Abstract
The late region of human neurotropic JC virus encodes a small 71-amino-acid agnoprotein that is also found in the polyomaviruses simian virus 40 and BK virus. Several functions of agnoprotein have been identified, including roles in regulating viral transcription and virion maturation. Earlier studies showed that agnoprotein expressed alone induced p21/WAF-1 expression and caused cells to accumulate in the G2/M stage of the cell cycle. Here we report that agnoprotein expression sensitized cells to the cytotoxic effects of the DNA-damaging agent cisplatin. Agnoprotein reduced the viability of cisplatin-treated cells and increased chromosome fragmentation and micronucleus formation. Whereas cisplatin-treated control cells accumulated in S phase, cells expressing agnoprotein did not, instead becoming aneuploid. Agnoprotein expression correlated with impaired double-strand-break repair activity in cellular extracts and reduced expression of the Ku70 and Ku80 DNA repair proteins. After agnoprotein expression, much of the Ku70 protein was located in the perinuclear space, where agnoprotein was also found. Results from binding studies showed an interaction of agnoprotein with Ku70 which was mediated by the N terminus. The ability of agnoprotein to inhibit double-strand break repair activity when it was added to cellular extracts was also mediated by the N terminus. We conclude that agnoprotein inhibits DNA repair after DNA damage and interferes with DNA damage-induced cell cycle regulation. Since Ku70 is a subunit of the DNA-dependent protein kinase that is responsible both for double-strand break repair and for signaling damage-induced cell cycle arrest, the modulation of Ku70 and/or Ku80 by agnoprotein may represent an important event in the polyomavirus life cycle and in cell transformation.
JC virus (JCV) is a human polyomavirus that was first isolated from the brain of a patient suffering from progressive multifocal leukoencephalopathy (PML) (26). Polyomavirus is a genus of nonenveloped DNA viruses with icosahedral capsids containing small, circular, double-stranded DNA genomes (4). JCV is the etiologic agent of PML. The virus is widespread throughout the population, with >80% of adults exhibiting JCV-specific antibodies. Infection is thought to take place during early childhood and is usually subclinical. However under conditions of immunosuppression, e.g., in patients with AIDS, JCV can emerge from latency to cause PML (12, 30). PML is a neurodegenerative disease of the central nervous system that is characterized by multiple regions of demyelination caused by a lytic infection of oligodendrocytes by JCV. The destruction of myelin-producing cells leads to brain lesions and death (30). The occurrence of PML was very rare until the advent of the AIDS pandemic, but now it is much more prevalent and affects ∼5% of human immunodeficiency virus-infected persons (2). JCV is one of the few opportunistic infections that continues to occur with some frequency in patients with AIDS despite the widespread use of highly active antiretroviral therapy (2, 3).
JCV can transform cells in culture and is oncogenic in laboratory animals (8, 19). The transforming ability of JCV appears to be limited to specific cell types, particularly those of neural origin, and this property maps to the noncoding regulatory sequence at the origin of DNA replication. JCV DNA sequences have been detected in several kinds of human cancer, including glial tumors (7), medulloblastoma (9), and colon cancer (10). The role of JCV in human malignancies has been reviewed recently (8, 19).
The genome of JCV is organized in a similar fashion to that of the other two primate polyomaviruses, simian virus 40 (SV40) and BK virus (BKV). The double-stranded circular DNA of JCV contains the following three functional regions: the early and late coding genes and the noncoding regulatory sequence (4). The early region encodes the large T and small t antigens, while the late region encodes the viral capsid proteins VP1, VP2, and VP3 and a small regulatory protein known as agnoprotein encoded near the 5′ end of the primary late transcript. Agnoprotein is produced late in the infectious cycle, although it is not incorporated into virions (4, 16). In cells infected by JCV, the 8-kDa agnoprotein is found mainly in the cytoplasm, especially in the perinuclear region, while a small amount may also be found in the nucleus (25). A similar localization was observed in cells transfected with a plasmid encoding agnoprotein (6). Agnoprotein may have regulatory roles in viral transcription and translation as well as in virion assembly and maturation, and these roles were reviewed recently (30). JCV agnoprotein can interact with the large T antigen and can downregulate viral gene expression and DNA replication (28). It also interacts with YB-1, a cellular transcription factor that contributes to JCV gene expression in glial cells, and negatively regulates YB-1-mediated JCV gene transcription (29, 31).
Previous studies demonstrated that the expression of JCV agnoprotein dysregulates cell cycle progression in the absence of other viral proteins (6). NIH 3T3 mouse fibroblasts that constitutively expressed JCV agnoprotein accumulated at the G2/M stage of the cell cycle, and a decline in cyclin A- and B-associated kinase activity was observed in these cells. Agnoprotein showed the ability to augment the activity of the p21/WAF-1 promoter and increased the level of p21/WAF-1 protein in cells. In addition, agnoprotein was shown to bind p53. The activation of p21/WAF-1 gene expression in cells expressing agnoprotein may be mediated, at least in part, through cooperation with p53 (6). Results for a p53 null cell line revealed that agnoprotein could induce p21/WAF-1 transcription, but to a much lesser extent than in p53-expressing cells, indicating the existence of a p53-independent mechanism for p21/WAF-1 activation by agnoprotein (6).
Since cell cycle progression is linked to the process of DNA repair through p53, we were interested in looking for a possible effect of agnoprotein in the response of cells to DNA damage. DNA damage was induced by the treatment of cultured cells with the cytotoxic antitumor drug cisplatin, which interacts with DNA to form DNA adducts, primarily intrastrand cross-linked adducts, which activate signaling pathways that arrest the cell cycle, including the p53 pathway (33). Data will be presented indicating that agnoprotein expression renders cells more prone to the effects of DNA damage and less able to signal cell cycle arrest.
MATERIALS AND METHODS
Cell culture, transfection, and plasmids.
NIH 3T3 cells expressing JCV agnoprotein (agnopositive) and control agnonegative NIH 3T3 cell lines have been described previously (6). These cells were stably transfected with a plasmid vector expressing agnoprotein from the cytomegalovirus (CMV) promoter (pCMV-agnoprotein) (28) or with an empty vector, respectively. Three independently isolated clonal cell lines that express agnoprotein were used for these experiments, and they were transfected in parallel with cultures that received the empty vector. All cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. U-87MG human glioblastoma cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and were transfected by the calcium phosphate precipitation method (13) with the pCMV-agnoprotein plasmid. Plasmids that express glutathione S-transferase (GST)-agnoprotein and GST-agnoprotein deletion mutants in bacteria have been described previously (28).
Antibodies.
Rabbit anti-Ku70 (AHP316) and rabbit anti-Ku80 (AHP317) antibodies were obtained from Serotec (Raleigh, N.C.), a mouse monoclonal anti-GRB2 antibody was obtained from BD Transduction laboratories (Lexington, Ky.), and a mouse monoclonal anti-Rad51 antibody (clone 3C10) was obtained from Upstate Biotechnology (Lake Placid, N.Y.). Members of our laboratory previously described a rabbit polyclonal antibody against JCV agnoprotein (9).
Cell viability assay.
NIH 3T3 cells expressing JCV agnoprotein and control agnonegative NIH 3T3 cells were plated in 100-mm-diameter dishes equally in triplicate in two sets (15,000 cells/dish). One set for each cell line was treated with 0.5 μg of cisplatin/ml in growth medium for 5 h and washed with saline, and the cell viability was evaluated by trypan blue exclusion. The percentage of cells in the treated set relative to the untreated set was determined for each cell line 4 and 5 days after treatment.
Clonogenic assay.
NIH 3T3 cells expressing JCV agnoprotein and control agnonegative NIH 3T3 cells were plated and treated with or without 0.5 μg of cisplatin/ml as described for the viability assay. After treatment, the cells were grown for 10 to 14 days, fixed, and stained with methylene blue. The plates were then scanned with an Epson Perfection 1650 scanner.
Metaphase spreads.
Cells were treated with 1.5 μg of cisplatin/ml for 18 h or were left without treatment. Two hours prior to the harvesting of cells, 100 ng of Colcemid/ml was added to each flask. After mitotic arrest with Colcemid, the cells were harvested, treated with a hypotonic solution of 0.075 M KCl, and fixed with methanol-acetic acid (3:1). The cell suspension was dropped onto a glass slide and allowed to air dry. The slides were stained with 0.4% Giemsa stain solution (Sigma, St. Louis, Mo.), soaked in xylene, and mounted in SP15-100 Permount (Fisher, Pittsburgh, Pa.). Photomicrographs were taken under an Olympus light microscope with a magnification of ×700.
Micronucleus formation assay.
Micronucleus formation was assayed by an adaptation of published methods (17, 18). Agnopositive and agnonegative cells were treated with 0, 0.125, or 0.25 μg of cisplatin/ml for 18 h. After the cisplatin was removed, cytochalasin B (1.2 μg/ml) was added to block cytoplasmic, but not nuclear, division. After 24 h, the plates were washed in 0.9% NaCl, fixed in methanol, allowed to air dry, and stained with 0.4% Giemsa stain for 15 min. The proportion of binucleated cells with micronuclei was determined. For each plate, 300 binucleated cells were scored as being either positive or negative for micronuclei. Cells containing one to four micronuclei were scored as positive. Cells with five or more micronuclei usually showed nuclear fragmentation and were excluded from the analysis. This was in accord with published criteria (11). A binucleated cell was defined as a cell with an intact cytoplasm and two nuclei of equal size. Micronuclei were defined as nonrefractile bodies that did not touch a nucleus and were morphologically identical to, but smaller than, the normal nuclei.
FACS analysis.
Cells were left untreated or were treated with 1.5 μg of cisplatin/ml for 18 h. The cells were harvested by centrifugation, washed with phosphate-buffered saline (PBS), and fixed in ice-cold 73% ethanol. After incubation for 24 h at −20°C, the cells were washed with PBS containing 1% bovine serum albumin, stained with propidium iodide (10 μg/ml) in PBS containing 250 μg of RNase A/ml, and incubated at 37°C for 30 min in the dark before analysis by fluorescence-activated cell sorting (FACS). Flow cytometry was performed with a Becton Dickinson FACScan flow cytometer. Data were analyzed with ModFitLT, v 2.0 (PMAC), software.
Nonhomologous end-joining (NHEJ) assay for double-strand break (DSB) DNA repair.
Cells were left untreated or were treated with 1 μg of cisplatin/ml for 32 h. Nuclear extracts were prepared as follows. Cells were harvested, washed three times with ice-cold PBS, and resuspended in ice-cold hypotonic lysis buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, and protease inhibitors). After 10 min on ice, the cells were mixed up and down with a pipette and cell lysis was verified by phase-contrast microscopy. Lysates were centrifuged for 5 min, and the cell pellets were resuspended in lysis buffer and placed on ice for 10 min. After centrifugation, the cell pellets were resuspended in ice-cold hypertonic nuclear lysis buffer (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 500 mM NaCl, 25% glycerol [vol/vol], 0.2 mM EDTA, and protease inhibitors). After 1 h on ice and three rapid cycles of freezing-thawing (with liquid nitrogen and a 37°C water bath), the nuclear lysates were cleared by centrifugation. The supernatants were dialyzed for 10 h against a buffer containing 25 mM Tris HCl (pH 7.5), l mM EDTA, 10% glycerol (vol/vol), and protease inhibitors.
NHEJ activity assays were performed by a modification of the method of Baumann and West (1). The DNA substrate for the reaction was made by cutting the vector plasmid pBlueScript II(SK) with the restriction enzymes SmaI and XhoI. The resulting 3-kb linear DNA fragment was purified in an agarose gel. End-joining reaction mixtures consisted of 25 mM Tris acetate (pH 7.5), 100 mM potassium acetate, 10 mM magnesium acetate, 1 mM dithiothreitol, 2 mM ATP, and a 200 μM concentration of each deoxynucleoside triphosphate. Nuclear extracts (50 μg) were incubated for 5 min at 37°C before the addition of the DNA substrate (400 ng). After incubation for 1 h at 37°C, the DNA products were deproteinized by treatment with 100 μg of proteinase K/ml and were analyzed by electrophoresis through a 0.7% agarose gel. For end-joining reactions containing recombinant proteins, GST fusion proteins (1 μM final concentration) were added to the reaction before the addition of DNA.
GST pull-down assays and Western blotting.
GST pull-down assays were performed as previously described (6, 28). Ten micrograms of either GST alone, GST-agnoprotein, or a deletion mutant immobilized on Sepharose beads was incubated with whole-cell extracts prepared from U-87MG cells. The beads were washed extensively with lysis buffer and boiled in Laemmli sample buffer, and the proteins were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) followed by Western blot analysis using an antibody against the Ku70 protein. Western blotting was performed as previously described (6). For Western blots with total cell protein, 50 μg of protein was resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted with a rabbit anti-Ku70 antibody. The primary antibody was used at a dilution of 1/5,000, and bound antibody was detected with an ECL detection kit (Amersham, Arlington Heights, Ill.) used according to the manufacturer's recommendations. The relative intensities of bands on autoradiographs were measured by densitometry in a Bio-Rad FX molecular imager. In the case of the Ku70 GST pull-down assay, the relative binding of Ku70 to each of the agnoprotein mutants was calculated as follows. The intensity of the GST-alone band (negative control) was subtracted from the intensity of each of the experimental lanes. The ratio of the intensity of each of the bands to that of the positive-control full-length agnoprotein band (residues 1 to 71) was calculated and expressed as a percentage.
Immunofluorescence.
U-87MG human glioblastoma cells were transfected with the pCMV-agnoprotein plasmid or with a control empty vector backbone plasmid as described above. After 16 h, the cells were seeded on polylysine-coated glass chamber slides at a low density and were treated with 1.0 μg of cisplatin/ml for 16 h. The cells were then fixed with cold acetone. Fixed cells were blocked with 5% bovine serum albumin in PBS for 2 h and incubated with an anti-agnoprotein rabbit polyclonal primary antibody for 1 h. The cells were then washed three times with PBS-0.01% Tween 20 at 10-min intervals and were incubated with an anti-rabbit secondary antibody labeled with Texas Red for 45 min. The cells were then blocked again with 5% bovine serum albumin in PBS for 2 h and incubated with anti-Ku70 rabbit polyclonal primary antibodies for 1 h, after which the cells were washed three times with PBS-0.01% Tween 20 at 10-min intervals and incubated with an anti-rabbit fluorescein isothiocyanate-conjugated secondary antibody for 45 min. Finally, the slides were washed three times with PBS, mounted, and examined by fluorescence microscopy.
GST fusion proteins.
The expression and purification of the GST-agnoprotein fusion protein and its deletion mutants have been described previously (6, 28). Briefly, the fusion protein constructs were transformed into Escherichia coli DH5α cells which were cultured and induced with isopropyl-β-d-thiogalactopyranoside. The fusion proteins were purified after lysis and sonication with glutathione-Sepharose beads.
RESULTS
Effect of cisplatin treatment on survival and clonogenic ability of agnopositive and agnonegative cells.
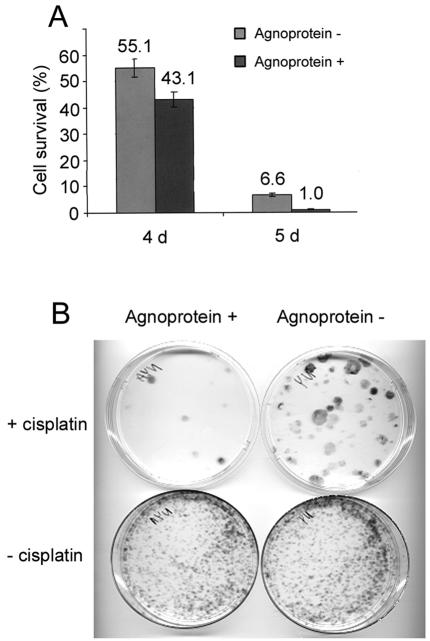
In our first experiments, we investigated whether cells expressing the JCV agnoprotein become more vulnerable to the DNA-damaging agent cisplatin. The ability of cells to survive after treatment with cisplatin was determined by a cell viability assay. Exponentially growing NIH 3T3 cells expressing JCV agnoprotein (agnopositive cells) and parallel cultures without agnoprotein (agnonegative cells) were treated with 0.5 μg of cisplatin/ml in growth medium for 5 h. The cells were counted 4 and 5 days after treatment and the numbers of cells were compared to those for untreated controls to calculate percentages of survival (Fig. 1A).
FIG. 1.
Effect of cisplatin treatment on survival and clonogenic ability of agnopositive and agnonegative cells. (A) Cell viability. NIH 3T3 cells expressing JCV agnoprotein and control agnonegative NIH 3T3 cells were treated with cisplatin as described in Materials and Methods. Sets of triplicates were done for each cell line under each condition (treated and untreated). Cell viability for each cell line was evaluated by counting the cells after 4 and 5 days of treatment. (B) Clonogenic ability. Cells were treated with or without cisplatin as described for panel A. After treatment, the cells were grown for 10 to 14 days, fixed, and stained with methylene blue.
Cell loss was observed in all cisplatin-treated cultures, but more so for the agnoprotein-expressing cells. After 5 days, there were more than six times as many surviving cisplatin-treated agnonegative cells than cisplatin-treated agnopositive cells. This experiment was performed on three independent occasions with independently isolated agnoprotein-expressing clonal cell lines, with similar results obtained each time.
To further investigate the effect of agnoprotein on cell survival, we performed a clonogenic assay. Agnopositive and agnonegative cells were plated as described above and treated with 0.5 μg of cisplatin/ml for 5 h. After treatment, the cisplatin was removed and the cells were cultured for 10 to 14 days, fixed, and stained with methylene blue (Fig. 1B). The number of colonies that formed after cisplatin treatment was much lower in plates with agnopositive cells than in those with agnonegative cells.
Induction of chromosome breaks and micronucleus formation after cisplatin treatment in agnopositive and agnonegative cell lines.
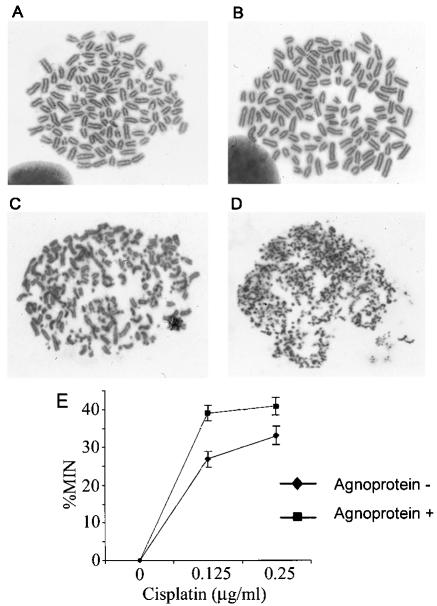
To find direct evidence of DNA damage, we analyzed metaphase spreads from cells treated with cisplatin to look for the presence of chromosome breaks. Agnopositive and agnonegative cell lines were plated and cultured in regular growth medium and then treated with 1.5 μg of cisplatin/ml for 18 h. For each cell line, a control group was left untreated. The cells were treated with Colcemid to induce mitotic arrest, harvested, and fixed, and metaphase spreads were examined as described in Materials and Methods (Fig. 2A to D). Non-cisplatin-treated cells exhibited a near tetraploid distribution. No chromatid or chromosomal gaps, breaks, rejoining, or complex configurations were noted (Fig. 2A and B). In the metaphase spreads from cisplatin-treated agnonegative cells, there was some degree of chromosomal breakage and rejoining, fragmentation, and complex configuration types of chromosomal aberrations (Fig. 2C). However, in cisplatin-treated cells expressing agnoprotein, extensive chromosomal structural abnormalities were found. Remarkably, virtually every chromosome was fragmented into many pulverized subchromosomal pieces (Fig. 2D). This experiment was performed twice with independently isolated agnoprotein-expressing clonal cell lines.
FIG. 2.
Induction of chromosome breaks and micronucleus formation after cisplatin treatment. NIH 3T3 cells expressing JCV agnoprotein and control agnonegative NIH 3T3 cells were treated with cisplatin, and metaphase spreads were prepared as described in Materials and Methods. All photographs were taken at a magnification of ×700. (A) Agnonegative, untreated cells; (B) agnopositive, untreated cells; (C) agnonegative, cisplatin-treated cells; (D) agnopositive, cisplatin-treated cells. (E) Agnopositive and agnonegative cells were treated with 0, 0.125, or 0.25 μg of cisplatin/ml, and micronucleus (MIN) formation was measured as described in Materials and Methods.
To further analyze DNA damage in these cells, we performed a micronucleus formation assay. This assay allows the detection of micronuclei that arise from chromosomal fragments that are not incorporated into daughter nuclei at mitosis and thus reflect genomic disruption (14, 17, 18). Agnopositive and agnonegative NIH 3T3 cells were treated with 0, 0.125, or 0.25 μg of cisplatin/ml for 18 h. After cisplatin removal, cytochalasin B was added to block cytoplasmic, but not nuclear, division. The proportion of binucleated cells with micronuclei was determined as described in Materials and Methods (Fig. 2E). After cisplatin treatment, micronuclei were formed more frequently in agnopositive cells, indicating a higher rate of chromosomal damage.
Accumulation of cells with aberrant DNA content after cisplatin treatment of agnopositive and agnonegative cell lines.
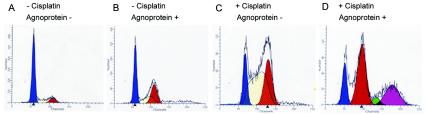
Cells with damaged DNA may delay the G2 phase to allow time for DNA repair. Previous studies have demonstrated that the expression of JCV agnoprotein dysregulates cell cycle progression. NIH 3T3 cells that constitutively expressed agnoprotein accumulated at the G2/M stage of the cell cycle (6). Thus, we were interested in analyzing whether agnoprotein affected the passage of cells through the cell cycle in response to cisplatin. FACS analysis was performed on agnopositive and agnonegative NIH 3T3 cells treated with 1.5 μg of cisplatin/ml for 18 h. Control groups were left untreated. The cells were stained with propidium iodide for analysis by flow cytometry (Fig. 3). For each FACS analysis, the relative numbers of diploid and aneuploid cells, as well as the percentages of diploid cells that were in the G0/G1, S, and G2/M phases of the cell cycle (Table 1), were determined. In the absence of cisplatin, most of the agnonegative cells were in G0/G1, with only 10% in G2/M, whereas 30% of the agnopositive cells were in G2/M, in concordance with earlier results (6). For cisplatin-treated, agnonegative cells, 50% of the cells were in S phase (Fig. 3C, yellow peak), with 32% in G2/M (orange peak), suggesting that the initiation of DNA repair had induced gatekeeper signaling that prevented cell division. No aneuploid cells were observed. For cisplatin-treated, agnopositive cells, 32% were aneuploid (Fig. 3D, purple peak), providing more evidence of a defect in DNA repair in these cells. It is unlikely that the purple peak represents tetraploid or polyploid cells given the extent of chromosome damage that is caused by cisplatin (Fig. 2D).
FIG. 3.
Accumulation of cells with aberrant DNA content after cisplatin treatment of agnopositive and agnonegative cell lines. Agnopositive and agnonegative cells were left untreated or treated with cisplatin and subject to FACS analysis as described in Materials and Methods. (A) Agnonegative, untreated cells; (B) agnopositive, untreated cells; (C) agnonegative, cisplatin-treated cells; (D) agnopositive, cisplatin-treated cells.
TABLE 1.
Cell cycle analysis of cisplatin-treated and untreated agnopositive and agnonegative cell lines
| Agnoprotein status | Cisplatin treatment | % Aneuploid cells | % Diploid cells | % Diploid cells in indicated phase
|
||
|---|---|---|---|---|---|---|
| G0/G1 | G2/M | S | ||||
| − | − | 0 | 100 | 74 | 10 | 16 |
| + | − | 0 | 100 | 49 | 32 | 21 |
| − | + | 0 | 100 | 19 | 32 | 50 |
| + | + | 32 | 68 | 27 | 73 | NDa |
ND, not determined.
Nuclear extract from agnopositive cells treated with cisplatin has reduced end-joining activity in NHEJ assay for DSB DNA repair.
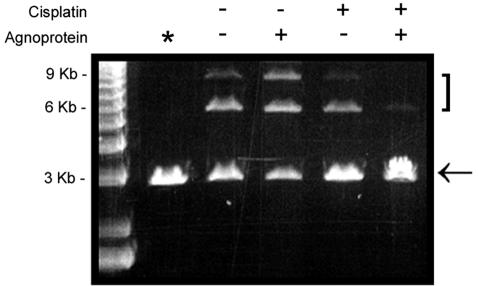
The two major pathways of DSB repair are NHEJ and homologous recombination (5, 15). In mammalian cells, the dominant mechanism of DNA repair is NHEJ, by which double-stranded DNA ends can be rejoined even if there is little or no base pairing at the site of the breaks. To assess the effect of agnoprotein on DSB repair after cisplatin treatment, we prepared nuclear extracts from cisplatin-treated (1 μg/ml) agnopositive and agnonegative cells and assayed them for NHEJ activity as described in Materials and Methods (Fig. 4). End joining of the 3-kb linearized double-stranded plasmid DNA was much reduced in the cisplatin-treated agnopositive cells, as evidenced by the lesser intensity of the 6-kb dimer band and the disappearance of the 9-kb trimer band (last lane). The NHEJ assay was performed multiple times with different DNA substrates and with nuclear extracts from two different independently isolated agnoprotein-expressing clonal cell lines.
FIG. 4.
NHEJ assay for DSB DNA repair. Agnopositive and agnonegative cells were left untreated or treated with cisplatin, and nuclear extracts were prepared as described in Materials and Methods. These were assayed by an NHEJ assay with a linear restriction-digested 3-kb plasmid. Lane *, control with no nuclear extract added. The arrow indicates the 3-kb monomeric linear plasmid substrate band. The 6- and 9-kb bands are the products of end joining and are indicated by a bracket.
Expression of NHEJ DNA repair proteins is downregulated in agnopositive cells.
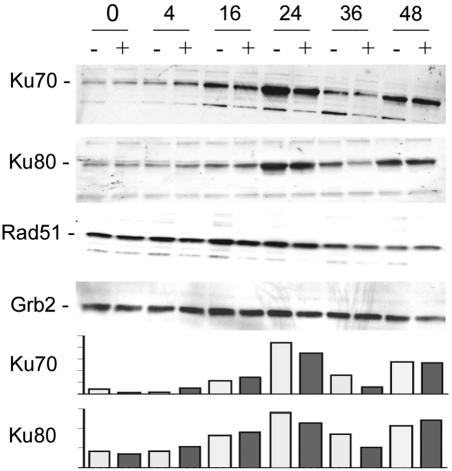
To investigate possible molecular mechanisms underlying the decrease in DSB repair activity in agnopositive cells, we examined the expression levels of Rad51, Ku70, and Ku80, which are key proteins involved in NHEJ DNA DSB repair. We found that the expression levels of Ku70 and Ku80 in cells that were negative for agnoprotein were noticeably higher than those in agnopositive cells after 24 and 36 h of cisplatin treatment, as determined by Western blot analysis (Fig. 5). These differences were verified by quantifying the intensities of the Ku70 and Ku80 bands with a densitometer (bottom panels). This experiment was performed twice, with the same results. The levels of Rad51 showed no differences. The levels of GRB2 protein served as a control for protein loading in this experiment. This Western blot analysis was performed twice with two independently isolated agnoprotein-expressing clonal cell lines.
FIG. 5.
Expression of NHEJ DNA repair proteins. Agnopositive (+) and agnonegative (−) cells were treated with cisplatin for 0, 4, 16, 24, 36, or 48 h, as indicated. Proteins were extracted and Western blotted with antibodies to Ku70, Ku80, Rad51, and Grb2 (loading control). The intensities of the Ku70 and Ku80 bands were measured by densitometry, and these data are shown in the lower panels.
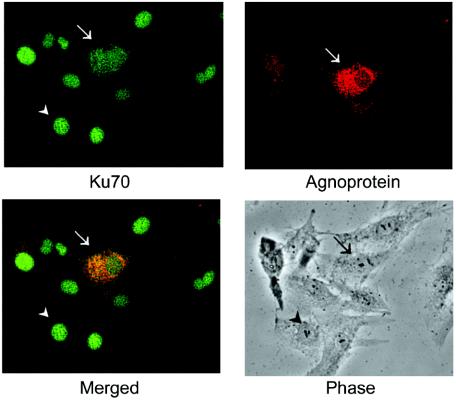
Agnoprotein expression is correlated with the appearance of Ku70 in the cytoplasm, where it colocalizes with agnoprotein in the perinuclear region.
U-87MG cells were transiently transfected with a plasmid expressing JCV agnoprotein from the CMV promoter. The cells were then treated with 1 μg of cisplatin/ml for 18 h and were double-stained for Ku70 (green) and agnoprotein (red), as shown in Fig. 6. This allowed for a direct observation of Ku70 subcellular localization in agnopositive cells (transfected) and agnonegative cells (untransfected) in the same culture. The cell in the center of the field that is indicated by an arrow is agnopositive. It shows bright red staining for agnoprotein that is predominantly perinuclear, as expected (6, 25). The other cells in the field, e.g., the cell indicated by the arrowhead, are agnonegative. These cells were untransfected and did not stain for agnoprotein. All cells show expression of Ku70 (green). In the untransfected cells, this staining is within the nucleus. However, in agnopositive cells, the staining is largely perinuclear. These data suggest that agnoprotein can sequester a significant proportion of cellular Ku70 in the perinuclear space.
FIG. 6.
Subcellular localization of agnoprotein and Ku70. U-87MG cells were transiently transfected with CMV-agnoprotein. The cells were then treated with 1 μg of cisplatin/ml for 18 h and double stained for Ku70 (green) and agnoprotein (red) as indicated. Arrows indicate an agnopositive cell; arrowheads indicate an agnonegative cell.
Functional and physical interaction of Ku70 and agnoprotein.
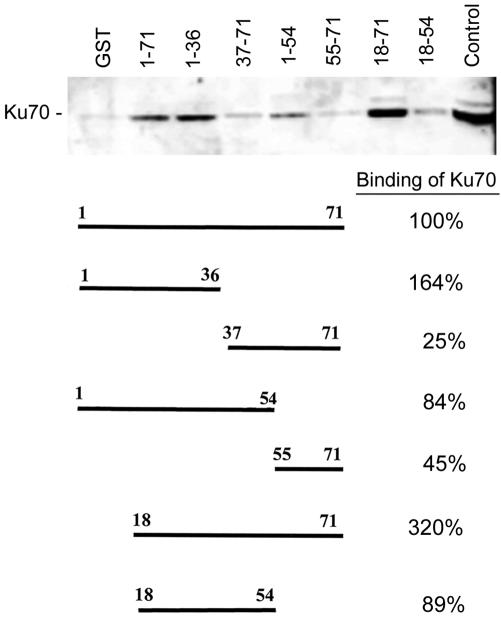
To determine if Ku70 and the JCV agnoprotein could directly interact, we performed binding assays with protein extracts from U-87MG human glioblastoma cells that were negative for agnoprotein. The protein extracts were incubated with GST, GST-agnoprotein, or GST-agnoprotein deletion mutants. Protein complexes were isolated by the use of glutathione-Sepharose 4B beads as described in Materials and Methods and were resolved by SDS-PAGE followed by Western blotting with an anti-Ku70 antibody (Fig. 7). The full-length agnoprotein (residues 1 to 71) bound Ku70 strongly. Deletion mutants of agnoprotein containing N-terminal residues 1 to 55 or 1 to 36 bound Ku70 well, whereas those containing residues 37 to 71 and 55 to 71 did not, suggesting that the Ku70-binding domain is in the N terminus of agnoprotein. The binding of the mutants containing residues 18 to 71 and 18 to 54 indicates that the region from residues 18 to 36 is involved.
FIG. 7.
Analysis of agnoprotein-Ku70 interaction by using GST-agnoprotein fusion proteins. To see if Ku70 and the JCV agnoprotein could directly interact, we performed binding assays with protein extracts from U-87MG human glioblastoma cells that were negative for agnoprotein. Protein complexes were isolated by using glutathione-Sepharose 4B beads as described in Materials and Methods and were resolved by SDS-PAGE, followed by Western blotting with an anti-Ku70 antibody. The top panel shows a Western blot of the Ku70 that was pulled down by each of the GST-agnoprotein deletion mutant fusion proteins. The structure of each deletion mutant and its relative percentage of Ku70 binding are shown in the bottom panel. The lane on the far left is a negative control with GST only; the lane on the far right is a positive control with an unfractionated U-87MG cell extract. The relative intensities of the bands on the autoradiograph were measured by densitometry and the relative binding of Ku70 to each of the agnoprotein mutants was calculated as follows. The intensity of the GST-alone band (negative control) was subtracted from the intensity of each of the experimental lanes. The ratio of each of the bands relative to the full-length positive-control lane (1-71) was calculated and expressed as a percentage.
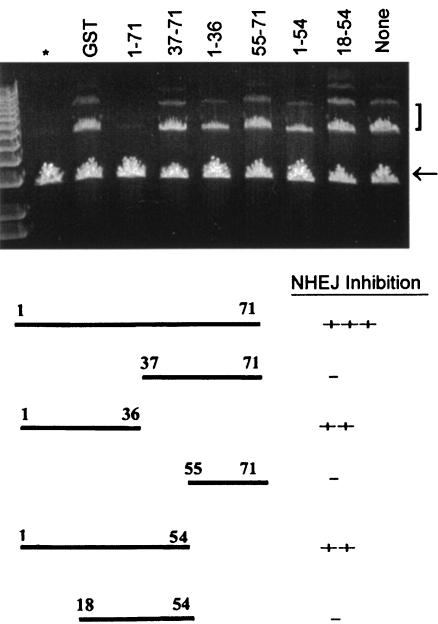
The effects of the addition of the full-length protein and of C-terminal deletion mutants of agnoprotein on the end-joining activity of agnonegative cell extracts were examined as follows. Nuclear extracts from agnonegative NIH 3T3 cells were prepared. The extracts were incubated with GST alone or with the various GST-agnoprotein fusion proteins (the full-length protein and various deletion mutants) and were assayed for NHEJ activity (Fig. 8). Full-length GST-agnoprotein (residues 1 to 71) gave a nearly total inhibition of NHEJ activity. Mutants containing N-terminal residues 1 to 55 or 1 to 36 caused inhibition, whereas those containing residues 37 to 71 and 55 to 71 did not, suggesting that the Ku70-binding domain is in the N terminus of agnoprotein. A mutant containing residues 18 to 54 did not inhibit NHEJ activity, indicating that the N-terminal 18 residues may be important.
FIG. 8.
Effects of addition of full-length and C-terminal deletion mutants of agnoprotein on the end-joining activity of agnoprotein-negative cell extracts. Nuclear extracts from agnoprotein-negative NIH 3T3 cells were prepared. The extracts were incubated with GST alone or with the various GST-agnoprotein fusion proteins (full-length protein and various deletion mutants, as indicated) and assayed for NHEJ activity as described in Materials and Methods. The lane on the far left (*) is a negative control without nuclear extract. The lane on the far right (none) is a positive control that has nuclear extract but no GST-protein addition. The arrow indicates the 3-kb monomeric linear plasmid substrate band. The 6- and 9-kb bands are the products of end joining and are indicated by a bracket.
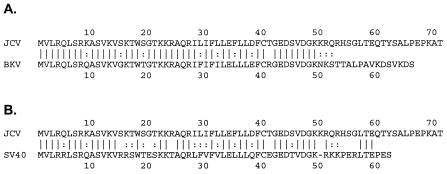
Sequence conservation between polyomavirus agnoproteins.
A question arose regarding whether the function of the JCV agnoprotein in the block of DNA repair is also found for SV40 and BKV agnoproteins. We compared the sequence of the region of the JCV agnoprotein that is involved in regulating DNA repair with the corresponding regions of the BKV and SV40 agnoproteins (Fig. 9). Of the 71 amino acid residues in the JCV agnoprotein, 41 are conserved in the BKV agnoprotein, 30 of which are in the N-terminal 36 amino acids (Fig. 9A). The N-terminal amino acids 1 to 36 of the JCV agnoprotein show 83% sequence identity with the BKV agnoprotein, while the C-terminal amino acids (residues 37 to 71) show only 31% identity. The six amino acid differences that occur in the region from residues 1 to 36 are all conservative. The high degree of homology in this region suggests that the DNA repair function is likely conserved in BKV. For SV40, the N-terminal amino acids 1 to 36 of the JCV agnoprotein show 61% sequence identity with the SV40 agnoprotein. Fourteen amino acid differences occur in the region from residues 1 to 36, 11 of which are conservative and 3 of which are not. Thus, the homology is only partial in this region and it is unclear whether the DNA repair function is conserved in SV40.
FIG. 9.
Comparison of amino acid sequence of JCV agnoprotein to BKV and SV40 agnoproteins. The amino acid sequence of the JCV agnoprotein was compared to the sequence of the agnoproteins of BKV (A) and SV40 (B). Sequence identity is indicated with parallel lines, and conservative sequence changes are indicated with colons.
DISCUSSION
With these studies, we have shown that agnoprotein expression has profound effects on the response of cells to DNA damage. Under conditions in which moderate chromosome damage was observed in control cells, severe chromosomal pulverization was seen in agnopositive cells. Cisplatin-treated agnonegative cells were mostly diploid G2/M or S-phase cells, while approximately one-third of the agnopositive cells became aneuploid, indicating a possible alteration of the signaling events that normally inhibit cell cycle progression when cells are undergoing DNA repair. NHEJ is thought to be the major mechanism of DSB DNA repair in vivo (5, 15). NHEJ activity was much reduced in cisplatin-treated agnopositive cells, indicating an impairment of DNA repair. The protein that mediates NHEJ DNA repair in the nucleus is a complex known as DNA-dependent protein kinase (DNA-PK), which consists of three subunits, Ku70, Ku80, and a 470-kDa protein (20-24). Ku70 and Ku80 form a heterodimer that binds to DSBs, protects the DNA ends from degradation, and orients the DNA strands for religation (24). The Ku70/Ku80 heterodimer also constitutes the regulatory subunit of DNA-PK that recruits the 470-kDa catalytic subunit. DNA-PK serine/threonine-specific protein kinase activity is required for the DNA ends to become ligated (23) and also is involved in an early stage of the signaling of DNA-damage-induced cell cycle arrest (24). The Ku70/Ku80 heterodimer localizes to the nucleus by virtue of nuclear localization signals that are present on both subunits (20, 22). An analysis of mutants of Ku70 and Ku80 in which the nuclear localization signal was deleted demonstrated that only one subunit needs to have a nuclear localization signal in order for the heterodimer to localize to the nucleus (22). Remarkably, we have found that agnoprotein causes much of the Ku70 to be localized within the cytoplasm, where it colocalizes with agnoprotein in the perinuclear space. There are only a few other reports of extranuclear Ku70. Ku70 is located in the cytoplasm during mitosis (21) and can localize on the plasma membranes of some cells, where it can act as an autoantigen (27). Oxidative stress induces the nuclear loss of Ku70 in pancreatic acinar cells (34), and it has been reported that the overexpression of Ku70 can suppress the apoptotic translocation of Bax to mitochondria due to Ku70 binding to Bax in the cytosol (32). Here we report that Ku70 bound to the JCV agnoprotein and caused a significant portion of the cellular Ku70 to be sequestered outside of the nucleus. The finding that a viral protein can inhibit cellular DNA repair by this mechanism is a novel one.
It is becoming clear that agnoprotein is a multifunctional protein that can bind multiple partners, as is the case for T antigen. Presumably, the small size of the genomes of polyomaviruses dictates that a few proteins must perform many tasks. Agnoprotein has been shown to bind to T antigen (28), the Y-box transcription factor (31), p53 (6), and now the Ku70 DNA repair protein. This binding interaction involves residues near the N terminus of agnoprotein and functionally inactivates Ku70. In this way, agnoprotein inhibits DNA repair and disrupts DNA-damage-induced cell cycle arrest. The ability of agnoprotein to circumvent the cell cycle checkpoint associated with DNA repair may be important in the lytic life cycle of polyomaviruses in a similar manner to the more well-known effects of the large T antigen on p53 and pRb (35). Cell cycle disruption and the impairment of DNA repair by JCV agnoprotein may contribute to the ability of JCV to transform cells and may be important in malignancies associated with JCV. Indeed, in a recent analysis of JCV in medulloblastoma tissue sections, some samples were found that expressed JCV agnoprotein but had no sign of T-antigen expression (9).
Acknowledgments
We thank past and present members of the Center for Neurovirology and Cancer Biology for their insightful discussions and for the sharing of ideas and reagents. We also thank C. Schriver for editorial assistance.
This work was supported by grants awarded by the NIH to K.K.
REFERENCES
- 1.Baumann, P., and S. C. West. 1998. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl. Acad. Sci. USA 95:14066-14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, J. R. 2003. Progressive multifocal leukoencephalopathy in acquired immuno-deficiency syndrome: explaining the high incidence and disproportionate frequency of the illness relative to other immunosuppressive conditions. J. Neurovirol. 9(Suppl. 1):38-41. [DOI] [PubMed] [Google Scholar]
- 3.Berger, J. R., and M. Concha. 1995. Progressive multifocal leukoencephalopathy: the evolution of a disease once considered rare. J. Neurovirol. 1:5-18. [DOI] [PubMed] [Google Scholar]
- 4.Cole, C. N. 1996. Polyomavirinae: the viruses and their replication, p. 917-946. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology, 3rd ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 5.Cromie, G. A., J. C. Connelly, and D. R. Leach. 2001. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell 8:1163-1174. [DOI] [PubMed] [Google Scholar]
- 6.Darbinyan, A., N. Darbinian, M. Safak, S. Radhakrishnan, A. Giordano, and K. Khalili. 2002. Evidence for dysregulation of cell cycle by human polyomavirus, JCV, late auxiliary protein. Oncogene 21:5574-5581. [DOI] [PubMed] [Google Scholar]
- 7.Del Valle, L., J. Gordon, M. Assimakopolou, S. Enam, J. F. Geddes, J. Varakis, C. Katsesos, S. E. Croul, and K. Khalili. 2001. Detection of JC virus DNA sequences and expression of the viral regulatory protein, T-antigen, in tumors of the central nervous system. Cancer Res. 61:4287-4293. [PubMed] [Google Scholar]
- 8.Del Valle, L., J. Gordon, P. Ferrante, and K. Khalili. 2001. JC virus in experimental and clinical brain tumorigenesis, p 409-430. In K. Khalili and G. L. Stoner (ed.), Human polyomaviruses: molecular and clinical perspective. Wiley-Liss Inc., New York, N.Y.
- 9.Del Valle, L., J. Gordon, S. Enam, S. Delbue, S. Croul, S. Abraham, S. Radhakrishnan, M. Assimakopoulou, C. D. Katsetos, and K. Khalili. 2002. Expression of human neurotropic polyomavirus JCV late gene product agnoprotein in human medulloblastoma. J. Natl. Cancer Inst. 94:267-273. [DOI] [PubMed] [Google Scholar]
- 10.Enam, S., L. Del Valle, C. Lara, D. D. Gan, C. Ortiz-Hidalgo, J. P. Palazzo, and K. Khalili. 2002. Association of human polyomavirus JCV with colon cancer: evidence for interaction of viral T-antigen and beta-catenin. Cancer Res. 2:7093-7101. [PubMed] [Google Scholar]
- 11.Fenech, M. 1993. The cytokinesis-block micronucleus technique: a detailed description of the method and its application to genotoxicity studies in human populations. Mutat. Res. 285:35-44. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, J., and K. Khalili. 1998. The human polyomavirus, JCV, and neurological diseases. Int. J. Mol. Med. 1:647-655. [DOI] [PubMed] [Google Scholar]
- 13.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 14.Heddle, J. A. 1973. A rapid in vivo test for chromosomal damage. Mutat. Res. 18:187-190. [DOI] [PubMed] [Google Scholar]
- 15.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 16.Jay, G., S. Nomura, C. W. Anderson, and G. Khoury. 1981. Identification of the SV40 agnogene product: a DNA binding protein. Nature 291:346-349. [DOI] [PubMed] [Google Scholar]
- 17.Kalweit, S., D. Utesch, W. von der Hude, and S. Madle. 1999. Chemically induced micronucleus formation in V79 cells—comparison of three different test approaches. Mutat. Res. 439:183-190. [DOI] [PubMed] [Google Scholar]
- 18.Kersten, B., P. Kasper, S. Y. Brendler-Schwaab, and L. Müller. 2002. Use of the photo-micronucleus assay in Chinese hamster V79 cells to study photochemical genotoxicity. Mutat. Res. 519:49-66. [DOI] [PubMed] [Google Scholar]
- 19.Khalili, K., L. Del Valle, J. Otte, M. Weaver, and J. Gordon. 2003. Human neurotropic polyomavirus, JCV, and its role in carcinogenesis. Oncogene 22:5181-5191. [DOI] [PubMed] [Google Scholar]
- 20.Koike, M. 2002. Dimerization, translocation and localization of Ku70 and Ku80 proteins. J. Radiat. Res. 43:223-236. [DOI] [PubMed] [Google Scholar]
- 21.Koike, M., T. Awaji, M. Kataoka, G. Tsujimoto, T. Kartasova, A. Koike, and T. Shiomi. 1999. Differential subcellular localization of DNA-dependent protein kinase components Ku and DNA-PKcs during mitosis. J. Cell Sci. 112:4031-4039. [DOI] [PubMed] [Google Scholar]
- 22.Koike, M., T. Shiomi, and A. Koike. 2001. Dimerization and nuclear localization of Ku proteins. J. Biol. Chem. 276:11167-11173. [DOI] [PubMed] [Google Scholar]
- 23.Kurimasa, A., S. Kumano, N. V. Boubnov, M. D. Story, C. S. Tung, S. R. Peterson, and D. J. Chen. 1999. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol. 19:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, S. H., and C. H. Kim. 2002. DNA-dependent protein kinase complex: a multifunctional protein in DNA repair and damage checkpoint. Mol. Cell 13:159-166. [PubMed] [Google Scholar]
- 25.Okada, Y., S. Endo, H. Takahashi, H. Sawa, T. Umemura, and K. Nagashima. 2001. Distribution and function of JCV agnoprotein. J. Neurovirol. 7:302-306. [DOI] [PubMed] [Google Scholar]
- 26.Padgett, B. L., D. L. Walker, G. M. ZuRhein, and R. J. Eckroade. 1971. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet i:1257-1260. [DOI] [PubMed] [Google Scholar]
- 27.Prabhakar, B. S., G. P. Allaway, J. Srinivasappa, and A. L. Notkins. 1990. Cell surface expression of the 70-kD component of Ku, a DNA-binding nuclear autoantigen. J. Clin. Investig. 86:1301-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Safak, M., R. Barucco, A. Darbinyan, Y. Okada, K. Nagashima, and K. Khalili. 2001. Interaction of JC virus Agno protein with T antigen modulates transcription and replication of the viral genome in glial cells. J. Virol. 75:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Safak, M., and K. Khalili. 2001. Physical and functional interaction between viral and cellular proteins modulates JCV gene transcription. J. Neurovirol. 7:288-292. [DOI] [PubMed] [Google Scholar]
- 30.Safak, M., and K. Khalili. 2003. An overview: human polyomavirus JC virus and its associated disorders. J. Neurovirol. 9(Suppl. 1):3-9. [DOI] [PubMed] [Google Scholar]
- 31.Safak, M., B. Sadowska, R. Barrucco, and K. Khalili. 2002. Functional interaction between JC virus late regulatory agnoprotein and cellular Y-box binding transcription factor, YB-1. J. Virol. 76:3828-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawada, M., W. Sun, P. Hayes, K. Leskov, D. A. Boothman, and S. Matsuyama. 2003. Ku70 suppresses the apoptotic translocation of Bax to mitochondria. Nat. Cell Biol. 5:320-329. [DOI] [PubMed] [Google Scholar]
- 33.Siddik, Z. H. 2003. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 22:7265-7279. [DOI] [PubMed] [Google Scholar]
- 34.Song, J. Y., J. W. Lim, H. Kim, T. Morio, and K. H. Kim. 2003. Oxidative stress induces nuclear loss of DNA repair proteins Ku70 and Ku80 and apoptosis in pancreatic acinar AR42J cells. J. Biol. Chem. 278:36676-36687. [DOI] [PubMed] [Google Scholar]
- 35.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]