Abstract
Ovine pulmonary adenocarcinoma (OPA) is an infectious lung tumor of sheep caused by Jaagsiekte sheep retrovirus (JSRV). To test the hypothesis that JSRV insertional mutagenesis is involved in the oncogenesis of OPA, we cloned and characterized 70 independent integration sites from 23 cases of OPA. Multiple integration sites were identified in most tumors. BLAST analysis of the sequences did not disclose any potential oncogenic motifs or any identical integration sites in different tumors. Thirty-seven of the integration sites were mapped to individual chromosomes by PCR with a panel of sheep-hamster hybrid cell lines. Integration sites were found on 20 of the 28 sheep chromosomes, suggesting a random distribution. However, four integration sites from four different tumors mapped to chromosome 16. By Southern blot hybridization, probes derived from two of these sites mapped to within 5 kb of each other on normal sheep DNA. These sites were found within a single sheep bacterial artificial chromosome clone and were further mapped to only 2.5 kb apart, within an uncharacterized predicted gene and less than 200 kb from a mitogen-activated protein kinase-encoding gene. These findings suggest that there is at least one common integration site for JSRV in OPA and add weight to the hypothesis that insertional mutagenesis is involved in the development of this tumor.
Jaagsiekte sheep retrovirus (JSRV) is the etiological agent of ovine pulmonary adenocarcinoma (OPA), a naturally occurring lung cancer of sheep (also known as sheep pulmonary adenomatosis or Jaagsiekte) (26, 39). OPA is characterized by a lung adenocarcinoma originating from the differentiated epithelial cells of the distal respiratory tract (8). It is one of the major infectious diseases of sheep, and lately it has also emerged as a unique large-animal model for lung cancer (11, 28). OPA is experimentally reproducible by the inoculation of either lung secretions obtained from OPA-affected animals (38, 40) or JSRV infectious molecular clones (9, 26).
JSRV is unique among replication-competent retroviruses in its ability to transform cells in vitro via the expression of its viral envelope (Env), which functions essentially as an oncoprotein (19). Transfection of rodent or chicken fibroblasts with expression plasmids or vectors for JSRV Env results in the appearance of classic foci of transformed cells in days or weeks after transfection. Several studies have investigated the mechanisms of JSRV Env-induced cell transformation in vitro (2, 3, 6, 7, 18, 29, 32, 41). Hyaluronidase-2, which is encoded by a putative tumor suppressor gene, is the cellular receptor for JSRV (32), and both receptor-dependent (7) and -independent (6) mechanisms of transformation have been suggested. Activation of the phosphatidylinositol 3-kinase/Akt pathway mediated by the JSRV Env transmembrane domain helps but is not essential for transformation of rodent or chicken fibroblasts (3, 18, 29, 41).
The mechanisms by which JSRV induces type II pneumocytes and Clara cells to become adenocarcinomas have not been studied in detail. Tumorigenesis is a multistep process, and the expression of JSRV Env might not be sufficient to transform these cells in vivo. The JSRV long terminal repeats (LTRs) are specifically active in the cells that are the target for viral transformation (21, 27). Abundant viral antigens are present in the tumor cells, and infectious virus is present in the lung secretions of OPA-affected animals (24, 31). Thus, JSRV replicates in type II pneumocytes and Clara cells, and this could allow the viral LTR to activate nearby genes through the classical mechanism of insertional activation used by most oncogenic retroviruses (4, 33). Insertional activation may consequently be part of or ultimately be the cause of JSRV-induced carcinogenesis in vivo.
Only two JSRV integration sites have previously been cloned (9, 26), both by screening of genomic DNA libraries. One, from an OPA tumor cell line, JS7, was in the pulmonary surfactant protein A gene. The other, from an OPA tumor, was uncharacterized because of repetitive elements in the clone. The analysis of the JSRV insertion sites is greatly complicated by the presence in the sheep genome of approximately 20 copies of endogenous retroviruses highly related to JSRV (10, 30).
In this study, to accelerate the identification and isolation of integration sites, we developed a multistep gene-walking technique, called low-stringency-high-stringency (ls/hs) PCR, with which we cloned 70 JSRV integration sites from 23 sheep and also the integration site from the JS7 OPA tumor cell line (15). The chromosomal locations of 37 of these integration sites were determined by PCR by using as the template DNA isolated from a panel of sheep-hamster somatic hybrid cells, each containing 1 or a few of the 28 sheep chromosomes. By this method, the two previously published integration sites also were mapped to individual chromosomes. Sequences aligning to the same chromosome were mapped further by Southern blotting on sheep genomic DNA. Our data suggest that there is a common integration site for JSRV on chromosome 16 in tumor DNA extracted from two sheep with OPA. We mapped this common integration site to chromosome 5q11.2 on the human genome map. This agrees with the somatic cell hybrid mapping of the integration sites, as sheep chromosome 16 is syntenic to HSA5. Further investigation is required to determine the importance of this site in OPA pathogenesis.
MATERIALS AND METHODS
Source of tumor tissue.
Nineteen sheep clinically affected with OPA were donated by farmers in Scotland. The diagnosis of OPA was confirmed by gross and histological examination of lungs. An additional four sheep had experimentally induced OPA (38). These animals had multiple smaller nodules, and disease was not clinically apparent. Kidney and lung tumor samples were collected with separate sterile instruments to avoid possible cross-contamination, snap-frozen in liquid nitrogen, and stored at −70°C.
Preparation of DNA.
DNA was prepared as described previously (35). Briefly, kidney and lung tissues were ground to a powder in liquid nitrogen, lysed by addition to TES (0.01 M Tris, 0.1 M EDTA, 0.5% sodium dodecyl sulfate [SDS]), digested with 100 μg of RNase per ml and 100 μg of proteinase K per ml, and extracted with phenol, pH 8.0. DNA was precipitated with sodium acetate and isopropanol, washed twice with 70% ethanol, and then resuspended in 0.01 M Tris-0.1 M EDTA. Cell lines were trypsinized, washed in 1× phosphate-buffered saline, lysed in TES, and then processed as described above.
ls/hs PCR.
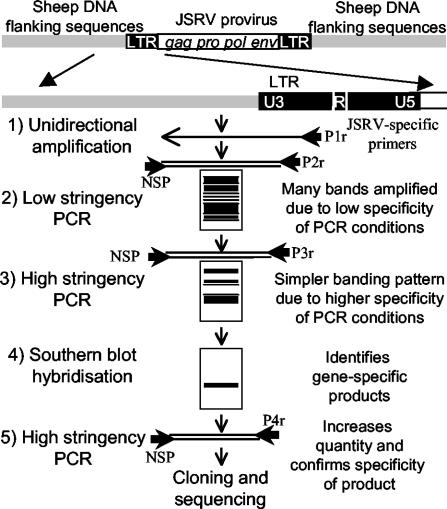
ls/hs PCR is a simple, multistep gene-walking PCR technique that can be used to amplify unknown flanking sequence even when other typical methods have been unsuccessful. Briefly, the target copy number was increased by unidirectional amplification with a sequence-specific primer DNA. Next, the target was subjected to primary amplification with a gene-specific primer in combination with an NSP under low-stringency conditions. This primary reaction mixture then underwent reamplification with the same NSP in conjunction with a second nested, gene-specific primer under conditions of high stringency. Amplification products were separated on an agarose gel, Southern blotted, and hybridized with a gene-specific probe to allow identification of bands that specifically correspond to extended sequence.
Nonspecific PCR primers (NSPs), i.e., primers not specific for sheep or viral sequences (donated by other projects at Moredun Research Institute) were used in conjunction with JSRV-specific primers (Table 1) that had been selected and optimized for specificity to ensure that they amplified JSRV and not JSRV-related endogenous virus sequences. For each PCR described below, the reaction mixtures contained 1 pmol of each primer (MWG-biotech), 50 mM KCl, 10 mM Tris-HCl (pH 9), 0.1% Triton X-100, 2 mM MgCl2, 200 μM each deoxynucleoside triphosphate (Roche), and 1.25 U of Taq polymerase (Promega) in a 50-μl volume. The ls/hs PCR technique was conducted as follows (Fig. 1). (i) Thirty cycles of unidirectional amplification were carried out with primer U3P1r with 100 ng of lung tumor DNA as the template. Cycles were 94°C for 30 s, 58°C for 30 s, and 72°C for 2 min. (ii) Five microliters of the product was used as the template for a series of PCRs with primer U3P2r together with one of 21 different NSPs. Conditions were 35 cycles of 94°C for 30 s, 30°C for 30 s, and 72°C for 2 min. (iii) One microliter of the product of this low-stringency PCR was used as the template for a heminested PCR with primer U3P3r plus the same NSP used in the previous round. Conditions were 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 2 min. (iv) A portion of the PCR product was run on a 1% agarose gel that was then Southern blotted and hybridized with a U3 probe generated by PCR with digoxigenin labeling mixture (Roche) with primers U3P5f and U3P4r. Conditions were 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 40 s. An alkaline phosphatase-nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (BCIP) detection system (Roche) was used to identify positive bands in accordance with the manufacturer's instructions. The rest of the product was run on a separate gel, and the positive bands were matched and recovered. (v) The specificity of the gel-purified amplimers was checked by U3 PCR (with primers U3P5f and U3P4r) and env-U3 PCR (with primers envP6f and U3P4r). Conditions for both were 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 40 s. (vi) Products that were U3 positive and env-U3 negative were reamplified with primer U3P4r together with the appropriate NSP and then cloned into pGemTeasy (Promega) in accordance with the manufacturers instructions. Plasmids were sequenced on an ABI 377 DNA sequencer (Applied Biosystems) with ABI Dye terminator chemistry. The sequences were compared to each other with the Vector NTI suite. Flanking region sequences, i.e., free of JSRV U3 or NSP sequences, were submitted for a BLASTn search (www.ncbi.nlm.nih.gov) both with and without repetitive sequences masked with RepeatMasker (http://ftp.genome.washington.edu/RM/RepeatMasker.html).
TABLE 1.
Primers used in this study
| Primer | Sequence |
|---|---|
| ls/hs PCR non-specific primers | |
| NSP1 | CTTTATAGCTTCATCATGCAG |
| NSP2 | CTTCCATGGAGGCGTGAA |
| NSP3 | GAGATCCTCGCATGGATC |
| NSP4 | GCTCGCCTGTGGTCCTGATA |
| NSP5 | GAAGACGCTTACAGACTACC |
| NSP6 | ATGAAATACTTGGTGCTGGC |
| NSP7 | CAGCGATGACAAGATACGCG |
| NSP8 | ATGAGGTACAACGTAGTTGC |
| NSP9 | CATCCATGTGGACATCATGG |
| NSP10 | AGGACGCTTACGAATTACCG |
| NSP11 | TTGACCTTCGGCTATGAGTA |
| NSP12 | GTTCAAGCTTGTCCAGGAATTC |
| NSP13 | CTAGTACTGGTACCATGTCTGC |
| NSP14 | TTAGCAATCCGGCGCCCT |
| NSP15 | AGCTCGAAATTAACCCTCAC |
| NSP16 | CTGATTGAGATCTGACGC |
| NSP17 | ACTCAAGCATTTCCTCACAAGGCAC |
| NSP18 | CTAGTACTGGTACCATGTCTGCAGCCAAGG |
| NP19 | TCGGTGAGCCGGTGAAA |
| NP20 | ATGTCCTCGCTGGACAC |
| NP21 | GGGAATTCAATGGTTGCAGCCATG |
| ls/hs PCR JSRV-specific primers | |
| U3P1r | CACCGGATTTTTACACAATCACCGG |
| U3P2r | CAATGCCAAAAGCAAACATCCG |
| U3P3r | ATTTCCTGRTATTTCTGTGAAGC |
| U3P4r | CAGCAAACATCCGAACCTTAAGAG |
| JSRV LTR probe- labeling PCR | |
| U3P5f | CGATCGATGCGGGGGACGACC |
| U3P4r | CAGCAAACATCCGAACCTTAAGAG |
| JSRV env-LTR PCR | |
| envP2f | CACTCCTTGGACTTTATGTCGAG |
| U3P4r | CAGCAAACATCCGAACCTTAAGAG |
| JSRV-specific LTR PCR | |
| U3P1f | TGGGAGCTCTTTGGCAAAAGCC |
| U3P1r | CACCGGATTTTTACACAATCACCGG |
| Flanking region probe- labeling PCRs | |
| 171/5af | AAATGCTGTGGGTGAGATTC |
| 171/5ar | TTGTGAGAGACATGGGAATAG |
| 175/10f | ACCAAATCTGAGAAAACGAGC |
| 175/10r | GAGTTGGGAAATTTTCGTG |
| 177/7f | GGAAACATTGAGTGTCTTGGG |
| 177/7r | CCAAAAAGACCTGCCTCCTTA |
| 178/3f | CTGCAATGGCCACACAGCGT |
| 178/3r | CCAACCAAGGAATCGAACC |
| Integration site PCR | |
| U3P4r | CAGCAAACATCCGAACCTTAAGAG |
| U3P1r | CACCGGATTTTTACACAATCACCGG |
| 177/7n1 | TCTCAACCTGACTCTGTGGTGC |
| 177/7n2 | CCAAAAAGACCTGCCTCCTTA |
| 175/10n1 | GAGTTGGGAAATTTTCGTG |
| 175/10n2 | CATTGTTTCCAGAGAGTTGG |
FIG. 1.
Schematic diagram of the ls/hs PCR technique used to amplify JSRV integration sites.
Integration site PCR on sheep-hamster somatic cell hybrid lines.
The integration site sequences were used to design primers to amplify a unique sequence within the sheep genome, avoiding repetitive sequences identified by RepeatMasker. Each primer pair was further optimized by gradient PCR (Mastercycler Gradient; Eppendorf). A 50-μl reaction mixture consisted of 350 ng of genomic DNA, 1× PCR buffer (Sigma), 200 μM each deoxynucleoside triphosphate (Sigma), 20 pmol of each primer, and 1.5 U of Taq polymerase (Sigma). The thermocycler was programmed for an initial cycle of 94°C for 3 min; 30 cycles of 94°C for 30 s, variable temperature depending on the primer pair for 30 s, and 72°C for 30 s; and then a final extension of 72°C for 5 min. PCR products of greater than 200 bp were separated in 1.5% agarose, and those of less than 200 bp were separated by 6% polyacrylamide gel electrophoresis in 1× Tris-borate-EDTA. The genomic DNA was prepared from a panel of 30 sheep-hamster somatic hybrid cell lines obtained from the Eleanor Roosevelt Institute, Denver, Colo. A full description of the cell lines and their selection media was published previously (5).
Southern hybridization.
For Southern blot analysis of the chromosome 16 integration sites, genomic DNA from the lung of an uninfected sheep was cleaved with the restriction endonuclease BamHI, EcoRI, PstI, or SacI (Fig. 2). Panels of digests (10 μg per lane) were separated on a 0.7% agarose gel and transferred to nylon membrane (MSI, Westborough, Mass.) with a PosiBlot 30-30 Pressure Blotter (Stratagene). [α-32P]dCTP-labeled DNA probes were generated by PCR with primer pairs specific for the flanking regions of clones 175/10, 177/7, 178/3, and 171/5 (Table 1). Each membrane was hybridized singly and in combination with the [α-32P]dCTP-labeled 175/10, 177/7, 178/3, and 171/5 probes with a probe (UltraHyb; Ambion) specific activity of 106 cpm/ml at 42°C. Following hybridization, the membranes were washed at 42°C twice for 5 min (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS) and then twice for 15 min (0.1% SSC, 0.1% SDS), followed by either exposure to a PhosphorImager screen (Kodak) or autoradiography. By the same methods, Southern blots of uninfected sheep lung DNA were hybridized with probes for the integration sites that mapped to chromosome 6 singly and then in combination. Southern blots of lung tumor and kidney DNAs from four different animals and of DNAs from two different tumor cell lines (15) were hybridized with the 177/7 probe essentially as described above except that hybridization and washes were performed at 62°C.
FIG. 2.
Southern blot analysis of sheep DNA hybridized with chromosome 16 integration sites 175/10 and 177/7 showing colocalization of these two sites. Each lane consists of 10 μg of genomic DNA digested with BamHI (B), EcoRI (E), PstI (P), or SacI (S). The first panel was hybridized with flank probe 175/10, the second was hybridized with 177/7, and the third was hybridized with both probes. The molecular weight (MW) standard is HindIII-digested phage lambda DNA.
Integration site PCR.
The primers used for integration site PCR were nested primers (n1 and n2 in Table 1) chosen from the integration site flank regions together with primer U3P4r. PCRs were performed with plasmid, kidney, or lung tumor DNA as the template. Touchdown PCR was done as follows: 94°C for 30 s, 65°C for 30 s (decreasing by 0.4°C per cycle), and 72°C for 30 s for 35 cycles. In order to confirm our results and rule out contamination, the PCR assays were repeated with an exogenous specific primer (U3P1r) from outside the cloned integration sites together with the nested flank primers. This PCR could only be positive for genuine integration sites and could not amplify our clones or previous PCR product.
Screening of an ovine BAC library.
An ovine bacterial artificial chromosome (BAC) library (12) was screened by three rounds of PCR with primers 175/10 and 177/7 as described previously. Briefly, the first PCR was of 78 superpools, each of which contained DNAs from eight plates of 96 BAC clones. In the second round, eight single pools, representing DNAs from each of the eight plates in the positive superpool, were screened. Finally, the pooled row and column DNAs from the positive plate were screened. The intersection of the row and column identified the location of the positive BAC clone. A single colony of the positive clone was used to inoculate a 50-ml culture in Luria broth containing 12.5 μg of chloramphenicol per ml. After incubation with shaking at 37°C for 18 h, BAC DNA was extracted by standard alkaline lysis (35) and resuspended in 50 μl of Tris-EDTA. A PCR product was generated with 1 μl of BAC DNA as the template and, for verification, sequenced by ABI Dye terminator chemistry.
The selected BAC was subcloned into pBluescript following digestion with PstI, and colonies were picked at random for shotgun sequencing. The sequence data were analyzed by BLASTn to anchor the BAC to the draft human sequence.
Nucleotide sequence accession numbers.
The nucleotide sequences greater than 50 bp long determined in this study have been submitted to the GenBank database and assigned accession no. AY322366 to AY322426.
RESULTS AND DISCUSSION
We developed a highly sensitive and specific multiround ls/hs PCR technique that enabled us to clone 70 JSRV integration sites from OPA tumor tissue of 23 different sheep (Table 2). The validity of the clones could be confirmed because they contained 152 bp of U3 sequence uniquely identifying JSRV. The length of the flanking region (host cell DNA) ranged from 12 to 671 bp. Sequences of greater than 50 bp were deposited in the GenBank database. The sequences less than 60 bp long were too short for mapping to individual chromosomes by PCR assay. Nevertheless, they were informative regarding the presence of multiple unique integration sites in individual tumors. Multiple integration sites were shown to occur in most (15 of 23) of the OPA tumors. This is the same as for mouse mammary virus (MMTV)-induced tumors, which contain multiple (2 to 20) exogenous MMTV proviruses at different sites in the genome (22). OPA tumors 179 and 180 were completely negative with NSPs 1 to 14. Subsequently, additional NSPs (15 to 21) were used that did facilitate the amplification of multiple integration sites from these tissues. Application of the ls/hs PCR and blotting technique to nontumor tissue (n = 3) did not result in any positive bands (data not shown), demonstrating the specificity of this method.
TABLE 2.
JSRV integration site clones obtained from OPA
| Integration site (animal/ NSP) | Flanking region size (bp) | BLAST result summarya | Chromosome |
|---|---|---|---|
| 24/7 | 142 | Rpt (Bov-tA SINE) | |
| 24/8 | 193 | Rpt (human L2) | 3 |
| 37/5 | 289 | Rpt (Bov-B LINE) | |
| 37/9 | 270 | No matches >23 | 10 |
| 39/7 | 300 | Human chr, 14 ∼220 | 18 |
| 39/8 | 671 | Rpt (Bov-tA & -A2 SINE) | 7 |
| 39/12 | 280 | Human ∼80 bp (>1 chr) | 2 |
| 68/3 | 406 | Rpt (Bov-B LINE) | |
| 168/1 | 169 | Human chr 7 ∼80 bp | 4 |
| 170/5a | 155 | Bos taurus ∼80 bp | CHO |
| 170/5b | 121 | No matches >20 bp | |
| 170/7 | 81 | Rpt (μ satellite) | 6 |
| 170/12 | 38 | Rpt | 7 |
| 171/1 | 367 | Rpt (human Mariner ∼120 bp) | 3 |
| 171/4 | 184 | No matches >30 bp | |
| 171/5a | 115 | Human ∼40 bp (>1 chr) | 16 |
| 171/5b | 105 | No matches >30 bp | |
| 171/5c | 75 | No matches >20 bp | 14 |
| 171/7a | 218 | No matches >30 bp | 6 |
| 171/7b | 175 | No matches >30 bp | 3 |
| 171/9a | 169 | Rpt seqs ∼50 bp | 13 |
| 171/9b | 27 | No matches >20 bp | |
| 171/11a | 161 | Human chr 12 (PCCX2 gene) ∼60 bp | 17 |
| 171/11b | 140 | Rpt (μ satellite) | |
| 171/11c | 56 | No matches >20 bp | |
| 172/1 | 386 | Human chr 10 ∼80 bp | 21 |
| 172/3 | 217 | No matches >20 bp | 12 or 15 |
| 172/4 | 53 | No matches >20 bp | |
| 172/6 | 108 | No matches >27 bp | CHO |
| 172/7b | 189 | Human ∼35 bp | 20 |
| 172/7c | 165 | Human ∼35 bp | 10 |
| 172/7d | 123 | No matches >20 bp | 6 |
| 172/10 | 78 | No matches >20 bp | |
| 172/12 | 73 | Human chr 14 ∼50 bp | 18 |
| 173/7 | 466 | Rpt (Bov-B) | |
| 173/10 | 339 | Rpt (Bov SINE) | |
| 174/3 | 87 | No matches >30 bp | 5 |
| 174/10 | 72 | No matches >20 bp | 1 |
| 175/7a | 12 | No matches >20 bp | |
| 175/7b | 84 | Rpt ∼30 bp (SINE) | 11 |
| 175/7c | 182 | Rpt (Bov-tA SINE) | |
| 175/9 | 233 | Rpt (Bov-A2 SINE) | X |
| 175/10 | 90 | Human chr 5 ∼60 bp | 16 |
| 177/5a | 22 | No matches >20 bp | |
| 177/5b | 131 | Rpt (human L1) | |
| 177/7 | 246 | No matches >20 bp | 16 |
| 178/3 | 268 | Rpt (Bov-tA SINE) | 16 |
| 179/18 | 144 | No matches >20 bp | |
| 179/19 | 165 | No matches >20 bp | |
| 179/20a | 430 | No matches >20 bp | |
| 179/20b | 316 | No matches >20 bp | |
| 180/17a | 30 | No matches >20 bp | |
| 180/17b | 69 | No matches >20 bp | |
| 180/19 | 87 | No matches >20 bp | |
| 180/20a | 129 | Rpt (μ satellite) | |
| 180/20b | 126 | No matches >20 bp | |
| 180/21 | 168 | No matches >30 bp | |
| 184/3 | 307 | Rpt (Bov-A2 SINE) | |
| 184/6 | 423 | Rpt (Bov-2 SINE) | 13 |
| 184/7 | 519 | Human chr 8 ∼110 bp | 26 |
| 184/11 | 351 | No matches >20 bp | 2 |
| 187/6a | 296 | Human ∼50 bp (>1 chr) | 17 |
| 187/6b | 342 | No matches >20 bp | 7 |
| 192/10 | 95 | No matches >20 bp | 1 |
| 506/4 | 47 | No matches >20 bp | |
| e158M/6 | 34 | No matches >20 bp | |
| e159M/5 | 42 | No matches >20 bp | |
| e138G/9 | 98 | No matches >20 bp | 6 |
| e139G/7 | 345 | Rpt (Bov-A2, -1D SINE) | 26 |
| e139G/10 | 352 | Rpt (Bov-2 SINE) | |
| JS7/18 | 37 | Surfactant protein A | 10 |
| JSRV21 | No matches | 6 |
Rpt, repetitive; chr, chromosome; seqs, sequences.
BLAST analysis of the 70 sequences amplified from LT DNA did not reveal any significant alignment with known oncogenic sequences. Of the 70 integration sites, 22 (31%) aligned with repetitive sequences. At least 30 to 40% of the sheep genome consists of repeat sequences, so this cannot be interpreted as a preference to integrate into these regions. There was no bias of any particular NSP for repetitive sequences. Thirty-eight of the sequences had no significant matches with any sequences in the databases (only segments of <35 bp), and 10 integration sites aligned with fairly short regions (50 to 100 bp) of human or bovine sequence. At the sheep genome-JSRV junction there was no apparent bias for any particular nucleotide or nucleotide pattern.
To localize each integration site, including JSRV JS7 and JSRV 21 (9, 26), to a specific sheep chromosome, a PCR assay was performed with integration site-specific primers and template DNA from each of 30 sheep-hamster somatic hybrid cell lines (5), as well as CHO cell and normal sheep DNAs as negative and positive controls, respectively. It could not be determined from the available chromosome distribution on cell lines whether 172/3 mapped to chromosome 12 or 15. Three sites, 171/5b, 172/10, and 173/7, were PCR positive on our normal sheep DNA but did not map to a specific chromosome. Therefore, these may represent sequences that are present in the source DNA and our control but vary at one or both primer sites in the genome from which the cell lines are derived. Alternatively, one or more chromosomes or particular chromosome fragments are not represented in the panel of hamster-sheep hybrids. Integration sites 170/5a and 172/6 mapped to both hamster and sheep chromosomes, suggesting that the PCR detects a sequence common to both genomes. Nine of the original integration sites were uninformative owing to repeat sequences throughout the clones. Even though 11 of 39 integration sites were mapped to just two chromosomes (namely, 6 and 16), statistical analysis by a likelihood ratio test did not confirm any preference for a particular chromosome. Indeed, the distribution of integration sites among the sheep chromosomes was similar to that predicted by the Poisson distribution (5 hits per chromosome, 0.5 predicted, 1 actual; 4 hits, 1.5 predicted, 1 actual; 3 hits, 3.8 predicted, 3 actual; 2 hits, 7 predicted, 5 actual; 1 hit, 9 predicted, 9 actual; 0 hits, 6 predicted, 8 actual).
Five of the six integration sites assigned to single human chromosomes by BLASTn were assigned to a syntenic sheep chromosome (14) by the somatic cell hybrid mapping. For example, site 39/7 aligned with human chromosome 14, which is syntenic with sheep chromosomes 7 and 18.
In order to identify potential common integration loci, probes from different integration sites were hybridized to normal sheep DNA. Chromosomes with more than three integration sites were selected, and by this approach, evidence for a common integration site on chromosome 16 was obtained. Blots of normal sheep genomic DNA were hybridized singly and in combination with probes of 175/10 and 177/7 integration sites (Fig. 2). The BamHI and PstI bands were identical, and both had the same-size EcoRI band, with 177/7 having an additional band of 5 kb, which is suspected to be a restriction fragment length polymorphism in this restriction site in the normal sheep DNA. The hybridization of both probes to a single PstI fragment of approximately 4.5 kb showed that these two integration sites map less than 4.5 kb apart. A SacI restriction site is apparently situated between the two integration sites, as they hybridize with different SacI fragments. The other two chromosome 16 integration sites did not seem to have the same banding pattern by Southern blot analysis and, as a result, were not further analyzed. The five integration sites on chromosome 6 did not have any identical bands, indicating that these integrations did not occur near each other (data not shown).
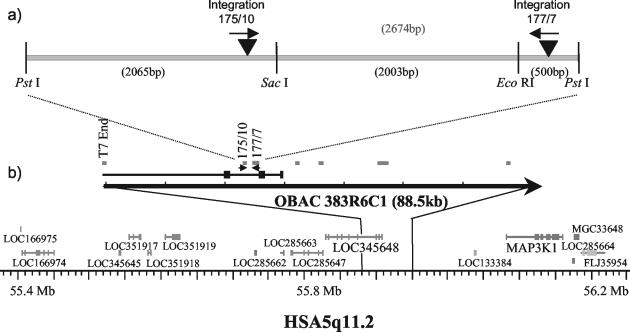
The proximity of integration sites 175/10 and 177/7 was confirmed by their presence in a single BAC clone selected from an ovine BAC library by PCR. Further, both sites were present in a 4.5-kb subfragment of this BAC clone (GenBank accession number AY523942). Sequence analysis showed that the integration sites were 2,535 bp apart (Fig. 3a), and the presence of PstI, EcoRI, and SacI restriction sites was in agreement with the information obtained by Southern hybridization (Fig. 2).
FIG. 3.
Ovine BAC (OBAC) clone 383R6C1 contains integration sites 175/10 and 177/7. (a) Schematic diagram of BAC subclone (GenBank accession no. AY523942) showing PstI, SacI, and EcoRI restriction sites in relation to the integration sites. Arrows indicate the orientation of the two integration sites. (b) Alignment of the BAC clone with the orthologous HSA5q11.2 genomic sequence represented according to Build 33 of the human genome sequence according to the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/mapview). Predicted genes are in dark grey, and confirmed genes are in light grey. Boxes indicate the relative positions of BLASTn hits from randomly sequenced BAC subclones.
A BLASTn analysis of sequences from subclones of the BAC clone aligned it against National Center for Biotechnology Information Build 33 of the human genome sequence to HSA5 (Fig. 3b). The 175/10 and 177/7 integration sites fall in a predicted gene, LOC345648, on human chromosome 5. No function has been assigned to this gene. Other predicted genes lie close to this region. Of particular interest, MAP3K1 lies less than 200 kb away from the integration sites. This is within the range of distances previously reported for enhancer activation of cellular oncogenes by retrovirus LTRs (16). In addition, mitogen-activated protein kinase activation has been demonstrated in OPA tumor cells in vivo by immunohistochemistry (M. De las Heras, personal communication, 2003).
In order to determine if integrations in this region of chromosome 16 occur in more tumors than 177 and 175, Southern blots of lung tumor and kidney DNAs from four sheep were hybridized with a probe corresponding to integration site 177/7. However, this did not reveal an additional band in lung tumor DNA compared to kidney DNA, which would have indicated a clonal population of cells. A novel 6-kb band hybridizing in HindIII-digested lung tumor, but not kidney, DNA of all four animals was found to be lung specific rather than tumor specific, suggesting that integration 177/7 is in a region specifically methylated in lung DNA and not kidney DNA. Digests of lung tumor DNA with six other restriction enzymes, including ones that are unaffected by methylation, did not reveal a second band, even in DNA from tumor 177. These findings suggested that the proportion of tumor cells that contained integrated JSRV was below the level of detection by Southern hybridization.
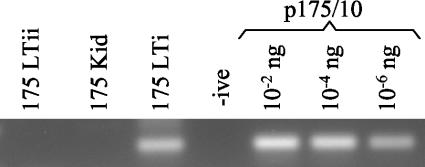
Subsequent PCR analysis showed that the 177/7 integration site was not present in DNA extracted from other parts of the same tumor. It could be amplified from plasmid, but unfortunately none of the original tumor DNA from which the clone was amplified remained. PCR amplification of integration site 175/10 revealed its presence only in the DNA from which the site was originally cloned (Fig. 4). Two other DNA samples from lung tumor of the same animal were negative. Thus, the PCRs showed that the integration sites were clonal only in a small area of the tumor. This result was verified with a primer in JSRV U3 downstream of any of the integration site clones to confirm that the positive results were not due to contamination with any previous PCR product or plasmid.
FIG. 4.
PCR assay specific for integration site 175/10 showing that it is present in a restricted region of the tumor only. A PCR assay was carried out with 250 ng of genomic DNA 175 LTi, from which integration site 175/10 was cloned; genomic DNA 175 LTii, made from a nearby (approximately 1 cm apart) piece of the same lung tumor; and genomic DNA 175 Kid, made from kidney tissue from the same animal. Plasmid p175/10, containing integration site 175/10, was used as the positive control. -ive, negative control.
In summary, two integration sites from different OPA cases were found only 2.5 kb apart on the genome, an event with a probability of approximately 10−6 if integration is completely random, compared to 1 in 20 in this study (i.e., 2 integrations of 40 mapped to individual chromosomes). This supports a role for insertional mutagenesis. In order to strengthen this hypothesis, further evidence of integration sites in this region is required, which requires generation and analysis of additional JSRV integration sites. In addition, comparative expression studies for the genes close to this region would be of great interest in assessing the role of these integrations.
Neither of these insertion sites was found to be clonal, suggesting that (i) the integration sites we have identified are not involved in tumorigenesis but are accumulated during tumor growth or viral replication or (ii) multiple tumors arise independently but nevertheless may contain integration sites in a preferred area, as occurs in MMTV-induced tumors. The discovery that JSRV tumors are polyclonal or oligoclonal does not argue against insertional mutagenesis, as multiclonality has also been reported for tumors induced by MMTV (36). It is clear that the host genetic background has a strong effect on the frequency with which a particular gene is insertionally activated in MMTV tumors. For instance, in CH3 mice wnt-1 is activated in 75% of tumors while in other mouse strains this value is 25 to 59% (20). OPA affects an outbred sheep population; therefore, analysis of greater numbers of tumors is required to identify additional preferred sites.
As has been shown for other retrovirus-induced tumors (1, 13, 17, 34), OPA development is likely to be a multistep process resulting from an accumulation of events that may not necessarily need to occur in a specific order. The recent data on the transforming activity of the transmembrane envelope protein in vitro (2, 3, 6, 7, 18, 29, 32, 41) suggest an important but not necessarily exclusive role for acute transformation by JSRV. In particular, an acute transformation mechanism does not fit with the long incubation periods between infection and tumor production seen in natural OPA, especially as no immune response to JSRV is detectable in infected animals (23, 25; C. Summers, personal communication). We suggest that proliferation of alveolar type II cells or a precursor target cell, induced by the JSRV envelope protein, may be a step in the tumorigenic pathway but that insertional mutagenic events also may be necessary.
Acknowledgments
This work was supported by grant CA59116 from the National Cancer Institute, National Institutes of Health. The BAC mapping was supported by NIH grant CA95706.
We are grateful to Dave Geyer and Dave Patterson at the Eleanor Roosevelt Institute for Cancer Research for making the sheep-hamster hybrid cell lines available to us. Many thanks to Daniel Soong, Linda May, Stephen Smith, and Colette Abbey for technical support, to Diane Redmond for introducing the ls/hs PCR method, to Tom Allen for helpful discussions, and to Jill Sales (BIOSS) for the statistical analysis.
REFERENCES
- 1.Aguilar-Cordova, E., R. Strange, L. J., Young, H. T. Billy, P. H. Gumerlock, and R. D. Cardiff. 1991. Viral Ha-ras mediated mammary tumor progression. Oncogene 6:1601-1607. [PubMed] [Google Scholar]
- 2.Alberti, A., C. Murgia, S.-L. Liu, M. Mura, C. Cousens, J. M. Sharp, A. D. Miller, and M. Palmarini. 2002. Envelope-induced cell transformation by ovine betaretroviruses. J. Virol. 76:5387-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, T. E., K. J. Sherrill, S. M. Crispell, M. R. Perrott, J. O. Carlson, and J. C. DeMartini. 2002. The Jaagsiekte retrovirus envelope gene induces transformation of the avian embryo fibroblast cell line DF-1 but does not require a conserved SH2 binding domain. J. Gen. Virol. 83:2733-2742. [DOI] [PubMed] [Google Scholar]
- 4.Brown, P. O. 1997. Integration, p. 106-203. In J. M. Coffin et al. (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 5.Burkin, D. J., T. E. Broad, M. R. Lambeth, H. R. Burkin, and C. Jones. 1998. New gene assignments using a complete, characterized sheep-hamster somatic cell hybrid panel. Anim. Genet. 29:48-54. [DOI] [PubMed] [Google Scholar]
- 6.Chow, Y. H., A. Alberti, M. Mura, C. Pretto, P. Murcia, L. M. Albritton, and M. Palmarini. 2003. Transformation of rodent fibroblasts by the Jaagsiekte sheep retrovirus envelope is receptor independent and does not require the surface domain. J. Virol. 77:6341-6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danilkovitch-Miagkova, A., F. M. Duh, I. Kuzmin, D. Angeloni, S. L. Liu, A. D. Miller, and M. I. Lerman. 2003. Hyaluronidase 2 negatively regulates RON receptor tyrosine kinase and mediates transformation of epithelial cells by Jaagsiekte sheep retrovirus. Proc. Natl. Acad. Sci. USA 100:4580-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De las Heras, M., L. Gonzalez, and J. M. Sharp. 2003. Pathology of ovine pulmonary adenocarcinoma. Curr. Top. Microbiol. Immunol. 275:25-54. [DOI] [PubMed] [Google Scholar]
- 9.DeMartini, J. C., J. V. Bishop, T. E. Allen, S. Brown, D. L. Knudson, F. A. Jassim, J. M. Sharp, D. R. Voelker, and J. O. Carlson. 2001. The Jaagsiekte sheep retrovirus clone JSRV JS7 induces ovine pulmonary carcinoma and is integrated into the surfactant protein A gene of the JS7 tumor cell line. J. Virol. 75:4239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeMartini, J. C., J. O. Carlson, C. Leroux, T. Spencer, and M. Palmarini. 2003. Endogenous retroviruses related to Jaagsiekte sheep retrovirus. Curr. Top. Microbiol. Immunol. 275:117-137. [DOI] [PubMed] [Google Scholar]
- 11.Fan, H., M. Palmarini, and J. C. DeMartini. 2003. Transformation and oncogenesis by Jaagsiekte sheep retrovirus. Curr. Top. Microbiol. Immunol. 275:139-177. [DOI] [PubMed] [Google Scholar]
- 12.Gill, C. A., S. K. Davis, J. F. Taylor, N. E. Cockett, and C. D. K. Bottema. 1999. Construction and characterization of an ovine bacterial artificial chromosome library. Mamm. Genome 10:1108-1111. [DOI] [PubMed] [Google Scholar]
- 13.Hoatlin, M. E., S. L. Kozak, F. Lilly, A. Chakraborti, C. A. Kozak, and D. Kabat. 1990. Activation of erythropoietin receptors by Friend viral gp55 and by erythropoietin and down-modulation by the murine Fv-2r resistance gene. Proc. Natl. Acad. Sci. USA 87:9985-9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iannuzzi, L., G. P. Di Meo, A. Perucatti, and D. Incarnato. 1999. Comparison of the human with the sheep genomes by use of human chromosome-specific painting probes. Mamm. Genome 10:719-723. [DOI] [PubMed] [Google Scholar]
- 15.Jassim, F. A. 1988. Identification and characterisation of transformed cells in Jaagsiekte a contagious lung tumor of sheep. Ph.D. thesis. University of Edinburgh, Edinburgh, United Kingdom.
- 16.Lazo, P. A., J. S. Lee, and P. N. Tsichlis. 1990. Long distance activation of the myc protooncogene by provirus insertion in Mlvi-1 or Mlvi-4 in rat T-cell lymphomas. Proc. Natl. Acad. Sci. USA 87:170-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, J. P., A. D. D'Andrea, H. F. Lodish, and D. Baltimore. 1990. Activation of cell growth by binding of Friend spleen focus-forming virus gp55 glycoprotein to the erythropoietin receptor. Nature 343(6260):762-764. [DOI] [PubMed] [Google Scholar]
- 18.Liu, S. L., M. I. Lerman, and A. D. Miller. 2003. Putative phosphatidylinositol 3-kinase (PI3K) binding motifs in ovine betaretrovirus Env proteins are not essential for rodent fibroblast transformation and PI3K/Akt activation. J. Virol. 77:7924-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maeda, N., M. Palmarini, C. Murgia, and H. Fan. 2001. Direct transformation of rodent fibroblasts by Jaagsiekte sheep retrovirus DNA. Proc. Natl. Acad. Sci. USA 98:4449-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchetti, A., J. Robbins, G. Campbell, F. Buttitta, F. Squartini, M. Bistocchi, and R. Callahan. 1991. Host genetic background effect on the frequency of mouse mammary tumor virus-induced rearrangements of the int-1 and int-2 loci in mouse mammary tumors. J. Virol. 65:4550-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGee-Estrada, K., M. Palmarini, and H. Fan. 2002. HNF-3β is a critical factor for the expression of the Jaagsiekte sheep retrovirus (JSRV) long terminal repeat in type II pneumocytes but not in Clara cells. Virology 292:87-97. [DOI] [PubMed] [Google Scholar]
- 22.Nusse, R., and H. E. Varmus. 1982. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 31:99-109. [DOI] [PubMed] [Google Scholar]
- 23.Ortín, A., E. Minguijon., P. Dewar, M. Garcia, L. M. Ferrer, M. Palmarini, L. Gonzalez, J. M. Sharp, and M. De las Heras. 1998. Lack of a specific immune response against a recombinant capsid protein of Jaagsiekte sheep retrovirus in sheep and goats naturally affected by enzootic nasal tumour or sheep pulmonary adenomatosis. Vet. Immunol. Immunopathol. 61:229-237. [DOI] [PubMed] [Google Scholar]
- 24.Palmarini, M., P. Dewar, M. De las Heras, N. F. Inglis, R. G. Dalziel, and J. M. Sharp. 1995. Epithelial tumour cells in the lungs of sheep with pulmonary adenomatosis are major sites of replication for Jaagsiekte retrovirus. J. Gen. Virol. 76:2731-2737. [DOI] [PubMed] [Google Scholar]
- 25.Palmarini, M., M. J. Holland, C. Cousens, R. G. Dalziel, and J. M. Sharp. 1996. Jaagsiekte retrovirus (JSRV) establishes a disseminated infection of the lymphoid tissues in sheep affected by pulmonary adenomatosis. J. Gen. Virol. 77:2991-2998. [DOI] [PubMed] [Google Scholar]
- 26.Palmarini, M., J. M. Sharp, M. De las Heras, and H. Fan. 1999. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J. Virol. 73:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmarini, M., S. Datta, R. Omid, C. Murgia, and H. Fan. 2000. The long terminal repeats of Jaagsiekte sheep retrovirus (JSRV) are preferentially active in type II pneumocytes. J. Virol. 74:5776-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmarini, M., and H. Fan. 2001. Retrovirus-induced ovine pulmonary adenocarcinoma, an animal model for lung cancer. J. Natl. Cancer Inst. 93:1603-1614. [DOI] [PubMed] [Google Scholar]
- 29.Palmarini, M., N. Maeda, C. Murgia, C. De-Fraja, A. Hofacre, and H. Fan. 2001. A phosphatidylinositol-3-kinase docking site in the cytoplasmic tail of the Jaagsiekte sheep retrovirus transmembrane protein is essential for envelope-induced transformation of NIH 3T3 cells. J. Virol. 75:11002-11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmarini, M., M. Mura, and T. E. Spencer. 2004. Endogenous betaretroviruses of sheep: teaching new lessons in retroviral interference and adaptation. J. Gen. Virol. 85:1-13. [DOI] [PubMed] [Google Scholar]
- 31.Platt, J. A., N. Kraipowich, F. Villafane, and J. C. DeMartini. 2002. Alveolar type II cells expressing Jaagsiekte sheep retrovirus capsid protein and surfactant proteins are the predominant neoplastic cell type in ovine pulmonary adenocarcinoma. Vet. Pathol. 39:341-352. [DOI] [PubMed] [Google Scholar]
- 32.Rai, S. K., F. M. Duh, V. Vigdorovich, A. Danilkovitch-Miagkova, M. I. Lerman, A. D. Miller. 2001. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for Jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc. Natl. Acad. Sci. USA 98:4443-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg, N., and P. Jolicoeur. 1997. Retroviral pathogenesis, p. 475-585. In J. M. Coffin, S. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 34.Ruscetti, S. K., N. J. Janesch, A. Chakraborti, S. T. Sawyer, and W. D. Hankins. 1990. Friend spleen focus-forming virus induces factor independence in an erythropoietin-dependent erythroleukemia cell line. J. Virol. 64:1057-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Sarkar, N. H. 1995. Clonal variations among multiple primary mammary tumors and within a tumor of individual mice: insertion mutations of int oncogenes. Virology 212:490-499. [DOI] [PubMed] [Google Scholar]
- 37.Scherdin, U., K. Rhodes, and M. Breindl. 1990. Transcriptionally active genome regions are preferred targets for retroviral integration. J. Virol. 64:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharp, J. M., K. W. Angus, E. W. Gray, and F. M. M. Scott. 1983. Rapid transmission of sheep pulmonary adenomatosis (Jaagsiekte) in young lambs. Arch. Virol. 78:89-95. [DOI] [PubMed] [Google Scholar]
- 39.Sharp, J. M., and J. C. DeMartini. 2003. Natural history of JSRV in sheep. Curr. Top. Microbiol. Immunol. 275:55-79. [DOI] [PubMed] [Google Scholar]
- 40.Verwoerd, D. W., A. L. Williamson, and E. M. De Villiers. 1980. Aetiology of Jaagsiekte: transmission by means of subcellular fractions and evidence for the involvement of a retrovirus. Onderstepoort J. Vet. Res. 47:275-280. [PubMed] [Google Scholar]
- 41.Zavala, G., C. Pretto, Y. H. Chow, L. Jones, A. Alberti, E. Grego, M. De las Heras, and M. Palmarini. 2003. Relevance of Akt phosphorylation in cell transformation induced by Jaagsiekte sheep retrovirus. Virology 312:95-105. [DOI] [PubMed] [Google Scholar]