Abstract
Several nonenveloped animal viruses possess an autolytic capsid protein that is cleaved as a maturation step during assembly to yield infectious virions. The 76-kDa major outer capsid protein μ1 of mammalian orthoreoviruses (reoviruses) is also thought to be autocatalytically cleaved, yielding the virion-associated fragments μ1N (4 kDa; myristoylated) and μ1C (72 kDa). In this study, we found that μ1 cleavage to yield μ1N and μ1C was not required for outer capsid assembly but contributed greatly to the infectivity of the assembled particles. Recoated particles containing mutant, cleavage-defective μ1 (asparagine → alanine substitution at amino acid 42) were competent for attachment; processing by exogenous proteases; structural changes in the outer capsid, including μ1 conformational change and σ1 release; and transcriptase activation but failed to mediate membrane permeabilization either in vitro (no hemolysis) or in vivo (no coentry of the ribonucleotoxin α-sarcin). In addition, after these particles were allowed to enter cells, the δ region of μ1 continued to colocalize with viral core proteins in punctate structures, indicating that both elements remained bound together in particles and/or trapped within the same subcellular compartments, consistent with a defect in membrane penetration. If membrane penetration activity was supplied in trans by a coinfecting genome-deficient particle, the recoated particles with cleavage-defective μ1 displayed much higher levels of infectivity. These findings led us to propose a new uncoating intermediate, at which particles are trapped in the absence of μ1N/μ1C cleavage. We additionally showed that this cleavage allowed the myristoylated, N-terminal μ1N fragment to be released from reovirus particles during entry-related uncoating, analogous to the myristoylated, N-terminal VP4 fragment of picornavirus capsid proteins. The results thus suggest that hydrophobic peptide release following capsid protein autocleavage is part of a general mechanism of membrane penetration shared by several diverse nonenveloped animal viruses.
The mechanisms by which nonenveloped animal viruses mediate membrane penetration to invade the host cytoplasm remain less well understood than the mechanisms of membrane fusion by enveloped viruses. Since nonenveloped viruses lack a membrane, they must traverse the cellular membrane barrier by a mechanism other than fusion. Nonenveloped animal viruses contain “penetration proteins, ” analogous to the fusion proteins of enveloped viruses, that accomplish membrane penetration, perhaps either by forming a membrane-spanning pore or by locally disrupting the membrane bilayer. For example, capsid proteins VP1 and VP4 of poliovirus, each present in 60 copies per virion, are thought to play the critical roles in membrane penetration by that nonenveloped virus (reviewed in reference 38).
In many viruses, the fusion or penetration proteins are primed to adopt their membrane-seeking forms by posttranslational cleavage, which is mediated in some cases by the proteins themselves (autocatalytic cleavage, or “autocleavage”). Cleavage may be necessary to remove conformational restraints, to expose terminal hydrophobic sequences for membrane insertion, or to allow peptide release. Examples of enveloped virus fusion proteins primed by cleavage include influenza virus HA (Orthomyxoviridae), Sendai virus F (Paramyxoviridae), and human immunodeficiency virus gp160 (Retroviridae) (reviewed in reference 37). Examples of nonenveloped virus penetration proteins primed by autocleavage include polio- and rhinovirus VP0 (Picornaviridae), flock house virus α (Nodaviridae), and Nudaurelia capensis omega virus α (Tetraviridae) (reviewed in reference 41). Polio- and rhinovirus VP0 proteins are cleaved into fragments VP4 and VP2, allowing VP4 release during cell entry (6, 24, 32, 46, 47). Similarly, flock house virus and N. capensis omega virus α proteins are cleaved into fragments β and γ, allowing γ release during cell entry (8, 35, 53, 59, 64, 68).
Mammalian orthoreoviruses (reoviruses), members of the Reoviridae family, are nonenveloped viruses comprising a 10-segment, double-stranded RNA genome surrounded by two concentric, icosahedral protein capsids. The 10 genome segments encode eight structural proteins, which constitute the T=1 inner and T=13 (laevo) outer capsid layers, and three nonstructural proteins not present in the mature virion. The inner capsid proteins possess the enzymatic activities necessary for viral transcription (reviewed in reference 63). In addition, the outer capsid protein λ2 is responsible for capping the 5′ end of each viral plus-strand RNA as it exits the particle during transcription (34; reviewed in reference 58). The other outer capsid proteins—μ1, σ3, and σ1—are involved in cell entry. The majority of the outer capsid lattice is formed by 600 copies of μ1, the putative membrane penetration protein (reviewed in reference 16), which are organized as trimers on the virus particle (29, 48). The σ3 protein, also present in 600 copies, closely associates with μ1 to coat the particle surface (29, 48). The remainder of the outer capsid is made up of 36 copies of the attachment protein σ1, arranged as trimers at the fivefold axes of symmetry (20, 22, 33; reviewed in reference 44). The double-layered nature of the reovirus capsid (diameter, ∼85 nm) makes it distinct from—and much larger than—the T=3 single capsids of poliovirus and flock house virus (diameter, 30 to 35 nm) (31, 39) and the T=4 single capsid of N. capensis omega virus (diameter, 40 to 45 nm) (53).
To initiate infection, the σ1 protein in reovirus virions binds to host cell surface receptors (reviewed in references 5 and 44), and the particles are then internalized, most likely by receptor-mediated endocytosis (10, 62). Within the endocytic vesicles, lysosomal proteases cleave the outer capsid proteins (30, 62) to generate particles very similar to the infectious subvirion particles (ISVPs) that can be generated in vitro by protease treatment (42, 54). In these particles, σ3 is degraded, leaving μ1 as the major surface protein (29). Through a process that we continue to dissect, ISVPs can undergo a structural transformation to yield a related particle form, the ISVP* (15, 42). Generation of the ISVP*, which contains an altered conformer of μ1 and has shed σ1, appears necessary for membrane penetration, promoting particle release into the host cytoplasm (10, 11, 15, 18, 43). Either during the process of membrane penetration or after release into the cytoplasm, a large piece of μ1, the central “δ” fragment (see below), is also lost from the particle (18). Furthermore, once in the cytoplasm, the now transcriptionally activated particle can produce the viral plus-strand RNAs that are used as templates for new protein synthesis and duplex RNA genome synthesis. This cytoplasmically delivered “payload” of reovirus—a partially uncoated particle with transcriptase activity—is distinct from those of poliovirus, flock house virus, and N. capensis omega virus, which are thought to enter the cytoplasm by delivering only their translation-competent, plus-sense RNA genomes across the membrane (reviewed in references 38 and 41).
The putative membrane penetration protein of reoviruses, μ1 (76 kDa; 708 amino acids), is known to undergo at least three different proteolytic cleavages in vitro and in vivo (Fig. 1A). Upon exposure to host proteases in the intestinal lumen (7) or in endocytic vesicles (30, 62), particle-bound μ1 is cleaved near amino acid 580 to generate two fragments, μ1δ and φ, that remain particle bound (54) (Fig. 1A). The cleavage of μ1 into μ1δ and φ has been shown to be dispensable for membrane permeabilization and transcriptase activation, as well as for infection, by ISVPs (17, 19). As recently identified (51; M. L. Nibert, unpublished data), removal of ∼10 amino acids from the C terminus of μ1 (i.e., from the φ region) can also accompany protease treatment, but the significance of this cleavage remains unclear. Lastly, μ1 is cleaved near its N terminus, between amino acids 42 and 43, to generate two fragments, a small N-terminal fragment, μ1N (4 kDa), and a large C-terminal fragment, μ1C (72 kDa), which are also both present in particles (55, 61) (Fig. 1A). No known protease has been linked to the μ1N/μ1C cleavage, and this cleavage is instead considered to be autocatalytic (48, 55). Interestingly, μ1N contains the N-myristoyl group of μ1 at its N terminus, making it further reminiscent of polio- and rhinovirus VP4, which has been implicated in membrane penetration by those nonenveloped viruses (23, 25, 32, 46, 47, 52).
FIG. 1.
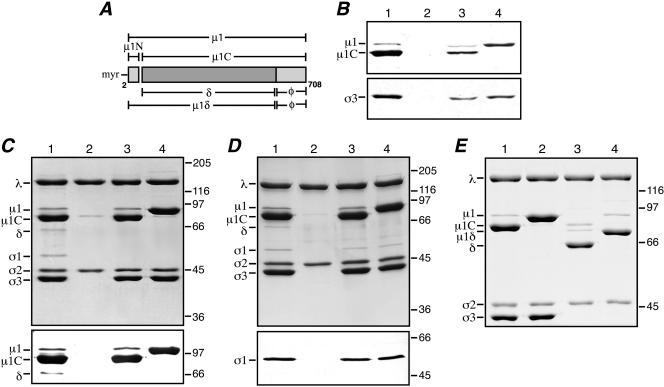
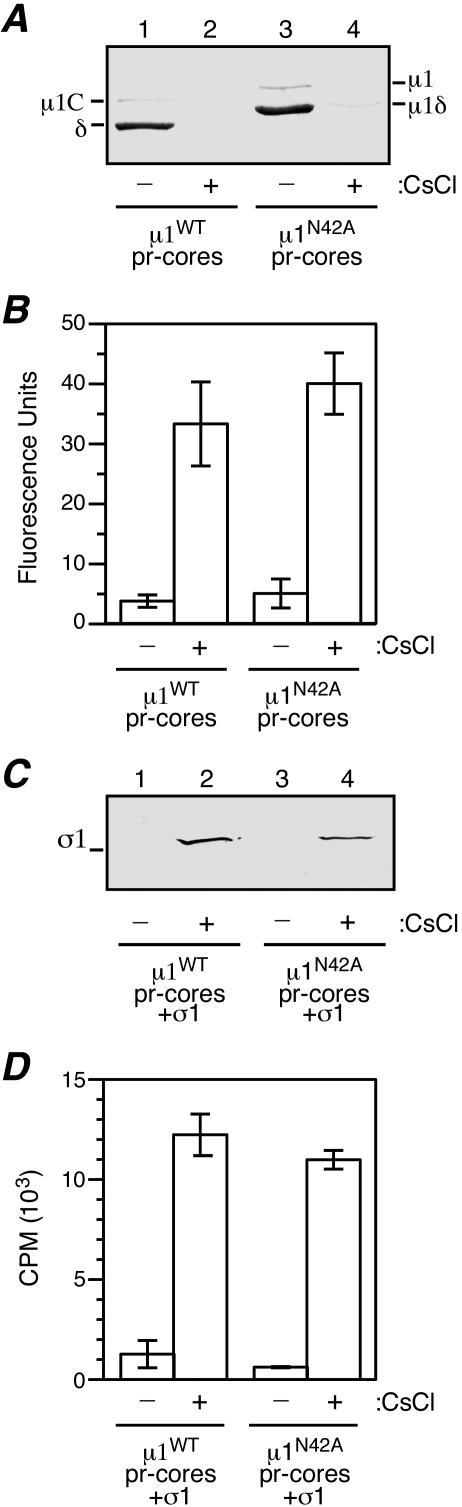
Expression of μ1N42A, generation of recoated particles, and protease treatment of recoated particles to generate ISVPs. (A) The μ1 protein and its cleavage products are diagrammed. The μ1 N-terminal and C-terminal amino acid positions are labeled 2 and 708, respectively. The autocleavage products μ1N and μ1C are indicated. The N-terminal myristoyl group (myr) is present in both full-length μ1 and μ1N. Cleavage of full-length μ1 by exogenous proteases (e.g., chymotrypsin or trypsin) yields the fragments μ1δ and φ. Cleavage of μ1C by exogenous proteases yields the fragments δ and φ. (B) SDS-PAGE and immunoblotting of insect cell lysates containing either coexpressed μ1WT and wild-type σ3 (lane 3) or μ1N42A and wild-type σ3 (lane 4) was performed. The samples were probed with either the μ1-specific mouse MAb 10H2 (top) or a σ3-specific rabbit polyclonal antiserum (bottom). Virions (lane 1) and cores (lane 2) were included for comparison. (C) Samples of purified μ1WT r-cores (lane 3) and μ1N42A r-cores (lane 4) were examined by SDS-PAGE followed by either Coomassie staining (top) or immunoblotting with μ1-specific MAb 10H2 (bottom). Virions (lane 1) and cores (lane 2) were included for comparison. (D) Samples of purified μ1WT r-cores plus σ1 (lane 3) and μ1N42A r-cores plus σ1 (lane 4) were examined by SDS-PAGE followed by either Coomassie staining (top) or immunoblotting with a σ1-specific rabbit polyclonal antiserum (bottom). Virions (lane 1) and cores (lane 2) were included for comparison. (E) Samples of chymotrypsin-generated μ1WT pr-cores (lane 3) and μ1N42A pr-cores (lane 4) were examined by SDS-PAGE and Coomassie staining. Untreated μ1WT r-cores (lane 1) and μ1N42A r-cores (lane 2) were included for comparison.
The significance of the μ1N/μ1C cleavage in reovirus infection has not been directly addressed, largely because of an inability to block this cleavage in viral particles. We overcame this obstacle by generating a point mutation in μ1 to prevent μ1N/μ1C cleavage and then studying its effects in “recoated cores” (r-cores), a versatile system developed for studies of reovirus outer capsid assembly and functions in cell entry (19, 20). The residue immediately N-terminal to the μ1N/μ1C cleavage site, Asn42, was specifically chosen for mutagenesis based on a previous report that it is required for the cleavage to occur (65). In this report, we describe our results with this approach and the new insights we obtained into the role of μ1 in membrane penetration by reoviruses. The findings suggest that hydrophobic peptide release following capsid protein autocleavage is part of a general mechanism of membrane penetration shared by several diverse nonenveloped viruses.
MATERIALS AND METHODS
Mouse and insect cells and native reovirus particles.
Spinner-adapted mouse L929 cells were grown in suspension in Joklik's modified minimal essential medium (Irvine) supplemented to contain 2% fetal and 2% neonatal bovine sera (HyClone), as well as 2 mM glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin (Irvine)/ml. Spodoptera frugiperda clone 21 (Sf21) and Trichoplusia ni High Five insect cells (Invitrogen) were grown in TC-100 medium (Invitrogen) supplemented to contain 10% heat-inactivated fetal bovine serum. Mv1Lu and CV1 cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented to contain 10% fetal bovine serum and 10 μg of gentamicin (Invitrogen)/ml. Purified reovirus type 1 Lang (T1L) virions, ISVPs, and top component particles (33), as well as cores (19), were obtained as previously described. The purified particles were stored at 4°C in virion buffer (150 mM NaCl, 10 mM MgCl2, 10 mM Tris, pH 7.5).
Generation of recombinant baculovirus to express mutant μ1.
Site-directed mutagenesis of the plasmid pBKS-M2L (19) was performed using the QuikChange protocol (Stratagene). The missense mutation that resulted in changing Asn42 to alanine (μ1N42A) was introduced using the complementary mutagenic primers 5′-CTATCACCTGGAATGTTAGCGCCTGGAGGAGTACCATGG-3′ and 5′-CCATGGTACTCCTCCAGGCGCTAACATTCCAGGTGATAG-3′ (the changes are underlined). The nucleotide changes also introduced a HaeII restriction site that was used to screen for plasmids containing the mutant clone. Mutant clones were sequenced from nucleotides 1 to 1120 to confirm that they contained the desired changes and to rule out any additional mutations. The NotI-Bsu36I restriction fragment (nucleotides 1 to 727) containing the desired changes was subcloned into pFb-M2L (19). Clones containing the subcloned insert were then further subcloned into pFbD-M2L/S4L (19) using the Bsu36I-SalI restriction sites (nucleotides 1 to 727). All restriction enzymes were obtained from New England BioLabs.
Expression of reovirus proteins from baculovirus vectors.
The recombinant baculovirus containing the T1L M2 and S4 genes used to coexpress wild-type μ1 (μ1WT) and wild-type σ3 was described previously (20). The recombinant baculovirus containing the T1L S1 gene used to express wild-type σ1 has been described (20, 21). To express these wild-type proteins or μ1N42A, T. ni High Five cells were infected at a multiplicity of infection of 5 to 10 PFU/cell with fourth-passage baculovirus stocks. Baculovirus-infected cells expressing σ1 or coexpressing μ1 and σ3 were harvested 65 h postinfection (p.i.), and cytoplasmic extracts were prepared as previously described (19, 20).
Protein gel electrophoresis and immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of reduced or nonreduced samples was carried out as previously described (56). Samples were reduced unless otherwise indicated in the text. For immunoblot analyses, proteins were transferred to a nitrocellulose membrane in 25 mM Tris-192 mM glycine (pH 8.3). The μ1-specific mouse monoclonal antibody (MAb) 10H2 (67), T1L σ1-specific rabbit polyclonal antiserum (15), and σ3-specific rabbit polyclonal antiserum (40) were each used at a 1:2,000 dilution. The μNS-specific rabbit polyclonal antiserum (14) was used at a 1:5,000 dilution. The binding of these primary antibodies was detected with alkaline phosphatase-coupled goat anti-mouse or goat anti-rabbit antibodies (Bio-Rad) and the colorimetric reagents p-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt (Bio-Rad).
R-cores.
R-cores and r-cores plus σ1 were generated as described previously (19, 20). Briefly, cytoplasmic insect cell extracts containing baculovirus-expressed μ1 and σ3 or μ1, σ3, and σ1 were incubated with purified T1L cores at 37°C for 2 to 4 h. Following incubation, the reaction mixtures were chilled and loaded atop a 14-ml CsCl step gradient (1.25 to 1.45 g/cm3) with a 2-ml sucrose cushion (20% [wt/vol]). The gradients were subjected to centrifugation at 25,000 rpm in a Beckman SW28 rotor for 2 to 20 h at 4°C. The resulting particles were harvested and concentrated on a second CsCl step gradient and centrifuged at 40,000 rpm in a Beckman SW50.1 rotor for 2 h at 4°C. The r-cores were harvested from the second gradient and dialyzed extensively in virion buffer. Particle concentrations were determined by densitometry of gels stained with Coomassie brilliant blue R250 (Sigma) as described previously (19).
Electron cryomicroscopy and image reconstruction.
Vitrified samples of μ1N42A r-cores were prepared on holey carbon films and maintained at −176°C for electron cryomicroscopy as described previously (4). Electron micrographs were recorded under low-dose conditions (∼20 e−/Å2) in an FEI-Philips CM300 FEG microscope at a calibrated magnification of ×47,440. The micrographs were digitized at 7-μm intervals on a Zeiss PHODIS scanner, and the data were then bin-averaged to yield pixels corresponding to 2.95 Å in the specimen. A total of 22 micrographs, recorded with defocus settings ranging between 1.9- and 3.6-μm underfocus, were selected for processing. From these micrographs, 973 particle images were masked and extracted using the program RobEM (http://bilbo.bio.purdue.edu/∼baker/programs/programs.html), and each particle image was stored as a 363- by 363-pixel array. Origin and orientation parameters for each particle were determined and refined using model-based procedures (3). Eleven cycles of refinement and data screening led to a three-dimensional reconstruction at 15.6-Å resolution, computed from 807 particle images. The effects of the microscope contrast transfer function were compensated for in part for each image (12). A difference density map was computed by subtracting the map of μ1N42A r-cores from an existing 17.6-Å map of μ1WT r-cores plus σ1 (20), after first reducing the resolution and scaling the magnification and contrast of the former against the latter using RobEM. High-spatial-frequency features in all maps were sharpened through the application of a 1/600-Å2 inverse temperature factor (36).
Infectivity and hemagglutination.
Determinations of numbers of PFU per milliliter of reovirus particle preparations were performed as previously described (33). Particle/PFU ratios were used to compare relative infectivities. Hemagglutination titers were determined as previously described (26).
Immunostaining and immunofluorescence (IF) microscopy.
For immunostaining, the following primary antibody dilutions were used: μ1-specific mouse MAb 10H2 (67) (1:2,000 dilution) and μNS-specific rabbit polyclonal antiserum (14) and core-specific rabbit polyclonal antiserum (19) (1:5,000 dilutions). Goat anti-mouse immunoglobulin G and goat anti-rabbit immunoglobulin G, each conjugated to either Alexa 488 or Alexa 594, were obtained from Molecular Probes. CV1 cells were plated directly on glass coverslips at a density of 1.2 × 104 cells/cm2 and allowed to adhere overnight. When indicated, cycloheximide (50 μg/ml; Sigma) was added at 37°C 1 h prior to infection. Cells were infected at 2 × 104 to 5 × 104 particles/cell for 1 h at 4°C. Following virus attachment, the cells were either fixed immediately in 2% paraformaldehyde for 10 min at room temperature or incubated at 37°C to allow infection to proceed and fixed in 2% paraformaldehyde at the appropriate time p.i. Unless otherwise indicated, the cells were permeabilized, blocked, incubated with antibodies and 4′,6-diamidino-2-phenylindole (Molecular Probes), and mounted as described previously (57). Microscopy images were collected digitally using a Nikon TE-300 inverted microscope equipped with phase and fluorescence optics and analyzed using Metamorph version 5.1 (Universal Imaging) as described previously (57). Images were processed and presented using Photoshop and Illustrator software (Adobe).
Detection of μNS by immunoblotting.
CV1 cells (2.5 × 106) were infected at 1,000 particles/cell. Lysates were collected at 0, 2, or 24 h p.i. by scraping the cells into 2 ml of phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.5). The cells were pelleted by centrifugation and resuspended in 30 μl of PBS. The cells were then lysed in sample buffer, boiled for 10 min, and subjected to SDS-PAGE. Following transfer to nitrocellulose, μNS protein was detected using a μNS-specific rabbit polyclonal antiserum (14) and an alkaline phosphatase-coupled goat anti-rabbit secondary antibody (Bio-Rad).
Flow cytometry.
Confluent monolayers of Mv1Lu cells were incubated with virus at 2 × 104 particles/cell for 1 h at 4°C to allow virus attachment to occur. Following attachment, 0-h p.i. time point specimens were collected by scraping the cells into 2 ml of PBS, pelleting the cells at 500 × g in a Beckman Allegra 6R centrifuge, and fixing the cells in 1 ml of 2% paraformaldehyde for 10 min at room temperature. Cells analyzed at later times p.i. were first incubated at 37°C to allow infection to proceed for the desired time and then processed as described above. Following fixation, the cells were washed with PBS containing 1% bovine serum albumin (PBSA), permeabilized with PBSA containing 0.1% Triton X-100, and incubated for 1 h at 4°C with either μNS-specific rabbit polyclonal antiserum (14) (1:5,000 dilution) or μ1-specific mouse MAb 10H2 (67) (1:2,000 dilution). The cells were then washed with PBSA to remove unbound antibody and incubated with goat anti-rabbit immunoglobulin G or goat anti-mouse immunoglobulin G, each conjugated to Alexa 488, to detect the rabbit or mouse primary antibodies, respectively. The cells were again washed with PBSA to remove unbound antibodies and resuspended in PBS. Flow cytometry was performed using a Becton Dickinson FACS Calibur.
Hemolysis and other assays of the ISVP → ISVP* transition.
Hemolysis experiments were performed as described elsewhere (17). μ1 protease sensitivity, bis-ANS (Molecular Probes) fluorescence, and σ1 release assays were performed as described previously (15). For all of the described experiments, the final particle concentration was in the range of 4 × 1012 to 7.5 × 1012 particles/ml. Cs+ was added to promote the ISVP → ISVP* transition and associated activities relative to Na+ in these types of experiments (15, 17-20). K+ also demonstrates this effect and may be physiologically relevant in this regard as the predominant IA cation inside cells (9-11, 15). Available data indicate that the effect of larger IA cations is to accelerate the rate of the transition, not to affect its outcome.
Transcriptase activation.
Transcription reactions containing 9 × 1010 particles in a total volume of 15 μl were performed as described elsewhere (27), with slight modifications. Briefly, 32P-labeled transcripts were generated by incubating NaCl-treated ISVPs (or chymotrypsin-treated r-cores with or without σ1) or CsCl-treated ISVPs (or chymotrypsin-treated r-cores with or without σ1) with a transcription reaction mixture (100 mM Tris, pH 8; 10 mM MgCl2; 1 mM [each] ATP, UTP, and CTP; 10 mM phosphoenolpyruvate; 0.4 U of pyruvate kinase σ; 0.33 μU of RNasin [Promega]; 2 mM dithiothreitol; and 2.5 μCi of [α-32P]GTP [Perkin-Elmer]) at 37°C for 105 min. Transcription levels were measured by trichloroacetic acid precipitation followed by scintillation counting.
α-Sarcin coentry.
The α-sarcin coentry assay was performed as previously described (18).
μ1N release.
ISVPs labeled with [3H]myristate (Perkin-Elmer) were generated as previously described (55). These particles were then incubated for 30 min at 32°C in buffer containing 50 mM Tris (pH 7.5), 0.5% Triton X-100, and either 300 mM NaCl or 300 mM CsCl. Following incubation, the samples were cooled on ice for 10 min and then layered atop a prechilled 1.25- to 1.45-g/cm3 CsCl density gradient. The gradients were subjected to centrifugation in a Beckman SW60 rotor at 50,000 rpm and 4°C for 2 h and then fractionated using a peristaltic pump (5 drops/fraction). Ready Safe scintillation fluid (20 ml; Beckman) was added to each fraction, and the radioactivity (in counts per minute) was measured using a Beckman LS 6500 scintillation counter.
RESULTS
Alanine substitution for Asn42 abrogates μ1N/μ1C cleavage.
Mutant μ1 containing an asparagine → alanine substitution at position 42 (μ1N42A) was expressed in insect cells using a recombinant baculovirus. This vector was designed to coexpress μ1 with its “protector” protein, σ3, which binds to μ1 in solution and is required for its assembly into particles (19, 60). Insect cell lysates containing either μ1N42A or μ1WT coexpressed with wild-type σ3 were analyzed by SDS-PAGE and immunoblotting to estimate the levels of protein expression and μ1N/μ1C cleavage. The levels of μ1 and σ3 expression with μ1N42A were comparable to those with μ1WT (Fig. 1B). However, μ1N42A exhibited no detectable μ1N/μ1C cleavage, as evidenced by the absence of μ1C; most of the mutant protein comigrated with uncleaved μ1 (Fig. 1B, lane 4).
μ1N/μ1C cleavage is not required for outer capsid assembly.
To test for outer capsid assembly with μ1N42A, insect cell lysates containing wild-type σ3 coexpressed with either μ1N42A or μ1WT were incubated with cores, and the resulting particles (μ1N42A and μ1WT r-cores, respectively) were isolated on CsCl gradients. In both cases, a single particle band was observed, at a density consistent with virion-like particles containing the μ1-σ3 outer capsid. SDS-PAGE and Coomassie staining (Fig. 1C, top) or immunoblotting (Fig. 1C, bottom) confirmed that μ1 and σ3 had been incorporated into the particles in both samples. As expected for the μ1WT particles (19), most μ1 underwent μ1N/μ1C cleavage (μ1C was visible in these gels), while only a small portion (∼5%) remained uncleaved. In the μ1N42A particles, however, μ1 remained wholly uncleaved (Fig. 1C, lane 4), suggesting that μ1N/μ1C cleavage is not required for outer capsid assembly. Densitometry of Coomassie-stained gels confirmed that recoating was largely complete, since the levels of μ1 and σ3 relative to other structural proteins in μ1N42A r-cores approximated those of μ1 plus μ1C and σ3 in native virions or μ1WT r-cores (data not shown). Use of insect cell lysates containing [3H]myristate confirmed that the μ1 protein in both μ1N42A and μ1WT r-cores was myristoylated (data not shown), as is that in native virions (55).
R-cores including the wild-type attachment protein σ1 (r-cores plus σ1) were also generated (20). When purified on CsCl gradients, both μ1N42A and μ1WT r-cores plus σ1 migrated at a density similar to that of virions. SDS-PAGE and Coomassie staining (Fig. 1D, top), followed by densitometry (data not shown), confirmed that both μ1N42A and μ1WT r-cores plus σ1 contained a nearly full complement of μ1 (or μ1 plus μ1C) and σ3. The amount of σ1 could not be easily quantified due to the usual low-copy number of this protein. However, the presence of σ1 was confirmed by immunoblotting (Fig. 1D, bottom), and those results suggested that the amounts of σ1 in both types of r-cores plus σ1 were similar to that in native virions.
Evidence for proper outer capsid assembly by μ1N42A.
To determine whether the outer capsid had been properly assembled, μ1N42A and μ1WT r-cores were treated with protease (trypsin [Fig. 1E] or chymotrypsin [data not shown]) to generate partially uncoated particles resembling ISVPs (pr-cores), which were then isolated on CsCl gradients. SDS-PAGE of the pr-cores (Fig. 1E, lanes 3 and 4) alongside untreated r-cores (Fig. 1E, lanes 1 and 2) indicated that σ3 was degraded and most μ1 was cleaved at the δ-φ junction, as expected for ISVPs (54). The cleavage of μ1N42A at the δ-φ junction generated two fragments: a large N-terminal fragment, μ1δ (Fig. 1E, lane 4), and a small C-terminal fragment, φ (not visible on this gel, but observed on others). This differs from native ISVPs (54) and μ1WT pr-cores (Fig. 1E, lane 3) (19), in which the larger μ1 cleavage product is primarily δ instead of μ1δ. Since μ1N/μ1C cleavage is necessary to convert μ1δ to δ (Fig. 1A), the presence of μ1δ confirmed that this cleavage had not occurred in the μ1N42A particles. Essentially the same results were observed with μ1N42A and μ1WT pr-cores plus σ1 as with pr-cores (data not shown). The finding that digestion of the outer capsid with protease generates ISVP-like particles from μ1N42A particles provides evidence that μ1N42A and its associated σ3 protein are conformationally similar to wild-type μ1 and σ3 in native virions and μ1WT r-cores. SDS-PAGE of μ1N42A r-cores disrupted under nonreducing conditions additionally revealed a disulfide bond between the φ regions of adjacent μ1 monomers (data not shown), as was also found in virions and μ1WT r-cores (56).
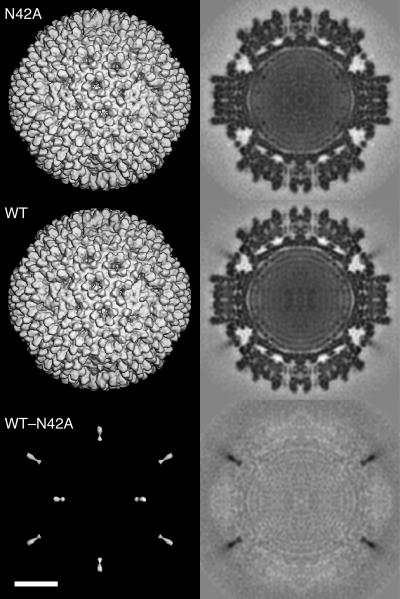
As a further test for proper outer capsid assembly, we subjected μ1N42A r-cores to electron cryomicroscopy and three-dimensional image reconstruction. In micrographs the individual particles were well dispersed and had an appearance similar to those of virions and μ1WT r-cores (data not shown). A final reconstruction, computed using data from 807 particles, was determined to be reliable to at least 15.6-Å resolution (4). The resolution was subsequently reduced to 17.6 Å (Fig. 2, top row) for quantitative comparisons with other reconstructed density maps. Compared to reconstructions of μ1WT r-cores (data not shown) (19) and μ1WT r-cores plus σ1 (Fig. 2, middle row) (20), few if any differences were apparent. In a difference map obtained by subtracting the density map of μ1N42A r-cores from that of μ1WT r-cores plus σ1, the only clear features were densities attributable to σ1 at the icosahedral vertices (Fig. 2, bottom row). We conclude that the μ1N42A mutation has minimal effects on outer capsid structure and assembly.
FIG. 2.
Three-dimensional-image reconstruction of μ1N42A recoated particles. Shown are surface-shaded representations (left) and central, density-coded sections (right) of μ1N42A r-cores (top row) and μ1WT r-cores plus σ1 (middle row) and a difference map obtained by subtracting the densities of the former from those of the latter (bottom row). Both reconstructions, as well as the difference map revealing σ1- but not μ1-related densities, are shown at 17.6-Å resolution. Scale bar, 200 Å.
μ1N42A particles are poorly infectious.
Consistent with previous findings (20), μ1WT r-cores plus σ1 were nearly as infectious as native virions in L929 cells (Table 1). In contrast, μ1N42A r-cores plus σ1 were much less infectious than either native virions or μ1WT r-cores plus σ1 (Table 1) (relative [per-particle] infectivity, ≈1/3,200 that of virions). This suggests that the μ1N/μ1C cleavage is critical for productive infection, although infection can still occur at very low efficiency (∼0.03% that of virions) in the absence of this cleavage. Similarly, μ1N42A pr-cores plus σ1 were much less infectious than native ISVPs or μ1WT pr-cores plus σ1 (Table 1) (relative infectivity, ≈1/1,600 that of ISVPs), suggesting that proteolytic removal of σ3 and cleavage of μ1 at the δ-φ junction are not sufficient to enhance the relative infectivity of μ1N42A r-cores plus σ1 to any substantial degree. Also consistent with previous findings (19), r-cores containing the outer capsid proteins μ1WT and σ3, but lacking the attachment protein σ1, were ∼200 times more infectious than core particles in L929 cells (Table 1). Although the addition of μ1WT and σ3 enhanced the relative infectivity of r-cores to this large extent, the μ1WT r-cores were still only 1/2,500 as infectious as virions (Table 1). This is consistent with previous evidence that the attachment protein σ1 is needed, in addition to μ1 and σ3, for r-cores to approach virion-like levels of infectivity (20). μ1N42A r-cores, in comparison, showed little if any enhancement in relative infectivity over cores and were only ∼1/100 as infectious as μ1WT r-cores and ∼1/27,500 as infectious as native virions. These findings suggest that the μ1N/μ1C cleavage is required to confer enhanced infectivity on r-cores. Experiments were next performed to identify the step(s) in infection at which μ1N42A particles are defective.
TABLE 1.
Relative infectivities of native and recoated particles
| Particle typea | Relative infectivity (log10)b |
|---|---|
| Cores | 0.0 |
| Virions | 5.7 ± 0.2 |
| R-cores plus σ1; μ1WT | 5.4 ± 0.3 |
| R-cores plus σ1; μ1N42A | 2.2 ± 0.3 |
| R-cores; μ1WT | 2.3 ± 0.2 |
| R-cores; μ1N42A | 0.2 ± 0.2 |
| ISVPs | 6.3 ± 0.3 |
| Pr-cores plus σ1; μ1WT | 5.4 ± 0.3 |
| Pr-cores plus σ1; μ1N42A | 3.1 ± 0.1 |
All viral particles and proteins were derived from reovirus T1L.
Infectivity levels are expressed relative to that of cores, as means ± standard deviations for three or more determinations each.
Barrier to infectivity of μ1N42A particles occurs after attachment but before or during protein synthesis.
The capacity of μ1N42A pr-cores plus σ1 to attach to cells was addressed by IF microscopy. CV1 cells were incubated with μ1N42A pr-cores plus σ1, μ1WT pr-cores plus σ1, or native ISVPs for 1 h at 4°C. The cells were then washed extensively to remove free viral particles and fixed. IF microscopy using the μ1-specific MAb 10H2 (67) revealed that binding of the μ1N42A pr-cores plus σ1 to CV1 cells (Fig. 3A, bottom left) was comparable to that of μ1WT pr-cores plus σ1 (Fig. 3A, top left) or ISVPs (data not shown). Essentially the same results were observed in L929 and Mv1Lu cells (data not shown). To compare the levels of binding more quantitatively, flow cytometry using Mv1Lu cells was performed. Mv1Lu cells were incubated with μ1N42A pr-cores plus σ1, μ1WT pr-cores plus σ1, or native ISVPs for 1 h at 4°C to allow attachment. The cells were then washed extensively to remove free particles, fixed, and stained using μ1 MAb 10H2. Cells incubated with any of the three particle types showed essentially identical levels of increased 10H2 staining in comparison to uninfected cells (data not shown), providing further evidence that the μ1N42A pr-cores plus σ1 are fully competent for attachment. We also used a hemagglutination assay (26) to test cell-binding actvity. μ1N42A r-cores plus σ1 displayed a relative (per-particle) capacity for hemagglutination very similar to those of μ1WT r-cores plus σ1 and native virions (data not shown), indicating that the μ1N42A r-cores plus σ1 are largely functional for binding to erythrocytes. In sum, these findings indicate that the infectivity barrier for μ1N42A particles follows attachment.
FIG. 3.
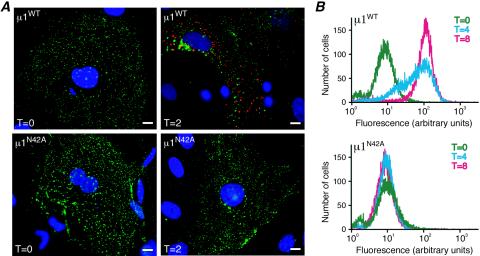
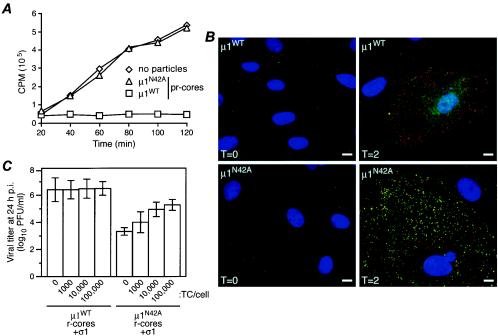
Attachment and protein synthesis during infection of cells by recoated particles. (A) CV1 cells were infected with μ1WT pr-cores (top row) or μ1N42A pr-cores (bottom row) (50,000 particles per cell) and then fixed at either 0 (left column) or 2 (right column) h p.i. The fixed cells were coimmunostained with a μNS-specific rabbit polyclonal antiserum followed by Alexa 594-conjugated goat anti-rabbit immunoglobulin G (red) and the μ1-specific mouse MAb 10H2 followed by Alexa 488-conjugated goat anti-mouse immunoglobulin G (green). Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (blue). Samples were examined by fluorescence microscopy. (B) Mv1Lu cells were infected with μ1WT pr-cores (top) or μ1N42A pr-cores (bottom) (20,000 particles per cell) and then fixed at either 0 (green), 4 (cyan), or 8 (magenta) h p.i. The fixed cells were stained with μNS-specific rabbit polyclonal antiserum followed by Alexa 488-conjugated goat anti-rabbit immunoglobulin G. Samples were examined by flow cytometry.
The synthesis of new viral proteins in infected cells was first evaluated by IF microscopy using a μNS-specific polyclonal antiserum (14). μNS was used as a marker of new protein synthesis, since it is a nonstructural protein that is produced during infection but is not present in input virions or r-cores plus σ1. In CV1 cells infected with μ1N42A pr-cores plus σ1 (Fig. 3A, bottom left), μ1WT pr-cores plus σ1 (Fig. 3A, top left), or native ISVPs (data not shown), no μNS was detected at 0 h p.i. By 2 h p.i., however, μNS was clearly evident in cells infected with μ1WT r-cores plus σ1 (Fig. 3A, top right) or ISVPs (data not shown), but not in cells infected with μ1N42A r-cores plus σ1 (Fig. 3A, bottom right). Essentially the same differences among these particle types were observed in L929 cells at 0 and 2 h p.i. (data not shown). Similarly, μNS was detected by immunoblotting in lysates harvested as early as 2 h p.i. from CV1 cells infected with μ1WT pr-cores plus σ1 but was not detected in lysates harvested as late as 24 h p.i. from CV1 cells infected with μ1N42A pr-cores plus σ1 (data not shown). Flow cytometry after staining with μNS-specific polyclonal antiserum revealed that Mv1Lu cells infected with native ISVPs (data not shown) or μ1WT pr-cores plus σ1 (Fig. 3B, top) showed increased levels of μNS at 4 h p.i and that nearly every cell was strongly μNS positive by 8 h p.i. In Mv1Lu cells infected with μ1N42A r-cores plus σ1, in contrast, μNS remained virtually undetectable at both 4 and 8 h p.i. (Fig. 3B, bottom). These findings suggest that the infectivity barrier for μ1N42A particles occurs before or during protein synthesis. We next attempted to pinpoint the step between attachment and protein synthesis at which μ1N42A r-cores are blocked.
μ1N42A particles fail to permeabilize membranes in vitro.
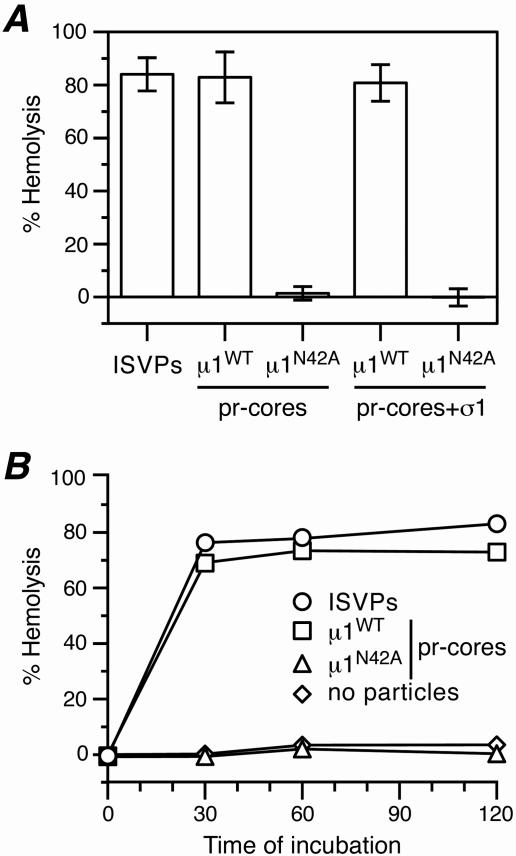
A membrane permeabilization defect of μ1N42A particles was demonstrated in vitro using a hemolysis assay (17, 19). When incubated with erythrocytes, ISVPs can permeabilize the cells' plasma membranes, leading to hemoglobin release and providing a useful model for membrane penetration during cell entry. Consistent with previous reports (15, 17-20), we found that both native ISVPs and μ1WT pr-cores (with or without σ1) displayed robust hemolytic activity (Fig. 4A). μ1N42A pr-cores (with or without σ1), in contrast, did not mediate hemolysis (Fig. 4A). Time course experiments confirmed the rapidity with which both ISVPs and μ1WT pr-cores (with or without σ1) mediated hemolysis, compared to the failure of μ1N42A pr-cores (with or without σ1) to mediate hemolysis over extended periods (Fig. 4B and data not shown). These findings suggest that μ1N/μ1C cleavage is an important step in the membrane permeabilization pathway and that the barrier to infectivity of μ1N42A particles may involve a specific defect in membrane penetration.
FIG. 4.
Hemolysis by μ1N42A recoated particles. (A) Samples of native ISVPs, μ1WT pr-cores (with or without σ1), or μ1N42A pr-cores (with or without σ1) were incubated with bovine calf erythrocytes (final concentration, 3% [vol/vol]) at 32°C for 30 min in the presence of 200 mM CsCl. The reactions were terminated by incubation on ice for 10 min, and the cells were pelleted by centrifugation at 300 × g for 5 min. The extent of hemolysis was determined by measuring the A405 of the supernatant and was expressed as a percentage (hemolysis by 1% Triton X-100 = 100%). Each bar represents the mean ± standard deviation from three trials. (B) Samples of μ1WT or μ1N42A pr-cores were evaluated for hemolysis as described for panel A, except that samples were harvested for analysis after different periods at 32°C. The results of a representative experiment are shown. Use of CsCl as a promoting agent is discussed in Materials and Methods.
μ1N42A particles exhibit other hallmarks of the ISVP → ISVP* transition in vitro.
ISVPs can transform into a distinct particle form, the ISVP*, that contains a protease-sensitive conformer of μ1, is more hydrophobic, has released σ1, and has been activated for synthesis of viral plus-strand RNAs (15). The transition from ISVP to ISVP* precedes or accompanies hemolysis (15) and appears to precede or accompany membrane penetration during productive cell entry as well (18). In an attempt to determine why μ1N42A particles are defective for hemolysis, we examined whether they exhibit other hallmarks of the ISVP → ISVP* transition in vitro.
(i) Increase in μ1 protease sensitivity.
The μ1 conformer in ISVP*s (μ1*) is highly sensitized to protease degradation at 4°C (15). Upon conversion to ISVP*-like particles, μ1WT pr-cores also contained a protease-sensitive conformer of μ1 (Fig. 5A), as should occur if pr-cores are mimicking the behaviors of native ISVPs (15). Interestingly, samples of μ1N42A pr-cores incubated under the same conditions to promote the ISVP → ISVP* transition also contained a protease-sensitive conformer of μ1 (Fig. 5A). None of the particle samples incubated under conditions that do not promote the ISVP → ISVP* transition showed increased protease sensitivity of μ1 (Fig. 5A), indicating that they had not been converted to ISVP*s. We performed identical experiments in the presence of erythrocytes and looked for μ1 protease sensitivity following a hemolysis reaction. After immunoblotting for μ1, we obtained essentially the same findings with regard to changes in μ1 protease sensitivity in the three particle types that we obtained in the absence of erythrocytes (data not shown). We conclude that μ1N42A particles can exhibit at least one structural hallmark of the ISVP → ISVP* transition, increased protease sensitivity of μ1, in the absence of μ1N/μ1C cleavage. Since the ISVP*-like μ1N42A particles are defective at hemolysis, however, the findings suggest that some missing element(s) of the transition in these particles is necessary for membrane permeabilization.
FIG. 5.
Capacity of μ1N42A recoated particles to undergo the ISVP → ISVP* transition. (A) Protease sensitivity of μ1. Samples of μ1WT pr-cores plus σ1 (lanes 1 and 2) or μ1N42A pr-cores plus σ1 (lanes 3 and 4) were incubated with bovine erythrocytes, and hemolysis was performed as for Fig. 4 in the presence of CsCl or NaCl (200 mM). Following incubation at 32°C and removal to ice, the samples were incubated with trypsin (100 μg/ml) for 45 min on ice. The trypsin was then inactivated by addition of soybean trypsin inhibitor (300 μg/ml). Samples were subjected to SDS-PAGE, followed by immunoblotting with the μ1-specific MAb 10H2. (B) Hydrophobicity. Samples of μ1WT or μ1N42A pr-cores plus σ1 were incubated in reaction buffer containing bis-ANS (25 μM) and either NaCl or CsCl (300 mM) for 30 min at 32°C. The levels of bis-ANS fluorescence were then measured on a fluorescence microplate reader (Spectramax; Molecular Dynamics) (excitation, 405 nm; emission, 485 nm). The results of a representative experiment are shown. (C) Release of σ1. Samples of μ1WT pr-cores plus σ1 (lanes 1 and 2) or μ1N42A pr-cores plus σ1 (lanes 3 and 4) were incubated in reaction buffer containing either CsCl or NaCl (300 mM) for 30 min at 32°C. The samples were removed to ice for 10 min and then loaded atop a sucrose cushion (20% [wt/vol]; 500 μl). Following centrifugation in a Beckman TLA 100.2 rotor (90,000 rpm for 1 h at 5°C), the top fraction (200 μl) of the sucrose cushion was removed, concentrated by precipitation with trichloroacetic acid, and subjected to SDS-PAGE, followed by immunoblotting with a σ1-specific rabbit polyclonal antiserum. (D) Transcriptase activation. Samples of μ1WT or μ1N42A pr-cores plus σ1 were incubated in reaction buffer containing either CsCl or NaCl (300 mM) for 30 min at 32°C. The samples were removed to ice for 10 min and then incubated in transcription reaction buffer (see Materials and Methods) containing [α-32P]GTP at 37°C for 105 min. RNA products were concentrated by precipitation with trichloroacetic acid, and the radioactivity in each sample was measured by scintillation counting. Each bar represents the mean ± standard deviation from three separate trials. Use of CsCl as a promoting agent is discussed in Materials and Methods.
(ii) Increase in particle hydrophobicity.
To address changes in hydrophobicity following the ISVP → ISVP* transition, we used a fluorescent probe, bis-ANS, that binds to exposed hydrophobic regions of proteins (15). Consistent with previous findings, ISVPs showed increased binding of bis-ANS upon conversion to ISVP*s (data not shown). Both μ1WT pr-cores and μ1N42A pr-cores showed similarly increased binding of bis-ANS as well (Fig. 5B). In contrast, the control samples held under conditions that do not promote the ISVP → ISVP* transition showed only limited increases in bis-ANS binding (Fig. 5B and data not shown), indicating they had not been converted to ISVP*s. We conclude that μ1N42A particles can exhibit a second structural feature of the ISVP → ISVP* transition, increased hydrophobicity, and that μ1N/μ1C cleavage is not required for this increase to occur.
(iii) Release of σ1.
To assay for loss of attachment protein σ1 that accompanies the ISVP → ISVP* transition, we subjected particles to centrifugation through a sucrose cushion, during which particle-associated proteins should pellet while released proteins remain near the top of the cushion (15). Following centrifugation, the top portion of the sucrose cushion was collected and analyzed by SDS-PAGE and immunoblotting to detect the presence of released σ1. Consistent with previous findings, σ1 was released from ISVPs upon conversion to ISVP*s (data not shown). Both μ1WT pr-cores and μ1N42A pr-cores also showed release of σ1 (Fig. 5C). In contrast, none of the control samples held under conditions that do not promote the ISVP → ISVP* transition showed σ1 release (Fig. 5C and data not shown), indicating that they had not been converted to ISVP*s. The δ or μ1δ fragment of μ1 remained particle associated in each of the ISVP* samples (data not shown), consistent with previous results (15). We conclude that μ1N42A particles can exhibit a third structural hallmark of the ISVP → ISVP* transition, release of σ1, and that μ1N/μ1C cleavage is not required for this release to occur.
(iv) Transcriptase activation.
μ1 must adopt the hydrophobic, protease-sensitive conformation now known to be associated with ISVP*s in order for the particle-associated transcriptases to be activated from their latent state and become capable of synthesizing the full-length viral plus-strand RNAs (9, 15, 27, 42). To test if μ1N/μ1C cleavage is required for transcriptase activation in vitro, samples of μ1N42A or μ1WT pr-cores plus σ1 were incubated under standard conditions either to promote or not to promote the ISVP → ISVP* transition. The particles were then added to an in vitro transcription reaction mixture that included [α-32P]GTP. The RNA products were isolated by trichloroacetic acid precipitation, and 32P incorporation was measured by scintillation counting. In this assay, after incubation under conditions to promote the ISVP → ISVP* transition, μ1N42A pr-cores plus σ1 produced RNA transcripts at high levels, similar to those of μ1WT pr-cores plus σ1 (Fig. 5D) or native ISVPs (data not shown). Only low levels of transcripts were produced, however, after each of the particle types was incubated under nonpromoting conditions (Fig. 5D and data not shown). Essentially the same results were obtained in experiments with μ1N42A and μ1WT pr-cores lacking σ1 (data not shown). We conclude that μ1N42A particles can exhibit a major functional hallmark of the ISVP → ISVP* transition, transcriptase activation, and thus that μ1N/μ1C cleavage is not required for this activation to occur.
In summary, the results indicate that μ1N42A pr-cores with or without σ1 (ISVP-like particles) are competent to undergo important elements of the ISVP → ISVP* transition in vitro but are nevertheless not functional for in vitro membrane permeabilization.
μ1N42A particles are also defective at membrane permeabilization during cell entry.
Since μ1N42A pr-cores (with or without σ1) failed to mediate membrane permeabilization in vitro, we hypothesized that the barrier to infectivity of μ1N42A particles may involve the membrane penetration step during cell entry. To test this hypothesis, we assessed the capacity of μ1N42A pr-cores plus σ1 to mediate cytoplasmic coentry of the ribonucleotoxin α-sarcin, which in turn inhibits both cellular and viral protein synthesis (18, 50). L929 cells were infected with μ1WT pr-cores plus σ1 or μ1N42A pr-cores plus σ1 in the presence of α-sarcin, and levels of protein synthesis were measured at 20-min intervals p.i. In cells infected with μ1WT pr-cores plus σ1 (Fig. 6A), inhibition of protein synthesis occurred within 20 min p.i. at 37°C, consistent with entry of the toxin along with penetrating viral particles and also consistent with previous results (18). Similar results were seen in cells infected with ISVPs (data not shown). In contrast, cells infected with μ1N42A pr-cores plus σ1 showed no decrease in protein synthesis relative to uninfected cells (Fig. 6A), supporting the conclusion that the μ1N42A particles are defective at membrane permeabilization, and α-sarcin-based intoxication, during cell entry.
FIG. 6.
Behaviors of μ1N42A recoated particles in cells. (A) Coentry of α-sarcin during viral infection. L929 cells were incubated with μ1WT or μ1N42A pr-cores plus σ1 at 4°C to allow attachment, washed with methionine-free medium containing [35S]methionine-cysteine with or without α-sarcin (50 μg/ml), and incubated at 37°C for different times, as indicated. The cells were then lysed, and macromolecules were concentrated by trichloroacetic acid precipitation. The radioactivity (in counts per minute) in the washed precipitates was measured by scintillation counting. The values shown represent the average of two experiments, each done in duplicate. (B) Antibody-detected structural changes in infected cells. CV1 cells were infected with μ1WT pr-cores plus σ1 (top row) or μ1N42A pr-cores plus σ1 (bottom row) (50,000 particles per cell) and then fixed at either 0 (left column) or 2 (right column) h p.i. The fixed cells were coimmunostained with a core-specific rabbit polyclonal antiserum followed by Alexa 594-conjugated goat anti-rabbit immunoglobulin G (red) and the μ1-specific mouse MAb 4A3 followed by Alexa 488-conjugated goat anti-mouse immunoglobulin G (green). Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (blue). Samples were examined by fluorescence microscopy. (C) Rescue of infectivity by top component. L929 cell monolayers were coinfected with μ1WT or μ1N42A r-cores plus σ1 (1,000 per cell) plus top component particles (0, 1,000, 10,000, or 100,000 per cell). After 24 h at 37°C, the cells were lysed, and infectious titers were determined by plaque assay. Samples of cells infected with each amount of top component particles alone were identically generated and analyzed in parallel. Within each experiment, the latter titers were subtracted from the respective former titers to correct for any residual infectivity of the top component. Each bar represents the mean ± standard deviation of the results from three separate experiments.
μ1N42A particles exhibit hallmarks of the ISVP → ISVP* transition in cells.
The μ1-specific MAb 4A3 (67) recognizes a conformer of the δ region of μ1 that is present in ISVP*s both in vitro and in infected cells at early times p.i (18). This or associated changes in particle structure also allow the recognition of particles by a polyclonal antiserum specific for core surface proteins (18). The capacity of the δ region of μ1 in μ1N42A pr-cores plus σ1 to undergo these conformation-specific changes in infected cells was addressed using IF microscopy. CV1 cells were incubated with μ1N42A or μ1WT pr-cores plus σ1 in the presence of cycloheximide for 1 h at 4°C. At 0 and 2 h p.i., the infected cells were washed to remove unattached virus and fixed prior to being immunostained. At 0 h p.i., little staining was observed with either particle type and either antibody reagent (Fig. 6B, left), consistent with prior evidence that neither of these antibody reagents is effective at binding to ISVP-like particles (18). Also consistent with prior evidence (18), at 2 h p.i., cells infected with μ1WT pr-cores plus σ1 displayed diffuse staining with the MAb 4A3 and punctate staining with the core-specific antiserum (Fig. 6B, upper right), indicating that the δ region of μ1 had undergone a conformational change, was now largely separated from the particles, and was probably free in the cytoplasm and nucleus. Results similar to these were obtained with native ISVPs in this (data not shown) and the previous study (18). In contrast, at 2 h p.i., cells infected with μ1N42A pr-cores plus σ1 displayed punctate staining with both MAb 4A3 and the core-specific antiserum in colocalizing spots (Fig. 6B, lower right). These findings provide evidence that μ1N42A pr-cores plus σ1 undergo structural changes associated with ISVP* formation during cell entry but that the conformationally altered δ region of μ1N42A fails to separate from particles in the same manner as μ1WT. Two possible explanations are that (i) the δ region of μ1N42A remains bound to particles and (ii) the δ region of μ1N42A dissociates from particles but both remain trapped within the same subcellular compartments (e.g., endocytic vesicles) so that their separation cannot be resolved by IF microscopy.
Infectivity of μ1N42A particles is rescued by coinfection with genome-deficient particles.
Since μ1N42A particles can be activated for transcription, we reasoned that other viral particles may be capable of supplying membrane penetration activity in trans, enhancing the infectivity of μ1N42A particles. For this purpose, we chose reovirus top component particles, which have protein contents and structures essentially identical to those in virions but which cannot replicate in cells because they lack the viral genome (28, 61). Moreover, top component particles are known to mediate membrane penetration in vivo (13, 49), as well as in the hemolysis assay in vitro (M. L. Nibert, unpublished data). L929 cells were coinfected with either μ1N42A or μ1WT r-cores plus σ1, along with increasing amounts of top component particles. The infections were allowed to proceed for 24 h, and viral growth was then measured by plaque assay. Cells infected with matching amounts of top component alone were analyzed in parallel to permit correction for any growth attributable to those particles. With increasing amounts of top component, growth attributable to μ1N42A r-cores plus σ1 was greatly enhanced while growth attributable to μ1WT r-cores plus σ1 was not affected (Fig. 6C). With the largest amount of top component tested, growth attributable to μ1N42A r-cores plus σ1 was enhanced by >100-fold and approached 10% of the levels attained by μ1WT r-cores plus σ1 (Fig. 6C). The finding that coinfections with top component particles greatly enhanced the infectivity of μ1N42A particles supports the hypothesis that μ1N42A particles are defective at the membrane penetration step but are otherwise functional for subsequent steps in the onset of viral replication.
Release of μ1N from ISVP*s.
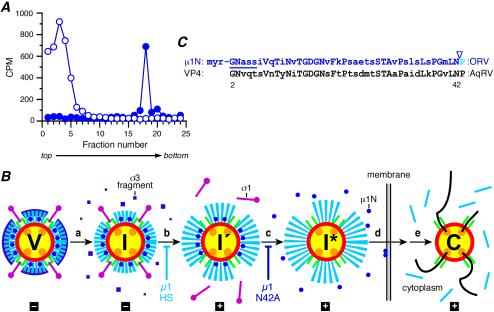
A likely possibility would be that μ1N/μ1C cleavage is critical to allow release of the N-myristoylated μ1N fragment (600 copies per virion) (29, 48, 55) before or during membrane penetration. We therefore tested for μ1N release from particles at the ISVP → ISVP* transition in vitro, which had not been addressed in previous studies. ISVPs labeled with [3H]myristate, which is specifically incorporated as the N-terminal myristoyl group of μ1 and μ1N (55, 65), were used to monitor the particle association of μ1N. Separate samples of ISVPs were incubated under conditions that either converted them to ISVP*s or retained them as ISVPs. Upon centrifugation in CsCl gradients at 4°C, the particles should migrate to their known densities (∼1.38 g/cm3) (15, 42) while released proteins remain near the top of the gradient. Following centrifugation, the gradients were fractionated, and scintillation counting was performed to identify fractions containing the radiolabel. In initial experiments, we found that the label could be efficiently recovered from ISVP* samples only when a nonionic detergent (e.g., Triton X-100) was included in the reaction mixture (data not shown). This was presumably due to the hydrophobic nature of μ1N, which caused it to stick to surfaces, such as that of the centrifuge tube, once released. In later experiments, we therefore included this detergent in both samples and consistently found that μ1N was retained in ISVPs but was indeed released from ISVP*s (Fig. 7A). This finding is consistent with the hypothesis that μ1N/μ1C cleavage is required during cell entry to allow the release of the myristoylated μ1N fragment from particles.
FIG. 7.
μ1N release from ISVP*s, model, and sequences. (A) μ1N release from ISVP*s. [3H]myristate-labeled ISVPs were incubated with either 300 mM NaCl (solid circles) or 300 mM CsCl (open circles) in the presence of 0.5% Triton X-100 for 30 min at 32°C. The particles were then purified on a 1.25- to 1.45-g/cm3 CsCl density gradient. The gradients were fractionated, and the radioactivity (in counts per minute) in each sample was measured by scintillation counting. The results of a representative experiment are shown. (B) Updated model of reovirus uncoating in vitro and during cell entry. Interactions of viral particles and proteins with cell surface receptors and possible localizations to specific subcellular compartments are not included in the diagram. The proposed uncoating intermediates and fates of the outer capsid proteins—μ1N (blue), μ1C (cyan), σ1 (magenta), and σ3 (purple)—are shown. (Step a) The virion (V) undergoes proteolytic processing to generate the ISVP (I). In ISVPs, the σ3 protein has been degraded, and its differently sized fragments have been released. (Step b) The ISVP then undergoes a major structural transition to the ISVP′ (I′). ISVP′s lack σ1 and contain an altered, hydrophobic conformer of μ1. Particles containing μ1HS are blocked at step b (18). (Step c) The ISVP′ then undergoes a further transition to the ISVP* (I*), during which the remaining μ1N/μ1C cleavage occurs and the μ1N peptide is released. As suggested in the diagram (by the increase in particle diameter), further conformational changes in μ1C seem likely to accompany this transition. Particles containing μ1N42A are blocked at step c. (Steps d and e) The released μ1N peptides, putatively in concert with other portions of μ1C remaining in the ISVP*, effects membrane penetration (step d), and transcriptionally active core particles (C) are ultimately released into the cytoplasm (step e). The capacity of each particle type to mediate viral transcription is indicated below the diagram (+, able to mediate; −, unable to mediate). Note that cleavage of μ1C at the δ-φ junction during generation of ISVPs is not shown in this diagram, because the fate of φ during and after membrane penetration remains unknown. The cyan lines accompanying the core after step e specifically represent the released δ fragment (18). (C) Sequence comparison of the N-terminal regions of orthoreovirus (ORV) μ1 and aquareovirus (AqRV) VP4 proteins. Amino acids 2 to 43 of μ1 are shown on top, with corresponding residues of VP4 aligned below. Identical residues are shown in uppercase. The myristoylation consensus sequence in both proteins is indicated by the line. The N-terminal N-myristoyl group of μ1 is labeled (myr), and the site of the putative autocleavage is indicated by the arrowhead. The myristoyl group plus amino acids 2 to 42 constitute the μ1N peptide.
DISCUSSION
Consistent with results of previous studies, in which Asn42 in μ1 was mutated to threonine, glutamine, glutamate, or histidine (65), we found that an alanine substitution at Asn42 completely blocked μ1N/μ1C cleavage. Autolytic capsid proteins from several other nonenveloped animal viruses also require an asparagine at the cleavage site (1, 46, 59, 64, 68; reviewed in reference 41), suggesting that this particular amino acid plays a key role in the cleavage mechanism. The current model for the cleavage mechanism of both flock house virus and N. capensis omega virus α proteins is that the nitrogen of the asparagine side chain nucleophilically attacks the carbonyl group of the peptide backbone immediately C-terminal to the asparagine residue (64). We are currently working to determine a crystal structure of the reovirus μ1N42A-σ3 heterohexamer (L. Zhang, A. L. Odegard, M. L. Nibert, and S. C. Harrison, unpublished data) in preparation for further studies to dissect the μ1N/μ1C cleavage mechanism.
This report further illustrates the utility of r-cores (19, 20) for studies of cell entry by reovirus. Using this system, we were able to show that μ1N42A can assemble into a largely complete outer capsid surrounding the core, despite not having undergone the μ1N/μ1C cleavage. Most of the previous literature would have led us not to predict this, since μ1N/μ1C cleavage appears to occur upon association of μ1 and σ3 and thus has been thought to be important for assembly (e.g., see references 45 and 65). Based partly on evidence in this study, we now believe that μ1N/μ1C cleavage must occur before or during membrane penetration, but not necessarily as a step in assembly. Other data indicate that in a fraction of the μ1 subunits, cleavage in fact does not occur during assembly but rather during disassembly of the μ1-σ3 heterohexamer (48) or the particle-associated outer capsid. After disruption of virions under certain conditions preceding SDS-PAGE, a much larger fraction of the μ1 subunits remain uncleaved at the μ1N/μ1C junction, suggesting that at least a portion of this cleavage accompanies disassembly (M. L. Nibert, A. L. Odegard, M. A. Agosto, K. Chandran, and L. A. Schiff, unpublished data). The coupling of μ1N/μ1C cleavage to disassembly provides a plausible explanation for why the cleavage-defective mutant μ1N42A can assemble normally into r-cores.
Since proteolytic cleavage is required to prime many enveloped virus fusion proteins (reviewed in reference 37) and nonenveloped virus penetration proteins (reviewed in references 38 and 41) for membrane interactions, we reasoned that the μ1N/μ1C cleavage may allow μ1 to adopt its membrane-seeking conformation during cell entry. In this study, we obtained multiple points of evidence that μ1N/μ1C cleavage is indeed critical for membrane permeabilization and penetration by reovirus particles, but not for some earlier steps in entry, such as attachment. If membrane penetration activity was supplied in trans by genome-deficient top component particles containing μ1WT, the μ1N42A particles displayed much higher levels of infectivity. The large number of top component particles needed to maximize this effect may reflect the inefficiency with which appropriate forms of the two particles were colocalized to the same subcellular compartment, from which membrane penetration could then occur. Thus, although we hypothesize that the defect in μ1N42A particles involves the membrane penetration step per se, we recognize that this mutation could alternatively or additionally affect particle delivery to the appropriate subcellular location.
Recent studies have proposed a model in which ISVPs undergo a series of structural and functional changes to yield a new particle form, the ISVP*, before or during membrane penetration (15, 18). Structurally, ISVP*s contain an altered conformer of μ1 and have lost the attachment protein σ1 (15, 18, 42). Functionally, these particles not only are associated with membrane permeabilization in vitro and in vivo but also have been activated to transcribe the viral plus-strand RNAs (15, 18). The evidence in this study that μ1N42A particles can progress only partially through the transition to ISVP*s leads us to propose a new uncoating intermediate, the ISVP′, at which particles are trapped in the absence of μ1N/μ1C cleavage (Fig. 7B). The existence of this intermediate is supported by evidence that in a fraction of the μ1 subunits, μ1N/μ1C cleavage directly accompanies the ISVP → ISVP* transition (Nibert et al., unpublished). The ISVP′ is thus proposed to be the particle form that immediately precedes this cleavage. Another mutant, μ1HS, has also recently been characterized using the r-core system (18) and shown to be blocked at an earlier step, between the ISVP and the newly defined ISVP′ (Fig. 7B). The results indicate that μ1HS particles do not mediate membrane permeabilization, but neither do they exhibit any hallmarks of the ISVP → ISVP* transition. Thus, through studies with r-cores containing different mutations in μ1, we appear to be dissecting the uncoating and membrane penetration processes of reoviruses into a series of discrete steps. Trapped uncoating intermediates obtained with these mutants may be useful for other biochemical and structural studies to characterize the changes in μ1 that allow it to interact with and alter the membrane for particle translocation.
Whether the released μ1N peptide plays an active role in membrane penetration or must simply be discarded before other regions of μ1 can act remains to be determined. The very low residual infectivity of μ1N42A particles (Table 1) is consistent with either possibility. For example, the μ1N peptide might be capable of playing an active role, but with greatly reduced efficiency, even if it remains tethered to the δ region of μ1C. If μ1N indeed plays an active role, there are several possible reasons why it must be cleaved and released for maximal activity. The freed C-terminal end of μ1N may be necessary for the released peptide to fold into a particular conformation for its activity. The cleavage and release may also be necessary to allow the peptide to diffuse farther from the particle than would be possible if it remained tethered to the δ region of μ1C (Fig. 1A). Yet another possibility is that the cleavage and release may be necessary to allow large numbers of μ1N peptides to associate, either in solution or in the membrane. One simple idea is that of a “μ1N bullet, ” which is ejected from the particle and interacts with the nearby membrane, directly participating in the penetration process that allows the particle to enter the cytoplasm. The N-terminal N-myristoyl group is an obvious candidate to play a key role in this proposed function, although the μ1N peptide sequence is itself quite hydrophobic (Fig. 7C). Preliminary evidence obtained with r-cores containing a myristoylation-defective μ1 protein suggest that these particles are indeed poorly infectious (I. S. Kim, M. A. Agosto, K. Chandran, and M. L. Nibert, unpublished data). Notably, the μ1N region (including the myristoylation signal and autocleavage site) is highly conserved with those parts of the 69-kDa outer capsid protein VP4 of aquareoviruses (Fig. 7C) and much more conserved than is the μ1C region with the rest of VP4 (2, 48), suggesting that μ1N and the N-terminal portion of the aquareovirus protein share conserved functions.
In several other nonenveloped animal viruses, small and sometimes N-myristoylated peptides are known or thought to play critical roles in cell entry. One well-known example is the autocleavage product VP4 of poliovirus. After binding of a poliovirus virion to its cell surface receptors, and either before or after uptake and acidification along the endocytic pathway, the 65- to 70-residue, N-terminally N-myristoylated VP4 peptide is released from the particle and the previously buried N terminus of VP1 is externalized (reviewed in reference 38). A favored hypothesis for polio- and rhinovirus membrane penetration is that the newly exposed N terminus of VP1, possibly in concert with VP4, interacts with the plasma or endosomal membrane to form a pore through which the genomic RNA is extruded into the cytoplasm (6, 24, 46, 47, 66). As with μ1N, the exact role of VP4 is not known. However, Danthi et al. and Moscufo et al. have described mutations in VP4 that severely impact infection even though the virus particle binds to the cell surface and undergoes all of its normal entry-related structural changes, including VP0 cleavage and VP4 release, suggesting that the released VP4 plays a direct role in entry (25, 52). Similarly, in flock house virus, the 44-residue autocleavage product, γ, is thought be directly involved in the membrane penetration process (8, 59; reviewed in reference 41). Mutations that block the autocleavage needed to generate γ render the virus very poorly infectious (59). In addition, an α-helical region of the γ peptide has been shown to interact with and permeabilize membranes, supporting the hypothesis that γ is directly involved in penetrating the endosomal membrane during cell entry (8). A similar role has been proposed for the 74-residue γ peptide of N. capensis omega virus (41, 53).
Further studies of these and other systems should provide additional comparisons for obtaining a more complete mechanistic description of how different nonenveloped viruses traverse the membrane barrier during cell entry. In such comparisons, it is important to note that reovirus virions are much larger than those of poliovirus, flock house virus, or N. capensis omega virus (85 versus 30 to 45 nm); that reovirus μ1 occupies a different structural position than does poliovirus VP0, flock house virus α, or N. capensis omega virus α (T=13 outer capsid versus T=3 or 4 single capsid); and that the entry payload of reovirus is substantially different from those of the other viruses (transcriptase particle versus plus-strand RNA). Despite these differences, these diverse nonenveloped viruses have now been suggested to share hydrophobic-peptide release following capsid-protein autocleavage as part of a general mechanism of membrane penetration during entry.
Acknowledgments
We thank H. Ploegh and his laboratory members for access to and assistance with flow cytometry instrumentation and experiments; E. C. Freimont and J. B. Dinoso for superb technical assistance; other members of our laboratory, D. Bubeck, M. Ehrlich, S. C. Harrison, J. M. Hogle, T. Kirchhausen, and L. Zhang, for helpful discussions; and M. A. Agosto, I. S. Kim, and K. S. Myers for comments on the manuscript.
This work was supported in part by NIH grants K08 AI52209 to J.S.L.P., R01 GM33050 to T.S.B., and R01 AI46440 to M.L.N.; a Keck Foundation award to the Purdue Structural Biology group for purchase of the CM300 FEG microscope; and a Purdue University reinvestment grant to the Structural Biology group. K.C. was additionally supported by a Fields postdoctoral fellowship made available to the Department of Microbiology and Molecular Genetics through the generosity of Ruth Peedin Fields.
REFERENCES
- 1.Ansardi, D. C., and C. D. Morrow. 1995. Amino acid substitutions in the poliovirus maturation cleavage site affect assembly and result in accumulation of provirions. J. Virol. 69:1540-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attoui, H., Q. Fang, F. M. Jaafar, J. F. Cantaloube, P. Biagini, P. De Micco, and X. De Lamballerie. 2002. Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of Golden shiner reovirus, Grass carp reovirus, Striped bass reovirus and golden ide reovirus (genus Aquareovirus, family Reoviridae). J. Gen. Virol. 83:1941-1951. [DOI] [PubMed] [Google Scholar]
- 3.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryo-electron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 4.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton, E. S., J. C. Forrest, J. L. Connolly, J. D. Chappell, Y. Liu, F. J. Schnell, A. Nusrat, C. A. Parkos, and T. S. Dermody. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441-451. [DOI] [PubMed] [Google Scholar]
- 6.Belnap, D. M., D. J. Filman, B. L. Trus, N. Cheng, F. P. Booy, J. F. Conway, S. Curry, C. N. Hiremath, S. K. Tsang, A. C. Steven, and J. M. Hogle. 2000. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 74:1342-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodkin, D. K., M. L. Nibert, and B. N. Fields. 1989. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J. Virol. 63:4676-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bong, D. T., C. Steinem, A. Janshoff, J. E. Johnson, and M. Reza Ghadiri. 1999. A highly membrane-active peptide in Flock House virus: implications for the mechanism of nodavirus infection. Chem. Biol. 6:473-481. [DOI] [PubMed] [Google Scholar]
- 9.Borsa, J., D. G. Long, M. D. Sargent, T. P. Copps, and J. D. Chapman. 1974. Reovirus transcriptase activation in vitro: involvement of an endogenous uncoating activity in the second stage of the process. Intervirology 4:171-188. [DOI] [PubMed] [Google Scholar]
- 10.Borsa, J., B. D. Morash, M. D. Sargent, T. P. Copps, P. A. Lievaart, and J. G. Szekely. 1979. Two modes of entry of reovirus particles into L cells. J. Gen. Virol. 45:161-170. [DOI] [PubMed] [Google Scholar]
- 11.Borsa, J., M. D. Sargent, P. A. Lievaart, and T. P. Copps. 1981. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology 111:191-200. [DOI] [PubMed] [Google Scholar]
- 12.Bowman, V. D., E. S. Chase, A. W. E. Franz, P. R. Chipman, X. Zhang, K. L. Perry, T. S. Baker, and T. J. Smith. 2002. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J. Virol. 76:12250-12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broering, T. J., J. Kim, C. L. Miller, C. D. S. Piggott, J. B. Dinoso, M. L. Nibert, and J. S. L. Parker. 2004. Reovirus nonstructural protein μNS recruits viral core surface proteins and entering core particles to factory-like inclusions. J. Virol. 78:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broering, T. J., J. S. L. Parker, P. L. Joyce, J. Kim, and M. L. Nibert. 2002. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J. Virol. 76:8285-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandran, K., D. L. Farsetta, and M. L. Nibert. 2002. Strategy for nonenveloped virus entry: a hydrophobic conformer of reovirus penetration protein μ1 mediates membrane disruption. J. Virol. 76:9920-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandran, K., and M. L. Nibert. 2003. Animal cell invasion by a large nonenveloped virus: reovirus delivers the goods. Trends Microbiol. 11:374-382. [DOI] [PubMed] [Google Scholar]
- 17.Chandran, K., and M. L. Nibert. 1998. Protease cleavage of reovirus capsid protein μ1/μ1C is blocked by alkyl sulfate detergents, yielding a new type of infectious subvirion particle. J. Virol. 72:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandran, K., J. S. L. Parker, M. Ehrlich, T. Kirchhausen, and M. L. Nibert. 2003. The δ region of outer-capsid protein μ1 undergoes conformational change and release from reovirus particles during cell entry. J. Virol. 77:13361-13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandran, K., S. B. Walker, Y. Chen, C. M. Contreras, L. A. Schiff, T. S. Baker, and M. L. Nibert. 1999. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J. Virol. 73:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandran, K., X. Zhang, N. O. Olson, S. B. Walker, J. D. Chappell, T. S. Dermody, T. S. Baker, and M. L. Nibert. 2001. Complete in vitro assembly of the reovirus outer capsid produces highly infectious particles suitable for genetic studies of the receptor-binding protein. J. Virol. 75:5335-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chappell, J. D., E. S. Barton, T. H. Smith, G. S. Baer, D. T. Duong, M. L. Nibert, and T. S. Dermody. 1998. Cleavage susceptibility of reovirus attachment protein σ1 during proteolytic disassembly of virions is determined by a sequence polymorphism in the σ1 neck. J. Virol. 72:8205-8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chappell, J. D., A. E. Porta, T. S. Dermody, and T. Stehle. 2002. Crystal structure of reovirus attachment protein σ1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 21:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chow, M., J. F. Newman, D. Filman, J. M. Hogle, D. J. Rowlands, and F. Brown. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327:482-486. [DOI] [PubMed] [Google Scholar]
- 24.Curry, S., M. Chow, and J. M. Hogle. 1996. The poliovirus 135S particle is infectious. J. Virol. 70:7125-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danthi, P., M. Tosteson, Q. H. Li, and M. Chow. 2003. Genome delivery and ion channel properties are altered in VP4 mutants of poliovirus. J. Virol. 77:5266-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dermody, T. S., M. L. Nibert, R. Bassel-Duby, and B. N. Fields. 1990. A σ1 region important for hemagglutination by serotype 3 reovirus strains. J. Virol. 64:5173-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drayna, D., and B. N. Fields. 1982. Activation and characterization of the reovirus transcriptase: genetic analysis. J. Virol. 41:110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dryden, K. A., D. L. Farsetta, G. Wang, J. M. Keegan, B. N. Fields, T. S. Baker, and M. L. Nibert. 1998. Internal structures containing transcriptase-related proteins in top component particles of mammalian orthoreovirus. Virology 245:33-46. [DOI] [PubMed] [Google Scholar]
- 29.Dryden, K. A., G. Wang, M. Yeager, M. L. Nibert, K. M. Coombs, D. B. Furlong, B. N. Fields, and T. S. Baker. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 122:1023-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebert, D. H., J. Deussing, C. Peters, and T. S. Dermody. 2002. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 277:24609-24617. [DOI] [PubMed] [Google Scholar]
- 31.Fisher, A. J., and J. E. Johnson. 1993. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature 361:176-179. [DOI] [PubMed] [Google Scholar]
- 32.Fricks, C. E., and J. M. Hogle. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furuichi, Y., S. Muthukrishnan, J. Tomasz, and A. J. Shatkin. 1976. Caps in eukaryotic mRNAs: mechanism of formation of reovirus mRNA 5′-terminal m7GpppGm-C. Prog. Nucleic Acid Res. Mol. Biol. 19:3-20. [DOI] [PubMed] [Google Scholar]
- 35.Gallagher, T. M., and R. R. Rueckert. 1988. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J. Virol. 62:3399-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Havelka, W. A., R. Henderson, and D. Oesterhelt. 1995. Three-dimensional structure of halorhodopsin at 7 Å resolution. J. Mol. Biol. 247:726-738. [DOI] [PubMed] [Google Scholar]
- 37.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 38.Hogle, J. M. 2002. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 56:677-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 40.Jané-Valbuena, J., L. A. Breun, L. A. Schiff, and M. L. Nibert. 2002. Sites and determinants of early cleavages in the proteolytic processing pathway of reovirus surface protein σ3. J. Virol. 76:5184-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson, J. E. 1996. Functional implications of protein-protein interactions in icosahedral viruses. Proc. Natl. Acad. Sci. USA 93:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joklik, W. K. 1972. Studies on the effect of chymotrypsin on reovirions. Virology 49:700-715. [DOI] [PubMed] [Google Scholar]
- 43.Joklik, W. K., and M. R. Roner. 1995. What reassorts when reovirus genome segments reassort? J. Biol. Chem. 270:4181-4184. [DOI] [PubMed] [Google Scholar]
- 44.Lee, P. W. K., and R. Gilmore. 1998. Reovirus cell attachment protein σ1: structure-function relationships and biogenesis. Curr. Top. Microbiol. Immunol. 233:137-154. [DOI] [PubMed] [Google Scholar]
- 45.Lee, P. W. K., E. C. Hayes, and W. K. Joklik. 1981. Characterization of anti-reovirus immunoglobulins secreted by cloned hybridoma cell lines. Virology 108:134-146. [DOI] [PubMed] [Google Scholar]
- 46.Lee, W.-M., S. S. Monroe, and R. R. Rueckert. 1993. Role of maturation cleavage in infectivity of picornaviruses: activation of an infectosome. J. Virol. 67:2110-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, Q., A. G. Yafal, Y. M. Lee, J. Hogle, and M. Chow. 1994. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of these sequences at physiological temperature. J. Virol. 68:3965-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liemann, S., K. Chandran, T. S. Baker, M. L. Nibert, and S. C. Harrison. 2002. Structure of the reovirus membrane-penetration, μ1, in a complex with its protector protein, σ3. Cell 108:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lucia-Jandris, P., J. W. Hooper, and B. N. Fields. 1993. Reovirus M2 gene is associated with chromium release from mouse L cells. J. Virol. 67:5339-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez, C. G., R. Guinea, J. Benavente, and L. Carrasco. 1996. The entry of reovirus into L cells is dependent on vacuolar proton-ATPase activity. J. Virol. 70:576-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendez, I. I., Y. M. She, W. Ens, and K. M. Coombs. 2003. Digestion pattern of reovirus outer capsid protein σ3 determined by mass spectrometry. Virology 311:289-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moscufo, N., A. G. Yafal, A. Rogove, J. Hogle, and M. Chow. 1993. A mutation in VP4 defines a new step in the late stages of cell entry by poliovirus. J. Virol. 67:5075-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munshi, S., L. Liljas, J. Cavarelli, W. Bomu, B. McKinney, V. Reddy, and J. E. Johnson. 1996. The 2.8 Å structure of a T = 4 animal virus and its implications for membrane translocation of RNA. J. Mol. Biol. 261:1-10. [DOI] [PubMed] [Google Scholar]
- 54.Nibert, M. L., and B. N. Fields. 1992. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 66:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nibert, M. L., L. A. Schiff, and B. N. Fields. 1991. Mammalian reoviruses contain a myristoylated structural protein. J. Virol. 65:1960-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Odegard, A. L., K. C. Chandran, S. Liemann, S. C. Harrison, and M. L. Nibert. 2003. Disulfide bonding among μ1 trimers in mammalian reovirus outer capsid: a late and reversible step in virion morphogenesis. J. Virol. 77:5389-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parker, J. S., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 59.Schneemann, A., W. Zhong, T. M. Gallagher, and R. R. Rueckert. 1992. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 66:6728-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shing, M., and K. M. Coombs. 1996. Assembly of the reovirus outer capsid requires μ1/σ3 interactions which are prevented by misfolded σ3 protein in temperature-sensitive mutant tsG453. Virus Res. 46:19-29. [DOI] [PubMed] [Google Scholar]
- 61.Smith, R. E., H. J. Zweerink, and W. K. Joklik. 1969. Polypeptide components of virions, top component and cores of reovirus type 3. Virology 39:791-810. [DOI] [PubMed] [Google Scholar]
- 62.Sturzenbecker, L. J., M. Nibert, D. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tao, Y., D. L. Farsetta, M. L. Nibert, and S. C. Harrison. 2002. RNA synthesis in a cage—structural studies of the reovirus polymerase λ3. Cell 111:733-745. [DOI] [PubMed] [Google Scholar]
- 64.Taylor, D. J., N. K. Krishna, M. A. Canady, A. Schneemann, and J. E. Johnson. 2002. Large-scale, pH-dependent, quaternary structure changes in an RNA virus capsid are reversible in the absence of subunit autoproteolysis. J. Virol. 76:9972-9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tillotson, L., and A. J. Shatkin. 1992. Reovirus polypeptide σ3 and N-terminal myristoylation of polypeptide μ1 are required for site-specific cleavage to μ1C in transfected cells. J. Virol. 66:2180-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tosteson, M. T., and M. Chow. 1997. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J. Virol. 71:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Virgin, H. W., IV, M. A. Mann, B. N. Fields, and K. L. Tyler. 1991. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J. Virol. 65:6772-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zlotnick, A., V. S. Reddy, R. Dasgupta, A. Schneemann, W. J. Ray, Jr., R. R. Rueckert, and J. E. Johnson. 1994. Capsid assembly in a family of animal viruses primes an autoproteolytic maturation that depends on a single aspartic acid residue. J. Biol. Chem. 269:13680-13684. [PubMed] [Google Scholar]