Abstract
Although Japan is classified as a country with a low prevalence of human immunodeficiency virus type 1 (HIV-1), domestic sexual transmission has been increasing steadily. Because 70% of the Japanese population expresses HLA-A24 (genotype HLA-A*2402), we wished to assess the effect of the dominant HLA type on the evolution and transmission of HIV-1 among the Japanese population. Twenty-three out of 25 A24-positive Japanese patients had a Y-to-F substitution at the second position [Nef138-10(2F)] in an immunodominant A24-restricted CTL epitope in their HIV-1 nef gene (Nef138-10). None of 12 A24-negative Japanese hemophiliacs but 9 out of 16 patients infected through unprotected sexual intercourse had Nef138-10(2F) (P < 0.01). Two of two A24-positive but none of six A24-negative Australians had Nef138-10(2F). Nef138-10(2F) peptides bound well to the HLA-A*2402 heavy chain; however, Nef138-10(2F) was expressed poorly on the cell surface from the native protein. Thus, HIV-1 with Nef138-10(2F) appears to be a cytotoxic-T-lymphocyte escape mutant and has been transmitted frequently by sexual contact among the highly A24-positive Japanese population.
While cytotoxic T lymphocytes (CTLs) exert immune pressure on human immunodeficiency virus type 1 (HIV-1) throughout the course of primary and chronic infection (4, 24, 30), HIV-1 escapes through a variety of immune evading mechanisms such as downregulation of HLA class I molecules by Nef (7, 32, 33, 36) and defects in differentiation and maturation of CTLs (2, 6, 27, 35). Viral mutation also plays a crucial role in immune escape, and CTL escape mutant viruses may appear early or late in the clinical course of infection (5, 14, 31). Mutations leading to CTL escape may occur at amino acid residues essential for major histocompatibility complex binding (8), for T-cell-receptor recognition (10), or in flanking regions that affect antigen processing (3, 26).
HIV-1 CTL escape mutants may be stable. One such example at the HLA-B27-restricted Gag epitope, which is related to slower disease progression in adults, could be transmitted vertically from mother to child (12). Although significant association between HLA alleles and polymorphism in reverse transcriptase sequences in a large cohort of patients indicated HIV-1 adaptation at a population level (28), direct horizontal transmission of CTL escape mutants is yet to be shown.
Japan is classified as a country of low HIV-1 prevalence; however, national HIV-1 and AIDS surveillance has shown a steady increase of HIV-1 and AIDS cases mainly through unprotected sexual intercourse (USI) (84% of HIV-1 patients and 71% of AIDS patients were infected through USI within the country) (1). The Japanese population is less polymorphic than other populations in that 70% express HLA-A24 (genotype HLA-A*2402) (13). We speculated that stable CTL escape mutants from HLA-A24 might be transmitted more frequently in Japan than in other countries where the prevalence of HLA-A24 is much lower. We postulated that Japanese hemophiliacs with HIV-1 infection might be a good comparator group since they were infected directly by contaminated blood products from abroad. We therefore examined an immunodominant CTL epitope in the nef gene (Nef138-10) in HLA-A24-positive and -negative hemophiliacs and compared the sequence with sequences from those patients infected through USI (13, 18). We included Caucasian Australians infected through USI as another control of transmission of CTL escape mutants in a country where HLA-A24 is less prevalent (19).
MATERIALS AND METHODS
Patient samples.
For sequence analysis, blood specimens were collected in EDTA. Plasmas were separated and preserved at −80°C until use. For enzyme-linked immunospot (ELISPOT) assay, peripheral blood mononuclear cells (PBMCs) were separated from heparinized whole blood and used on the day of the assay. Patient HLA was typed serologically. In selected patients, HLA genotype was determined after written informed consent was obtained. The study was approved by institutional review boards. All patients serologically typed as A24 positive proved to be positive for HLA-A*2402.
RNA extraction and reverse transcription.
Viral RNA was extracted from 140 μl of plasma from patients by using the QIAamp viral RNA Mini kit (QIAGEN) and subjected to reverse transcription according to the manufacturer's protocol with SuperScript II RNase H− reverse transcriptase (Invitrogen) and 5 μM random primers (Takara).
PCR amplification and sequencing.
Fifteen microliters of cDNA (a one-sixth volume of the reverse transcription reaction) was subjected to the first PCR. One-tenth of the first PCR was subjected to the nested PCR. PCR was performed by using Ex-Taq (Takara) with 35 cycles of 30 s at 94°C, 30 s at 58°C, 30 s at 72°C, and a final extension for 7 min at 72°C. The primer sets are as follows (all nucleotide positions are in accordance with the HIV-1 SF2 strain). For the env V3 region, first PCR primer set 1, primers CBE297P (5′-GGTAGAACAGATGCATGAGGAT-3′) (consensus B env, nucleotides [nt] 297 to 318) and E7668 M (5′-TTCTCCAATTGTCCCTCATATCTCCTCCTCCA-3′) (SF2, nt 7668 to 7636) were used; and for the second PCR primer set 1, primers E6554P (5′-ATCAGTTTATGGGATCAAAGCC-3′) (SF2, nt 6554 to 6575) and E7353 M (5′-ACAATTTCTGGGTCCCCTCCTGAGGA-3′) (SF2, nt 7353 to 7328) were used. For the first PCR primer set 2, primers E6984P (5′-ACATGGAATTAGGCCA-3′) (SF2, nt 6984 to 7000) and E7395 M (5′-TTACAGTAGAAAAATTCCCC-3′) (SF2, nt 7395 to 7375) were used; and for the second PCR primer set 2, primers E7028P (5′-GGCAGTCTAGCAGAAGAAGA-3′) (SF2, nt 7028 to 7047) and E7353 M (5′-ACAATTTCTGGGTCCCCTCCTGAGGA-3′) (SF2, nt 7353 to 7328) were used. For the first PCR primer set 3, primers P6951 (5′-GACCATGTACAAATGTCAGC-3′) (SF2, nt 6951 to 6970) and M7592 (5′-CTCTTGTTAATAGCAGCCCT-3′) (SF2, nt 7592 to 7573) were used; and for the second PCR primer set 3, primers E6984P (5′-ACATGGAATTAGGCCA-3′) (SF2, nt 6984 to 7000) and E7353 M (5′-ACAATTTCTGGGTCCCCTCCTGAGGA-3′) (SF2, nt 7353 to 7328) were used.
For the Nef138-10 epitope, first PCR primer set 1, primers n226p (5′-CTCAGGTACCTTTAAGACCAATG-3′) (nt 9028 to 9050) and n650m (5′-GAAAGTCCCCAGCGGAAAGTCCC-3′) (nt 9474 to 9452) were used; and for the second PCR primer set 1, primers n296p (5′-GGGACTGGAAGGGCTAATTTGGT-3′) (nt 9098 to 9120) and n564m (5′-GAAATGCTAGTTTGCTGTCAAAC-3′) (nt 9387 to 9365) were used. For the first PCR primer set 2, primers P8923 (5′-TGGAAAAACATGGAGCAATCA-3′) (nt 8923 to 8944) and M9290 (5′-TCCTTCATTGGCCTCTTCTAC-3′) (nt 9290 to 9270) were used; and for the second PCR primer set 2, primers P8924 (5′-GGAAAAACATGGAGCAATCAC-3′) (nt 8924 to 8945) and M9288 (5′-CTTCATTGGCCTCTTCTACCT-3′) (nt 9288 to 9268) were used. For the first PCR primer set 3, primers P8923 (5′-TGGAAAAACATGGAGCAATCA-3′) (nt 8923 to 8944) and n694m (5′-CAGCATCTGAGGGACGCCAC-3′) (nt 9525 to 9506) were used; and for the second PCR primer set 3, primers n226p (5′-CTCAGGTACCTTTAAGACCAATG-3′) (nt 9028 to 9050) and n532m (5′-TCTCCGCGTCCTCCATCCCA-3′) (nt 9345 to 9326) were used.
The PCR products were electrophoresed through agarose gels and purified with a Minielute gel extraction kit (QIAGEN) before sequencing. Purified PCR products were directly sequenced. When sequence ambiguities resulted, DNA fragments were subcloned into the pGEM-T vector (Promega) and sequenced. DNA sequencing was performed by using an ABI Prism dye terminator cycle sequencing ready reaction kit (Applied Biosystems) on a Perkin-Elmer ABI-377 sequencer.
Cells and media.
T2-A24, a kind gift from K. Kuzushima, was cultured in RPMI 1640 (Sigma) supplemented with 10% heat-inactivated fetal calf serum (FCS) (Sigma) and 0.8 mg of G418 (Invitrogen)/ml (25). We transformed PBMCs from an HLA-A*2402-positive person with human T-cell leukemia virus type 1 (HTLV-1) and established an HLA-A*2402- and CD4-positive-T-cell line (KWN-T4). KWN-T4 was cultured with RPMI 1640 supplemented with 25 U of interleukin-2 (Wako)/ml, 100 U of penicillin/ml, 100 U of streptomycin (Invitrogen)/ml, and 10% heat-inactivated FCS (JRH Bioscience). We also established Nef138-10-specific CTL clones as previously described (22). CTL clones were cultured with RPMI 1640 supplemented with 50 U of interleukin-2/ml, 100 U of penicillin/ml, 100 U of streptomycin/ml, and 10% heat-inactivated FCS.
Peptides.
Synthetic peptides Nef138-10 (RYPLTFGWCF), 2F (RFPLTFGWCF), 5C (RYPLCFGWCF), and 2F5C (RFPLCFGWCF) were purchased from Sigma-Genosys. All peptides were more than 95% pure as determined by high-performance liquid chromatography and mass spectroscopy.
Peptide binding assays.
Peptide binding to HLA-A*2402 was quantified by using a T2-A24 stabilization assay as previously described (25). T2-A24 cells were incubated at 26°C for 16 h, and then 2 × 105 cells were incubated with peptides at concentrations from 10−4 to 10−9 M for 1 h at 4°C. After incubation for 3 h at 37°C, the cells were stained with anti-HLA-A24 monoclonal antibody, A11.1 M (11), and an R-phycoerythrin (RPE)-conjugated F(ab′)2 fragment of anti-mouse immunoglobulin (DAKO). The mean fluorescence intensity was measured by FACSCalibur (Becton Dickinson).
ELISPOT assay and functional avidity analysis.
Freshly prepared PBMCs (20,000 to 50,000) were added to 96-well multiscreen plates (Millipore) which had been precoated with 100 μl of 5 μg of anti-gamma interferon (IFN-γ) monoclonal antibody 1-D1K (Mabtech)/ml at room temperature for 3 h and blocked with RPMI 1640 medium containing 10% FCS for 1 h. The cells were cultured with synthetic peptide Nef138-10 or its derivatives at concentrations from 10−5 to 10−11 M for 18 h. After the plates were washed, 100 μl of 1 μg of biotinylated anti-IFN-γ monoclonal antibody 7-B6-1 (Mabtech)/ml was added and incubated at room temperature for 90 min. After the plates were washed again, 100 μl of 1:1,000-diluted streptavidin-alkaline phosphatase conjugate (Mabtech) was added and incubated at room temperature for 60 min. Spots were developed with an alkaline phosphatase conjugate substrate kit (Bio-Rad) and counted with a KS ELISPOT compact (Carl Zeiss). The IFN-γ responses to peptide dilutions were expressed as a percentage of the maximal IFN-γ response seen in each individual assay.
Expression of recombinant Nef protein.
Mutations were introduced into nef derived from HIV-1 strain SF2 by site-directed mutagenesis based on overlap extension (16). Four proline residues in the Nef proline-rich domain that are important for HLA class I down-regulation were replaced by alanine as described previously (36). The wild type and various nef mutants were tagged by His6 and introduced into a Sendai virus vector (SeV) as previously described (36). For Western blot analysis, KWN-T4 cells were infected with various SeVs at a multiplicity of infection of 10 and lysed 20 h after infection. Western blot analysis was performed according to the standard procedure. Anti-His6 antibody (QIAGEN) and anti-SeV mouse antiserum were used to detect Nef and SeV proteins, respectively.
51Cr release assay.
Cytotoxicity was measured with a standard 51Cr release assay as previously described (21). Briefly, KWN-T4 was labeled with 100 μCi of Na251CrO4 for 2 h and washed three times with R10. Labeled cells (2 × 103) were added to a 96-well round-bottom microtiter plate with a corresponding amount of peptide. After 1 h of incubation, Nef138-10-specific CTL clones were added and incubated for 4 h. When SeV-infected cells were used as target cells, the cells were infected with SeVs at a multiplicity of infection of 10, 20 h before adding the CTLs.
The supernatants were collected and analyzed with a microbeta counter. Spontaneous 51Cr release was determined by measuring counts per minute in the supernatant of wells containing only target cells (cpmspn). The maximum release (cpmmax) was determined by measuring the release of 51Cr from target cells in the presence of 2% Triton X-100. Specific lysis was determined as follows: specific lysis = (cpmexp − cpmspn)/(cpmmax − cpmspn) × 100, where cpmexp represents the counts per minute in the supernatant of wells containing target and effector cells.
RESULTS
Sexual transmission of HIV-1 with stereotypic amino acid substitution among the Japanese population.
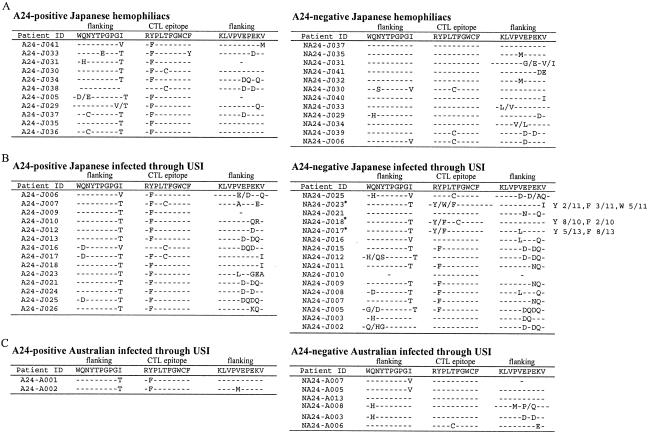
Only patients infected with virus subtyped as B by phylogenic comparison of envelope sequences were included to avoid potential bias introduced by sequence differences across subtypes (data not shown). We extensively sequenced the Nef138-10 epitope and its flanking region from plasma HIV-1 RNA of 23 Japanese hemophiliacs (11 A24-positive and 12 A24-negative individuals) and 30 Japanese (14 A24-positive and 16 A24-negative individuals) and 8 Caucasian Australians (2 A24-positive and 6 A24-negative individuals) infected through USI (Table 1). Ten out of 11 A24-positive but none of A24-negative Japanese hemophiliacs had a Y-to-F amino acid substitution at the second position [Nef138-10(2F)] (Fig. 1A) (P < 0.01), suggesting that HLA-A24 selected for Nef138-10(2F). In the case of patients infected through USI, 13 out of 14 A24-positive and 9 out of 16 A24-negative Japanese patients had Nef138-10(2F) by direct sequencing (Fig. 1B) (data not significant). The frequency of Nef138-10(2F) was significantly higher in Japanese A24-negative patients infected through USI than A24-negative hemophiliacs (P < 0.01). Two out of two A24-positive but none of six A24-negative Caucasian Australians had Nef138-10(2F) (Fig. 1C). The frequency of Nef138-10(2F) in A24-negative patients infected through USI was significantly higher for Japanese patients than for Australian patients (P < 0.05), suggesting that sexual transmission of the variant was more frequent among the Japanese population, which has a higher prevalence of HLA-A*2402.
TABLE 1.
Patient profilea
| Patient ID | Sex | HLA type | No. of CD4 cells/μl | Viral load (copies/ml) | Sample date (mo/day/yr) | HIV subtype |
|---|---|---|---|---|---|---|
| A24-positive Japanese hemophiliacs | ||||||
| A24-J037 | M | A24/26, B35/51 | 207 | 180,000b | 03/09/95 | B |
| A24-J041 | M | A24/26, B44/61 | 261 | 7,500bd | 03/09/95 | B |
| A24-J033 | M | A24/26, B46/52 | 27 | 200,000b | 03/27/95 | B |
| A24-J035 | M | A24, B40/48 | 148 | 360,000 | 04/10/95 | B |
| A24-J031 | M | A24/31, B51/60 | 29 | 180,000b | 10/23/95 | B |
| A24-J030 | M | A11/24, B13/62 | 3 | 380,000bd | 02/26/96 | B |
| A24-J029 | M | A24/31, B35/61 | 38 | ND | 04/01/96 | B |
| A24-J036 | M | A2 /24, B35/51 | 60 | 74,000b | 05/13/96 | B |
| A24-J034 | M | A24, B46/52 | 180 | 74,000bd | 05/20/96 | B |
| A24-J038 | M | A2 /24, B51/62 | 356 | 29,000b | 03/03/97 | B |
| A24-J005 | M | A24, B52/70 | 39 | 220,000b | 06/19/97 | B |
| A24-negative Japanese hemophiliacs | ||||||
| NA24-J037 | M | A26, B40 | 8 | >1,600,000bd | 06/08/95 | B |
| NA24-J035 | M | A11/26, B54/56 | 342 | 100,000b | 09/07/95 | B |
| NA24-J031 | M | A2/26, B51/61 | 521 | 130,000b | 09/18/95 | B |
| NA24-J041 | M | A26, B39/54 | 12 | 700,000bd | 10/05/95 | B |
| NA24-J032 | M | A2/11, B46/54 | 1d | 150,000b | 11/10/95 | ND |
| NA24-J030 | M | A31/33, B44/51 | 363 | 65,000b | 03/21/96 | B |
| NA24-J040 | M | A2/33, B17/54 | 101 | 74,000b | 03/21/96 | ND |
| NA24-J033 | M | A26, B61 | 143 | 140,000b | 04/18/96 | B |
| NA24-J029 | M | A11/33, B44/51 | 401 | <10,000 | 07/15/96 | B |
| NA24-J034 | M | A11/33, B17/56 | 38 | 81,000b | 08/15/96 | B |
| NA24-J039 | M | A11/26, B51/62 | 3 | 88,000b | 09/01/97 | B |
| NA24-J006 | M | A2/26, B39/61 | 335 | 9,200 | 10/30/00 | B |
| A24-positive Japanese infected through USI | ||||||
| A24-J006 | M | A2/24, B7/54 | 212 | 33,000 | 09/19/97 | B |
| A24-J007 | M | A24/26, B17/56 | 103 | 120,000 | 11/06/97 | B |
| A24-J009 | M | A24, B48/52 | 278 | 4,500 | 01/19/98 | B |
| A24-J010 | M | A24, B52 | 393 | 18,000 | 03/09/98 | B |
| A24-J024 | M | A24, B35/61 | 274 | 110,000 | 10/27/98 | B |
| A24-J012 | M | A24/26, B46/60 | 253 | 24,000 | 07/19/99 | B |
| A24-J013 | M | A24/26, B35/48 | 168 | 15,000 | 9/20/99 | B |
| A24-J016 | M | A11/24, B7/55 | 245 | 150,000 | 05/15/00 | B |
| A24-J017 | M | A1/24, B54/70 | 255 | 70,000 | 10/17/00 | B |
| A24-J018 | M | A24/31, B37/61 | 185 | 8,300 | 01/04/01 | B |
| A24-J025 | M | A24, B51/52 | 282 | 130,000 | 06/07/01 | B |
| A24-J023 | M | A2/24, B51/54 | 856d | 17,000d | 08/06/01 | B |
| A24-J021 | M | A2/24, B46/52 | 344 | 35,000 | 11/26/01 | B |
| A24-J026 | M | A2/24, B13/51 | 381 | 110,000 | 11/28/01 | B |
| A24-negative Japanese infected through USI | ||||||
| NA24-J025 | M | A2/31, B51/61 | 352 | 18,000b | 03/23/95 | B |
| NA24-J023 | M | A11/26, B35/51 | 23 | 5,000b | 04/01/96 | ND |
| NA24-J021 | M | A26, B52/54 | 9 | 44,000 | 08/04/97 | B |
| NA24-J018 | M | A2, B39/60 | 378 | 72,000 | 04/06/98 | B |
| NA24-J017 | M | A11/31, B51/56 | 197 | 72,000 | 04/16/98 | B |
| NA24-J016 | M | A3/31, B51/58 | 257 | 200,000 | 05/25/98 | B |
| NA24-J015 | M | A2/26, B51/62 | 543 | 13,000 | 06/26/98 | B |
| NA24-J012 | M | A31, B13/51 | 268 | 26,000 | 10/19/98 | B |
| NA24-J011 | M | A2, B55/60 | 408 | 12,000 | 10/22/98 | B |
| NA24-J010 | M | A2/26, B51/61 | 206 | 16,000 | 12/17/98 | B |
| NA24-J009 | M | A2, B52/60 | 115 | 850,000 | 05/24/99 | B |
| NA24-J008 | M | A11/33, B44/60 | 312 | 2,600 | 07/08/99 | ND |
| NA24-J007 | M | A26, B7/52 | 396 | 450 | 08/09/00 | B |
| NA24-J005 | M | A2/31, B48/52 | 604 | 17,000 | 01/18/01 | B |
| NA24-J003 | M | A31/33, B44/51 | 308 | 20,000 | 06/04/01 | B |
| NA24-J002 | M | A2/33, B44/46 | 496 | 14,000 | 09/27/01 | ND |
| A24 positive Australian infected through USI | ||||||
| A24-A001 | M | A3/24, B7 | 255 | 38,000 | 08/16/96 | ND |
| A24-A002 | M | A24/30, B13 | 598 | 21,700 | 03/22/01 | B |
| A24-negative Australian infected through USI | ||||||
| NA24-A007 | M | A2/3, B7 | 704 | NDc | 11/02/95 | B |
| NA24-A005 | M | A1/3, B8/70 | 620 | 7,700 | 05/26/96 | B |
| NA24-A013 | M | A32, B13/64 | 851 | 23,200 | 09/28/98 | B |
| NA24-A008 | M | A2/3, B39/44 | 543 | 52,836 | 01/04/99 | B |
| NA24-A003 | M | A2, B18/62 | 575 | 19,400 | 11/06/99 | B |
| NA24-A006 | M | A3/26, B18/27 | 594 | 18,200 | 04/13/00 | B |
ND, not determined.
Data were obtained by Branch DNATM version 1.0.
Nearest data were 17,000 with CD4 counts of 638.
Nearest data were within 6 months of sample collection.
FIG. 1.
Nef138-10 epitope and its flanking sequences. Amino acid sequences deduced from the direct DNA sequencing of Nef138-10 CTL epitope and both flanking regions are presented. Wild-type sequences (HIV-1 strain SF2) are presented on the top. Dashes indicate the same amino acid as that of the wild type. Sequence substitutions are presented by single amino acid characters. Where a mixture of two or three amino acids was plausible, two or three amino acids were shown together separated by a shill. A single dash indicates that the sequences could not be determined by ambiguities. (A) Sequences from A24-positive and -negative Japanese hemophiliacs. (B) Sequences from A24-positive and -negative Japanese patients infected through USI. Asterisks indicate samples for which sequence ambiguities were found by direct sequence analysis. We cloned these PCR fragments into the pGEM-T vector and sequenced each 10 to 13 colonies. All amino acid sequences are indicated. (C) Sequences from A24-positive and -negative Australians infected through USI.
Nef138-10(2F) accompanied a particular amino acid substitution in the N-terminal flanking region. We detected an I-to-T substitution at the −1 position (−1T) in 32 flanking sequences out of 34 accompanying Nef138-10(2F) sequences (94%), while others were two I-to-V substitutions (Fig. 1). The −1T substitution was quite unusual in the flanking region of the wild-type Nef138-10 CTL epitope in our cohort (Fig. 1).
Reversion of CTL escape mutants.
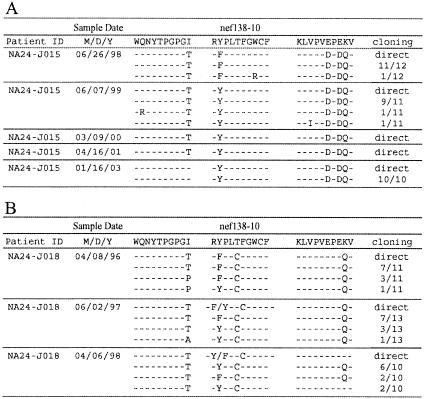
Since three acutely infected A24-positive patients (A24-J023, A24-J024, and A24-J025) had Nef138-10(2F) in their earliest plasma samples available, we could not demonstrate the evolution of Nef138-10(2F) from the wild type under the selective pressure of HLA-A*2402 (data not shown). However, we could analyze serial samples from chronically infected A24-negative patients who had been followed without treatment over years. All the 12 cloned sequences in the earliest plasma samples available from NA24-J015 had F at the second position; however, 11 out of 11 clones displayed wild-type sequence within a year (Fig. 2A). It is interesting that the −1T substitution in the flanking region was present for at least a further two years before reverting to the wild type (Fig. 2A). In another chronically infected A24-negative patient (NA24-J018), we observed that the proportion of Nef138-10(2F) decreased gradually but persisted in the plasma for almost two years after the start of the follow-up (Fig. 2B). This patient had a T-to-C substitution at the fifth position with [Nef138-10(2F5C)] or without [Nef138-10(5C)] a substitution at the second position (Fig. 2B). Interestingly, the ratio of Nef138-10(2F5C) to Nef138-10(5C) decreased as time went by (Fig. 2B), suggesting that Nef138-10(5C) is more stable than Nef138-10(2F5C). Actually, we observed Nef138-10(5C) in both A24-positive and -negative patients (Fig. 1).
FIG. 2.
Serial Nef138-10 epitope and its flanking sequences in two A24-negative patients. Data are shown as described in the legend to Fig. 1. Fractional numbers in the right-most column indicate clone numbers with the sequences shown in the numerator and total clone numbers sequenced shown in denominator. “Direct” indicates the result of direct sequencing. (A) Patient NA24-Jo15. (B) Patient NA24-J018.
In order to elucidate the higher stability of the 5C rather than the 2F substitution, we examined the codon usage at these positions (data not shown). The wild-type codon for the second tyrosine (Y) residue in Nef138-10 was coded by TAT or TAC in 23 (77%) and 12 (40%) out of 30 patients, respectively. Five patients (17%) had a mixture of TAT and TAC for the codon (data not shown). Mutated nucleotide triplet TTT or TTC was responsible for the Y-to-F amino acid substitution in 27 (80%) and 9 (26%) out of 34 patients, respectively (data not shown). In two patients (6%) Nef138-10(2F) was coded by a mixture of HIV-1 using TTT and TTC for the codon. It appeared that at least one point mutation was necessary for the Y-to-F amino acid substitution. The wild-type codon for the fifth threonine (T) residue in Nef138-10 was coded by ACC or ACT in 49 (98%) and 2 (4%) out of 50 patients. One patient (2%) had a mixture of ACC and ACT. Mutated nucleotide triplet TGC or TGT was responsible for the T-to-C amino acid substitution in 5 (45%) and 6 (55%) out of 11 patients, respectively (data not shown). It appeared that at least two nucleotides had to be mutated for the T-to-C amino acid substitution, although we could not exclude the possibility of a three-nucleotide mutation for the amino acid substitution. Therefore, a Y-to-F amino acid substitution, or vice versa, at the second position required less nucleotide mutations than did the T-to-C substitution at the fifth position.
Peptide-based analysis of Nef138-10 and its variants.
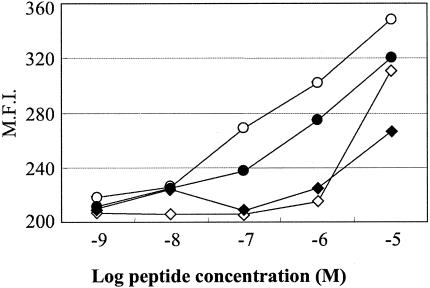
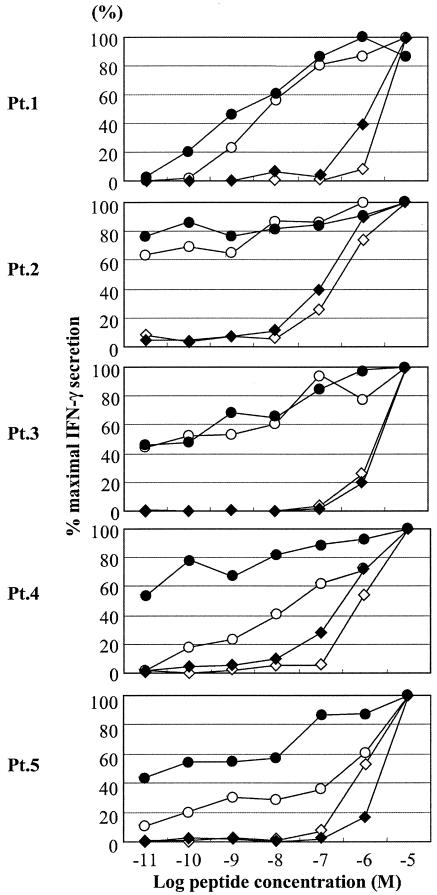
We measured the binding affinities of Nef138-10 and its variants to HLA-A*2402 (Fig. 3). Although a Y-to-F substitution occurred at the amino acid crucial for peptide affinity with the binding groove, Nef138-10(2F) binding to the HLA-A*2402 heavy chain was essentially preserved. However, the acquisition of a T-to-C substitution at the fifth position, such as Nef138-10(2F5C) and Nef138-10(5C), greatly reduced the binding affinity (Fig. 3). A functional avidity assay in which PBMCs from five patients were used confirmed the results of the binding assay (Fig. 4). Namely, the patients' PBMCs recognized Nef138-10(2F) at a very low concentration (one-half maximum response <1 nM) and had equivalent or even higher functional avidity than did the wild-type peptide. On the contrary, patients' PBMCs showed very low functional avidity against Nef138-10(2F5C) and Nef138-10(5C) (one-half maximum response >100 nM).
FIG. 3.
Binding of the wild-type and mutant peptides to HLA*2402 molecules. Peptide binding to HLA-A*2402 was quantified by using a T2-A24 stabilization assay. Symbols: ○, wild type; •, 2F; ◊, 5C; ⧫, 2F5C. M.F.I., mean fluorescence intensity.
FIG. 4.
Functional avidity assay. The reactivity of peptide-specific cells in PBMCs from five patients against log-fold dilutions of peptide was determined. Symbols: ○, wild type. •, 2F; ◊, 5C; ⧫, 2F5C.
Epitope presentation from native Nef protein.
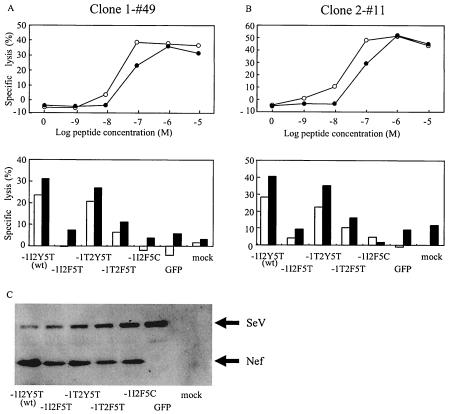
Strong selection for Nef138-10(2F) in the presence of CTLs with high in vivo functional avidity against the peptide prompted us to examine the processing and presentation of the Nef138-10 CTL epitope from the native protein. Native Nef proteins containing wild-type or variant CTL epitopes were expressed in an HLA-A*2402-positive-T-cell line (KWN-T4) via SeV. CTL epitope presentation was examined by two CTL clones established from A24-positive patients outside these cohorts. Although the two CTL clones were established by stimulation with the wild-type peptide (Nef138-10), they killed the target cells pulsed with Nef138-10(2F) peptides almost as well as the wild type (Fig. 5A and B). Both CTL clones efficiently killed the target cells expressing either wild-type Nef or Nef with −1T substitution in the flanking region (−1T2Y5T). However, these CTL clones failed to kill the target cells infected with vectors expressing Nef138-10(2F) with (−1T2F5T) or without (−1I2F5T) the −1T substitution in the flanking region. As expected, the CTL clones did not kill the target cells infected with a vector coding Nef138-10(2F5C), a nonbinding mutant (−1I2F5C) (Fig. 5A and B). Western blot analysis revealed that Nef proteins with wild-type or variant CTL epitopes were expressed abundantly in the target cells. Taken together, these data indicate that a Y-to-F substitution within the CTL epitope itself but not the −1T substitution in the flanking region resulted in the poor antigen presentation against CTL, which resulted in the escape.
FIG. 5.
Killing activity of clone 1-#49 (A) against KWN-T4 target cells pulsed with log-fold dilutions of peptide (top) and expressing native Nef proteins containing wild-type sequences (−1I2Y5T), a Y-to-F substitution at the second position of the CTL epitope (−1I2F5T), an I-to-T substitution at the −1 flanking position (−1T2Y5T), double substitutions at the −1 and second positions (−1T2F5T), and double substitutions at the second and fifth positions (−1I2F5C) (bottom). The effector-versus-target ratio was 1:1 (□) or 2.5:1 (▪) in panel A and 1:1 (□) or 4:1 (▪) in panel B. Killing activity against KWN-T4 cells infected with control vector expressing green fluorescent protein (GFP) and mock infected (mock) are also shown. (C) Western blot analysis of intracellular expression of various Nef mutants in KWN-T4 target cells. KWN-T4 target cells expressing native Nef proteins containing wild-type sequences (−1I2Y5T), a Y-to-F substitution at the second position of the CTL epitope (−1I2F5T), an I-to-T substitution at the −1 flanking position (−1T2Y5T), double substitutions at the −1 and second positions (−1T2F5T), and double substitutions at the second and fifth positions (−1I2F5C) were examined. KWN-T4 cell lysates infected with control vector expressing GFP and mock infected are also shown. An aliquot (3 μg) of the same KNW-T4 target cells used for the killer assay in the upper panel was used for the Western blot. Symbols: ○, Nef138-10; •, Nef138-10(2F).
DISCUSSION
We showed a significantly higher prevalence of a stereotypic amino acid substitution [Nef138-10(2F)] at an A24-restricted CTL epitope in Nef among A24-positive Japanese hemophiliacs compared with A24-negative counterparts. The origin of their HIV-1 infection was from the plasma collected and processed in Western countries where HLA-A*2402 was less prevalent (19). It is inferred that Nef138-10(2F) might be rare in a population where HLA-A*2402 is not prevalent but that it has a selective advantage in the presence of HLA-A*2402. Our findings with Australians are consistent with this notion. Although we examined only two HIV-1-infected A24-positive Caucasian Australians, both had Nef138-10(2F). On the other hand, Nef138-10(2F) was rare in A24-negative Australians. Japanese and Australians are distinctly different in the frequency of HLA-A*2402 within their respective populations (allele frequency of HLA-A24 is 35.1 and 7.8%, respectively) (19). Nef138-10(2F) was also positively selected among Japanese patients who were infected through USI. Interestingly, we detected Nef138-10(2F) frequently among A24-negative Japanese who were infected through USI. The result suggests that HIV-1 that went through selective pressure by HLA-A*2402 is actually circulating among the Japanese population because of the high prevalence of HLA-A24. Although we showed the reversion of Nef138-10(2F) to the wild type, it occurred very slowly over years, allowing the horizontal spread via sexual contact. In this study, we showed that HIV-1 with Nef138-10(2F) is actually a CTL escape mutant. Although the stereotypic Y-to-F substitution occurred at an anchor residue, Nef138-10(2F) peptide did bind to HLA-A*2402 heavy chain with almost the same efficiency as did the wild type (Fig. 3). This result is consistent with the algorithm prediction of the published binding motif (http://hiv-web.lanl.gov/content/immunology/motif_scan/motif.html). When native Nef proteins with or without a substitution were overexpressed in the A24-positive target cells via SeV, the Y-to-F substitution at the second position of the CTL epitope virtually abolished the killing by the CTL clones. The substitution in the flanking region did not affect the killing substantially. Therefore, the mechanism for the CTL escape appeared to reside in the processing of native Nef proteins and subsequent antigen presentation rather than HLA binding. A proteosomal cleavage prediction program, NetChop (23), suggested the possibility that the Y-to-F substitution in the second position creates a new cleavage site at the fifth T residue in the CTL epitope. Proteolytic cleavage within the epitope could be the cause of poor antigen presentation.
Although we could not show the process of positive selection for Nef138-10(2F), Nef138-10(2F5C), and Nef138-10(5C), the high prevalence of Nef138-10(2F) in A24-positive patients and the reversion in A24-negative patients suggested that one point mutant, Nef138-10(2F), was selected first, and then two or three point mutants, Nef138-10(2F5C), evolved. Once the T-to-C amino acid substitution at the fifth position is acquired, the binding capacity of the CTL epitope to the HLA-A*2402 heavy chain is abolished (Fig. 3), and the Y-to-F substitution at the second position may become dispensable even in the presence of HLA-A*2402.
In our cohort of patients, Nef138-10(2F) accompanied a −1T substitution in the flanking region very frequently. We observed sequential reversion in the CTL epitope and flanking region at least in one patient with an A24-negative background. As of 11 October 2003, the HIV-1 sequence database showed that the 2F substitution (74 sequences) accompanied the −1T substitution frequently (64.9%) but accompanied the wild-type residue (I) only rarely (9.5%). On the other hand, the wild-type residue (Y) in the second position of the CTL epitope (195 sequences) accompanied wild-type (I) residue more frequently (57.4%) than the −1T substitution (20.5%). Although the function of the region surrounding Nef138-10 has not been elucidated, there seems to be a compensation between these two residues.
In simian immunodeficiency virus infection, CTLs with high functional avidity select for escape variants (29). However, we found CTLs with high functional avidity not only against the wild type but also against Nef138-10(2F) in five patients studied. It is not known how these CTLs against Nef138-10(2F) are maintained in vivo. Very recently, new insights into the exogenous pathway for antigen presentation to CTLs have been elucidated (15, 17). Cross presentation by professional antigen-presenting cells such as dendritic cells may be responsible. Our study underlines the difficulties in evaluating the effective CTL responses in vivo by CTL assays in which peptides are used, such as ELISPOT.
For example, a CTL escape variant of Epstein-Barr virus was demonstrated in a highly A11-positive population in New Guinea (9). HLA-restricted CTL responses appear to be driving HIV-1 evolution at a population level (20). As far as we know, this is the first direct demonstration of horizontal transmission of CTL escape mutants of HIV-1 at a population level. We previously reported stereotypic amino acid substitutions in HIV-1 at some CTL epitopes restricted by HLA-B35 (21). Stereotypically selected HIV-1 may become dominant through transmission where certain HLA types are highly prevalent. Recently, a rare HLA supertype was shown to have a selective advantage for the prognosis of HIV-1 infection (34). In a population with less diverse HLA types, such as that of Japan, HLA types may have a large impact on HIV-1 evolution and escape. Our study may prove to have important implications for understanding viral pathogenesis and vaccine development.
Acknowledgments
This work was partly supported by grants for AIDS research from the Ministry of Health, Labor, and Welfare of Japan, a grant-in-aid for Scientific Research (A) from the Japan Society of the Promotion of Science, and the Japan Health Sciences Foundation.
REFERENCES
- 1.Anonymous. 2003. HIV/AIDS in Japan, 2002. Infect. Agents Surveill. Rep. 24:203-204. [Google Scholar]
- 2.Appay, V., P. R. Dunbar, M. Callan, P. Klenerman, G. M. A. Gillespie, L. Papagno, G. S. Ogg, A. King, F. Lechner, C. A. Spina, S. Little, D. V. Havlir, D. D. Richman, N. Gruener, G. Pape, A. Waters, P. Easterbrook, M. Salio, V. Cerundolo, A. J. McMichael, and S. L. Rowland-Jones. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 8:379-385. [DOI] [PubMed] [Google Scholar]
- 3.Beekman, N. J., P. A. van Veelen, T. van Hall, A. Neisig, A. Sijts, M. Camps, P. M. Kloetzel, J. J. Neefjes, C. J. Melief, and F. Ossendorp. 2000. Abrogation of CTL epitope processing by single amino acid substitution flanking the C-terminal proteasome cleavage site. J. Immunol. 164:1898-1905. [DOI] [PubMed] [Google Scholar]
- 4.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 6.Champagne, P., G. S. Ogg, A. S. King, C. Knabenhans, K. Ellefsen, M. Nobile, V. Appay, G. P. Rizzardi, S. Fleury, M. Lipp, R. Forster, S. Rowland-Jones, R. P. Sekaly, A. J. McMichael, and G. Pantaleo. 2001. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature 410:106-111. [DOI] [PubMed] [Google Scholar]
- 7.Collins, K. L., B. K. Chen, S. A. Kalams, B. D. Walker, and D. Baltimore. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397-401. [DOI] [PubMed] [Google Scholar]
- 8.Couillin, I., B. Culmann-Penciolelli, E. Gomard, J. Choppin, J. P. Levy, J. G. Guillet, and S. Saragosti. 1994. Impaired cytotoxic T lymphocyte recognition due to genetic variations in the main immunogenic region of the human immunodeficiency virus 1 NEF protein. J. Exp. Med. 180:1129-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Campos-Lima, P. O., R. Gavioli, Q. J. Zhang, L. E. Wallace, R. Dolcetti, M. Rowe, A. B. Rickinson, and M. G. Masucci. 1993. HLA-A11 epitope loss isolates of Epstein-Barr virus from a highly A11+ population. Science 260:98-100. [DOI] [PubMed] [Google Scholar]
- 10.Douek, D. C., M. R. Betts, J. M. Brenchley, B. J. Hill, D. R. Ambrozak, K. L. Ngai, N. J. Karandikar, J. P. Casazza, and R. A. Koup. 2002. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J. Immunol. 168:3099-3104. [DOI] [PubMed] [Google Scholar]
- 11.Foung, S. K., B. Taidi, D. Ness, and F. C. Grumet. 1986. A monoclonal antibody against HLA-A11 and A24. Hum. Immunol. 15:316-319. [DOI] [PubMed] [Google Scholar]
- 12.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 13.Goulder, P. J., A. Edwards, R. E. Phillips, and A. J. McMichael. 1997. Identification of a novel HLA-A24-restricted cytotoxic T-lymphocyte epitope within HIV-1 Nef. AIDS 11:1883-1884. [DOI] [PubMed] [Google Scholar]
- 14.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 15.Guermonprez, P., L. Saveanu, M. Kleijmeer, J. Davoust, P. Van Endert, and S. Amigorena. 2003. ER-phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature 425:397-402. [DOI] [PubMed] [Google Scholar]
- 16.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 17.Houde, M., S. Bertholet, E. Gagnon, S. Brunet, G. Goyette, A. Laplante, M. F. Princiotta, P. Thibault, D. Sacks, and M. Desjardins. 2003. Phagosomes are competent organelles for antigen cross-presentation. Nature 425:402-406. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda-Moore, Y., H. Tomiyama, K. Miwa, S. Oka, A. Iwamoto, Y. Kaneko, and M. Takiguchi. 1997. Identification and characterization of multiple HLA-A24-restricted HIV-1 CTL epitopes: strong epitopes are derived from V regions of HIV-1. J. Immunol. 159:6242-6252. [PubMed] [Google Scholar]
- 19.Imanishi, T., T. Akaza, A. Kimura, K. Tokunaga, and T. Gojobori. 1992. Allele and haplotype frequencies for HLA and complement loci in various ethnic groups, p. 1065-1220. In K. Tsuji, M. Aizawa, and T. Sasazuki (ed.), HLA 1991, vol. 1. Oxford University Press. [Google Scholar]
- 20.Jost, S., M. C. Bernard, L. Kaiser, S. Yerly, B. Hirschel, A. Samri, B. Autran, L. E. Goh, and L. Perrin. 2002. A patient with HIV-1 superinfection. N. Engl. J. Med. 347:731-736. [DOI] [PubMed] [Google Scholar]
- 21.Kawana, A., H. Tomiyama, M. Takiguchi, T. Shioda, T. Nakamura, and A. Iwamoto. 1999. Accumulation of specific amino acid substitutions in HLA-B35-restricted human immunodeficiency virus type 1 cytotoxic T lymphocyte epitopes. AIDS Res. Hum. Retrovir. 15:1099-1107. [DOI] [PubMed] [Google Scholar]
- 22.Kawana-Tachikawa, A., M. Tomizawa, J. Nunoya, T. Shioda, A. Kato, E. E. Nakayama, T. Nakamura, Y. Nagai, and A. Iwamoto. 2002. An efficient and versatile mammalian viral vector system for major histocompatibility complex class I/peptide complexes. J. Virol. 76:11982-11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kesmir, C., A. K. Nussbaum, H. Schild, V. Detours, and S. Brunak. 2002. Prediction of proteasome cleavage motifs by neural networks. Protein Eng. 15:287-296. [DOI] [PubMed] [Google Scholar]
- 24.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuzushima, K., N. Hayashi, H. Kimura, and T. Tsurumi. 2001. Efficient identification of HLA-A*2402-restricted cytomegalovirus-specific CD8(+) T-cell epitopes by a computer algorithm and an enzyme-linked immunospot assay. Blood 98:1872-1881. [DOI] [PubMed] [Google Scholar]
- 26.McMichael, A. J., and R. E. Phillips. 1997. Escape of human immunodeficiency virus from immune control. Annu. Rev. Immunol. 15:271-296. [DOI] [PubMed] [Google Scholar]
- 27.Migueles, S. A., A. C. Laborico, W. L. Shupert, M. S. Sabbaghian, R. Rabin, C. W. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, and M. Connors. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061-1068. [DOI] [PubMed] [Google Scholar]
- 28.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 29.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 30.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279:2103-2106. [DOI] [PubMed] [Google Scholar]
- 31.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz, O., V. Marechal, S. Le Gall, F. Lemonnier, and J. M. Heard. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2:338-342. [DOI] [PubMed] [Google Scholar]
- 33.Tomiyama, H., H. Akari, A. Adachi, and M. Takiguchi. 2002. Different effects of Nef-mediated HLA class I down-regulation on human immunodeficiency virus type 1-specific CD8+ T-cell cytolytic activity and cytokine production. J. Virol. 76:7535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trachtenberg, E., B. Korber, C. Sollars, T. B. Kepler, P. T. Hraber, E. Hayes, R. Funkhouser, M. Fugate, J. Theiler, Y. S. Hsu, K. Kunstman, S. Wu, J. Phair, H. Erlich, and S. Wolinsky. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9:928-935. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe, N., M. Tomizawa, A. Tachikawa-Kawana, M. Goto, A. Ajisawa, T. Nakamura, and A. Iwamoto. 2001. Quantitative and qualitative abnormalities in HIV-1-specific T cells. AIDS 15:711-715. [DOI] [PubMed] [Google Scholar]
- 36.Yamada, T., N. Kaji, T. Odawara, J. Chiba, A. Iwamoto, and Y. Kitamura. 2003. Proline 78 is crucial for human immunodeficiency virus type 1 Nef to down-regulate class I human leukocyte antigen. J. Virol. 77:1589-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]