Abstract
Murid herpesvirus 4 (commonly called MHV-68) is closely related to Kaposi's sarcoma-associated herpesvirus (KSHV) and provides an excellent model system for investigating gammaherpesvirus-associated pathogenesis. MHV-76 is a naturally occurring deletion mutant of MHV-68 that lacks 9,538 bp of the left end of the unique portion of the genome encoding nonessential pathogenesis-related genes. The KSHV K1 protein has been shown to transform rodent fibroblasts in vitro and common marmoset T lymphocytes in vivo. Using homologous recombination techniques, we successfully generated recombinants of MHV-76 that encode green fluorescent protein (MHV76-GFP) and KSHV K1 (MHV76-K1). The replication of MHV76-GFP and MHV76-K1 in cell culture was identical to that of MHV-76. However, infection of BALB/c mice via the intranasal route revealed that MHV76-K1 replicated to a 10-fold higher titer than MHV76-GFP in the lungs at day 5 postinfection (p.i.). We observed type 2 pneumocyte proliferation in areas of consolidation and interstitial inflammation of mice infected with MHV76-K1 at day 10 p.i. MHV76-K1 established a 2- to 3-fold higher latent viral load than MHV76-GFP in the spleens of infected mice on days 10 and 14 p.i., although this was 10-fold lower than that established by wild-type MHV-76. A salivary gland tumor was present in one of four mice infected with MHV76-K1, as well as an increased inflammatory response in the lungs at day 120 p.i. compared with that of mice infected with MHV-76 and MHV76-GFP.
Murid herpesvirus 4 (commonly called MHV-68) infection of laboratory mice provides an excellent system for the study of gammaherpesvirus pathogenesis (2, 7, 19, 23, 24, 33). After intranasal infection of mice, MHV-68 replicates initially in the lungs (27). This infection is cleared by days 10 to 14 postinfection (p.i.) by CD8+ T cells (8), although the virus persists in a latent form in epithelial cells at this site (25). MHV-68 spreads to the spleen, where it infects B lymphocytes, macrophages, and dendritic cells (9, 28, 30, 34), causing splenomegaly driven by CD4+ T cells (29). Life-long virus persistence occurs predominantly in surface immunoglobulin D-negative B cells after the resolution of splenomegaly (35).
MHV-76 was isolated from the yellow-necked mouse (Apodemus flavicollis) in Slovakia and was recently shown by genome sequencing to be identical to MHV-68 except for a 9,538-bp deletion at the left end of the unique region (17). This deletion is nonessential, encompassing four protein-coding genes (M1, M2, M3, and M4) and eight viral tRNA-like genes, none of which are found in other gammaherpesviruses. The roles of M1, M4, and the viral tRNAs during MHV-68 infection are currently unknown. M2 is expressed in B cells during latency (18). Although it is nonessential for the development of latency, M2 is responsible for much of the MHV-68-induced peak of viral latency in the spleen during splenomegaly (11, 18), and the M2 protein is a target for the host immune response (10). M3 encodes a secreted chemokine binding protein that is expressed during both acute infections and the persistence of MHV-68 in mice (21, 31). Although the replication of MHV-76 is identical to that of MHV-68 in cell culture, after infections of mice MHV-76 is cleared more rapidly from the lungs (17). This is probably due to an enhanced inflammatory response in the lungs against MHV-76. Splenomegaly is also significantly reduced after MHV-76 infection, consistent with a much reduced latent viral load in the spleen (17).
Since the start of the AIDS pandemic, Kaposi's sarcoma (KS) has become one of the most prevalent tumors in men under 60 years old (1). Long before AIDS, KS was thought to be caused by an infectious agent. The large relative risk of KS among certain categories of AIDS patients and the clustering of KS in different populations have strengthened this view. DNA sequences from a novel gammaherpesvirus, termed KS-associated herpesvirus (KSHV), were identified in an AIDS-associated KS biopsy (5). KSHV has since been found in all epidemiological forms of KS and also in primary effusion lymphomas (4). This new herpesvirus is related to an oncogenic herpesvirus from monkeys (herpesvirus saimiri [HVS]) and to a well-characterized human herpesvirus (Epstein-Barr virus) which is associated with several human cancers (20, 22).
At a position equivalent to that of the main transforming proteins of HVS (saimiri transforming protein [STP]) and Epstein-Barr virus (latent membrane protein 1), KSHV contains a distinct open reading frame (ORF) called K1 (12). K1 is a 46-kDa transmembrane glycoprotein (12). While the amino-terminal extracellular domain of K1 is extremely variable between isolates of KSHV, the carboxy-terminal short cytoplasmic tail is relatively well conserved (36). This carboxy-terminal cytoplasmic tail contains a functional immunoreceptor tyrosine-based activation motif (ITAM) (15). The ITAM is capable of transducing signals to induce cellular activation, calcium mobilization, and tyrosine phosphorylation, which are all indicative of lymphocyte activation (14, 15). However, unlike other ITAM-based signal transduction events that require a ligand-receptor interaction, K1 signaling appears to occur constitutively (14). The K1 protein has been shown to interact with several cellular signal transduction proteins, including Vav, p85, and Syk kinase (15), and to induce the activity of nuclear factor of activated T cells (14). ITAM-dependent signaling by K1 has been shown to augment the lytic replication of KSHV in B cells (13). In addition to the transformation of rodent fibroblasts, K1 can also functionally replace STP in HVS for the immortalization of common marmoset T lymphocytes to interleukin-2-independent growth and for the induction of lymphomas in common marmosets (16). Prakash et al. (21a) produced transgenic mice expressing K1 under the control of a simian virus 40 (SV40) promoter. These mice showed enhanced NF-κB activity in nonmalignant lymphocytes and tumors in 2 of 13 14-month-old mice. One of these tumors was a plasmoblastic malignant lymphoma and the other was a spindle-cell sarcomatoid tumor (21a).
The aim of this study was to ascertain the function of the KSHV K1 protein in vivo. There is no amenable animal model system for KSHV. While MHV-68 has been informative about aspects of gammaherpesvirus biology, it does not contain an equivalent of K1 (32). To assess the function of K1 in vivo, we therefore adopted the approach of generating an MHV-68 recombinant containing K1. We surmised that MHV-76 was the best vector to use for this since it is missing several pathogenesis-related genes (17). There was therefore less chance of the biological effects of K1 being masked by a duplicate MHV-68 function. We successfully generated two recombinants of MHV-76 that encode green fluorescent protein (GFP) and KSHV K1. The pathogenesis of these recombinant viruses in the context of infection of BALB/c mice was investigated.
MATERIALS AND METHODS
Cell lines and viruses.
MHV-76 and recombinant viruses were grown and titrated on BHK-21 cells (in Glasgow's medium supplemented with 10% [vol/vol] newborn calf serum, 10% [vol/vol] tryptose phosphate buffer, penicillin [60 μg/ml], streptomycin [100 μg/ml], and 2 mM glutamine) as described previously. MHV-76 was originally isolated during field studies from the yellow-necked mouse, A. flavicollus (3). It was subsequently plaque purified on BHK-21 cells and characterized as described previously (17).
Plasmid construction.
KSHV ORF K1 was amplified from BCP1 DNA with the oligonucleotides KLA (GTACGAGCTCGCTAGCAAGATGTTCCTGTATGTTGTCTG) and KLB (GTACGCGGCCGCGGTACCAATCCACTGGTTGCG) and Advantage cDNA polymerase (BD Clontech). The PCR product was cloned into the SacI and NotI sites of pBluescript KS-Myc (has a Myc epitope tag between the NotI and XbaI sites of pBluescript KS). The K1-Myc sequence was then subcloned into the SacI and KpnI sites of pUC18. The encephalomyocarditis virus (EMCV) internal ribosome entry sequence (IRES) (from pBVIRESEGFP) was inserted at the EcoRI and SacI sites upstream of K1-Myc. The IRES-K1Myc sequence was inserted into the SalI and XmaI 3′ multiple cloning site of pHygEGFP (BD Clontech). BglII linkers (NEB) were then added at the BsaBI site and a BglII fragment (CMV-HygEGFP-IRES-K1Myc-POLYA) was then cloned into the BamHI site of pBS76LHE (contains nucleotides 9539 to 12569 of MHV-68). An enhanced GFP (EGFP) control plasmid was constructed by inserting the CMV-HygEGFP-IRES-POLYA sequence into the BamHI site of pBS76LHE.
Construction of recombinant viruses.
All recombinant virus work was performed at the University of Edinburgh under United Kingdom Advisory Committee on Genetic Modification safety level 2 conditions and with notification of the work to the Health and Safety Executive. MHV-76 genomic DNA was prepared from purified virions as described previously (17). MHV-76 DNA (5 μg) and 10 μg of linearized plasmid were transfected by electroporation into 2 × 106 BHK-21 cells by using a double-pulse setting (high-voltage setting = 600 V, 25 μF, 99 Ω; low-voltage setting = 260 V, 1,500 μF, 329 Ω; 0.1-s interpulse delay) on an EasyJect electroporator (EquiBio). Electroporated cells were cultured in a six-well plate. Green fluorescent plaques were picked after 5 days and subjected to four rounds of plaque purification by limiting dilution (in 96-well plates) with hygromycin selection (100 μg/ml). Each round of purification was verified by a PCR to amplify MHV-76 ORF74 and K1 or EGFP. A Southern blot analysis was performed to ensure that the recombinant viruses were free from contamination with wild-type MHV-76. The Southern blots were probed with a 1-kb BamHI-XmaI fragment of pBS-76LHE and a 1-kb SacII-XmaI fragment of pHygEGFP (Fig. 1 and 2).
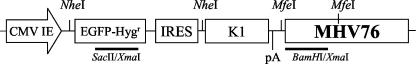
FIG. 1.
Schematic diagram of plasmid used to generate recombinant MHV76-K1 virus. The expression cassette consists of the CMV-IE promoter and the EGFP-Hygr gene from pEGFPHyg (Clontech), followed by the IRES element from EMCV. Cloned immediately downstream from the IRES is the KSHV K1 ORF followed by the SV40 poly(A) signal (pA). These elements were cloned into a BamHI site next to the MHV-76 genomic DNA (nucleotides 9539 to 12569 of MHV-68) in pBluescript (Stratagene). The MHV76-GFP virus was generated by using an identical plasmid lacking the KSHV K1 gene. The positions of the probes used for Southern hybridization (BamHI-XmaI and SacII-XmaI) and the restriction sites for NheI and MfeI are indicated.
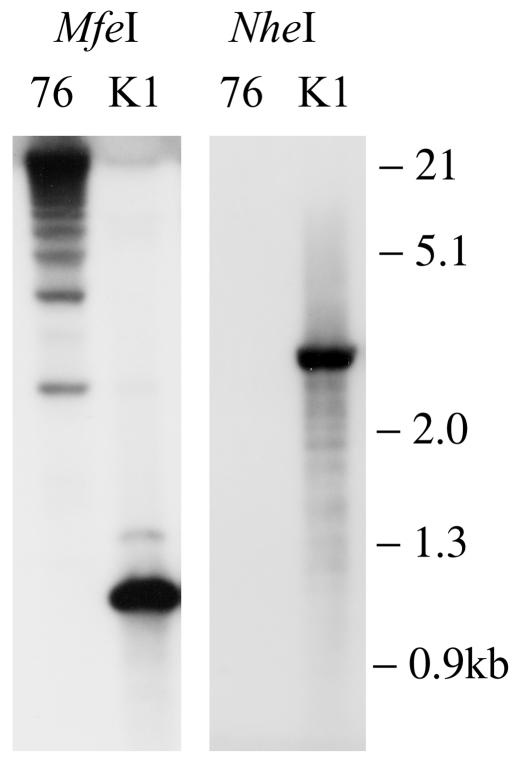
FIG. 2.
Southern blot showing purified MHV76-K1 recombinant virus compared with parental MHV-76. The left panel shows MHV-76 and MHV76-K1 digested with MfeI and probed with the BamHI-XmaI probe (Fig. 1). The right panel shows MHV-76 and MHV76-K1 digested with NheI and probed with the SacII-XmaI probe (Fig. 1).
In vitro infections.
In vitro one-step growth curves were obtained by infecting subconfluent BHK-21 cells at a multiplicity of infection of 5 and allowing the virus to adsorb for 1 h at 37°C. The cells were then washed three times to remove unbound virus, and fresh medium was added. At various times p.i., the cells were harvested by scraping into the medium. Viruses were released from the cells by three freeze-thaw cycles, and infectious virus was quantified by a plaque assay. All experiments were performed in duplicate.
Infection of mice and analysis of tissues.
All animal experiments were performed at the University of Edinburgh under United Kingdom Home Office License number 60/2429. Female BALB/c mice were obtained from Harlan Olac and were infected when they were 3 to 4 weeks old. Mice were anesthetized with halothane and infected intranasally with 4 × 105 PFU of virus in 40 μl of sterile phosphate-buffered saline (PBS). At various times p.i., mice were sacrificed by CO2 asphyxiation and tissues were harvested for analysis. Infectious virus was detected by a plaque assay, and an infective-center assay was used to detect latent virus as described previously (28).
The expression of KSHV K1 was detected by immunoprecipitation and Western blotting. Tissue samples from MHV76-K1- and MHV76-GFP-infected mice were disrupted by Dounce homogenization in RIPA buffer (PBS, 1% [vol/vol] Nonidet P-40, 0.5% [wt/vol] sodium deoxycholate, 0.1% [wt/vol] sodium dodecyl sulfate). The cell lysate was immunoprecipitated with a mouse monoclonal antibody (9E10) recognizing the Myc epitope tag (Santa Cruz Biotechnology). Precipitated proteins were separated in a sodium dodecyl sulfate-12% (wt/vol) polyacrylamide gel and transferred to a nitrocellulose membrane. The Western blot was probed with a biotin-conjugated anti-Myc-tag monoclonal antibody (Santa Cruz Biotechnology) and streptavidin-conjugated alkaline phosphatase (Sigma). The blot was developed with 4-nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) (Roche).
Histopathology.
Mice were sacrificed by CO2 asphyxiation. Lungs were perfused in situ via the trachea with 10% neutral buffered Formol saline and then processed into paraffin-embedded sections. Five-micrometer-thick sections were cut and stained with hematoxylin and eosin for examination by light microscopy.
Immunohistochemistry.
Frozen sections were fixed on slides in 4% (wt/vol) paraformaldehyde for 1 h, washed in PBS for 5 min, and blocked for 30 min (PBS, 1% [wt/vol] bovine serum albumin, 0.1% [vol/vol] Triton, 0.1% [wt/vol] sodium azide, and 5% [vol/vol] normal goat serum). The primary antibody was a rabbit polyclonal anti-MHV-68 antibody used at 1:500 for 2 h. The slides were washed twice in PBS for 5 min each time. The secondary antibody was either goat anti-rabbit-tetramethyl rhodamine isocyanate used at 1:400 or goat anti-rabbit-Alexa Fluor 594 used at 1:200 for 30 min. The slides were washed twice in PBS for 5 min each time before being mounted with fluorescent mounting medium (Dako). All antibody incubations were carried out in a humidified chamber at room temperature.
For cytokeratin immunohistochemistry, sections of fixed tissue were cut onto Biobond (British Biocell Ltd.)-coated microscope slides. Sections were dewaxed and rehydrated, and endogenous peroxidase activity was blocked with 3% (vol/vol) hydrogen peroxide in distilled water for 10 min. The slides were washed in PBS, and antigen retrieval was performed by incubating the slides in distilled water containing 0.1% trypsin and 0.1% calcium (pH 7.8) for 30 min at 37°C. The slides were washed in PBS and then incubated with an anti-cytokeratin antibody for 1 h at room temperature (mouse monoclonal NCL-L-Pan-CK diluted 1:100; Novocastra Laboratories). The detection of bound antibody was done by the use of a commercially available kit (M.O.M. peroxidase kit; Vector Laboratories) and visualization was performed with 3,3′-diaminobenzidine tetrahydrochloride (DAB) (Vector Laboratories).
Statistical analysis.
Data were analyzed with GraphPad Prism software (San Diego, Calif.). In all cases, a two-way analysis of variance with Bonferroni's posttest was used.
RESULTS
Generation of MHV-76 recombinants expressing EGFP and K1.
Two recombinant MHV-76 viruses were produced by homologous recombination in BHK cells. The region chosen for insertion was the left end of the unique portion of the genome. This is the region in which MHV-76 has a deletion relative to MHV-68 and was therefore the region least likely to cause a disruption of viral function (17). The first virus (MHV76-GFP) encoded an EGFP-hygromycin resistance (EGFP-Hygr) gene expressed from the cytomegalovirus immediate-early promoter (CMV-IE). The IRES from EMCV was inserted downstream of the EGFP-Hygr gene, followed by the SV40 polyadenylation signal. The second virus (MHV76-K1) included the coding sequence of the KSHV K1 gene inserted downstream of the EMCV IRES. The vectors used are illustrated in Fig. 1. Recombinant viruses were purified by plaque purification and limiting dilution on BHK cells. Once a pure population of green fluorescent plaques was obtained, the genomic structure was checked by Southern blotting. These data (Fig. 2) showed that the MHV76-K1 virus had the insert in the predicted position on the MHV-76 genome and that there were no DNA rearrangements. The multiple high-molecular-weight bands observed in the MHV-76 lane probed with the Bam-Xma probe were due to variability in the numbers of terminal repeats.
In vitro growth curve.
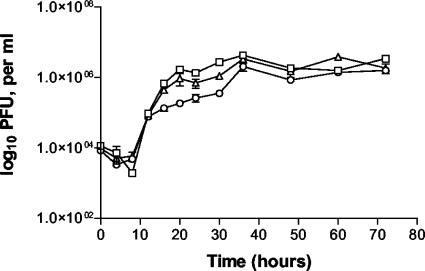
The growth kinetics of MHV76-GFP and MHV76-K1 were compared with those of wild-type MHV-76 in an in vitro one-step growth curve. BHK cells were infected at a multiplicity of infection of five; viruses were harvested over a period of 72 h and titrated. As shown in Fig. 3, the growth kinetics of MHV76-GFP and MHV76-K1 were not significantly different from those of MHV-76, reaching maximal titers at about 60 h p.i.
FIG. 3.
In vitro single-step growth curves of MHV-76 (□), MHV76-GFP (▵), and MHV76-K1 (○) on BHK-21 cells at a multiplicity of infection of 5. Data are shown as mean log10 virus titers ± standard errors and are representative of two separate experiments, with each performed in duplicate.
In vivo infection of mice.
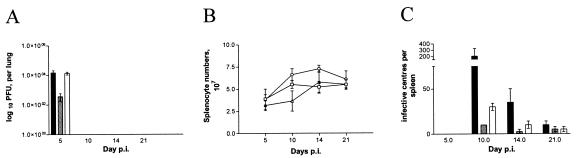
Groups of 20 BALB/c mice were infected intranasally with 4 × 105 PFU of MHV-76, MHV76-GFP, or MHV76-K1. The virus titers in the lungs were determined at various times p.i. by plaque assays for groups of four mice. The data in Fig. 4A show that the titers of MHV-76 and MHV76-K1 reached a similar level on day 5 p.i. but that the MHV76-GFP recombinant produced significantly lower infectious virus titers (P < 0.01). In all three cases, virus was effectively cleared from the lungs by day 10 p.i.
FIG. 4.
(A) Viral replication in lungs of BALB/c mice infected intranasally with 4 × 105 PFU of MHV-76 (black bars), MHV76-GFP (gray bars), or MHV76-K1 (white bars). The mean log10 virus titer ± standard error for four mice per group is shown for each time point. MHV76-GFP had a significantly lower titer than MHV-76 or MHV76-K1 on day 5 p.i. (P < 0.01). (B) Numbers of spleen cells during intranasal infection of BALB/c mice with 4 × 105 PFU of MHV-76 (□), MHV76-GFP (▵), or MHV76-K1 (○). The mean total number of splenocytes ± standard error for four mice per group is shown for each time point. MHV76-K1-infected mice had a significantly higher splenocyte number than MHV76-GFP- or MHV-76-infected mice on day 14 p.i. (P < 0.05). (C) Latent virus in spleens of BALB/c mice infected intranasally with 4 × 105 PFU of MHV-76 (black bars), MHV76-GFP (gray bars), or MHV76-K1 (white bars), as determined by an infective-center assay. The mean number of infective centers per spleen ± standard error for four mice per group is shown for each time point. MHV76-K1 had a significantly higher latent viral load than MHV76-GFP on day 10 p.i. (P < 0.001).
To examine the extent of splenomegaly after infection, we determined the total numbers of splenocytes in mice infected with MHV76-K1 and MHV76-GFP. As determined previously, infection with MHV-76 does not induce a significant increase in splenocyte numbers compared with infection by MHV-68 (6, 17). This was also shown to be the case for mice infected with MHV76-GFP (Fig. 4B). However, we observed an increase in the numbers of splenocytes in mice infected with MHV76-K1 on days 10 and 14 p.i. compared with those in mice infected with either MHV76-GFP or MHV-76. This difference was significant (P < 0.05) on day 14 p.i.
To investigate the ability of MHV76-GFP and MHV76-K1 to establish latency in vivo, we determined the latent virus load in the spleen by using an infective-center assay. The data in Fig. 4C show that infective centers peaked around day 10 for all three viruses but that the numbers of cells that were latently infected with MHV76-GFP and MHV76-K1 were significantly smaller than those of cells that were latently infected with MHV-76. This indicates that both MHV76-K1 and MHV76-GFP are able to establish latency in the spleen cell population, but at much reduced levels compared to MHV-76. We noted that although the levels were low, the numbers of latently MHV76-K1-infected cells were two- to threefold higher than those of latently MHV76-GFP-infected cells on days 10 and 14 p.i. This difference was found to be significant (P < 0.001) on day 10 p.i.
Pathology.
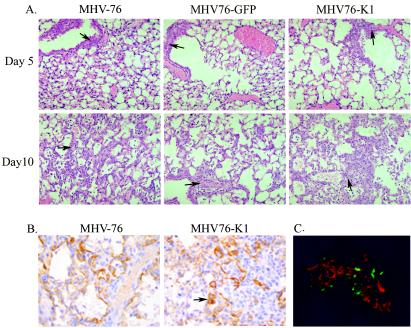
The lungs of mice were examined on days 5 and 10 p.i. for histopathological changes associated with virus infection. At day 5 p.i., all viruses produced a significant inflammatory response in the lungs, as shown by perivascular inflammatory cell infiltrates (dominated by lymphocytes) and edema (Fig. 5A), which persisted until day 10 p.i. with the development of an associated proliferative response (Fig. 5B). These data are consistent with those observed previously for MHV-76 infections and are in contrast with the limited inflammatory response seen at day 5 p.i. in the lungs of MHV-68-infected mice.
FIG. 5.
(A) Hematoxylin and eosin staining of lung sections of mice infected with MHV-76, MHV76-GFP, or MHV76-K1 at days 5 and 10 p.i., showing perivascular inflammatory cell infiltrates (arrows). (B) Cytokeratin staining of lung sections of mice infected with MHV-76 or MHV76-K1 at day 10 p.i., showing strong staining of type 2 pneumocytes in chronic proliferative inflammatory foci (arrow). (C) Mice infected with MHV76-K1 express GFP in the lungs at day 5 p.i. MHV-76 antigens were stained with polyclonal antisera and an anti-rabbit tetramethyl rhodamine isocyanate conjugate (red).
The immunohistochemical detection of cytokeratin was used to confirm type 2 pneumocyte proliferation in the lungs (Fig. 5B). At day 5 p.i., occasional areas of type 2 pneumocyte proliferation were present in MHV76-K1-infected mice. At day 10 p.i., strong staining of type 2 pneumocytes in chronic proliferative inflammatory foci in MHV-76- and MHV76-K1-infected mice was identified (Fig. 5B), but this change was more severe for MHV76-K1-infected mice. These data confirm the presence of epithelial hyperproliferation in association with inflammation.
The lungs of MHV-76-, MHV76-GFP-, and MHV76-K1-infected mice were also examined for the expression of GFP and immunostained for MHV-76 antigens at days 5 and 10 p.i. On day 5 p.i., the expression of GFP was clearly visible as foci of viral infection in the lungs of MHV76-K1-infected mice (Fig. 5C). Similarly, GFP expression was observed for MHV76-GFP-infected mice, but it was not observed for MHV-76-infected mice (data not shown). We noted that GFP expression was not observed in all foci of viral replication and that GFP expression was limited to cells that did not express the late viral antigens to which the antisera were directed. This may be due to the silencing of the CMV-IE promoter during productive viral replication. On day 10 p.i., there was no detectable GFP or viral antigen expression in any of the lung samples examined, consistent with the clearance of viral infection from the lungs at this time p.i. (data not shown).
The lungs of mice infected with MHV76-K1 were immunostained for expression of the K1 protein with an antibody that recognized a Myc epitope at the C terminus, but the protein was found to be below the level of detection of this technique (data not shown).
Long-term infected mice.
Groups of four BALB/c mice were infected with MHV-76, MHV76-GFP, or MHV76-K1 and sacrificed on day 120 p.i. Their organs (lungs, livers, spleens, kidneys, and blood) were removed for histopathological examination and an analysis of viral antigen expression. The livers of MHV76-K1-infected mice showed focal mixed zone 1 vasculitis/perivasculitis compared with a mild chronic periportal inflammation in mice infected with MHV-76 or MHV76-GFP (data not shown). No abnormalities were detected in the kidneys or spleens of infected mice (data not shown). An increased inflammatory response was observed in the lungs of MHV76-K1-infected mice at day 120 p.i. compared with those of both MHV-76- and MHV76-GFP-infected mice (data not shown).
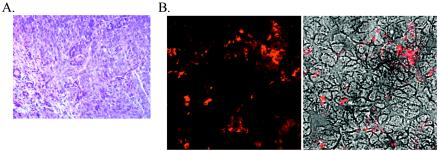
One of four mice infected with MHV76-K1 developed a salivary gland adenocarcinoma at day 120 p.i. (Fig. 6A). Immunostaining of the tumor with polyclonal anti-MHV-68 sera confirmed the presence of the virus (Fig. 6B). The tumor was also immunostained for expression of the K1 protein with an antibody that recognized a Myc epitope at the C terminus, but again the protein was found to be below the level of detection of this technique (data not shown).
FIG. 6.
(A) Hematoxylin and eosin staining of the salivary gland adenocarcinoma present in an MHV76-K1-infected mouse at day 120 p.i. (B) Expression of MHV-76 antigens in the salivary gland adenocarcinoma detected with rabbit polyclonal antisera. The left panel shows the immunofluorescent image, and the right panel shows a phase-contrast image merged with the immunofluorescent image.
Expression of K1 in MHV76-K1-infected mice.
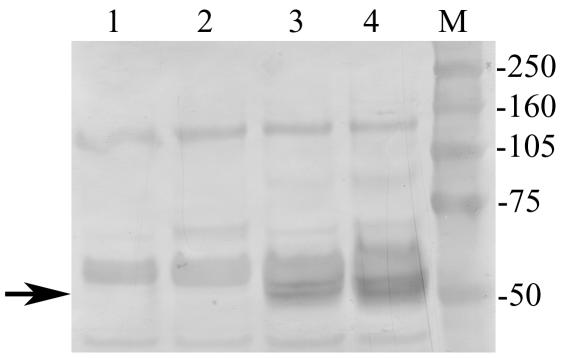
Although immunostaining of tissue sections for the expression of K1 proved negative, we were able to show the expression of K1 in MHV76-K1-infected mice by immunoprecipitation with a monoclonal antibody against the Myc epitope tag. The Western blot in Fig. 7 shows the presence of a 50-kDa protein in cell lysates from both the lung (day 5 p.i.) and salivary gland adenocarcinoma. The migration of this protein species was consistent with the predicted molecular mass of the glycosylated K1 protein (14).
FIG. 7.
Western blot of immunoprecipitated proteins showing the expression of Myc-tagged K1 in the lungs of MHV76-K1-infected mice at day 5 p.i. (lane 3) and in a salivary gland adenocarcinoma at day 120 p.i. (lane 4). K1 expression was not detected in either the lungs (lane 1) or salivary glands (lane 2) of MHV76-GFP-infected mice. The position of the K1 protein at ∼50 kDa is indicated with an arrow.
DISCUSSION
Investigations of the mechanisms through which KSHV is involved in tumorigenesis are hampered by the lack of useful animal model systems. We have made use of the well-characterized murine gammaherpesvirus MHV-76 to generate recombinants expressing both EGFP and the KSHV ORF K1. MHV-76 is a naturally occurring deletion variant which, similar to MHV-68, grows to high titers and forms plaques in monolayer cell cultures, thus facilitating the generation of virus recombinants (6, 17). The recombinant viruses were generated by homologous recombination at the left end of the unique portion of the MHV-76 genome and resulted in the insertion of a CMV-IE promoter expressing a bicistronic transcript. This transcript encoded EGFP, an IRES element, and the K1 gene. Recombinant viruses could be identified by virtue of their expression of EGFP, and the K1 gene was expressed through the IRES. The in vitro growth kinetics of viruses expressing EGFP alone (MHV76-GFP) or EGFP and K1 (MHV76-K1) were compared with those of MHV-76 and were found to be similar. However, after the infection of BALB/c mice by the intranasal route, we observed a 10-fold increase in the titer of MHV76-K1 compared with that of MHV76-GFP in the lungs. A histological examination of lung sections revealed a minor hyperproliferative response of the lung epithelial cells in response to the MHV76-K1 infection which appeared to be quantitatively different but qualitatively similar to that in response to MHV-76 infection. It is possible that this was due to the transient expression of the K1 gene promoting a proliferative response in the infected cell; however, further studies would be required to investigate this hypothesis further. One function of K1 during KSHV infection might therefore be to induce cellular proliferation and increase the pool of infected cells, which would be reflected in the increased titers observed in the lungs. Alternatively, it has been demonstrated that ITAM-dependent signaling by K1 can act to augment lytic reactivation in KSHV-infected B cells (13). The enhancement of lytic replication of the MHV76-K1 recombinant virus by K1 expression would also result in the observed increase in lung titers.
The increased replication of MHV76-K1 in the lungs (compared with MHV76-GFP) was not reflected in the number of latently infected cells in the spleen. The latent virus load peaked at day 10 p.i. but was found to be 10-fold lower for mice infected with MHV76-K1 than for mice infected with MHV-76. This may have been due to an increased immune response to the EGFP or K1 gene product or possibly to interference of the inserted DNA with latency determinants within the MHV-76 genome. This region of the genome is known to be important for the establishment of latency since MHV-76 has a 50-fold reduction in latent virus load compared with MHV-68 (17). Although the number of splenocytes that were latently infected with MHV76-K1 was low, it was approximately threefold higher than that for MHV76-EGFP-infected mice on days 10 and 14 p.i. (P < 0.001 on day 10 p.i.). We also observed a small increase (two- to threefold) in the number of splenocytes in mice infected with MHV76-K1 compared with that in mice infected with MHV-76 or MHV76-GFP (P < 0.05 on day 14 p.i.). Thus, as for productively infected cells, the function of K1 during infection might be to induce cellular proliferation, resulting in an expansion of the pool of infected B cells.
A salivary gland adenocarcinoma was observed in one of a group of four mice infected with MHV76-K1 at day 120 p.i. This is comparable with the low rate of tumor development (2 of 13 mice at 14 months) observed in transgenic mice expressing K1 under the control of the SV40 promoter (21A). Immunostaining of sections of the tumor with polyclonal MHV-68 antisera revealed the presence of viral antigens. Although we were unable to detect the expression of K1 in the tumor by immunostaining, K1 expression in the tumor was confirmed by immunoprecipitation and Western blot analysis (Fig. 7). Long-term infection with MHV-68 can result in the formation of lymphomas (26). However, we have never previously observed salivary gland tumors developing in mice infected with MHV-68 or MHV-76. These results suggest that K1 is a contributing factor in KSHV-induced tumors. Lee et al. (16) have reported the construction of a recombinant HVS in which STP was replaced with KSHV K1. This virus was shown to induce T-cell lymphomas in two of two common marmosets. Although the level of tumor induction in mice infected with MHV76-K1 was lower, this may reflect the lower natural tumorigenic potential of MHV-76 than that of HVS. A small proportion of mice infected with MHV-68 for >12 months or in the context of immunosuppression have been observed to develop B-cell lymphomas (26). Since HVS is an inherently more tumorigenic virus, we cannot discount the contribution of other viral genes to the tumor development observed with the HVS-K1 recombinant.
In conclusion, we have presented the development of a small-animal model for the study of the pathogenicity of KSHV-encoded gene products. The infection of mice with recombinant MHV-76 expressing KSHV K1 induced a transient hyperproliferation of lung epithelial cells and a salivary gland tumor in one of four infected mice.
Acknowledgments
This work was funded by an Association for Internal Cancer Research (AICR) grant to S.J.T. and J.P.S. and a Medical Research Council (MRC) career establishment grant to S.J.T. J.P.S. is a Royal Society University Research Fellow.
We thank L. Bieleski for help with statistical analysis.
REFERENCES
- 1.Beral, V., and R. Newton. 1998. Overview of the epidemiology of immunodeficiency-associated cancers. J. Natl. Cancer Inst. Monogr. 1998:1-6. [DOI] [PubMed] [Google Scholar]
- 2.Blackman, M. A., E. Flano, E. Usherwood, and D. L. Woodland. 2000. Murine gamma-herpesvirus-68: a mouse model for infectious mononucleosis? Mol. Med. Today 6:488-490. [DOI] [PubMed] [Google Scholar]
- 3.Blaskovic, D., M. Stancekova, J. Svobodova, and J. Mistrikova. 1980. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 24:468. [PubMed] [Google Scholar]
- 4.Boshoff, C., and R. A. Weiss. 2001. Epidemiology and pathogenesis of Kaposi's sarcoma-associated herpesvirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:517-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 6.Clambey, E. T., H. W. Virgin, and S. H. Speck. 2002. Characterization of a spontaneous 9.5-kilobase-deletion mutant of murine gammaherpesvirus 68 reveals tissue-specific genetic requirements for latency. J. Virol. 76:6532-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doherty, P. C., J. P. Christensen, G. T. Belz, P. G. Stevenson, and M. Y. Sangster. 2001. Dissecting the host response to a gamma-herpesvirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:581-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehtisham, S., N. P. Sunil-Chandra, and A. A. Nash. 1993. Pathogenesis of murine gammaherpesvirus infection in mice deficient in CD4 and CD8 T cells. J. Virol. 67:5247-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flano, E., S. M. Husain, J. T. Sample, D. L. Woodland, and M. A. Blackman. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J. Immunol. 165:1074-1081. [DOI] [PubMed] [Google Scholar]
- 10.Husain, S. M., E. J. Usherwood, H. Dyson, C. Coleclough, M. A. Coppola, D. L. Woodland, M. A. Blackman, J. P. Stewart, and J. T. Sample. 1999. Murine gammaherpesvirus M2 gene is latency-associated and its protein a target for CD8(+) T lymphocytes. Proc. Natl. Acad. Sci. USA 96:7508-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacoby, M. A., H. W. Virgin, and S. H. Speck. 2002. Disruption of the M2 gene of murine gammaherpesvirus 68 alters splenic latency following intranasal, but not intraperitoneal, inoculation. J. Virol. 76:1790-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagunoff, M., and D. Ganem. 1997. The structure and coding organization of the genomic termini of Kaposi's sarcoma-associated herpesvirus. Virology 236:147-154. [DOI] [PubMed] [Google Scholar]
- 13.Lagunoff, M., D. M. Lukac, and D. Ganem. 2001. Immunoreceptor tyrosine-based activation motif-dependent signaling by Kaposi's sarcoma-associated herpesvirus K1 protein: effects on lytic viral replication. J. Virol. 75:5891-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagunoff, M., R. Majeti, A. Weiss, and D. Ganem. 1999. Deregulated signal transduction by the K1 gene product of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 96:5704-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, H., J. Guo, M. Li, J. K. Choi, M. DeMaria, M. Rosenzweig, and J. U. Jung. 1998. Identification of an immunoreceptor tyrosine-based activation motif of K1 transforming protein of Kaposi's sarcoma-associated herpesvirus. Mol. Cell. Biol. 18:5219-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, H., R. Veazey, K. Williams, M. Li, J. Guo, F. Neipel, B. Fleckenstein, A. Lackner, R. C. Desrosiers, and J. U. Jung. 1998. Deregulation of cell growth by the K1 gene of Kaposi's sarcoma-associated herpesvirus. Nat. Med. 4:435-440. [DOI] [PubMed] [Google Scholar]
- 17.Macrae, A. I., B. M. Dutia, S. Milligan, D. G. Brownstein, D. J. Allen, J. Mistrikova, A. J. Davison, A. A. Nash, and J. P. Stewart. 2001. Analysis of a novel strain of murine gammaherpesvirus reveals a genomic locus important for acute pathogenesis. J. Virol. 75:5315-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macrae, A. I., E. J. Usherwood, S. M. Husain, E. Flano, I. J. Kim, D. L. Woodland, A. A. Nash, M. A. Blackman, J. T. Sample, and J. P. Stewart. 2003. Murid herpesvirus 4 strain 68 M2 protein is a B-cell-associated antigen important for latency but not lymphocytosis. J. Virol. 77:9700-9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. 2001. Natural history of murine gamma-herpesvirus infection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parry, C. M., J. P. Simas, V. P. Smith, C. A. Stewart, A. C. Minson, S. Efstathiou, and A. Alcami. 2000. A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J. Exp. Med. 191:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Prakash, O., Z. Y. Tang, X. Peng, R. Coleman, J. Gill, G. Farr, and F. Samaniego. 2002. Tumorigenesis and aberrant signaling in transgenic mice expressing the human herpesvirus-8 K1 gene. J. Natl. Cancer Inst. 94:926-935. [DOI] [PubMed] [Google Scholar]
- 22.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevenson, P. G., J. M. Boname, B. de Lima, and S. Efstathiou. 2002. A battle for survival: immune control and immune evasion in murine gamma-herpesvirus-68 infection. Microbes Infect. 4:1177-1182. [DOI] [PubMed] [Google Scholar]
- 24.Stewart, J. P. 1999. Of mice and men: murine gammaherpesvirus 68 as a model. Epstein-Barr Virus Rep. 6:31-35. [Google Scholar]
- 25.Stewart, J. P., E. J. Usherwood, A. Ross, H. Dyson, and T. Nash. 1998. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J. Exp. Med. 187:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sunil-Chandra, N. P., J. Arno, J. Fazakerley, and A. A. Nash. 1994. Lymphoproliferative disease in mice infected with murine gammaherpesvirus 68. Am. J. Pathol. 145:818-826. [PMC free article] [PubMed] [Google Scholar]
- 27.Sunil-Chandra, N. P., S. Efstathiou, J. Arno, and A. A. Nash. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 73:2347-2356. [DOI] [PubMed] [Google Scholar]
- 28.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 29.Usherwood, E. J., A. J. Ross, D. J. Allen, and A. A. Nash. 1996. Murine gammaherpesvirus-induced splenomegaly: a critical role for CD4 T cells. J. Gen. Virol. 77:627-630. [DOI] [PubMed] [Google Scholar]
- 30.Usherwood, E. J., J. P. Stewart, K. Robertson, D. J. Allen, and A. A. Nash. 1996. Absence of splenic latency in murine gammaherpesvirus 68-infected B cell-deficient mice. J. Gen. Virol. 77:2819-2825. [DOI] [PubMed] [Google Scholar]
- 31.van Berkel, V., K. Preiter, H. W. Virgin IV, and S. H. Speck. 1999. Identification and initial characterization of the murine gammaherpesvirus 68 gene M3, encoding an abundantly secreted protein. J. Virol. 73:4524-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Virgin, H. W., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Virgin, H. W., and S. H. Speck. 1999. Unraveling immunity to gamma-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr. Opin. Immunol. 11:371-379. [DOI] [PubMed] [Google Scholar]
- 34.Weck, K. E., S. S. Kim, H. W. Virgin IV, and S. H. Speck. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol. 73:3273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willer, D. O., and S. H. Speck. 2003. Long-term latent murine gammaherpesvirus 68 infection is preferentially found within the surface immunoglobulin D-negative subset of splenic B cells in vivo. J. Virol. 77:8310-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zong, J. C., D. M. Ciufo, D. J. Alcendor, X. Wan, J. Nicholas, P. J. Browning, P. L. Rady, S. K. Tyring, J. M. Orenstein, C. S. Rabkin, I. J. Su, K. F. Powell, M. Croxson, K. E. Foreman, B. J. Nickoloff, S. Alkan, and G. S. Hayward. 1999. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi's sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J. Virol. 73:4156-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]