Abstract
Hepatitis C virus (HCV) infection causes hepatitis, hepatocellular carcinoma, and B-cell lymphomas in a significant number of patients. Previously we have shown that HCV infection causes double-stranded DNA breaks and enhances the mutation frequency of cellular genes, including proto-oncogenes and immunoglobulin genes. To determine the mechanisms, we studied in vitro HCV infection of cell culture. Here we report that HCV infection activated the immunologic (type II) isoform of nitric oxide (NO) synthase (NOS), i.e., inducible NOS (iNOS), thereby inducing NO, which in turn induced DNA breaks and enhanced the mutation frequencies of cellular genes. Treatment of HCV-infected cells with NOS inhibitors or small interfering RNA specific for iNOS abolished most of these effects. Expression of the core protein or nonstructural protein 3 (NS3), but not the other viral proteins, in B cells or hepatocytes induced iNOS and DNA breaks, which could be blocked by NOS inhibitors. The core protein also enhanced the mutation frequency of cellular genes in hepatocytes derived from HCV core transgenic mice compared with that in control mice. The iNOS promoter was activated more than fivefold in HCV-infected cells, as revealed by a luciferase reporter assay driven by the iNOS promoter. Similarly, the core and NS3 proteins also induced the same effects. Therefore, we conclude that HCV infection can stimulate the production of NO through activation of the gene for iNOS by the viral core and NS3 proteins. NO causes DNA breaks and enhances DNA mutation. This sequence of events provides a mechanism for HCV pathogenesis and oncogenesis.
Hepatitis C virus (HCV) infects more than 170 million people in the world. The importance of HCV infection in hepatocellular carcinomas (HCC) and non-Hodgkin's B-cell lymphomas has been well documented (6, 29), indicating that HCV is a unique nonretroviral oncogenic RNA virus. From the genetic perspective, cancer represents the phenotypic consequence of the accumulation of mutations or deletions of DNA involving the inactivation of tumor suppressor genes and the activation of oncogenes by a stepwise process. We have recently demonstrated that HCV infection induces DNA damage that leads to a mutator phenotype (15).
HCV-induced inflammation and oxidative stress have been implicated as risk factors for liver damage and cancer development (13, 21, 22). HCV infection induces the production of total nitric oxide (NO), i.e., NOX which includes both nitrites (NO2−) and nitrates (NO3−); there is an association between the viral load and the level of NOX in the serum of HCV-infected patients (17). In several human gastrointestinal neoplasms, including HCV-associated HCC, the immunologic (type II) isoform of NO synthase (NOS), i.e., inducible NOS (iNOS), which generates NO from l-arginine in inflamed tissues, is elevated (24). The enhanced levels of iNOS in HCV-associated T lymphocytes correlated with the high level of expression of HCV proteins (32). Therefore, one of the means by which HCV exerts its effects upon infected cells is likely oxidative stress, including NO production.
NO plays an important role in many physiological and pathological conditions, serving as an intercellular and intracellular messenger and antimicrobial agent (9, 18, 19). Overproduction of NO in HCV infection is likely the result of activation of iNOS. Cloning and functional analysis of the human iNOS gene promoter have identified several copies of nuclear factor κB (NF-κB) response elements and several copies of activator protein 1 (AP-1) binding sites (16). The plasmid DNA and cellular genes exposed to exogenous NO gave rise to mutations when replicated in Escherichia coli or in mammalian cells (20, 27). In addition, NO can kill cells through both necrotic and apoptotic pathways; acute enhancement of the NO level produces killing through necrosis, while chronic NO produces predominantly apoptotic features (12). The DNA damage and apoptosis induced by NO may result in genome instability in HCV-infected cells. Furthermore, NO production in macrophages has been reported to induce DNA mutations (38).
These observations suggested that NO production by HCV infection plays an important role in the initiation and promotion or progression of cancers arising from HCV-infected tissues. Despite the reported clinical association, the molecular mechanisms linking HCV infection to the induction of iNOS and malignant transformation remain obscure. To answer these questions, we examined whether the HCV-induced DNA damage and mutations depend on the activation of iNOS expression and production of NO. For this purpose, we used a recently established in vitro HCV infection system (34) and a transgenic mouse model. We assessed the effects of HCV infection on the iNOS-induced genetic instability in human B cells and hepatocytes.
MATERIALS AND METHODS
Cell culture.
Raji (an established human B lymphoblastoma cell line) cells were obtained from the American Type Culture Collection and grown in RPMI 1640 medium (Invitrogen) containing 20% fetal bovine serum. Raji cells were further cloned by single-cell dilution; individual cell clones were selected and infected in vitro with the culture supernatant from an HCV-producing lymphoma cell line (SB cells) (34) and maintained for various periods of time in culture. The JT cell line was established by in vitro infection of peripheral blood mononuclear cells from a healthy donor with Epstein-Barr virus and HCV from SB cells simultaneously (34). The immortalized cells were further cloned by single-cell dilution. A control infection with UV-irradiated SB culture supernatant was included in both cases. Mouse embryonic fibroblasts (MEFs) were established from HCV core transgenic mice (unpublished data).
To induce human iNOS, cells were treated with a mixture of recombinant human cytokines, including gamma interferon (Boehringer Mannheim) at 100 U/ml, interleukin-1β at 0.5 ng/ml, and tumor necrosis factor alpha (Pharmingen) at 10 ng/ml.
Mice.
For animal studies, mice expressing HCV core gene genotype 1b under control of the human elongation factor 1a promoter were generated and bred at the University of Southern California transgenic mouse facility (unpublished data). The primary mouse fibroblast cultures were prepared from both core transgenic mouse and littermate embryos by trypsinizing the embryonic tissue and plating the dissociated cells.
Plasmids.
The various expression plasmids were constructed by inserting the HCV core, E2, NS3, NS4B, NS5A, and NS5B cDNAs behind the cytomegalovirus (CMV) major immediate-early promoter/enhancer in pCDNA3.1 (Invitrogen). E1 expression vector was kindly provided by T. Miyamura, Tokyo, Japan. The reporter plasmid used for analyzing iNOS promoter activity, pGL3-NOS 8.3 (16), contains 8.3 kb of the human iNOS promoter cloned into the pGL3-basic luciferase reporter gene vector (Promega). Plasmids pGL3-336, -911, -2483, -3665, and -5774, which contain various fractions of the iNOS promoter, were kindly provided by Joel Moss and Arnold S. Kristof, National Institutes of Health, Bethesda, Md. (16).
Cloning and sequencing.
Genomic DNA was extracted by standard methods from the various cell lines and hepatocytes from HCV core transgenic mice. PCR amplification was performed with Pfu Turbo DNA polymerase (Stratagene) and the reported primers for VH (from VH Framework 1 to constant region 1 of heavy-chain genes) (8) and p53 (exons 5 to 8) (5, 33). Pfu Turbo DNA polymerase has an expected error rate of about 4 × 10−5 mutations after 30 cycles of PCR amplification (about 1.3 × 10−6 changes/base/cycle). The purified PCR products were further incubated with Taq polymerase (Roche) and 0.2 mM dATP for 15 min at 72°C. The PCR products were ligated into the TOPO cloning vector (Invitrogen), and individual clones containing an insert of the expected size were sequenced by Laragen, Inc. (Los Angeles, Calif.), with T7 and T3 primers or p53 sequencing primers (5). For each PCR product, 20 to 40 individual clones were sequenced. PCR products made from DNA of mouse tumors were purified with a QIAquick gel extraction kit (QIAGEN) and used directly for sequencing.
Sequences of PCR products were compared to the corresponding germ line sequences with DNASIS-Mac v3.0 (Hitachi Software Engineering Co.). Nucleotide changes corresponding to the reported polymorphism or present in normal DNA from the same control cell line were excluded from the calculation of mutation frequencies. The identical base changes in colinear DNA sequence in different clones, which were presumably derived from the same mutational events (shared mutations), were counted only once in the census of somatic point mutations. The common heterozygous mutations in the p53 gene (codon 213 CGA→CAA and codon 234 TAC→CAC) of Raji cells were excluded from the calculation.
Reverse transcription (RT)-PCR.
Raji cells were lysed with TRI reagent (Molecular Research Center). The extracted RNA was reverse transcribed with oligo(dT) primers (New England Biolabs) and Superscript II enzyme (Invitrogen) in accordance with the manufacturer's protocol. The cDNA product was diluted fivefold with water sequentially twice; 1 μl each of these dilutions was used in a PCR. Taq polymerase (Roche) was used for amplification with primers specific for iNOS (10) and β-actin (36), yielding 289- and 600-bp PCR products, respectively. Amplification was performed for 22 to 32 cycles. HCV RNA was detected by a previously described procedure (34). The PCR products were analyzed by electrophoresis on 2% agarose gel.
Transfection and luciferase assay.
Cells were transiently transfected with FuGENE transfection reagent (Roche) or gene pulser II (Bio-Rad) with a construct containing the luciferase reporter gene. Briefly, at 50% confluence, cells were transfected with a mixture of 1 μg of DNA of the reporter plasmids and 3 μl of FuGENE reagent and incubated for 6 h at 37°C; cells were then washed, and fresh medium was added. Cells were harvested at the indicated time points, and luciferase activity was determined with a Luciferase Assay System Kit (Promega). Briefly, cells were washed twice in phosphate-buffered saline and lysed by adding 200 μl of a lysis buffer (Promega). After 15 min at room temperature, the lysate was removed and centrifuged. To 20 μl of supernatant, 100 μl of a luciferase assay reagent was added, and firefly luciferase activity was measured in a Lumat LB9501 luminometer (Berthold, Wildbad, Germany) in accordance with the manufacturer's guide.
Ligation-mediated PCR (LM-PCR) analysis of DSBs.
double-strand DNA breaks (DSBs) were identified by a modification of the previously reported procedure (31). Detailed descriptions of the methods used have been published previously (15).
Determination of NOX by Griess reaction.
NOX was determined by measuring the formation of both stable oxidation products of NO, namely, nitrites (NO2−) and nitrates (NO3−). The NOX concentration in 100 μl of culture supernatant was determined by Griess reagent reaction (Promega) as described before (3). The samples were incubated at room temperature for 10 min before their optical A550 was measured. With a standard curve, the absorbance of the samples was converted to micromolar NO.
RNA interference with siRNA.
The small interfering RNAs (siRNAs) used (synthesized by the University of Southern California Microchemical Core) were selected in accordance with the guidelines of Elbashir et al. (4) with the following sequences: NOS-S sense strand, 5′-ACAACAGGAACCUACCAGCTT-3′, and NOS-AS antisense strand, 5′-GCUGGUAGGUUCCUGUUGUTT-3′. The sense and antisense strands of the RNA were annealed at a concentration of 80 mM in 10 mM Tris (pH 7.7)-1 mM EDTA-100 mM NaCl by heating to 90°C for 1 min and then cooling in a thermocycler at a rate of 0.1°C/s until 22°C was reached. To transfect Raji cells with siRNAs, 2 × 105 cells were washed, resuspended in 50 μl of serum- and antibiotic-free RPMI medium, and cultured in a 96-well tissue culture dish. A preincubated mixture of 100 pmol of siRNA and 0.8 μl of Oligofectamine (total volume of 50 μl; Invitrogen) was added to Raji cells, and the mixture was incubated overnight at 37°C as previously described (35). The transfection efficiency was determined with control (nonsilencing) siRNA labeled with rhodamine (QIAGEN) to be more than 80%. Cells were resuspended in 200 μl of RPMI medium containing 20% fetal bovine serum and infected with HCV-containing SB culture supernatant (34). To prolong the effects of siRNA, the HCV-infected, siRNA-transfected cells were retransfected with the same siRNA at day 4. The samples were harvested at various time points after infection. Nonfunctional siRNAs (Ambion) were used as controls.
Western blot assay.
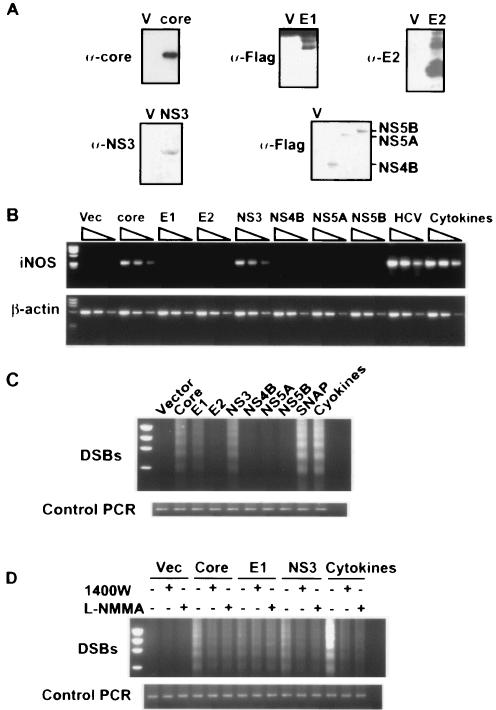
The expression of the core, Flag-E1, E2, NS3, Flag-NS4B, Flag-NS5A, and Flag-NS5B proteins was analyzed by Western blotting. Exponentially growing cells were harvested with a lysis buffer containing 50 mM Tris-HCl (pH 8), 150 mM NaCl, 1% Nonidet P-40, and protease inhibitor cocktail (Roche). Proteins were resolved by electrophoresis in sodium dodecyl sulfate-polyacrylamide gels and electrophoretically transferred onto nitrocellulose membranes (Amersham Biosciences). The membrane was incubated with core (Anogen)-, E2 (Biodesign)-, NS3 (Novocastra Laboratories)- or Flag-specific monoclonal antibodies (Sigma) and then reacted with a peroxidase-conjugated secondary antibody. Immunoreactivity was visualized by an enhanced chemiluminescence detection system (Amersham Biosciences).
Statistical analysis.
Statistical analysis of the data in the tables was performed by the χ2 test. Values of P < 0.05 were considered to be statistically significant.
Nucleotide sequence accession number.
The sequence of p53 has been submitted to the GenBank database and assigned accession number U94788.
RESULTS
HCV infection induces iNOS expression.
To examine whether NO is involved in HCV-induced pathogenesis, we first determined the level of human iNOS mRNA in HCV-infected Raji cells by RT-PCR as a semiquantitative analysis, with the human β-actin mRNA as an endogenous internal standard. HCV-infected Raji cells showed a higher level of iNOS mRNA at day 8 postinfection than that in their uninfected counterparts (Fig. 1A). The iNOS expression level in HCV-infected cells was almost the same as that in the cells treated with a mixture of stimulatory cytokines that have been shown to induce iNOS expression (16). We had determined that the iNOS and β-actin cDNA amplification under this condition occurred in a linear range, ensuring accurate determination of the ratio of iNOS mRNA to β-actin mRNA (data not shown). We further followed the kinetics of iNOS mRNA activation at various time points following HCV infection. The activation of iNOS was confirmed at several time points postinfection; the iNOS mRNA level started to increase as early as day 2 postinfection, progressively increased, and was maintained at the high level at least until day 28 postinfection (Fig. 1B).
FIG. 1.
Detection and kinetic analysis of iNOS mRNA expression. (A) cDNA from Raji cells infected with HCV and control cells was serially diluted (0-, 5-, and 25-fold) and used for PCR amplification to detect iNOS mRNA (32 PCR cycles). β-Actin mRNA levels (22 PCR cycles) were used as a control. These experiments were repeated three times with similar results. (B) Kinetics of iNOS expression in HCV-infected Raji cells were followed from day 2 up to day 28 postinfection (p.i.). HCV RNA was detected by RT-PCR assay. (C) NO3−-NO2− production in cell culture medium in the presence or absence of 1400W or L-NMMA. For a positive control, cells were treated with a mixture of cytokines (Cyt) (interleukin-1β at 0.5 ng/ml; gamma interferon at 100 U/ml, and tumor necrosis factor alpha at 10 ng/ml) or 0.3 mM SNAP, releasing endogenous and exogenous NO, respectively. HCV (−), cells treated with UV-inactivated HCV.
To confirm that the iNOS induction seen is functionally significant, we determined the nitrite and nitrate levels in the culture supernatant of infected cells. The amount of nitrites and nitrates produced from HCV-infected cells was significantly higher than that from mock-infected cells or cells incubated with the UV-inactivated culture supernatant (Fig. 1C). Treatment of HCV-infected Raji cells with the iNOS-specific inhibitor 1400W or the general NOS inhibitor NG-monomethyl-l-arginine (L-NMMA) suppressed the elevation of the nitrite and nitrate levels in the supernatant of HCV-infected cell culture (Fig. 1C). As a positive control, cells treated with the stimulatory cytokines or the NO donor SNAP (S-nitroso-N-acetylpenicillamine) also produced higher levels of nitrites and nitrates. These results demonstrated that HCV infection enhanced the production of nitrites and nitrates as a result of iNOS activation.
HCV-induced DNA damage is NO dependent.
Since NO has been reported to induce DNA damage (10) and we recently reported that HCV infection induced DNA damage (15), we next determined whether the DNA damage associated with HCV infection was caused by iNOS activation. We detected DSBs by LM-PCR assay. As a positive control, NO donor SNAP or cytokine mixtures induced strong DSBs (Fig. 2A), confirming that NO induces DNA breaks. Significantly, HCV infection of Raji cells induced detectable DSBs whereas mock-infected cells or cells incubated with UV-irradiated HCV supernatant did not generate detectable DSBs. DSBs were also detected in HCV-infected JT cells that were established by Epstein-Barr virus-induced transformation (34) (Fig. 2B). The generation of DSBs induced both by HCV infection and by the stimulatory cytokines was substantially inhibited by the iNOS inhibitor 1400W or L-NMMA, indicating that the generation of DSBs occurred mainly through the production of NO as a result of iNOS induction (Fig. 2A and B).
FIG. 2.
HCV infection induces NO-dependent DNA damage. (A and B) DSBs in HCV-infected and uninfected Raji (A) or JT (B) cells in the presence or absence of NOS inhibitors were detected by LM-PCR and separated by gel electrophoresis as previously described (15, 31). A control PCR assay was done with β-actin. (C and D) The region-specific DSBs were detected with p53- and VH-specific primers (15). (E) HCV RNA was detected by RT-PCR assay. (F) Control PCR: amplification of the VH gene of genomic DNA as an internal control.
We have previously demonstrated that HCV-associated DSBs could be induced in both the p53 and VH genes, probably by different mechanisms (15). We therefore examined the effects of iNOS inhibitors on DSBs observed in these two loci. The results showed that DSBs could be detected in both regions in HCV-infected cells but not in uninfected cells (Fig. 2C and D). Interestingly, the generation of DSBs in the p53 region of HCV-infected cells was substantially, but not completely, inhibited by the NOS inhibitor 1400W or L-NMMA (Fig. 2C). In comparison, the generation of DSBs in the VH gene was also reduced, but to a lesser extent than the reduction observed in the p53 gene, by the iNOS inhibitor 1400W or L-NMMA (Fig. 2D). These results indicate that NO production is largely responsible for the HCV-induced DSBs, but other mechanisms are also involved, particularly in the case of the VH gene. HCV RNA was detected by RT-PCR in cells infected with HCV (Fig. 2E), confirming that the cells were indeed infected. Figure 2F serves as a loading control for input DNA. The ability of NOS inhibitors to provide substantial protection against the generation of HCV-induced DSBs implies that HCV-induced iNOS and the resulting NO formation contributes to DSBs in HCV-infected cells.
iNOS-specific siRNA lowers the incidence of HCV-induced DSBs.
To further establish that HCV-induced iNOS is responsible for the generation of DSBs, we next examined the effects of the iNOS-specific siRNA. We first evaluated the activity of an siRNA targeted to all iNOS transcripts. The iNOS-specific siRNA was transfected into Raji cells; 2 days later, the cells were infected with HCV. The iNOS protein or mRNA was examined at day 8 postinfection. Introduction of the iNOS-specific siRNA almost completely eliminated the expression of iNOS protein in HCV-infected cells (Fig. 3A). Correspondingly, the iNOS mRNA level in HCV-infected cells was also substantially reduced (Fig. 3B). The control siRNA did not have any effects on the expression levels of iNOS protein or mRNA (Fig. 3A and B). The expression of iNOS siRNA did not prevent viral infection, as HCV RNA could be detected in the treated cell culture (Fig. 3A). The expression levels of β-actin protein (Fig. 3A) or mRNA (Fig. 3B) were not affected by the iNOS siRNA. As shown above, neither iNOS protein nor iNOS mRNA could be detected in the uninfected cells. We next determined whether the introduction of siRNAs affected the generation of DSBs in HCV-infected Raji cells. Introduction of the iNOS-specific siRNA caused a significant reduction of DSBs in HCV-infected cells compared to that caused by the control siRNA (Fig. 3C). These results together established that activation of iNOS is largely responsible for HCV-induced DSBs.
FIG. 3.
iNOS-specific siRNA lowers HCV-induced DSBs. (A and B) Effect of siRNA on levels of iNOS protein (A) and RNA (B) in HCV-infected cells. (A) Western blot analysis shows iNOS protein expression at day 8 postinfection in HCV-infected cells. A Western blot assay of β-actin was used as a loading control. HCV RNA was detected by RT-PCR. (B) iNOS and β-actin mRNAs in serial dilutions from Raji cells transfected with iNOS or control siRNA. (C) DSBs in cells transfected with iNOS or control siRNA. DSBs were examined at day 8 postinfection.
The iNOS inhibitor partially reduces the mutation frequency of cellular genes.
We have recently found that HCV infection induces increases in the mutation frequency of several cellular genes (15). To address whether the HCV-induced NO was responsible for these mutagenic effects, we determined the mutation frequency of cellular genes in HCV-infected Raji cells in the presence or absence of iNOS inhibitors. We chose p53 for this analysis. The iNOS inhibitor was added at the beginning of HCV infection and maintained throughout the study period. The mutation frequency was determined by sequencing multiple PCR clones of p53 genes (exons 5 to 8) (5). The results showed that the treatment of HCV-infected Raji cells with iNOS inhibitors reduced the mutation frequency of the p53 gene to nearly the level of the uninfected cells at 8 days postinfection (Table 1). This result indicates that HCV-induced NO production is largely responsible for the enhancement of mutations in p53. The same treatment only partially reduced the mutation frequency of the VH gene, suggesting that the enhancement of VH mutations was caused by both NO production and other additional mechanisms.
TABLE 1.
Effects of iNOS inhibitor 1400W on mutation frequencies of p53 and VH in HCV-infected human B-cell linesa
| Locus | HCV infection | iNOS inhibitor 1400W | % of clones mutatedb | Mutation frequencyc (mutations/bp, 104) |
|---|---|---|---|---|
| p53 | − | − | 0 (0/35) | 0 |
| − | + | 3 (1/32) | 0.8 | |
| + | − | 24 (7/42) | 4.2 | |
| + | + | 2 (1/42) | 0.6d | |
| VH | − | − | 16 (5/31) | 4.1 |
| − | + | 14 (4/29) | 4.7 | |
| + | − | 77 (24/31) | 20.9 | |
| + | + | 29 (10/34) | 7.4d |
Raji cells were infected with HCV-containing culture supernatant of SB cells (34) in the presence or absence of 1400W. Cells were harvested for DNA sequence analysis on day 8 postinfection. Identical mutations in different plasmid clones from the same individual Raji cells were counted only once unless genealogies indicated that the mutations were unique.
The number of PCR clones containing at least a mutation versus the total number of PCR clones sequenced shown in parenthesis.
Mutation frequency was calculated as the number of mutations versus the total number of base pairs sequenced.
P < 0.05.
The core and NS3 proteins are responsible for HCV-associated iNOS induction.
To determine which viral genes are responsible for the activation of iNOS, the occurrence of DNA damage, and the enhanced mutation of cellular genes, we performed transient transfection of various HCV genes individually into Raji cells. Following transient transfection of the expression vectors, the expression of the various HCV proteins in the transfected cells was confirmed by Western blot analysis with specific antibodies (Fig. 4A). The ability of each viral protein to induce iNOS mRNA was examined at 48 h posttransfection. The results showed that of all of the HCV proteins examined, the core and NS3 proteins induced iNOS mRNA expression, whereas none of the other proteins, including E1, E2, NS4B, NS5A, and NS5B, elevated the iNOS mRNA level in Raji cell cultures (Fig. 4B). To examine whether there is a correlation between iNOS expression and DSBs, these transfected cells were studied for DSBs. Of the viral proteins examined, core, NS3 and, to a lesser extent, E1 proteins, enhanced DSBs (Fig. 4C). None of the other viral proteins caused DSBs. These results concerning the core and NS3 proteins were consistent with the iNOS induction by these two proteins. However, the generation of DSBs associated with E1 protein expression was unexpected. To determine whether DNA damage associated with the expression of the core and NS3 proteins were caused by iNOS expression, we added iNOS inhibitors to these transfected cells. The DSBs induced by the core and NS3 proteins were significantly, but not completely, reduced by the inhibitors of iNOS, but the E1-induced DSBs, which were not as pronounced as those induced by either the core or NS3 protein, were not affected by the iNOS inhibitors (Fig. 4D). These results suggest that core and NS3 protein-induced DSBs occurred mainly through the generation of NO, whereas E1-induced DSBs were caused by other mechanisms that are independent of iNOS induction. This conclusion is in agreement with the finding that E1 did not induce iNOS (Fig. 4B).
FIG. 4.
Core or NS3 protein induces NO-dependent DNA damage. (A) Raji cells were transfected with various expression plasmids. Expression of HCV proteins was determined by Western blot analysis with specific antibodies. V, vector-transfected cells. (B) iNOS mRNA expression in cells transfected with viral proteins was determined by a semiquantitative RT-PCR assay in Raji cell expressing various HCV proteins. Serial dilutions (0-, 5-, and 25-fold) of each cellular cDNA were used. (C) DSB formation was determined by LM-PCR in Raji cells expressing various HCV proteins. Vector, vector control; SNAP, Raji cells treated with the NO donor SNAP; Cytokines, Raji cells treated with a cytokine mixture. (D) DSBs in Raji cells transfected with various HCV proteins in the presence or absence of NOS inhibitors (1400W and L-NMMA). Vec, vector.
The core and NS3 proteins induce iNOS and DSBs in hepatocytes.
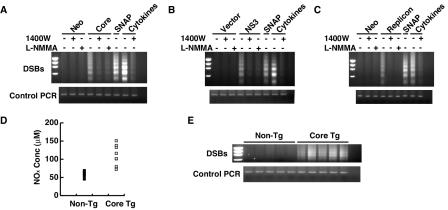
To determine whether the core and NS3 proteins induced iNOS expression and caused DSBs in cells other than B lymphocytes, we studied a HepG2 cell line stably expressing the core protein and an Huh7 cell line harboring an HCV subgenomic replicon (NS3-NS5B). An Huh7 cell line expressing a neomycin resistance-encoding gene was used as a control. The results showed that the core protein-expressing HepG2 cells had significantly enhanced DSBs compared to the neomycin-resistant control cells (Fig. 5A). The core protein-induced DSBs could be inhibited by iNOS inhibitors almost completely, suggesting that the core protein caused DSBs mainly by iNOS induction. Similarly, transient expression of NS3 in Huh7 cells also caused significantly more pronounced DSBs compared to those in the vector-transfected cells (Fig. 5B). Again, these DSBs were inhibited by treatment with iNOS inhibitors. In addition, cells harboring an HCV subgenomic replicon (NS3-NS5B) also had pronounced DSBs that were inhibited by the iNOS inhibitors (Fig. 5C). In contrast, overexpression of the NS4B, NS5A, and NS5B proteins alone did not elevate iNOS expression and did not generate a detectable level of DSBs in Huh7 cells (data not shown), similar to the finding in Raji cells. These results indicate that the core and NS3 proteins induced DSBs through iNOS activation not only in Raji cells but also in hepatocytes.
FIG. 5.
DSBs in hepatocytes expressing various HCV proteins. DSBs were determined by LM-PCR assay. (A) Core protein-expressing (Core) or neomycin-resistant (Neo) HepG2 cells. For some of the experiments indicated, the iNOS inhibitors 1400W and L-NMMA were added to the cell culture 5 days before the cells were harvested. (B) Huh7 cells transiently expressing NS3 or the vector plasmid. Cells were examined 2 days after transfection. (C) Huh7 cells with the HCV replicon. (D) NOX concentration in serum from HCV core transgenic (Tg) mice and littermates. (E) DSB formation in hepatocytes from HCV core transgenic mice. In panels D and E, each symbol and each lane represents an individual animal.
To determine whether the HCV core protein induces iNOS expression in vivo, we analyzed the nitrite and nitrate concentrations in the serum of core transgenic mice in comparison to those of their littermates. The serum of HCV core transgenic mice had a significantly higher level of nitrites and nitrates than did that of their nontransgenic littermates (P < 0.05, Fig. 5D). Correspondingly, hepatocytes from core transgenic mice had significantly enhanced DSBs compared to those of their nontransgenic littermates (Fig. 5E). We also found that MEFs of the core transgenic mice exhibited a much higher level of DSBs than did MEFs of their age-matched littermates (data not shown). These results indicate that the core protein induced iNOS expression and DSBs through an NO-dependent mechanism in hepatocytes, both in vitro and in vivo.
Expression of the HCV core and NS3 proteins enhances the frequency of mutations in tumor suppressor genes.
The data in Table 1 suggested that HCV-induced iNOS was most likely responsible for the increased mutations in cellular genes associated with HCV infection (15). Since the core and NS3 proteins were sufficient to induce iNOS in Raji cells or hepatocytes, we next examined whether the expression of these proteins alone could enhance mutations of cellular genes. Huh7 cells were transiently transfected with the core and NS3 proteins, and the p53 gene (exons 5, 6, 7, and 8) was amplified by PCR at 5 days posttransfection. Multiple PCR clones of each sample were sequenced to detect possible sequence mutations. The results showed that cells transiently transfected with the core or NS3 protein had a significantly higher mutation frequency (6- to 10-fold higher) than that detected in cells with the neomycin resistance transgene or the vector plasmid (P < 0.05) (Table 2). The mutation frequency in the cells transfected with the neomycin resistance gene or the vector plasmid was similar to that in the untransfected Huh7 cells. However, coexpression of the core and NS3 proteins did not show any additive effect over that of the core or NS3 protein alone (Table 2). We also performed a similar analysis of HepG2 cells stably transfected with the core gene (37). These cells also showed a mutation frequency (4.9 × 10−4) higher than that of the cells transformed with the neomycin resistance gene (Table 2).
TABLE 2.
Mutation frequencies in the p53 gene in various cellsa
| Cell line | Transgene | NT/Tb | % of sequences mutated | Mutation frequency (mutations/bp, 104) |
|---|---|---|---|---|
| Huh7 | Vector | 4 (1/24) | 0.9 | |
| Neo | 5 (1/22) | 1.0 | ||
| Core | 32 (6/19) | 8.2c | ||
| NS3 | 35 (7/20) | 8.9c | ||
| Core + NS3 | 30 (6/20) | 6.7c | ||
| HepG2 | Neo | 0 (0/19) | 0 | |
| Core | 17 (13/18) | 4.9c | ||
| Mouse | None | NT | 3 (2/58) | 0.8 |
| Core | NT | 21 (20/96) | 4.6d | |
| Core | T | 23 (10/44) | 5.1d |
Huh7 cells were transfected with the various plasmids, and DNA was harvested from cells at 5 days posttransfection. HepG2 cells were stable transformants of the core protein or neomycin resistance gene selected with neomycin. Tissues were derived from two different lineages of transgenic mice carrying the HCV core transgene.
Nontumor (NT) and tumor (T) tissues of livers derived from HCV core transgenic mice and their littermates were examined.
P < 0.05.
P < 0.01.
We further confirmed the mutagenic effects of the core protein in hepatocytes of core transgenic mice. Hepatocytes derived from these animals had a mutation frequency approximately fivefold higher than those of the wild-type, age-matched mice (Table 2). There was no significant difference between the mutation frequencies of the tumor and nontumor regions of the liver from the core transgenic mice, indicating that the mutations alone are not sufficient to cause tumors.
HCV proteins activate the iNOS promoter.
To understand the mechanism of induction of iNOS mRNA by HCV infection and the individual core and NS3 proteins, we used a reporter plasmid containing a luciferase gene under the control of the iNOS promoter to examine whether these effects were due to regulation at the transcription level. The reporter plasmid was transfected into HCV-infected and mock-infected Raji cells. Luciferase activity was at least fivefold higher in HCV-infected Raji cells than in uninfected cells (Fig. 6), suggesting that HCV can transactivate the iNOS promoter in B cells. When the reporter plasmid was cotransfected with the core or NS3 protein into uninfected Raji cells, a three- to fivefold induction of luciferase activity, compared to that of the cells transfected with the vector plasmid, was also observed. These results demonstrate that the transactivation of the iNOS promoter by HCV is most likely mediated by the core and NS3 proteins. To define the minimal iNOS promoter required for activation by HCV or by the core or NS3 protein, Raji cells infected with HCV or mock infected were transfected with a series of iNOS promoter truncation mutants. Alternatively, Raji cells were cotransfected with the core or NS3 protein expression plasmids and one of the iNOS promoter truncation mutants. After normalization of the protein amounts, it was observed that the serial truncation of the iNOS promoter proportionally lowered the levels of iNOS promoter activation. The smallest truncation mutant, pGL3-336, which contains only one NF-κB binding site, was still able to respond to the activation by HCV infection or core or NS3 transfection to at least a small extent. The extents of transactivation were comparable for the core and NS3 proteins, although the core protein appeared to have slightly higher transactivation activity. Furthermore, the levels of transactivation of the various iNOS promoter truncation mutants were proportional to the copy number of the binding sites for NF-κB or AP-1, suggesting that core or NS3 transactivation of the iNOS promoter was mediated by NF-κB and AP-1. These results together indicate that HCV caused upregulation of the gene for iNOS through the core and NS3 proteins at the transcriptional level.
FIG. 6.
Assays of iNOS promoter activity in HCV-infected cells and in cells transfected with the core or NS3 protein. (Top two panels) Mock-infected or HCV-infected Raji cells were transfected with the indicated luciferase reporter constructs containing different fractions of the iNOS promoter at 8 days postinfection of HCV. Luciferase activity was determined from the cell lysates at 2 days posttransfection. (Bottom three panels) Uninfected Raji cells were transfected with the vector or the core- or NS3-expressing plasmid and one of the luciferase reporter controls as described on the left. Luciferase activity was assayed at 2 days posttransfection. Data are the mean values of three experiments with assays done in triplicate, expressed as the luciferase activity (relative light units [RLU]) normalized for total protein content. Positions of NF-κB and AP-1 binding sites in the promoter are depicted.
DISCUSSION
This study, which was based mainly on an in vitro culture infection system and partially confirmed in transgenic mice, demonstrates that HCV infection induces iNOS expression and production of NO, which in turn causes DNA damage, resulting in enhanced mutations of cellular genes (Fig. 7). This putative sequential pathway can be blocked by treatment of HCV-infected cells with iNOS inhibitors or by the introduction of an iNOS-specific siRNA. Furthermore, the HCV core or NS3 protein alone can activate these sequential events. Therefore, the core and NS3 proteins may play a key role in HCV pathogenesis and oncogenesis. However, we cannot rule out the possible involvement of other viral proteins in the pathogenesis and oncogenesis of HCV infection. For example, E1 also induces DSBs through a mechanism not involving iNOS activation. These results combined suggested that the observed elevation of the mutation frequency of cellular genes associated with HCV infection was the consequence of early genetic events in HCV infection. Our results are consistent with the reports that the HCV core protein can activate the NF-κB pathway (11); it stands to reason that the core protein can activate the iNOS promoter, which includes several NF-κB binding sites. It has been shown recently that NS3 inhibits a cellular signaling molecule (interferon regulatory factor 3) and that the protease activity is responsible for this inhibition (7). Our results presented here have shown that NS3 has opposite effects on the activation of NF-κB and AP-1 transcription factors. We do not know whether the protease activity is required for this activation. Our results confirmed and further extended our previous findings that HCV infection causes DSBs and enhances mutations of cellular genes (15). We showed that the viral core and NS3 proteins alone can induce DSBs and enhance mutations of cellular DNAs. These effects were demonstrated not only in the transient transfection of these proteins in Raji cells and hepatocytes but also in the stable transformants of HepG2 cells expressing the core proteins and in the core transgenic mice. Furthermore, the core transgenic mice produce a higher NOX level in their serum. These results add further weight to the association of HCV infection with DSBs and enhanced mutations of cellular genes; i.e., HCV induces a mutator phenotype (15). It is significant that both the core and NS3 proteins have been shown to be capable of transforming cells under in vitro conditions (25, 30). Our findings here may have provided a mechanism for these transforming activities. Nevertheless, DNA mutations alone cannot fully account for tumor formation, as the tumor and nontumor regions of the livers of core transgenic mice had similar mutation frequencies (Table 2). Thus, additional potentiating events are necessary for triggering tumor formation. We have recently shown that HCV-associated B-cell lymphomas and HCC had amplification of mutations in several proto-oncogenes (15).
FIG. 7.
Postulated mechanism of HCV-induced DNA damage.
NO has been shown to cause predominantly transitional nucleotide substitutions of cellular genes (20). Interestingly, we have recently shown that HCV infection causes predominantly transition mutations in cellular genes (15). NO and reactive NO species, such as the strong oxidant peroxynitrite anion (ONOO−) and dinitrogen trioxide peroxynitrite, may cause DNA damage through three chemical mechanisms (9, 14, 19, 20). The first is the direct reaction of reactive NO species with DNA, which induces nitrosative or oxidative deamination of DNA bases by metabolically activated N-nitrosamines, leading to DNA strand breakage under cell-free conditions (20, 27). Indeed, addition of NO to TK6 cells produced a 40- to 50-fold increase in hypoxanthine and xanthine in cellular DNA (20). It has also been shown that gaseous NO can induce predominantly transitional mutations (28). Thus, HCV-induced NO has the ability to modify genomic DNA with transitional substitutions. The second is inhibition of DNA repair processes (10). The third is increased production of genotoxic species such as alkylating agents and hydrogen peroxide. Thus, production of NO may explain most of the mutations of cellular genes, such as p53, associated with HCV infection (15). Indeed, most of the mutations found in the p53 gene of HCV-infected cells were blocked by iNOS inhibitors (Table 1). At low physiological concentrations, NO inhibits apoptosis, but at higher concentrations, NO may be proapoptotic as a result of activation of caspases (Fig. 7) (12). Although NO has been reported to induce cell death (12), a low level of NO does not kill the cells but may activate angiogenesis and cell survival (19). Therefore, HCV-induced NO may either inhibit or promote cell survival. We have previously separated nonapoptotic from apoptotic cells with HCV-infected and uninfected B cells by Annexin V staining (15). The HCV-infected nonapoptotic cells had a significantly higher level of DSBs than did their uninfected counterparts, confirming the occurrence of spontaneous DNA strand breakage even in the absence of apoptosis (15).
A variety of viruses, including herpes simplex virus type 1, influenza virus, Sendai virus, coxsackieviruses, simian immunodeficiency virus, hepatitis B virus, and human immunodeficiency virus type 1, have been reported to induce iNOS, leading to NO production (2). HCV-infected patients also have higher iNOS levels (24, 32). NO production has been shown to play a role in viral clearance (2), immunopathology (26), and cognitive dysfunction (1). The mutagenic effects of NO reported here are a novel phenomenon associated with viral infection.
In contrast to those found in the p53 gene, the mutations found in the VH gene were not completely blocked by iNOS inhibitors, indicating that other mechanisms also contributed to DNA damage in these genes. We have shown that HCV activates AID (15), which also causes transition mutations in the RGYW motif of DNA (23). The viral protein responsible for the activation of AID has not been identified.
Our observations suggest that a large viral load, maybe even transiently, would increase the risk of generation of DNA damage, which leads to mutations of cellular genes. NO is a potent antimicrobial effector molecule capable of nitrating tyrosine residues of proteins into nitrotyrosine to exhibit antiviral activity against a wide range of viruses in rodents (19). Conceivably, NO may have an antiviral activity against HCV; this may explain the observed low levels of virus replication detectable in vivo even in the presence of significant liver damage. However, in this study, we did not see a significant change in the amount of HCV RNA in cells treated with iNOS inhibitors or iNOS-specific siRNA. Nevertheless, more quantitative studies are needed to examine the effects of NO on viral replication.
In conclusion, we have demonstrated that HCV induces the production of NO in hepatocytes and B cells by activating the iNOS promoter, which is mediated by two HCV proteins. Data from this model system support the notion that elaboration of a mutator phenotype is a relatively early event in HCV pathogenesis. The potential linkage of iNOS induction to HCV-associated dysfunction implies that inhibitors of iNOS could have therapeutic effects.
Acknowledgments
We thank Joel Moss and Arnold S. Kristof (National Institutes of Health, Bethesda, Md.) for iNOS reporter constructs and T. Miyamura (National Institute of Infectious Diseases, Tokyo, Japan) for E1 expression vector.
This project was supported by NIH research grants AI 40038 and CA108302.
REFERENCES
- 1.Adamson, D. C., B. Wildemann, M. Sasaki, J. D. Glass, J. C. McArthur, V. I. Christov, T. M. Dawson, and V. L. Dawson. 1996. Immunologic NO synthase: elevation in severe AIDS dementia and induction by HIV-1 gp41. Science 274:1917-1921. [DOI] [PubMed] [Google Scholar]
- 2.Akaike, T., and H. Maeda. 2000. Nitric oxide and virus infection. Immunology 101:300-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding, Y., and R. S. Rana. 1998. Nitric oxide does not initiate but potentiates glucose-induced insulin secretion in pancreatic beta-cells. Biochem. Biophys. Res. Commun. 251:699-703. [DOI] [PubMed] [Google Scholar]
- 4.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 5.Farrell, P. J., G. J. Allan, F. Shanahan, K. H. Vousden, and T. Crook. 1991. p53 is frequently mutated in Burkitt's lymphoma cell lines. EMBO J. 10:2879-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferri, C., F. Caracciolo, A. L. Zignego, L. La Civita, M. Monti, G. Longombardo, F. Lombardini, F. Greco, E. Capochiani, A. Mazzoni, et al. 1994. Hepatitis C virus infection in patients with non-Hodgkin's lymphoma. Br. J. Haematol. 88:392-394. [DOI] [PubMed] [Google Scholar]
- 7.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 8.Ivanovski, M., F. Silvestri, G. Pozzato, S. Anand, C. Mazzaro, O. R. Burrone, and D. G. Efremov. 1998. Somatic hypermutation, clonal diversity, and preferential expression of the VH 51p1/VL kv325 immunoglobulin gene combination in hepatitis C virus-associated immunocytomas. Blood 91:2433-2442. [PubMed] [Google Scholar]
- 9.Jaffrey, S. R., and S. H. Snyder. 1995. Nitric oxide: a neural messenger. Annu. Rev. Cell Dev. Biol. 11:417-440. [DOI] [PubMed] [Google Scholar]
- 10.Jaiswal, M., N. F. LaRusso, L. J. Burgart, and G. J. Gores. 2000. Inflammatory cytokines induce DNA damage and inhibit DNA repair in cholangiocarcinoma cells by a nitric oxide-dependent mechanism. Cancer Res. 60:184-190. [PubMed] [Google Scholar]
- 11.Kato, N., H. Yoshida, S. Kioko Ono-Nita, J. Kato, T. Goto, M. Otsuka, K. Lan, K. Matsushima, Y. Shiratori, and M. Omata. 2000. Activation of intracellular signaling by hepatitis B and C viruses: C-viral core is the most potent signal inducer. Hepatology 32:405-412. [DOI] [PubMed] [Google Scholar]
- 12.Kim, P. K., R. Zamora, P. Petrosko, and T. R. Billiar. 2001. The regulatory role of nitric oxide in apoptosis. Int. Immunopharmacol. 1:1421-1441. [DOI] [PubMed] [Google Scholar]
- 13.Lai, M. M. 2002. Hepatitis C virus proteins: direct link to hepatic oxidative stress, steatosis, carcinogenesis and more. Gastroenterology 122:568-571. [DOI] [PubMed] [Google Scholar]
- 14.Lyons, C. R. 1995. The role of nitric oxide in inflammation. Adv. Immunol. 60:323-371. [DOI] [PubMed] [Google Scholar]
- 15.Machida, K., K. T. Cheng, V. M. Sung, S. Shimodaira, K. L. Lindsay, A. M. Levine, M.-Y. Lai, and M. M. Lai. 2004. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc. Natl. Acad. Sci. USA 101:4262-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marks-Konczalik, J., S. C. Chu, and J. Moss. 1998. Cytokine-mediated transcriptional induction of the human inducible nitric oxide synthase gene requires both activator protein 1 and nuclear factor κB-binding sites. J. Biol. Chem. 273:22201-22208. [DOI] [PubMed] [Google Scholar]
- 17.Mihm, S., A. Fayyazi, and G. Ramadori. 1997. Hepatic expression of inducible nitric oxide synthase transcripts in chronic hepatitis C virus infection: relation to hepatic viral load and liver injury. Hepatology 26:451-458. [DOI] [PubMed] [Google Scholar]
- 18.Moncada, S., and A. Higgs. 1993. The l-arginine-nitric oxide pathway. N. Engl. J. Med. 329:2002-2012. [DOI] [PubMed] [Google Scholar]
- 19.Nathan, C. 1992. Nitric oxide as a secretory product of mammalian cells. FASEB J. 6:3051-3064. [PubMed] [Google Scholar]
- 20.Nguyen, T., D. Brunson, C. L. Crespi, B. W. Penman, J. S. Wishnok, and S. R. Tannenbaum. 1992. DNA damage and mutation in human cells exposed to nitric oxide in vitro. Proc. Natl. Acad. Sci. USA 89:3030-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohshima, H., and H. Bartsch. 1994. Chronic infections and inflammatory processes as cancer risk factors: possible role of nitric oxide in carcinogenesis. Mutat. Res. 305:253-264. [DOI] [PubMed] [Google Scholar]
- 22.Okuda, M., K. Li, M. R. Beard, L. A. Showalter, F. Scholle, S. M. Lemon, and S. A. Weinman. 2002. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology 122:366-375. [DOI] [PubMed] [Google Scholar]
- 23.Petersen-Mahrt, S. K., R. S. Harris, and M. S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418:99-103. [DOI] [PubMed] [Google Scholar]
- 24.Rahman, M. A., D. K. Dhar, E. Yamaguchi, S. Maruyama, T. Sato, H. Hayashi, T. Ono, A. Yamanoi, H. Kohno, and N. Nagasue. 2001. Coexpression of inducible nitric oxide synthase and COX-2 in hepatocellular carcinoma and surrounding liver: possible involvement of COX-2 in the angiogenesis of hepatitis C virus-positive cases. Clin. Cancer Res. 7:1325-1332. [PubMed] [Google Scholar]
- 25.Ray, R. B., L. M. Lagging, K. Meyer, and R. Ray. 1996. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J. Virol. 70:4438-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reiss, C. S., and T. Komatsu. 1998. Does nitric oxide play a critical role in viral infections? J. Virol. 72:4547-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Routledge, M. N. 2000. Mutations induced by reactive nitrogen oxide species in the supF forward mutation assay. Mutat. Res. 450:95-105. [DOI] [PubMed] [Google Scholar]
- 28.Routledge, M. N., D. A. Wink, L. K. Keefer, and A. Dipple. 1993. Mutations induced by saturated aqueous nitric oxide in the pSP189 supF gene in human Ad293 and E. coli MBM7070 cells. Carcinogenesis 14:1251-1254. [DOI] [PubMed] [Google Scholar]
- 29.Saito, I., T. Miyamura, A. Ohbayashi, H. Harada, T. Katayama, S. Kikuchi, Y. Watanabe, S. Koi, M. Onji, and Y. Ohta. 1990. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 87:6547-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakamuro, D., T. Furukawa, and T. Takegami. 1995. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J. Virol. 69:3893-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlissel, M., A. Constantinescu, T. Morrow, M. Baxter, and A. Peng. 1993. Double-strand signal sequence breaks in V(D)J. recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 7:2520-2532. [DOI] [PubMed] [Google Scholar]
- 32.Schweyer, S., S. Mihm, H. J. Radzun, H. Hartmann, and A. Fayyazi. 2000. Liver infiltrating T lymphocytes express interferon gamma and inducible nitric oxide synthase in chronic hepatitis C virus infection. Gut 46:255-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sotillo, R., P. Dubus, J. Martin, E. de la Cueva, S. Ortega, M. Malumbres, and M. Barbacid. 2001. Wide spectrum of tumors in knock-in mice carrying a Cdk4 protein insensitive to INK4 inhibitors. EMBO J. 20:6637-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung, V. M., S. Shimodaira, A. L. Doughty, G. R. Picchio, H. Can, T. S. Yen, K. L. Lindsay, A. M. Levine, and M. M. Lai. 2003. Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection. J. Virol. 77:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Surabhi, R. M., and R. B. Gaynor. 2002. RNA interference directed against viral and cellular targets inhibits human immunodeficiency virus type 1 replication. J. Virol. 76:12963-12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zan, H., A. Cerutti, P. Dramitinos, A. Schaffer, and P. Casali. 1998. CD40 engagement triggers switching to IgA1 and IgA2 in human B cells through induction of endogenous TGF-β: evidence for TGF-β- but not IL-10-dependent direct Sμ→Sα and sequential Sμ→Sγ, Sγ→Sα DNA recombination. J. Immunol. 161:5217-5225. [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu, N., C. F. Ware, and M. M. Lai. 2001. Hepatitis C virus core protein enhances FADD-mediated apoptosis and suppresses TRADD signaling of tumor necrosis factor receptor. Virology 283:178-187. [DOI] [PubMed] [Google Scholar]
- 38.Zhuang, J. C., C. Lin, D. Lin, G. N. Wogan, and C. Ma. 1998. Mutagenesis associated with nitric oxide production in macrophages. Proc. Natl. Acad. Sci. USA 95:8286-8291. [DOI] [PMC free article] [PubMed] [Google Scholar]