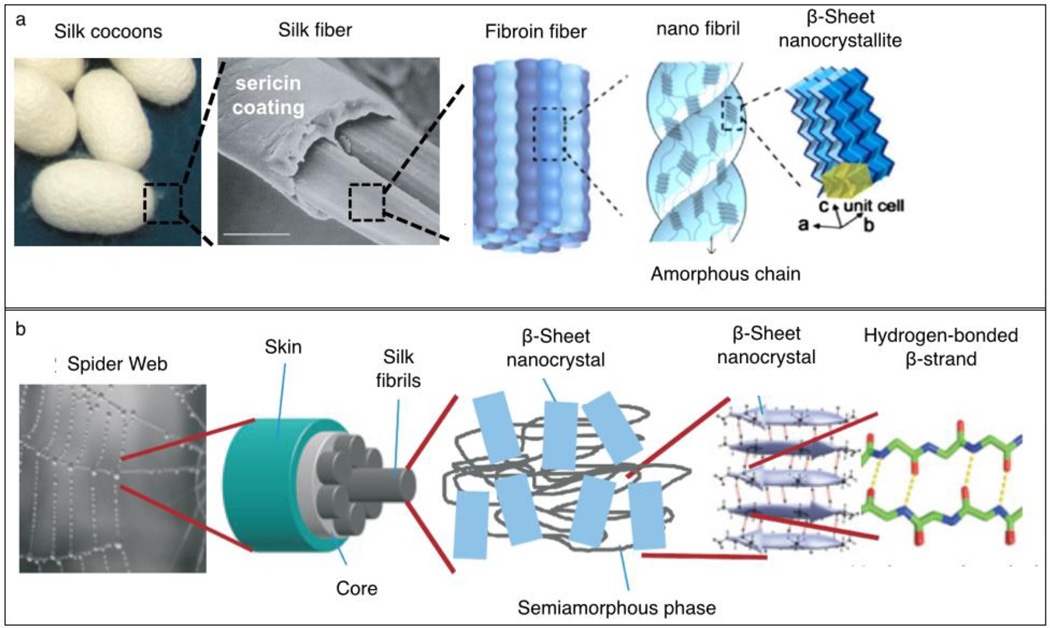
Silks are a diverse class of protein biopolymers that are produced for use outside the physical environment of the organism and exhibit structure-dependent functional properties.44 The silk proteins most extensively studied for regenerative medicine include materials naturally produced by spiders and silkworms, as well as engineered proteins whose sequences were inspired by such native silks45–47 (Figure 2). The major structural protein of silkworm (Bombyx mori) silk, is fibroin, an amphiphilic block copolymer with alternating repetitive hydrophobic and hydrophilic domains.48 During fiber spinning, fibroin proteins assemble into semicrystalline structures composed of β-sheet crystallites and less-structured (or amorphous) regions.49 Fibroins are spun by the silkworm into fibers of ~10–25-µm diameter (Figure 2a).50 These fibroin-based fibers are coated and cemented together by a second family of gluelike proteins called sericins to produce a cocoon that provides a protective environment for the silkworm to undergo its metamorphosis into a moth. Purification of stable aqueous fibroin solutions from these cocoons is straightforward and allows for the generation of regenerated fibroin materials in a variety of forms, including films, fibers, and foams.50
Figure 2.
Hierarchical organization of silk fibers. (a) Structure of silk fibers comprising silk cocoons produced by the silkworm (B. mori).The silk fiber is composed of fibers of fibroin, a structural protein, held together by sericins, gluelike proteins. The fibroin fibers are produced from smaller-diameter nanofibrils, made from assembled fibroin proteins. The fibroin proteins fold into a semicrystalline morphology during spinning, being organized into highly crystalline β sheets and less-ordered domains. (b) Structure of spider dragline silk. The spider dragline thread is composed of small silk fibrils. The fibrils are composed of structural proteins (spidroins) that assemble into β-sheet nanocrystals and a semiamorphous phase during spinning. Reproduced with permission from Reference 45 © 2002 Nature Publishing Group, 46 © 2014 Royal Society of Chemistry, and 47 © 2010 Nature Publishing Group.
The silk dragline fiber produced by spiders contains several high-molecular-weight proteins (spidroins) that are organized in three domains: a large central repetitive core and two nonrepetitive terminal domains (Figure 2b).51 The overall structure of the central core of these spidroins is reminiscent of the semicrystalline block co-polymer organization of B. mori silk, but there are differences between the two materials. Spidroins contain sequence motifs that fold into less-ordered helical and β-spiral confirmations that contribute to the extensibility and, thus, toughness of the protein fiber.52 Poly-alanine blocks form crystalline β-sheet structures that serve as strong intermolecular crosslinks and the crystalline content and crystallite size correlate with the strength of the silk fiber.53 Nonrepetitive N- and C-terminal domains have a globular structure and play a critical role in the storage of the proteins and the assembly of fibers during spinning.11 Significant advances in recombinant production of spidroin-inspired proteins have facilitated the generation of designer, silk-like proteins for biomedical research.54,55
Although the structures of collagen, fibronectin, and silk proteins are distinct, each biopolymer contains repetitive sequences and domains stabilized by noncovalent bonds that contribute to the bulk mechanical properties of the fibers or networks they form. These properties enable proteins to assemble into hierarchical structures with functions that depend on their ordered arrangements. Their abilities to self-assemble in defined ways, respond to specific environmental stimuli, and adapt to mechanical loads have inspired the development of supramolecular polymers that mimic core “design principles” of structural proteins for a variety of applications.
Bioinspired supramolecular polymers: structure and assembly
The hierarchical organization of proteins remains an ongoing inspiration for scientists seeking new methods for obtaining elaborate molecular assemblies. Supramolecular polymerization, which exploits noncovalent interactions including hydrogen-bonding or π−π interactions56, 57 to direct the self-assembly of molecular units, is a powerful approach to constructing such assemblies. The noncovalent interactions make this class of polymers unique, as it allows the rapid transformation of the material by (locally) breaking and re-forming the noncovalent bonds between monomers. Moreover, the materials can be molecularly engineered to achieve specific features such as biodegradability or bioactivity. As with biopolymers, the organization of supramolecular polymers starts with a molecular design that encodes the properties of the final assemblies (Figure 1, right). The noncovalent bonds between neighboring monomers result in the formation of a stable nucleus. The nuclei grow further to form an ensemble of many molecules, through the process of self-assembly, resulting in polymer growth. On the next hierarchical level, these polymers can interact with one another to form higher-order hierarchical assemblies. As with proteins, although the local interactions between the basic subunits are often simple, these interactions lead to complex architectures over various length scales.
Supramolecular polymers can be divided into two major classes: random-coil and ordered.57 The first class is reminiscent of an unfolded protein but formed by noncovalent interactions. Random-coil supramolecular polymers were studied by Meijer and co-workers, yielding an interesting class of materials with properties similar to those of regular polymers but with the dynamics of supramolecular assemblies.58 The second class involves the formation of one-dimensional nanoarchitectures with a high degree of internal order (ordered supramolecular polymers) and was developed and extensively studied in the laboratories of Stupp,59–62 Aida,63 and others.64,65 Ordered supramolecular polymers all share a common feature: The self-assembly of the monomers is driven by at least one type of anisotropic interaction, typically hydrogen-bonding or π−π interactions. These anisotropic interactions are the driving force to form one-dimensional structures, thereby creating polymer-like assemblies. The design of peptide-based ordered supramolecular polymers relies mainly on using conserved amino acids sequences that retain many of the molecular characteristics of the native protein,66 or using peptide sequences that adopt secondary structures, such as the β-sheet.67
The use of protein-derived peptides enables mimicking the structural properties of the native protein while using shorter peptide sequences. For example, collagen-mimetic peptides that contain the triad amino acid repeats Gly-Hyp-X or Gly-X-Hyp (where X is usually Pro)68 were employed to elucidate the triple-helix structure and the stabilization effect of different amino acid residues,69 similar to native collagen. Hartgerink and co-workers designed and synthesized self-assembling peptides that form collagen-like triple helixes with sticky overhangs that direct their assembly into longer fibers and eventually self-supporting hydrogels, recapitulating the hierarchical self-assembly of natural collagen.66 In addition to collagen-mimetic peptides, silk-mimetic peptides bearing the morphological features of the natural protein have been synthesized and used to study the secondary structural propensities and folding into an elongated confirmation.70
A different category of ordered supramolecular polymers includes short peptide sequences where their self-assembly is driven by electrostatic interactions and β-sheet formation.71 For instance, the RADA16 motif, a 16-fold repetition of the four amino acids arginine (R), alanine (A), aspartic acid (D) and alanine (A), has an alternating cationic–hydrophobic–anionic–hydrophobic peptide sequence.72–74 Complexation of the opposing charges allows the formation of strong β sheets, resulting in the formation of ordered supramolecular polymers (Figure 3a). These fibers are able to entangle and form hydrogels under physiological conditions, rendering them useful in the context of regenerative medicine.75 In addition to RADA16 and collagen-mimetic peptides, many other underivatized self-assembling peptides exist and are covered in various reviews.76
Figure 3.
Self-assembly of supramolecular polymers. (a) Molecular design of RADA16. The alternating cationic–hydrophobic–anionic–hydrophobic sequence forces assembly into ribbons, as evidenced by AFM (inset). Reproduced with permission from Reference 73 © 2013 American Chemical Society, and 74 © 2007 PloS one. (b) Molecular design of peptide amphiphiles. A hydrophobic tail and β-sheet domain drive self-assembly into supramolecular polymers. The charged domain ensures solubility of the fibers. Peptide amphiphile (PA) supramolecular polymers can be complexed with an anionic biopolymer to form hierarchically organized constructs. Reproduced with permission from Reference 62 © 2013 Royal Society of Chemistry. (c) Molecular design of Fmoc-YL-OMe and Fmoc-YL-OH, where Fmoc is the N-(fluorenyl-9-methoxycarbonyl) protecting group, Y is the amino acid tyrosine (Tyr), and L is the amino acid leucine (Leu). The hydrophobic Fmoc group and uncharged peptide renders Fmoc-YL-OMe insoluble in water. Only upon hydrolysis to Fmoc-YL-OH is the charge balance favorable for assembly into fibers, as evidenced by atomic force microscopy (AFM) (inset). Reproduced with permission from Reference 83 © 2010 Nature Publishing Group.
A prominent example of ordered supramolecular polymers was developed by Stupp and co-workers. Adding a hydrophobic tail to one end of a peptide sequence resulted in the formation of a peptide amphiphile (PA) (Figure 3b).59–62 This hydrophobic tail renders the peptide insoluble in water, and as a result, the monomers aggregate upon dispersion in aqueous solutions. Two domains in the peptide sequence prevent the aggregates from forming amorphous precipitates. The first domain consists of amino acids with a high β-sheet-forming propensity and is directly attached to the hydrophobic tail. These anisotropic interactions ensure that the molecules assemble into supramolecular polymers.77 The second domain contains charged amino acids and is attached to the β-sheet-forming domain. These charges render the fibers water-soluble. A fourth peptide domain can be attached to the charged domain to give the fiber functionality, such as a cell-binding domain78 or a catalytically active domain.79
Another example of functionalizing peptide sequences to drive self-assembly involves the addition of hydrophobic aromatic domains, such as the N-(fluorenyl-9-methoxycarbonyl) (Fmoc) protecting group. This hydrophobic group can drive the self-assembly of extremely short peptide sequences80 to form fibers.81 The addition of Fmoc dramatically decreases the concentration needed to form supramolecular polymers or hydrogels,82 making it more suitable as a biomaterial. In addition, the use of non-natural amino acids sequences allows for the creation of materials with functionalities that are not natively available in biology. For instance, Hirst et al. coupled the self-assembly of Fmoc-Tyr-Leu (where Tyr is tyrosine and Leu is leucine) to the hydrolytic activity of subtilisin (Figure 3c).83 This enzyme can hydrolyze the methyl esters of Fmoc-Tyr-Leu-OMe, thereby creating Fmoc-Tyr-Leu, which subsequently assembles into supramolecular polymers. Not only does this allow for the external triggering of self-assembly, it also creates a system in which the self-assembly rate can be controlled by the amount of enzyme present. The rate of self-assembly influenced the morphology of the resulting supramolecular polymer and, thereby, its material properties.
Using supramolecular polymers, researchers have studied the relationship between molecular design and self-assembled structure and their effect on the material properties. For instance, Stupp and co-workers showed that the rigidity of a supramolecular polymer84 or the cohesiveness of the fibers85 can be tuned by introducing minor mutations in the peptide domain of PA molecules. Better understanding of such molecular design and structure relationships is crucial for the resulting functional properties, and can induce differnet cell behaviors and fates86
Similar to native protein folding across multiple length scales to form macroscopic protein networks, supramolecular polymers can also be organized on higher length scales. For example, PA molecules were hierarchically assembled in perpendicularly aligned monodomain gels.87 The ordering within this hierarchical construct takes place over several length scales. A different hierarchically ordered supramolecular system involved the ordering of a hybrid system of PA nanofibers and an oppositely charged biopolymer88 (Figure 3b). A mixture of these two components resulted in the formation of nanofiber bundles aligned perpendicular to the diffusion barrier at the interface of the two solutions. The final structure was a hierarchically ordered membrane with a thickness on the order of 2–20 µm that was organized on two different levels. First, PA molecules were assembled into supramolecular polymers, and second, the supramolecular polymers were aligned perpendicular to the membrane.89 The length scale of such constructs was not limited to macroscopic sacs or membranes but rather that the structures could be downscaled by injecting microdroplets of biopolymer solution into a bath of supramolecular polymers using a picospray setup.90 This processing technique resulted in cell-sized hierarchically organized membranes, rather than macroscopic structures. Moreover, using a microemulsion technique, the length scale could be decreased even further to supramolecular-polymer-decorated particles with a diameter of only hundreds of nanometers, ideal for drug-delivery purposes.91
Peptide-derived supramolecular polymers offer great flexibility for applications in regenerative medicine, as the self-assembled structures provide the main structural components and various amounts of surface decoration with bioactive or signaling ligands can be doped into the structure. Control over the amino acid sequence allows the formation of domain structures with predisposed assembly at scales well beyond the length of the amino acid blocks. Controlling the peptide sequence or modifying the amino acids with different motifs enables the formation of fibers at the multinanometer scale and gels or higher hierarchical structures at micron and larger length scales. The dimensions of the resulting supramolecular assemblies enable them to mimic some of the components of the ECM and direct cell behavior.
Protein biopolymers and supramolecular polymers as biomaterials for regenerative medicine
Regenerative medicine aims at developing therapies for the repair or replacement of tissues and organs, thereby restoring function impaired by congenital defects, diseases, trauma, or aging.92 One of the goals in this field is the development of functional scaffolds that provide the microenvironment for the growth of cells and tissues. Cells in their natural environment are constantly signaled by surrounding factors to adhere, migrate, proliferate, or differentiate. These signaling molecules can be loosely divided into two groups: soluble factors, including growth factors and small molecules, and insoluble signaling cues that are covalently or noncovalently attached to the extracellular matrix. Therefore, in addition to providing physical support, an ideal engineered scaffold would regulate the delivery of bioactive factors and participate in signaling to control cell behavior and support tissue structure, growth, and function. In this context, protein biopolymers and bioinspired supramolecular polymers are ideal candidates to fulfill these requirements because of their biocompatibility, degradability, mechanical properties, and signaling capability.
The engineering of delivery matrices composed of biopolymers or supramolecular polymers has attracted extensive research efforts because of the critical role of growth factors in controlling cellular functions and their ability to elicit tissue regeneration. Precise control over the signaling of these factors in space and time could allow control of a regenerative process.29 To this end, the natural interactions between ECM proteins and growth factors, specifically the growth-factor-binding sites of fibronectin,93,94 enable the use of these domains as a generic approach for delivering growth factors. For example, a multifunctional recombinant fragment of fibronectin was engineered to integrate a fibrin-binding domain and a growth-factor-binding domain. This multifunctional domain enabled the co-delivery of vascular endothelial growth factor, VEGF,95 and platelet-derived growth factor, PDGF-BB, and induced angiogenesis at low doses, whereas growth factors delivered without the fibronectin fragment had no significant effect.94
Alternatively, different biopolymers including fibronectin, collagen, and silk have been used to encapsulate or immobilize growth factors by exploiting electrostatic or other secondary interactions between the biopolymers and the growth factors. For instance, silk has been studied as a potential material to deliver a growth factor able to stimulate the nervous system, thereby facilitating the treatment of peripheral nerve injury (PNI).96 For successful nerve regeneration after PNI, the axons from the severed nerves need to bridge the gap between the two stumps and restore the original connections.97 To increase the success rate, the axon growth can be stimulated by neural growth factors (NGFs) and guided through physical (contact guidance) and chemical (neurotropism) mechanisms. However, the inherent instability of growth factors in vivo poses a challenge. They are also sensitive to degradation during processing and formulation of scaffolds. Therefore, protection and the sustained release of growth factors from silk fibroin scaffolds have been studied to overcome this challenge.
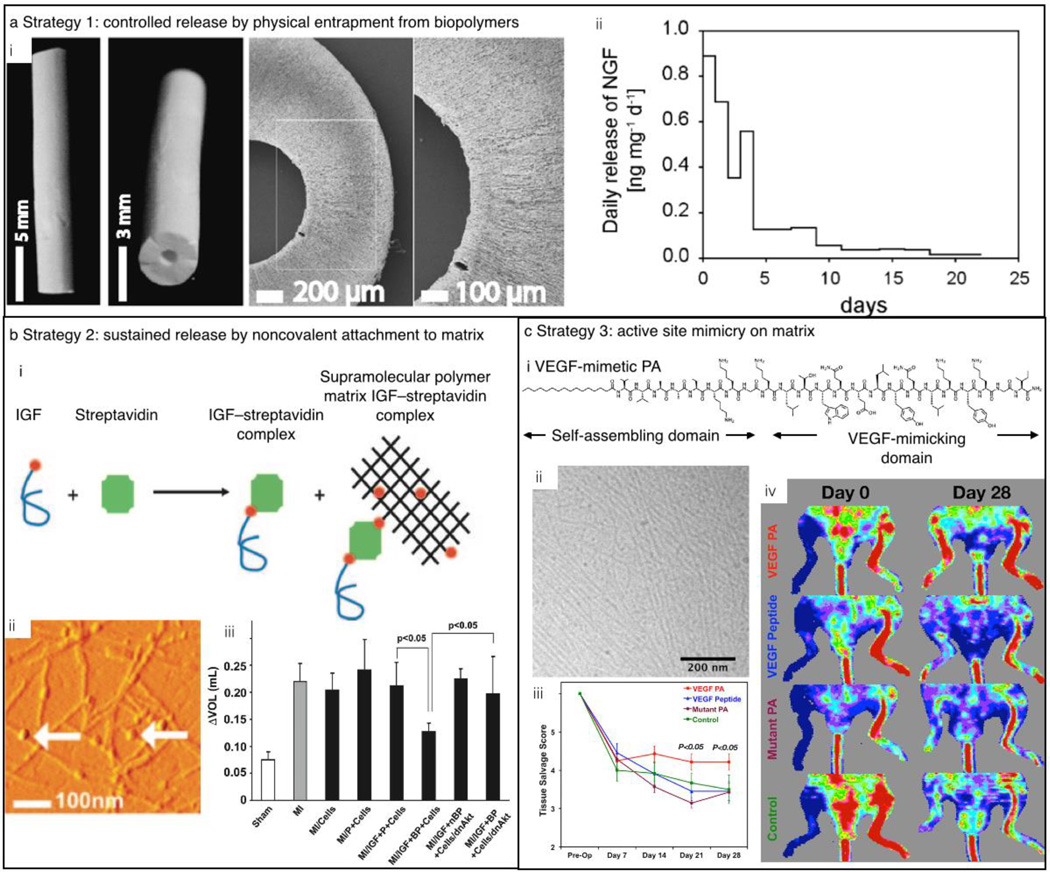
A great advantage of using fibroin to create such biomaterials is its water solubility and ability to form insoluble scaffolds under relatively mild conditions.98,99 Such benign, aqueous processing allows for the production of relatively sophisticated materials, such as fibroin nerve-guiding conduits loaded with NGF for peripheral nerve repair (Figure 4a). NGF-loaded fibroin matrices exhibit sustained release of the growth factor for weeks and support the adhesion of PC12 cells. Furthermore, silk fibroin scaffolds, loaded with NGF, can support adhesion and promote neurite outgrowth of dorsal root ganglion neurons.100 In addition to NGF, silk scaffolds have been studied for the controlled release of insulin-like growth factor I (IGF-I) and bone morphogenetic protein 2, resulting in the in vitro differentiation of human mesenchymal stem cells into chondrocytes and osteoblasts, respectively.101
Figure 4.
Strategies for the release of growth factors. (a) Growth-factor release by physical entrapment in a nerve conduit. (i) Photographs and scanning electron microscopy (SEM) images of the nerve conduit. (ii) Sustained release of neural growth factor (NGF) from the scaffolds. (b) Sustained release of proteins through noncovalent interactions between the scaffold and the growth factor. (i) Scheme of the supramolecular interaction among biotinylated insulin-like growth factor (IGF), streptavidin, and a biotinlyated peptide as evidenced by (ii) AFM. (iii) Ventricular dilation, as measured by the difference in ventricular volume between days 1 and 21 after an induced myocardial infarction, was not observed for rats with cells embedded in the IGF-nanofiber construct. (c) Sustained growth-factor efficacy by mimicking the active site of the growth factor. (i) Molecular design of VEGF-mimetic peptide. (ii) Cryogenic transmission electron microscopy (Cryo-TEM) image of the supramolecular polymers formed by the VEGF-mimetic PA. (iii) Tissue salvage score according to the hindlimb ischemia model showing a significantly higher tissue salvage for animals treated with VEGF-mimetic PA. (iv) Laser Doppler perfusion imaging shows significantly higher perfusion ratios for the VEGF-mimetic PA as compared to controls. Reproduced with permission from 99 © 2007 Elsevier (a), 105 © 2006 National Academy of Sciences (b), and 109 © 2012 National Academy of Sciences (c).
Besides biopolymers, supramolecular polymers are also attractive for the delivery of biological factors. Delivery of bioactive factors can be achieved by chemically immobilizing or physically encapsulating them into supramolecular polymer matrices, preventing their denaturation. Their release can be controlled by the degradation rate of the matrices,102 their diffusion through the polymer construct,103 or external triggers.104 For example, the controlled release of a growth factor from a protected reservoir has been explored which relies upon biotin / streptavidin interactions..105 In this approach, peptides were biotinylated and the resulting biotin-decorated supramolecular polymers then facilitated the binding of tetravalent streptavidin molecules that could, in turn, coordinate biotinylated IGF-I (Figure 4b). The binding of growth factors to the supramolecular polymers protected them from enzymatic degradation and allowed for prolonged growth-factor delivery and activity. To demonstrate the efficacy of this strategy, the supramolecular polymers were tested in a cell-based therapy using a myocardial infarction mouse model. IGF-I tethered to the supramolecular polymer was injected together with cardiomyocytes (heart muscle cells) into the infarct zone and showed a significantly improved systolic (contractile) function of the heart as compared to untethered growth factor. This demonstrated that prolonged release of growth factors by noncovalent binding to a supramolecular polymer can support cell therapies.
Although growth factor delivery matrices show promise in controlling different cellular and regeneration processes, the clinical applications of these proteins are hindered by short half-lives, immune-related side effects, and high costs. An attractive strategy for overcoming some of these challenges is to identify the peptide sequence of the growth factor that binds the cell receptors and attach it to the supramolecular polymer. The peptide domain activates the targeted membrane receptors in the proximity of the scaffold and avoids the use of growth factors. For instance, D’Andrea et al. designed a peptide sequence based on the crystal structure of VEGF bound to its transmembrane receptor.106 This binding sequence was coupled to different supramolecular polymers107,108 and the resulting scaffolds were able to mimic the action of VEGF without using the actual growth factor. Stupp et al showed that when the VEGF-mimetic sequence is attached to a PA, the survival, proliferation, and migration of human umbilical vein endothelial cells (HUVECs) was significantly improved, as compared to the unmodified supramolecular polymer or the peptide by itself.109 The potential of the VEGF-mimetic PA as a therapy for ischemic disease was evaluated using the mouse hindlimb ischemia model (Figure 4c). An improvement in tissue salvage as well as enhanced motor function and blood perfusion was found after intramuscular injection of these supramolecular polymers, as compared to injection of just the bioactive peptide.
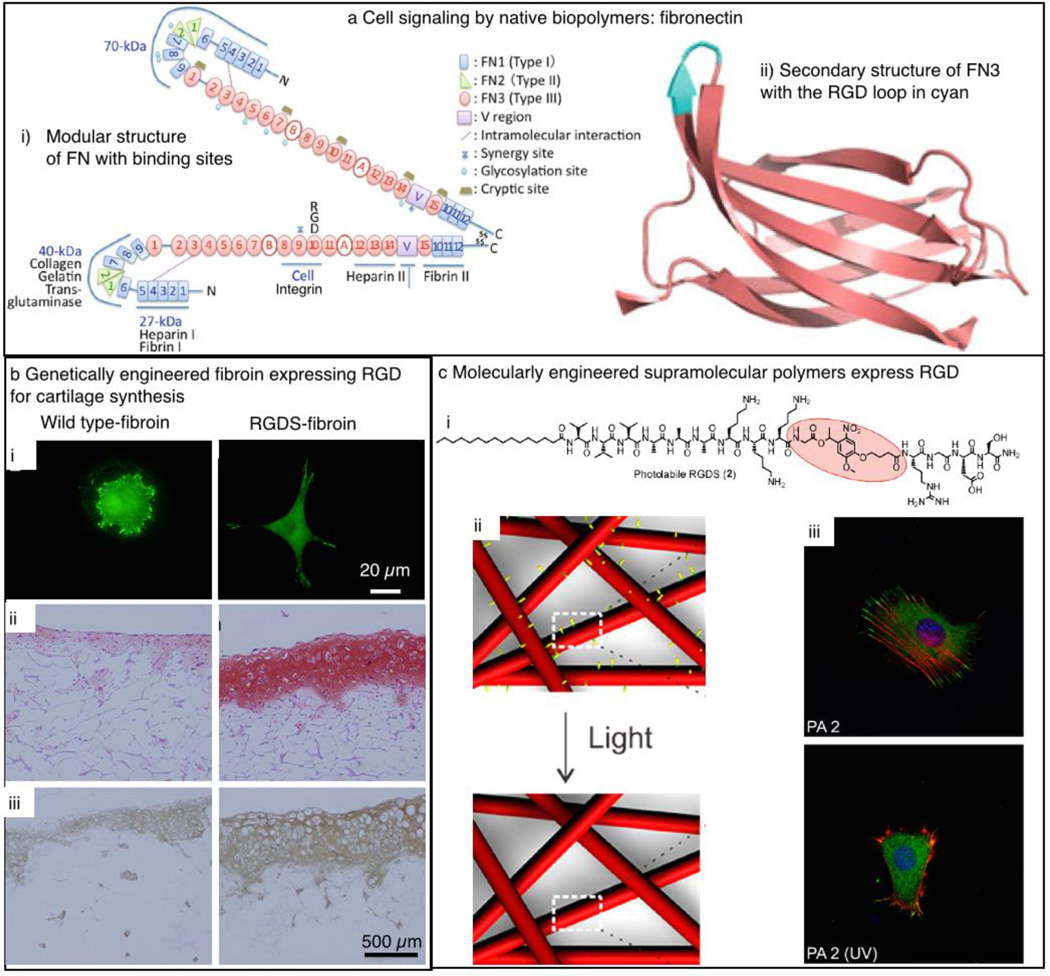
Beyond the ability of biopolymers and supramolecular polymers to deliver soluble signals to the environment, biomaterials for regenerative medicine need to be able to signal cells to undergo different processes such as adhesion, proliferation, migration, and differentiation. Cell adhesion is the biomimetic function most commonly incorporated in biomaterial design. Initially, this was achieved by merely coating scaffolds with biopolymers known to promote cell adhesion and spreading.110,111 Biopolymers such as fibronectin,112 laminin,113 vitronectin,114 tenascin,115 and certain collagens116 are known to facilitate cell adhesion and spreading through conserved amino acid sequences such as RGDS (Arg-Gly-Asp-Ser, where Arg is arginine, Gly is glycine, Asp is aspartic acid, and Ser is serine) that can be recognized by the transmembrane integrin proteins. In the case of fibronectin, integrin recognition and polymer self-assembly are reciprocal: Whereas cell adhesion depends on interaction with the fibronectin matrix, fibronectin matrix formation can occur only after soluble fibronectin interacts with integrins on the cell surface (Figure 5a).117 Although this process is not fully understood, fibronectin polymerization can be divided into two phases: nucleation and subsequent elongation. It is hypothesized that the binding of the RGDS sequence in fibronectin drives nucleation, as recombinant fibronectin that lacks the sequence cannot form fibrils.118 Integrin binding to the RGDS sequence initiates an intracellular cascade resulting in the formation of focal adhesions (cell–matrix adhesion points) allowing cells to attach and exert force on the surrounding matrix.119 This feature is extremely useful, as it permitted the incorporation of these sequences into synthetic systems, thus rendering them bioactive.
Figure 5.
Strategies to signal cells using biopolymers or supramolecular polymers. (a) Native cell-signaling peptides can be identified from extracellular matrix proteins, such as the RGD (Arg-Gly-Asp, where Arg is arginine, Gly is glycine, and Asp is aspartic acid) sequence in fibronectin. (b) (i) The RGD cell-signaling domain can be genetically engineered to be expressed by silkworms in the fibroin L-chain to significantly enhance cell adhesion compared to that in wild-type fibroin. Photographs of regenerated cartilage stained by (ii) Safranin-O or (iii) collagen type I immunostaining show enhanced cartilage regeneration on fibroin with RGD. (c) RGD can also be covalently engrafted on supramolecular polymers, such as (i) a PA construct. In this specific case, RGDS is attached via a photocleavable linker. (ii) Upon irradiation with UV light, the fibrous network of PA supramolecular polymers sheds their RGDS functional groups rendering them biologically inactive. (iii) Cells immobilized on the PA construct before (PA2) and after (PA2 (UV)) exposure with UV as imaged by confocal microscopy. Reproduced with permission from 40 © 2011 Springer (a), 129 © 2010 Elsevier (b), and 136 © 2012 American Chemical Society (c).
The most commonly applied peptide is the RGDS sequence, which has been incorporated into various polymers to render them bioactive.120 For the scope of this article, we focus only on examples of bioactivation of silk and supramolecular polymers with peptide cues. In the context of silk, Kaplan and co-workers showed that fibroin films can be chemically derivatized with RGD. These films were found to significantly upregulate bone formation as compared to controls without RGD.121 It was also shown that porous silk scaffolds functionalized with RGD can induce osteogenesis of human mesenchymal stem cells (differentiation into bone-forming osteoblasts).122 In conjunction with RGD functionalization, the surface patterning of silk facilitates the control of cell alignment on scaffolds.123 Such work shows the power of bioactivation and the potential of silk as a biomaterial for tissue reconstruction.
Scaffolds fabricated from recombinant silk proteins have also produced encouraging results in enabling attachment and growth of cells in vitro.124 The recombinant production of silk-inspired proteins allows for flexibility in the engineering of the protein sequence. For example, new proteins can be designed utilizing repetitive domains drawn from silkworm or spider silks in combination with sequences such as RGD.125–128 Kambe et al. genetically fused the RGD sequence into the silk fibroin light chain.129 To study whether the RGD was accessible for cells, the researchers found more attached chondrocytes (cartilage producing cells) on the RGD-fibroin as compared to wild-type fibroin (Figure 5b). The success in the application of silk fibroin in tissue engineering and the biomedical field in general has seeded considerable interest in the further development of these materials for medical devices and therapeutic treatments. Silk fibroin mesh structures (e.g., SERI Surgical Scaffold, from Allergan) are currently in clinical use as bioresorbable surgical scaffolds. In addition to their use as a fiber, a number of companies are pursuing silk fibroin solutions or silk-based materials for soft-tissue repair, the treatment of osteoarthritis, cartilage repair (currently in clinical trials), vascular grafts, and nerve regeneration. Recombinant spider silk proteins are also being commercially developed for tissue engineering and wound healing applications (e.g., AMSilk and Spiber Technologies).
Bioactivation of materials by incorporating bioactive cues can also be applied to supramolecular polymers. For instance, the above mentioned RGDS peptide has been applied on self-assembling PAs,130 Fmoc-peptide-based supramolecular polymers,131 self-assembling DNA nanotubes,132 and many others. Moreover, additional bioactive peptide sequences derived from collagen,133 laminin,78 or non-ECM proteins134,135 can also be used to increase the bioactivity of supramolecular polymers.
One goal when developing supramolecular polymers to mimic the ECM is to recapitulate the dynamic features of the native environment. Even though the ECM might seem static, ECM fibers are constantly broken down by enzymatic degradation, and new fibers are secreted by the cells. This homeostasis allows the tissue to change the composition of the ECM when necessary, for instance, in the case of wound healing or development. Although some supramolecular-polymer-based materials have been developed to mimic the native ECM dynamics,136,137 this feature remains a challenge. For instance, Sur et al. bioactivated a supramolecular polymer with an RGDS cue using a photocleavable linker, Figure 5c. Fibroblast cells recognized the RGDS cue, resulting in spreading, as expected. However, upon irradiation of the supramolecular polymers with light, the cues were shed, resulting in a loss of bioactivity.136 Although the dynamics in these examples and others138,139 are in stark contrast to the dynamics found in vivo, these strategies open up pathways to materials that display bioactivity on demand.
Conclusions and outlook
The selection of a bioactive biomaterial for tissue engineering and regenerative medicine is of great importance since its design can determine the functionality of the tissue formed, and when used as a scaffold seeded with cells it will greatly influence engraftment with host tissues. Overall, for a material to serve as a synthetic ECM, a few basic requirements are necessary. Key features are biodegradability and biocompatibility. The degradation kinetics has to be compatible with the rate of tissue regeneration and restoration of function, and materials used need to have the necessary mechanical integrity. The scaffold should also integrate and interact with the surrounding host tissues without eliciting an immunological response. Biomaterials and scaffolds must also allow easy diffusion of nutrients and cellular waste products, and allow cell penetration and tissue ingrowth/outgrowth depending on the application.. Biomaterials for regeneration should guide the appropriate cell response, deliver the required soluble factors, recruit endogenous cells, and in most cases induce vascularization. Finally, in order to translate biomaterials from the laboratory to the clinic they should be easy to handle, manufacture, store, sterilize, package and transport to the clinic.
Protein, peptide, and supramolecular polymers have emerged as attractive candidates for the fabrication of scaffolds for tissue engineering and regenerative medicine. They exhibit beneficial interactions with cells and have great potential as hydrogels. The mechanical properties, patternability, and biochemistry of ECM- and silk-derived proteins make these biomolecules ideal for the direction of cell behavior including adhesion, proliferation, migration, and differentiation. Scaffolds based on collagen and silk proteins have been shown to induce minimal immune and inflammatory responses when implanted in the body,140–142 are mechanically robust, and can be proteolytically degraded into nontoxic amino acids.
Silk fibroin derived from B. mori cocoons is a US Food and Drug Administration-approved biomaterial that is relatively inexpensive and widely available. The ability of silk fibroin to stabilize and deliver different molecules (e.g., growth factors, antibiotics, and vaccines) is of particular interest and holds tremendous promise for tissue engineering and other biomedical applications.143 The properties of silk and collagen make these materials attractive for further advances in the manufacturing of tissue engineering scaffolds, including additive manufacturing. Indeed, fibroin/collagen solutions have been successfully applied as the base materials in the bioprinting of chondrocyte cells.144 These advances hint at the possibilities of collagen- and silk-based clinical tissue regeneration work and commercialization in the future. The recombinant production of collagen- and spidroin-inspired proteins has progressed significantly and allows for the engineering of designer proteins with multiple functionalities. However, the high relative costs of these recombinant proteins currently limit their availability to the greater research community.
As biomaterials, supramolecular polymers are versatile materials that can be designed to mimic specific aspects of biopolymers by utilizing noncovalent interactions to direct the self-assembly of molecular building blocks. Specifically, self-assembling peptides offer numerous advantages as versatile and efficient biomaterials. Inspired by biological signaling, supramolecular polymers have been decorated with different bioactive peptide sequences for a variety of regenerative medicine applications. Furthermore, peptide biomaterials can biodegrade rapidly by hydrolytic and enzymatic processes into natural amino acids. Furthermore, the hierarchical assembly of peptide-based materials enables the crafting of materials organized from molecular to macroscopic scales. As an example, Stupp and co-workers’ monodomain gels can molecularly instruct cells to elicit a specific biological response by using peptide epitopes, but they can also instruct cells at the macroscopic scale to migrate along a specific direction.87
Multiple-length-scale signaling to cells is of crucial importance in regenerative medicine, especially when functional recovery is about spatial control as it is for the regeneration of nervous tissue. Two important examples are spinal-cord injury145–146 and the regeneration of peripheral nerves.147–149 The number of hierarchically organized materials remains limited so far, mainly because of a lack of design rules for the formation of hierarchical structures. Although great progress has been made on rules for the self-assembly of molecules into nanostructures, we currently do not have (with some exceptions150) adequate design rules that predict their assembly into hierarchically ordered structures across various length scales.
Another horizon in the field of supramolecular polymers is the development of stimuli-responsive dynamic materials. Self-assembled supramolecular architectures are optimal candidates for creating dynamic materials because the building blocks are not covalently fixed and the supramolecular forces that hold them together can be reconfigured to change their properties. Engineering dynamic triggers that allow changes in the physical structure and chemical composition of a synthetic matrix would be highly advantageous in mimicking the dynamic features of the ECM. This field will continue to expand to create such functionally powerful materials, possibly combining macromolecules and supramolecular structures.
Acknowledgments
Experimental work in the Stupp laboratory related to bioactive and bioinspired materials was supported by grants from the US National Institutes of Health (National Institute of Biomedical Imaging and Bioengineering, NIBIB/BRP Award 3R01EB003806-09S1;, National Heart, Lung, and Blood Institute, NHLBI/BRP Awards 5R01HL116577-02 and NHLBI/PPG Award 5P01HL108795-04; and National Institute of Dental and Craniofacial Research, NIDCR Award 5R01DE015920-09); the Department of Energy Office of Basic Energy Sciences under Award DE-FG02-00ER45810; the Center for Bio-Inspired Energy Science, an Energy Frontier Research Center funded by the US Department of Energy, Office of Science, Basic Energy Sciences (BES), under Award DE-SC0000989; and the National Science Foundation under Award DMR-1006713-004.
M.D. and R.R.N. thank Kristi M. Singh [Air Force Research Laboratory (AFRL)] for assistance in the editing of this manuscript. M.D. and R.R.N. gratefully acknowledge support from AFRL and Air Force Office of Scientific Research (AFOSR).
References
- 1.Frantz C, Stewart KM, Weaver VM. J Cell Sci. 2010;123:4195. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Järveläinen H, Sainio A, Koulu M, Wight TN, Penttinen R. Pharmacol Rev. 2009 Jun;61(2):198–223. doi: 10.1124/pr.109.001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaefer L, Schaefer RM. Cell Tissue Res. 2010;339:237. doi: 10.1007/s00441-009-0821-y. [DOI] [PubMed] [Google Scholar]
- 4.Barker TH. Biomaterials. 2011;32:4211. doi: 10.1016/j.biomaterials.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 5.Glowacki J, Mizuno S. Biopolymers. 2008;89:338. doi: 10.1002/bip.20871. [DOI] [PubMed] [Google Scholar]
- 6.Song JJ, Ott HC. Trends Mol. Med. 2011;17:424. doi: 10.1016/j.molmed.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Roy DC, Hocking DC. Tissue Eng., Part A. 2013;19:558. doi: 10.1089/ten.tea.2012.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadler KE, Holmes DF, Trotter JA, Chapman JA. Biochem. J. 1996;316:1. doi: 10.1042/bj3160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hynes RO. Cell. 1992;69:11. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 10.Garcia AJ, Vega MD, Boettiger D. Mol. Biol. Cell. 1999;10:785. doi: 10.1091/mbc.10.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasoju N, Bora U. Adv. Healthcare Mater. 2012;1:393. doi: 10.1002/adhm.201200097. [DOI] [PubMed] [Google Scholar]
- 12.Heim M, Keerl D, Scheibel T. Angew. Chem., Int. Ed. 2009;48:3584. doi: 10.1002/anie.200803341. [DOI] [PubMed] [Google Scholar]
- 13.Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, Lu H, Richmond J, Kaplan DL. Biomaterials. 2003;24:401. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 14.Charras G, Sahai E. Nat. Rev. Mol. Cell Biol. 2014;15:813. doi: 10.1038/nrm3897. [DOI] [PubMed] [Google Scholar]
- 15.Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. 2001;294:1708. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 16.Shaub A. Nat. Cell Biol. 1999;1:E173. doi: 10.1038/15608. [DOI] [PubMed] [Google Scholar]
- 17.Gulik-Krzywicki T, Fouquey C, Lehn JM. Proc. Natl. Acad. Sci. U.S.A. 1993;90:163. doi: 10.1073/pnas.90.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sijbesma RP, Beijer FH, Brunsveld L, Folmer BJB, Hirschberg JHKK, Lange RFM, Lowe JKL, Meijer EW. Science. 1997;278:1601. doi: 10.1126/science.278.5343.1601. [DOI] [PubMed] [Google Scholar]
- 19.Fogleman EA, Yount WC, Xu J, Craig SL. Angew. Chem., Int. Ed. 2002;41:4026. doi: 10.1002/1521-3773(20021104)41:21<4026::AID-ANIE4026>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 20.Park T, Zimmerman SC. J Am. Chem. Soc. 2006;128:13986. doi: 10.1021/ja064116s. [DOI] [PubMed] [Google Scholar]
- 21.Chen CC, Dormidontova EE. J Am. Chem. Soc. 2004;126:14972. doi: 10.1021/ja047521x. [DOI] [PubMed] [Google Scholar]
- 22.Rowan SJ, Beck JB. Polym. Prepr. 2005;46:1164. [Google Scholar]
- 23.Noro A, Matsushima S, He XD, Hayashi M, Matsushita Y. Macromolecules. 2013;46:8304. [Google Scholar]
- 24.Burattini S, Greenland BW, Merino DH, Weng W, Seppala J, Colquhoun HM, Hayes W, Mackay ME, Hamley IW, Rowan SJ. J Am. Chem. Soc. 2010;132:12051. doi: 10.1021/ja104446r. [DOI] [PubMed] [Google Scholar]
- 25.Miyauchi M, Harada A. J Am. Chem. Soc. 2004;126:11418. doi: 10.1021/ja046562q. [DOI] [PubMed] [Google Scholar]
- 26.Miyauchi M, Takashima Y, Yamaguchi H, Harada A. J Am. Chem. Soc. 2005;127:2984. doi: 10.1021/ja043289j. [DOI] [PubMed] [Google Scholar]
- 27.Yebeutchou RM, Tancini F, Demitri N, Geremia S, Mendichi R, Dalcanale E. Angew. Chem., Int. Ed. 2008;47:4504. doi: 10.1002/anie.200801002. [DOI] [PubMed] [Google Scholar]
- 28.Barnard A, Smith DK. Angew. Chem., Int. Ed. 2012;51:6572. doi: 10.1002/anie.201200076. [DOI] [PubMed] [Google Scholar]
- 29.Boekhoven J, Stupp SI. Adv. Mater. 2014;26:1642. doi: 10.1002/adma.201304606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Busseron E, Ruff Y, Moulin E, Giuseppone N. Nanoscale. 2013;5:7098. doi: 10.1039/c3nr02176a. [DOI] [PubMed] [Google Scholar]
- 31.Campbell BF, Chance MR, Friedman JM. Science. 1987;238:373. doi: 10.1126/science.3659921. [DOI] [PubMed] [Google Scholar]
- 32.Van der Rest M, Garrone R. FASEB J. 1991;5:2814. [PubMed] [Google Scholar]
- 33.Buehler MJ. J Mech. Behav. Biomed. Mater. 2008;1:59. doi: 10.1016/j.jmbbm.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Schulz RM, Bader A. Eur. Biophys. J. 2007;36:539. doi: 10.1007/s00249-007-0139-1. [DOI] [PubMed] [Google Scholar]
- 35.Gautieri A, Vesentini S, Redaelli A, Buehler MJ. Nano Lett. 2011;11:757. doi: 10.1021/nl103943u. [DOI] [PubMed] [Google Scholar]
- 36.Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM. J Mol. Biol. 2002;319:433. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- 37.Klotzsch E, Smith ML, Kubow KE, Muntwyler S, Little WC, Beyeler F, Gourdon D, Nelson BJ, Vogel V. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19267. doi: 10.1073/pnas.0907518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao Y, Schwarzbauer JE. Matrix Biol. 2005;24:389. doi: 10.1016/j.matbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Sottile J, Hocking DC. Mol. Biol. Cell. 2002;13:3546. doi: 10.1091/mbc.E02-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J, Mosher D. In: The Extracellular Matrix: An Overview. Mecham RP, editor. Berlin: Springer; 2011. pp. 41–75. [Google Scholar]
- 41.Engvall E, Ruoslahti E. Int. J. Cancer. 1977;20:1. doi: 10.1002/ijc.2910200102. [DOI] [PubMed] [Google Scholar]
- 42.Schwarzbauer JE, Sechler JL. Curr. Opin. Cell Biol. 1999;11:622. doi: 10.1016/s0955-0674(99)00017-4. [DOI] [PubMed] [Google Scholar]
- 43.Wierzbicka-Patynowski I, Schwarzbauer JE. J Cell Sci. 2003;116:3269. doi: 10.1242/jcs.00670. [DOI] [PubMed] [Google Scholar]
- 44.Heim M, Romer L, Scheibel T. Chem. Soc. Rev. 2010;39:156. doi: 10.1039/b813273a. [DOI] [PubMed] [Google Scholar]
- 45.Shao Z, Vollrath F. Nature. 2002;418:741. doi: 10.1038/418741a. [DOI] [PubMed] [Google Scholar]
- 46.Xu G, Gong L, Yang Z, Liu XY. Soft Matter. 2014;10:2116. doi: 10.1039/c3sm52845f. [DOI] [PubMed] [Google Scholar]
- 47.Keten S, Xu Z, Ihle B, Buehler MJ. Nat. Mater. 2010;9:359. doi: 10.1038/nmat2704. [DOI] [PubMed] [Google Scholar]
- 48.Asakura T, Suzuki Y, Nakazawa Y, Yazawa K, Holland GP, Yarger JL. Prog. Nucl. Magn. Reson. Spectrosc. 2013;29:23. doi: 10.1016/j.pnmrs.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 49.Hardy JG, Romer LM, Scheibel TR. Polymer. 2008;49:4309. [Google Scholar]
- 50.Rockwood DN, Preda RC, Yucel T, Wang X, Lovett ML, Kaplan DL. Nat. Protoc. 2011;6:1612. doi: 10.1038/nprot.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rising A, Widhe M, Johansson J, Hedhammar M. Cell. Mol. Life Sci. 2011;68:169. doi: 10.1007/s00018-010-0462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis RV. Chem. Rev. 2006;106:3762. doi: 10.1021/cr010194g. [DOI] [PubMed] [Google Scholar]
- 53.Eisoldt L, Smith A, Scheibel T. Mater. Today. 2011;14:80. [Google Scholar]
- 54.Tokareva O, Michalczechen-Lacerda VA, Rech EL, Kaplan DL. Microb. Biotechnol. 2013;6:651. doi: 10.1111/1751-7915.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heidebrecht A, Scheibel T. Adv. Appl. Microbiol. 2013;82:115. doi: 10.1016/B978-0-12-407679-2.00004-1. [DOI] [PubMed] [Google Scholar]
- 56.Brunsveld L, Folmer BJB, Meijer EW, Sijbesma RP. Chem. Rev. 2001;101:4071. doi: 10.1021/cr990125q. [DOI] [PubMed] [Google Scholar]
- 57.Aida T, Meijer EW, Stupp SI. Science. 2012;335:813. doi: 10.1126/science.1205962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dankers PY, Harmsen MC, Brouwer LA, van Luyn MJ, Meijer EW. Nat. Mater. 2005;4:568. doi: 10.1038/nmat1418. [DOI] [PubMed] [Google Scholar]
- 59.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 60.Zubarev ER, Pralle MU, Sone ED, Stupp SI. J Am. Chem. Soc. 2001;123:4105. doi: 10.1021/ja015653+. [DOI] [PubMed] [Google Scholar]
- 61.Hartgerink JD, Beniash E, Stupp SI. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5133. doi: 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stupp SI, Zha RH, Palmer LC, Cui H, Bitton R. Faraday Discuss. 2013;166:9. doi: 10.1039/c3fd00120b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamamoto Y, Fukushima T, Suna Y, Ishii N, Saeki A, Seki S, Tagawa S, Taniguchi M, Kawai T, Aida T. Science. 2006;314:1761. doi: 10.1126/science.1134441. [DOI] [PubMed] [Google Scholar]
- 64.Ghadiri MR, Granja JR, Milligan RA, McRee DE, Khazanovich N. Nature. 1993;366:324. doi: 10.1038/366324a0. [DOI] [PubMed] [Google Scholar]
- 65.Hartgerink JD, Granja JR, Milligan RA, Ghadiri MR. J Am. Chem. Soc. 1996;118:32. [Google Scholar]
- 66.O’Leary LER, Fallas JA, Bakota EL, Kang MK, Hartgerink JD. Nat. Chem. 2011;3:821. doi: 10.1038/nchem.1123. [DOI] [PubMed] [Google Scholar]
- 67.Aggeli A, Bell M, Boden N, Keen JN, Knowles PF, Mcleish TCB, Pitkeathly M, Radford SE. Nature. 1997;386:259. doi: 10.1038/386259a0. [DOI] [PubMed] [Google Scholar]
- 68.Di Lullo GA, Sweeney SM, Korkko J, Ala-Kokko L, San Antonio JD. J Biol. Chem. 2002;277:4223. doi: 10.1074/jbc.M110709200. [DOI] [PubMed] [Google Scholar]
- 69.Pires MM, Przybyla DE, Chmielewski J. Angew. Chem., Int. Ed. 2009;28:7813. doi: 10.1002/anie.200902375. [DOI] [PubMed] [Google Scholar]
- 70.Suzuki Y, Yamazaki T, Aoki A, Shindo H, Asakura T. Biomacromolecules. 2014;15(1):104. doi: 10.1021/bm401346h. [DOI] [PubMed] [Google Scholar]
- 71.Zhang S, Rich A. Proc. Natl. Acad. Sci. U.S.A. 1997;94:23. [Google Scholar]
- 72.Yokoi H, Kinoshita T, Zhang S. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8414. doi: 10.1073/pnas.0407843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cormier AR, Pang X, Zimmerman MI, Zhou HX, Paravastu AK. ACS Nano. 2013;7:7562. doi: 10.1021/nn401562f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horii A, Wang X, Gelain F, Zhang S. PLoS One. 2007;2:e190. doi: 10.1371/journal.pone.0000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ellis-Behnke RG, Liang YX, You SW, Tay DK, Zhang S, So K-F, Schneider GE. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5054. doi: 10.1073/pnas.0600559103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao X, Zhang S. Chem. Soc. Rev. 2006;35:1105. doi: 10.1039/b511336a. [DOI] [PubMed] [Google Scholar]
- 77.Velichko YS, Stupp SI, de la Cruz MO. J Phys. Chem. B. 2008;112:2326. doi: 10.1021/jp074420n. [DOI] [PubMed] [Google Scholar]
- 78.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 79.Guler MO, Stupp SI. J Am. Chem. Soc. 2007;129:12082. doi: 10.1021/ja075044n. [DOI] [PubMed] [Google Scholar]
- 80.Fleming S, Ulijn RV. Chem. Soc. Rev. 2014;43:8150. doi: 10.1039/c4cs00247d. [DOI] [PubMed] [Google Scholar]
- 81.Jayawarna V, Ali M, Jowitt TA, Miller AF, Saiani A, Gough JE, Ulijn RV. Adv. Mater. 2006;18:611. [Google Scholar]
- 82.Reches M, Gazit E. Science. 2003;300:625. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- 83.Hirst AR, Roy S, Arora M, Das AK, Hodson N, Murray P, Marshall S, Javid N, Sefcik J, Boekhoven J, van Esch JH, Santabarbara S, Hunt NT, Ulijn RV. Nat. Chem. 2010;2:1089. doi: 10.1038/nchem.861. [DOI] [PubMed] [Google Scholar]
- 84.Pashuck ET, Cui H, Stupp SI. J Am. Chem. Soc. 2010;132:6041. doi: 10.1021/ja908560n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Newcomb CJ, Sur S, Ortony JH, Lee O-S, Matson JB, Boekhoven J, Yu JM, Schatz GC, Stupp SI. Nat. Commun. 2014;5:3321. doi: 10.1038/ncomms4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 87.Zhang S, Greenfield MA, Mata A, Palmer LC, Bitton R, Mantei JR, Aparicio C, Olvera de la Cruz M, Stupp SI. Nat. Mater. 2010;9:594. doi: 10.1038/nmat2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Capito RM, Azevedo HS, Velichko YS, Mata A, Stupp SI. Science. 2008;319:1812. doi: 10.1126/science.1154586. [DOI] [PubMed] [Google Scholar]
- 89.Carvajal D, Bitton R, Mantei JR, Velichko YS, Stupp SI, Shull KR. Soft Matter. 2010;6:1816. [Google Scholar]
- 90.Rożkiewicz DI, Myers BD, Stupp SI. Angew. Chem., Int. Ed. 2011;123:6448. doi: 10.1002/anie.201100821. [DOI] [PubMed] [Google Scholar]
- 91.Boekhoven J, Zha RH, Tantakitti F, Zhuang E, Zandi R, Newcomb CJ, Stupp SI. RSC Adv. 2015;5:8753. doi: 10.1039/C4RA16593D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chien KR. Nature. 2008;453:302. doi: 10.1038/nature07037. [DOI] [PubMed] [Google Scholar]
- 93.Lin F, Ren X-D, Pan Z, Macri L, Zong W-X, Tonnesen MG, Rafailovich M, Bar-Sagi D, Clark RAF. J Invest. Dermatol. 2011;131:84. doi: 10.1038/jid.2010.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wijelath ES, Murray J, Rahman S, Patel Y, Ishida A, Strand K, Aziz S, Cardona C, Hammond WP, Savidge GF, Rafii S, Sobel M. Circ. Res. 2002;91:25. doi: 10.1161/01.res.0000026420.22406.79. [DOI] [PubMed] [Google Scholar]
- 95.Lee K, Silva EA, Mooney DJ. JR Soc. Interface. 2011;8:153. doi: 10.1098/rsif.2010.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guena S, Perroteau I, Los P, Battsiton B, editors. Tissue Engineering of the Peripheral Nerve: Biomaterials and Physical Therapy. New York: Elsevier; 2013. [Google Scholar]
- 97.Horch K. J Neurophysiol. 1979;42:1437. doi: 10.1152/jn.1979.42.5.1437. [DOI] [PubMed] [Google Scholar]
- 98.Nazarov R, Jin HJ, Kaplan DL. Biomacromolecules. 2004;5:718. doi: 10.1021/bm034327e. [DOI] [PubMed] [Google Scholar]
- 99.Uebersax L, Mattotti M, Papaloïzos M, Merkle HP, Gander B, Meinel L. Biomaterials. 2007;28:4449. doi: 10.1016/j.biomaterials.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 100.Benfenati V, Stahl K, Gomis-Perez C, Toffanin S, Sagnella A, Torp R, Kaplan DL, Ruani G, Omenetto FG, Zamboni R, Muccini M. Adv. Funct. Mater. 2012;22:1871. doi: 10.5301/JABFM.2012.10448. [DOI] [PubMed] [Google Scholar]
- 101.Wenk E, Merkle HP, Meinel L. J Controlled Release. 2011;150:128. doi: 10.1016/j.jconrel.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 102.Matson JB, Stupp SI. Chem. Commun. 2011;47:7962. doi: 10.1039/c1cc12570b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kisiday J, Jin M, Kurz B, Hung H, et al. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9996. doi: 10.1073/pnas.142309999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boekhoven J, Koot M, Wezendonk TA, Eelkema R, van Esch JH. J Am. Chem. Soc. 2012;134:19098. doi: 10.1021/ja3051876. [DOI] [PubMed] [Google Scholar]
- 105.Davis ME, Hsieh PC, Takahashi T, Song Q, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8155. doi: 10.1073/pnas.0602877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.D’Andrea LD, Iaccarino G, Fattorusso R, Sorriento D, Carannante C, Capasso D, Trimarco B, Pedone C. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14215. doi: 10.1073/pnas.0505047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang X, Horii A, Zhang S. Soft Matter. 2008;4:2388. [Google Scholar]
- 108.Liu X, Wang X, Horii A, Wang X, et al. Nanoscale. 2012;4:2720. doi: 10.1039/c2nr00001f. [DOI] [PubMed] [Google Scholar]
- 109.Webber MJ, Tongers J, Newcomb CJ, Marquardt KT, et al. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9220. [Google Scholar]
- 110.Klebe RJ. Nature. 1974;250:248. doi: 10.1038/250248a0. [DOI] [PubMed] [Google Scholar]
- 111.Pearlstein E. Science. 1976;262:497. doi: 10.1038/262497a0. [DOI] [PubMed] [Google Scholar]
- 112.Pierschbacher MD, Ruoslahti E. Nature. 1984;309:30. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 113.Ruoslahti E, Pierschbacher MD. Science. 1987;238:491. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 114.Hayman EG, Pierschbacher MD, Suzuki S, Ruoslahti E. Exp. Cell Res. 1985;160(2):245. doi: 10.1016/0014-4827(85)90173-9. [DOI] [PubMed] [Google Scholar]
- 115.Lots MM, Burdsal CA, Erickson HP, McClay DR. J Cell Biol. 1989;109:1795. doi: 10.1083/jcb.109.4.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kleiman HK, Klebe RJ, Martin GR. J Cell Biol. 1981;88:473. doi: 10.1083/jcb.88.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johansson S, Svineng G, Wennerberg K, Armulik A, Lohikangas L. Front. Biosci. 1997;2:d126. doi: 10.2741/a178. [DOI] [PubMed] [Google Scholar]
- 118.Sechler JL, Takada Y, Schwarzbauer JE. J Cell Biol. 1996;134:573. doi: 10.1083/jcb.134.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. J Biol. Chem. 2000;275:21785. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- 120.Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 121.Sofia S, McCarthy MB, Gronowicz G, Kaplan DL. J Biomed. Mater. Res. 2001;54:139. doi: 10.1002/1097-4636(200101)54:1<139::aid-jbm17>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 122.Meinel L, Karageorgiou V, Hofmann S, Fajardo R, Snyder B, Li C, Zichner L, Langer R, Vunjak-Novakovic G, Kaplan DL. J Biomed. Mater. Res., Part A. 2004;71:25. doi: 10.1002/jbm.a.30117. [DOI] [PubMed] [Google Scholar]
- 123.Gil ES, Mandal BB, Park SH, Marchant JK, Omenetto FG, Kaplan DL. Biomaterials. 2010;31:8953. doi: 10.1016/j.biomaterials.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Widhe M, Johansson J, Hedhammar M. Biopolymers. 2012;97:468. doi: 10.1002/bip.21715. [DOI] [PubMed] [Google Scholar]
- 125.Wohlrab S, Muller S, Schmidt A, Neubauer S, Kessler H, Leal-Egana A, et al. Biomaterials. 2012;33:6650. doi: 10.1016/j.biomaterials.2012.05.069. [DOI] [PubMed] [Google Scholar]
- 126.Yang M, Tanaka C, Yamauchi K, Ohgo K, Kurokawa M, Asakura T. J Biomed. Mater. Res., Part A. 2008;84:353. doi: 10.1002/jbm.a.31348. [DOI] [PubMed] [Google Scholar]
- 127.Bini E, Foo CW, Huang J, Karageorgiou V, Kitchel B, Kaplan DL. Biomacromolecules. 2006;7:3139. doi: 10.1021/bm0607877. [DOI] [PubMed] [Google Scholar]
- 128.Tanaka C, Asakura T. Biomacromolecules. 2009;10:923. doi: 10.1021/bm801439t. [DOI] [PubMed] [Google Scholar]
- 129.Kambe Y, Yamamoto K, Kojima K, Tamada Y, Tomita N. Biomaterials. 2010;31:7503. doi: 10.1016/j.biomaterials.2010.06.045. [DOI] [PubMed] [Google Scholar]
- 130.Webber MJ, Tongers J, Renault MA, Roncalli JG, et al. Acta Biomater. 2010;6:3. doi: 10.1016/j.actbio.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhou M, Smith AM, Das AK, Hodson NW, et al. Biomaterials. 2009;30:2523. doi: 10.1016/j.biomaterials.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 132.Stephanopoulos N, Freeman R, North HA, Sur S, et al. Nano Lett. 2015;15:603. doi: 10.1021/nl504079q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Saunders RL, Hammer DA. Cell. Mol. Bioeng. 2010;3:60. doi: 10.1007/s12195-010-0112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zha RH, Sur S, Boekhoven J, Shi HY, Zhang M, Stupp SI. Acta Biomater. 2015;12:1. doi: 10.1016/j.actbio.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shah RN, Shah NA, Del Rosario Lim MM, Hsieh C, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:3293. doi: 10.1073/pnas.0906501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sur S, Matson JB, Webber MJ, Newcomb CJ, Stupp SI. ACS Nano. 2012;6(12):10776. doi: 10.1021/nn304101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Khan S, Sur S, Dankers PY, da Silva RM, et al. Bioconjugate Chem. 2014;25:707. doi: 10.1021/bc400507v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Boekhoven J, Rubert Pérez CM, Sur S, Worthy A, Stupp SI. Angew. Chem., Int. Ed. 2013;52(46):12077. doi: 10.1002/anie.201306278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.An Q, Brinkmann J, Juskens H, Krabbenborg S, et al. Angew. Chem., Int. Ed. Engl. 2012;51:12233. doi: 10.1002/anie.201205651. [DOI] [PubMed] [Google Scholar]
- 140.Charriere G, Bejot M, Schnitzler L, Ville G, Hartmann DJ. J Am. Acad. Dermatol. 1989;21:1203. doi: 10.1016/s0190-9622(89)70330-3. [DOI] [PubMed] [Google Scholar]
- 141.Eaglstein WH, Alvarez OM, Auletta M, Leffel D, Rogers GS, Zitelli JA, Norris JE, Thomas I, Irondo M, Fewkes J, Hardin-Young J, Duff RG, Sabolinski ML. Dermatol. Surg. 1999;25:195. doi: 10.1046/j.1524-4725.1999.08186.x. [DOI] [PubMed] [Google Scholar]
- 142.Badylak SF, Gilbert TW. Semin. Immunol. 2008;20:109. doi: 10.1016/j.smim.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pritchard EM, Dennis PB, Omenetto F, Naik RR, Kaplan DL. Biopolymers. 2012;97:279. doi: 10.1002/bip.22026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Das S, Pati F, Choi YJ, Rijal G, Shim JH, Kim SW, et al. Acta Biomater. 2015;22:233. doi: 10.1016/j.actbio.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 145.Berns EJ, Sur S, Pan L, Goldberger JE, Suresh S, Zhang S, Kessler JA, Stupp SI. Biomaterials. 2014;35:185. doi: 10.1016/j.biomaterials.2013.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pan L, North HA, Sahni V, Jeong SJ, McGuire TL, Berns E, Stupp SI, Kessler JA. PLoS One. 2014;9:e104335. doi: 10.1371/journal.pone.0104335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Angeloni N, Bond CW, Harrington D, Stupp SI, Podlasek CA. J Sex. Med. 2013;10:1240. doi: 10.1111/j.1743-6109.2012.02930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bond CW, Angeloni N, Harrington D, Stupp SI, Podlasek CA. J Sex. Med. 2013;10:730. doi: 10.1111/jsm.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li A, Hokugo A, Yalom A, Berns EJ, Stephanopoulos N, McClendon MT, Segovia LA, Spigelman I, Stupp SI, Jarrahy R. Biomaterials. 2014;35:8780. doi: 10.1016/j.biomaterials.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cui H, Pashuck ET, Velichko YS, Weigand SJ, Cheetham AG, Newcomb CJ, Stupp SI. Science. 2010;327:555. doi: 10.1126/science.1182340. [DOI] [PMC free article] [PubMed] [Google Scholar]