Abstract
H9N2 influenza viruses are panzootic in domestic poultry in Eurasia and since 1999 have caused transient infections in humans and pigs. To investigate the zoonotic potential of H9N2 viruses, we studied the evolution of the viruses in live-poultry markets in Hong Kong in 2003. H9N2 was the most prevalent influenza virus subtype in the live-poultry markets between 2001 and 2003. Antigenic and phylogenetic analysis of hemagglutinin (HA) showed that all of the 19 isolates found except one belonged to the lineage represented by A/Duck/Hong Kong/Y280/97 (H9N2). The exception was A/Guinea fowl/NT184/03 (H9N2), whose HA is most closely related to that of the human isolate A/Guangzhou/333/99 (H9N2), a virus belonging to the A/Chicken/Beijing/1/94-like (H9N2) lineage. At least six different genotypes were recognized. The majority of the viruses had nonstructural (and HA) genes derived from the A/Duck/Hong Kong/Y280/97-like virus lineage but had other genes of mixed avian virus origin, including genes similar to those of H5N1 viruses isolated in 2001. Viruses of all six genotypes of H9N2 found were able to replicate in chickens and mice without adaptation. The infected chickens showed no signs of disease, but representatives of two viral genotypes were lethal to mice. Three genotypes of virus replicated in the respiratory tracts of swine, which shed virus for at least 5 days. These results show an increasing genetic and biologic diversity of H9N2 viruses in Hong Kong and support their potential role as pandemic influenza agents.
Aquatic birds are the natural reservoir of influenza A viruses (23, 27). After transfer to new avian or mammalian hosts, the viruses evolve rapidly and cause mild or occasionally severe respiratory disease (1, 11, 16). In 1997, 18 humans were infected with avian H5N1 influenza virus, and six died, refocusing global attention on the potential role of avian influenza viruses as precursors of human pandemic influenza virus strains (25, 28). Genetic characterization of avian influenza viruses circulating in China in 1997 revealed that the H5N1 influenza viruses isolated from humans and poultry in Hong Kong were probably reassortants that had obtained internal gene segments from either avian H9N2 (A/Quail/Hong Kong/G1/97 [Qa/HK/G1/97]-like) or H6N1 (A/Teal/Hong Kong/W312/97-like) viruses (6, 7, 14).
H9N2 viruses have become panzootic in Eurasia over the last decade and have been isolated from different types of terrestrial poultry worldwide (2, 9, 13). Two distinct lineages of H9N2 viruses, represented by the prototype viruses A/Duck/Hong Kong/Y280/97 (Dk/HK/Y280/97) and Qa/HK/G1/97, have become established in terrestrial poultry in Asia, predominantly in chickens and quail, respectively (6, 13). Qa/HK/G1/97-like viruses are thought to have been involved in the generation of the highly pathogenic H5N1 virus in 1997 (7). Dk/HK/Y280/97-like viruses were predominantly associated with infections of chickens that caused mild clinical signs (6). H9N2 viruses of both lineages have been isolated from humans (14, 20), and a virus of the Dk/HK/Y280/97-like lineage has been isolated from pigs in southern China (19). These H9N2 influenza viruses have acquired receptor-binding specificity for sialic acid with a terminal α2-6Gal linkage, as on human cells, as well as for sialic acid with an α2-3Gal linkage, as on avian cells (17). Between 2000 and 2001, H9N2 influenza viruses in southern China were transmitted from terrestrial poultry back to domestic ducks, in which multiple genotypes were generated through reassortment (13).
The prevalence of H9N2 viruses throughout Asia, along with their demonstrated ability to infect mammals, puts them high on the list of influenza viruses with pandemic potential. Therefore, it is imperative to understand the evolution and properties of these viruses. Here we describe the genetic and biologic properties of H9N2 viruses isolated from the live-poultry markets of Hong Kong in 2003.
MATERIALS AND METHODS
Sampling and virus isolation.
Influenza virus strains were isolated from poultry in live-poultry markets in Hong Kong, Special Administrative Region, during 2003. Tracheal swabs and fecal samples were collected into tissue culture medium containing penicillin G (2 × 106 U/liter), polymyxin B (2 × 106 U/liter), gentamicin (250 mg/liter), nystatin (0.5 × 106 U/liter), ofloxacin HCl (60 mg/liter), and sulfamethoxazole (0.2 g/liter). Virus isolates were subtyped with a panel of reference antisera recommended by the World Health Organization (http://www.who.int/csr/resources/publications/en/#influenza). Nineteen avian H9 viruses, including 15 viruses isolated from chickens, 2 viruses isolated from guinea fowl, 1 virus isolated from a pheasant, and 1 virus isolated from a pigeon, were chosen for detailed analysis (Table 1).
TABLE 1.
H9N2 influenza A viruses testeda
| Virus | Sampling date (mo/day/yr) | Location/type of market sampled | Genotypeb |
|---|---|---|---|
| A/Ck/HK/CSW153/03 | 1/16/03 | Kow/wholesale | D |
| A/Ck/HK/CSW291/03 | 1/16/03 | Kow/wholesale | D |
| A/Ck/HK/CSW304/03 | 1/16/03 | Kow/wholesale | D |
| A/Ck/HK/FY23/03 | 1/22/03 | Kow/retail | C |
| A/Ck/HK/SF1/03 | 1/23/03 | Kow/retail | C |
| A/Ck/HK/BD90/03 | 2/13/03 | Border | D |
| A/GF/HK/NT101/03 | 2/17/03 | Kow/retail | D |
| A/Ck/HK/AP45/03 | 2/18/03 | HK/retail | D |
| A/Ck/HK/TP38/03 | 2/20/03 | Newt/retail | F |
| A/Pg/HK/WF53/03 | 2/20/03 | Newt/retail | E |
| A/Ph/HK/WF54/03 | 2/20/03 | Newt/retail | D |
| A/Ck/HK/NT142/03 | 3/20/03 | Kow/retail | B |
| A/GF/HK/NT184/03 | 4/15/03 | Kow/retail | A |
| A/Ck/HK/WF126/03 | 4/22/03 | Newt/retail | C |
| A/Ck/HK/WF120/03 | 4/22/03 | Newt/retail | C |
| A/Ck/HK/YU427/03 | 5/19/03 | Newt/retail | C |
| A/Ck/HK/SSP418/03 | 6/23/03 | Kow/retail | C |
| A/Ck/HK/NT366/03 | 7/17/03 | Kow/retail | C |
Abbreviations: Pg, pigeon; Ph, pheasant; Newt, New Territories; Kow, Kowloon.
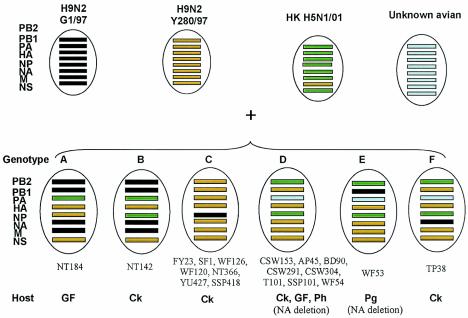
Genotypes are determined by the phylogenetic relationships shown in Fig. 1
Antigenic analysis.
A panel of monoclonal antibodies (MAbs) to the hemagglutinin (HA) of H9N2 strains was used in hemagglutination inhibition (HI) assays as previously described (18). We used three MAbs to Qa/HK/G1/97, three MAbs to A/Chicken/Hong Kong/G9/97 (Ck/HK/G9/97), and 10 MAbs to Dk/HK/Y280/97.
Genetic and phylogenetic analysis.
Viral gene sequencing and analysis were carried out as described previously (3, 5). For comparison, the phylogenetic analysis included sequences from H9N2 virus lineages established in southern China since the mid-1990s (including Dk/HK/Y280/97-like and Qa/HK/G1/97-like viruses), sequences from A/Goose/Guangdong/1/96-like virus (H5N1), and sequences from H5N1 viruses isolated in Hong Kong in 2001 (4). The regions used in the phylogenetic analyses were as follows: for the PB2 gene, nucleotides 10 to 1262; for the PB1 gene, nucleotides 66 to 1368; for the PA gene, nucleotides 8 to 1290; for the HA gene, nucleotides 1 to 1680; for the NP gene, nucleotides 55 to 962; for the NA gene, nucleotides 41 to 1393; for the M gene, nucleotides 1 to 889; and for the NS gene, nucleotides 1 to 842.
In vivo growth characteristics.
Virus replication in chickens (specific-pathogen-free white leghorn broilers), mice (BALB/c), and outbred pigs was measured after intranasal inoculation with virus-infected allantoic fluid (containing 106 50% tissue culture infective doses [TCID50] of virus), as previously described (6). Tracheal swabs were collected from chickens on days 3 through 7 postinoculation (p.i.), and virus was titrated in 10-day-old embryonated chicken eggs. Ten chickens were inoculated with each virus. The body weights of the inoculated mice were measured daily on days 0 through 5 p.i., at which time the animals were killed and virus in the lung tissue was titrated. Two 4-week-old, influenza virus antibody-free pigs were inoculated intranasally with 106 TCID50 of A/Guinea fowl/Hong Kong/WF10/99 (GF/HK/WF10/99), Ck/HK/CSW153/03, Ck/HK/WF126/03, and GF/HK/NT184/03 virus. Nasal swabs were collected on days 2 through 7 p.i. for virus titration in embryonated eggs. On days 5 and 7 p.i., a pig from each infection group was euthanized for viral titration in organs and for pathological examination.
RESULTS
Antigenic analysis.
H9N2 viruses continue to be the most abundant influenza viruses isolated in the Hong Kong live-poultry market system. Between 2001 and 2003, H9N2 viruses accounted for approximately 75% of all influenza viruses isolated. Antigenically distinct lineages of H9N2 viruses have been reported to be circulating in Asia. To investigate the antigenic properties of the H9N2 viruses isolated in 2003, we performed HI assays with a panel of anti-H9 MAbs (Table 2). Most of the H9N2 viruses isolated in 2003 reacted well with MAbs to Ck/HK/G9/97 and Dk/HK/Y280/97 but less well with MAbs to Qa/HK/G1/97. One isolate, GF/HK/NT184/03, showed little cross-reaction (HI titer, <200) with MAbs to Ck/HK/G9/97, Dk/HK/Y280/97, and Qa/HK/G1/97. These results demonstrated that most but not all of the H9N2 viruses circulating in the live-poultry markets of Hong Kong in 2003 were antigenically similar to Dk/HK/Y280/97-like viruses.
TABLE 2.
Cross-reactivity (HI-determined titers) of the H9N2 viruses isolated in 2003 with MAbs to H9N2 viruses
| Virusa | Virus titer determined with indicated MAb to strainb:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ck/HK/G9/97
|
Qa/HK/G1/97 lineage
|
Dk/HK/Y280/97
|
|||||||||||||
| G9-6 | G9-25 | 1073-9 | 26 | 29 | 7B10 | 8C4 | 15F1 | 18G4 | 3D11 | 4G3 | 19A10 | 18B10 | 2F4 | 18B1 | |
| Ck/HK/G9/97 | ≥ | ≥ | <200 | <200 | 3,200 | ≥ | 6,400 | ≥ | 3,200 | ≥ | 6,400 | 3,200 | 3,200 | ≥ | ≥ |
| Qa/HK/G1/97 | <200 | <200 | ≥ | ≥ | ≥ | 200 | <200 | <200 | <200 | <200 | <200 | <200 | <200 | <200 | <200 |
| HK/1074/99 | <200 | <200 | ≥ | ≥ | ≥ | <200 | 200 | <200 | <200 | <200 | <200 | <200 | <200 | <200 | <200 |
| Dk/HK/Y280/97 | 3,200 | 6,400 | <200 | <200 | 3,200 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ |
| Ck/HK/CSW153/03 | ≥ | ≥ | 3,200 | 1,600 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ |
| Ck/HK/AP45/03 | ≥ | ≥ | ≥ | 6,400 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ |
| Ck/HK/BD90/03 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ |
| Ck/HK/CSW291/03 | ≥ | ≥ | 1,600 | 1,600 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 200 | ≥ | ≥ |
| Ck/HK/CSW304/03 | ≥ | ≥ | 3,200 | 1,600 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 6,400 | ≥ | ≥ |
| Ck/HK/FY23/03 | ≥ | ≥ | ≥ | 6,400 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ |
| GF/HK/NT101/03 | ≥ | ≥ | ≥ | 3,200 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ |
| Ck/HK/NT142/03 | ≥ | ≥ | <200 | 800 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 6,400 | ≥ | ≥ |
| Ck/HK/SF1/03 | ≥ | ≥ | ≥ | 6,400 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ |
| Ck/HK/SSP101/03 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ |
| Ck/HK/TP38/03 | ≥ | ≥ | 3,200 | 1,600 | ≥ | ≥ | 800 | ≥ | ≥ | ≥ | ≥ | ≥ | 800 | ≥ | ≥ |
| Ck/HK/WF126/03 | ≥ | ≥ | 1,600 | 3,200 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 3,200 | ≥ | ≥ | ≥ | ≥ |
| Pg/HK/WF53/03 | ≥ | 640 | 6,400 | 6,400 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 800 | ≥ | ≥ | ≥ | ≥ |
| Ph/HK/WF54/03 | ≥ | ≥ | ≥ | 6,400 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 3,200 | ≥ | ≥ | ≥ | ≥ |
| GF/HK/NT184/03 | <200 | <200 | <200 | <200 | <200 | 800 | 1,600 | <200 | <200 | <200 | <200 | <200 | <200 | <200 | <200 |
| Ck/HK/WF120/03 | ≥ | ≥ | ≥ | 640 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ |
| Ck/HK/YU427/03 | ≥ | ≥ | 640 | 640 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ |
| Ck/HK/SSP418/03 | ≥ | ≥ | 320 | 320 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 6,400 | ≥ | ≥ |
| Ck/HK/NT366/03 | ≥ | ≥ | 160 | 320 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 6,400 | ≥ | ≥ |
Abbreviations: Pg, pigeon; Ph, pheasant;
≥, titer of ≥12,800.
Phylogenetic analysis.
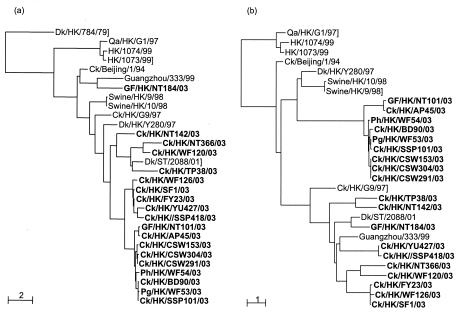
Antigenic analysis suggested that 18 of the 19 H9N2 viruses isolated in 2003 were similar to Dk/HK/Y280/97. To determine the genetic diversity among these viruses, we compared partial sequences of the eight gene segments of each virus. We identified six different genotypes, designated A, B, C, D, E, and F, among the 19 viruses (Fig. 1 and Table 1). Phylogenetic analysis of the HA1 region of the H9 gene showed that, as predicted by antigenic comparison, all of the H9N2 viruses isolated in 2003 except GF/HK/NT184/03 belonged to the Dk/HK/Y280/97-like lineage (Fig. 2a). GF/HK/NT184/03 clustered with A/Chicken/Beijing/1/94 (Ck/Beijing/1/94)-like viruses, which caused an outbreak of disease in chickens in Guangdong Province in 1994, and with Guangzhou/333/99, a human isolate. No Qa/HK/G1/97-like HA genes were found.
FIG. 1.
To determine the genetic diversity of the 19 virus isolates, we compared partial sequences of their eight gene segments. We identified six different genotypes, designated A, B, C, D, E, and F. Ph, pheasant.
FIG. 2.
(a) Phylogenetic analysis of the HA1 region of the H9 gene of the 19 H9N2 viruses isolated in 2003. As predicted by antigenic comparison, all but GF/HK/NT184/03 belonged to the Dk/HK/Y280/97-like lineage. (b) The N2 genes of the 19 isolates clustered into two main sublineages. Nine viruses clustered with Dk/HK/Y280/97-like viruses (which contain a three-amino-acid deletion in the stalk region), and 10 viruses grouped with Ck/HK/G9/97-like viruses (which do not contain a stalk deletion). Isolates in bold type represent isolates sequenced in this study. Ph, pheasant.
The N2 genes of the viruses isolated in 2003 formed two main sublineages. Nine viruses clustered with Dk/HK/Y280/97-like viruses (which contain a three-amino-acid deletion in the stalk region), and 10 viruses grouped with Ck/HK/G9/97-like viruses (which do not contain a stalk deletion) (Fig. 2b). As with the HA genes, no Qa/HK/G1/97-like NA genes were detected.
Phylogenetic analysis of the six internal genes revealed multiple viral genotypes and showed that reassortment had occurred. The M and PB1 genes of the H9N2 viruses isolated in 2003 separated into two groups, the Dk/HK/Y280/97-like and Qa/HK/G1/97-like lineages (data not shown). There were also two groups based on the NP gene sequence: one was more similar to Dk/HK/Y280/97-like viruses and the other was closer to the H5N1 viruses responsible for outbreaks in chickens in Hong Kong in 2001. Also included in this H5N1-like cluster were H9N2 viruses isolated from ducks between 2000 and 2001 (13). The PA genes also showed similarities to those of H5N1 viruses isolated in 2001.
The PB2 polymerase genes of the recent H9N2 viruses originated from a number of lineages. The PB2 genes of Ck/HK/NT184/03 (genotype A) and GF/HK/NT142/03 (genotype B) belonged to the Qa/HK/G1/97-like group. Seven viruses (genotype C) were part of the Dk/HK/Y280/97-like group. However, most PB2 genes of the H9N2 viruses isolated in 2003 (genotypes D, E, and F) were similar to those of the H5N1-like viruses isolated in 2001, as were those of H9N2 viruses isolated from ducks in 2000 and 2001. The NS genes of the H9N2 viruses isolated in 2003 formed a single group, since all were of Dk/HK/Y280/97 origin.
Genetic characteristics of the HA gene.
Matrosovich and colleagues (17) recently described avian H9N2 viruses with human virus-like receptor specificity. To determine the proportion of the viruses isolated in 2003 that had this trait, we compared the HA amino acid sequences (deduced from the nucleotide sequences) of these viruses with those of representative strains. Eighteen of the 19 H9N2 viruses possessed leucine at position 226, a feature characteristic of mammalian viruses as described by Matrosovich et al. GF/HK/NT184/03 had valine at position 190 and glutamine at position 226, as did avian H9 viruses isolated from ducks in the 1970s and the earliest viruses of the Dk/HK/Y280/97 lineage, such as Ck/Beijing/1/94 and Ck/HK/739/94. All of the recent H9N2 isolates maintained the Arg-Ser-Ser-Arg motif at the connecting peptide of their HA, which is characteristic of H9 viruses of land-based poultry (13).
In vivo growth properties.
The increased genetic diversity of influenza viruses in the Hong Kong live-bird markets is a cause for some concern. To determine whether this genetic heterogeneity is accompanied by biologic heterogeneity, we compared the levels of replication of six of the H9N2 viruses isolated in 2003 (GF/HK/NT184/03, Ck/HK/NT142/03, Ck/HK/WF126/03, Ck/HK/CSW153/03, A/Pigeon/Hong Kong/WF53/03 [Pg/HK/WF53/03], and Ck/HK/TP38/03), each of which represented one of the genotypes A, B, C, D, E, or F in avian and mammalian hosts. GF/HK/WF10/99 (H9N2) (Qa/HK/G1/97-like lineage) was used for comparison.
Growth in chickens.
None of the H9N2 viruses tested induced signs of disease in inoculated chickens over the duration of the experiment (7 days). However, all of the viruses replicated in the respiratory tracts of the infected birds; titers were ≥2.3 log10 50% egg infective doses (EID50)/0.1 ml in tracheal swabs at day 3 p.i. (Table 3). Chickens inoculated with Ck/HK/TP38/03 (genotype F) had a higher lung virus titer (5.3 log10 EID50/0.1 ml) on day 4 p.i. than did those inoculated with other genotypes (3.7 to 4.3 log10 EID50/0.1 ml). Virus was detected in tracheal swabs for at least 6 days p.i. Although most of the tracheal swabs were negative on day 7 p.i., traces of virus (1.5 log10 EID50/0.1 ml) were found in birds inoculated with GF/HK/NT184/03 (genotype A).
TABLE 3.
Replication of H9N2 influenza viruses in tissues from inoculated animalsa
| Virusb | Genotype | Virus titer [log10 EID50/0.1 ml (SD)] in:
|
Virus titer in mouse lung [log10 EID50/0.1 ml (SD)] at 5 dpi | No. of dead mice/total no. inoculated
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chickens
|
Swine
|
|||||||||||||
| Trachea
|
Lung
|
Nasal swab
|
Lung
|
|||||||||||
| 3 dpi | 4 dpi | 5 dpi | 4 dpi | 7 dpi | 2 dpi | 3 dpi | 4 dpi | 5 dpi | 5 dpi | 3 dpi | 5 dpi | |||
| GF/HK/WF10/99 | Qa/HK/G1/97-like | 2.7 (0.3) | 3.7 (0.4) | 2.5 (0.4) | 4.0 (0.3) | 1.3 (0.1) | 2.3 (0.3) | 2.7 (0.1) | 2.3 (0.3) | 1.0 (0.1) | 2.3 (0.3) | 5.3 (0.4) | 1/10 | 5/10 |
| GF/HK/NT184/03 | A | 3.3 (0.3) | 3.7 (0.1) | 2.3 (0.1) | 4.3 (0.3) | 1.3 (0.1) | 2.3 (0.1) | 2.5 (0.1) | 2.3 (0.3) | 0.7 (0.1) | 2.7 (0.4) | 5.0 (0.3) | 0/10 | 0/10 |
| Ck/HK/NT142/03 | B | 2.3 (0.3) | 3.3 (0.3) | 2.0 (0.4) | 4.0 (0.3) | 1.0 (0.1) | NT | NT | NT | NT | NT | 4.7 (0.4) | 0/10 | 0/10 |
| Ck/HK/WF126/03 | C | 2.3 (0.2) | 3.3 (0.3) | 2.3 (0.2) | 4.0 (0.1) | 1.0 (0.1) | 2.3 (0.3) | 3.3 (0.4) | 2.5 (0.3) | 1.7 (0.1) | 3.7 (0.3) | 5.0 (0.3) | 0/10 | 0/10 |
| Ck/HK/CSW153/03 | D | 3.3 (0.2) | 3.7 (0.3) | 2.5 (0.4) | 4.0 (0.4) | 1.5 (0.1) | 3.0 (0.3) | 3.5 (0.1) | 3.5 (0.3) | 1.5 (0.1) | 2.3 (0.3) | 5.3 (0.3) | 1/10 | 4/10 |
| Pg/HK/WF53/03 | E | 2.3 (0.3) | 3.3 (0.3) | 2.3 (0.1) | 3.7 (0.1) | 1.0 (0.1) | NT | NT | NT | NT | NT | 4.3 (0.3) | 0/10 | 0/10 |
| Ck/HK/TP38/03 | F | 2.7 (0.2) | 3.3 (0.2) | 2.3 (0.1) | 5.3 (0.3) | 1.3 (0.1) | NT | NT | NT | NT | NT | 5.3 (0.1) | 0/10 | 0/10 |
The dose of inoculum was 106 TCID50. Chicken were inoculated intranasally and intratracheally; BALB/c mice and swine were inoculated intranasally. dpi, days postinoculation; NT, not tested.
Pg, pigeon.
Growth in mice.
All genotypes of H9N2 viruses were able to replicate well in mice without prior adaptation. Virus titers in lungs ranged from 4.3 to 5.3 log10 EID50/0.1 ml on day 5 p.i. Death was observed after inoculation of mice with GF/HK/WF10/99 (Qa/HK/G1/97-like) or GF/HK/NT184/03 viruses (Table 3). Mice inoculated with the other genotypes lost body weight over the first 5 days p.i.
Growth in swine.
Qa/HK/G1/97-like viruses have been isolated from humans (14, 20), and a virus of the Dk/HK/Y280/97-like lineage has been isolated from pigs in southern China (19). To investigate the ability of our isolates to infect swine, we intranasally inoculated two 4-week-old influenza virus antibody-free pigs with GF/HK/NT184/03 (genotype A), Ck/HK/WF126/03 (genotype C), Ck/HK/CSW153/03 (genotype D), or GF/HK/WF10/99 virus. All viruses replicated well in the respiratory tract; the titers on day 2 p.i. were ≥2.3 log10 EID50/0.1 ml in nasal swabs, and virus was shed for at least 5 days (Table 3). Virus titers of nasal swabs peaked on day 3 p.i. at 2.5 to 3.5 log10 EID50/0.1 ml. Mild cough and elevated body temperature were observed on days 2 through 4 p.i. in all inoculated pigs. Ck/HK/WF126/03 and Ck/HK/CSW153/03 caused moderate interstitial pneumonia, as observed on days 5 and 7 p.i., whereas the other viruses caused less severe gross lung damage. No virus was detected in spleens, kidneys, livers, or intestines.
DISCUSSION
H9N2 influenza viruses are evolving and circulating widely in the Hong Kong live-poultry markets. The most recent study of H9N2 viruses from these markets, in 1999, detected two genotypes: Qa/HK/G1/97-like viruses isolated primarily from quail and Dk/HK/Y280/97-like viruses isolated primarily from chickens (6). We were unable to identify any Qa/HK/G1/97-like viruses circulating in the markets in 2003. Live quail have been banned from the poultry markets since the 1999 study (21), and the Qa/HK/G1/97-like viruses have apparently disappeared. It is encouraging to note that such changes in marketing practices can significantly reduce the presence of specific viruses. However, individual gene segments (from the PB2, PB1, NP, NA, and M genes) similar to those of the Qa/HK/G1/97-like viruses were found in numerous isolates, although no isolate contained more than four of these genes. Our identification of multiple genotypes of H9N2 viruses in 2003 was markedly unlike the findings of the 1999 study but was similar to those of a recent study of ducks in poultry markets in China's Guangdong province (13). Despite the multiple genotypes identified in this study and by Li et al., there did not appear to be genotypes of virus common to both studies; this finding suggests a high degree of diversity among the H9N2 viruses in the region. It is also worth noting that all six of the virus genotypes identified in this study were isolated within a 4-month period of 2003; therefore, they were cocirculating rather than appearing in a stepwise fashion. The suggestion by Li and colleagues that a two-way transmission occurs between ducks and poultry is a plausible explanation for the emergence of multiple genotypes. It is likely that reassortment events would be more frequent in the aquatic bird reservoir, where the virus burden is greater. Such reassortants could then transmit back to domestic poultry species and create the diversity of H9N2 viruses seen in the present study. This genetic heterogeneity is of some concern in view of the prevalence of H9N2 viruses in Hong Kong and Asia. H9N2 viruses have infected humans, but there has been little evidence of human-to-human transmission (26). Although we do not know which viral factors are necessary for successful human-to-human spread, reassortment is certain to increase the chance that a transmissible virus will be generated. The different phenotypic characteristics of the H9N2 genotypes support this premise. Although some early Ck/Beijing/1/94-like H9N2 viruses induced mortality rates as high as 80% in infected birds (9), none of the viruses tested in this study caused signs of disease in chickens, not even GF/HK/NT184/03, which has an HA protein that is 96.6% homologous to that of Ck/Beijing/1/94. The amount of virus shed by H9N2-infected chickens was similar to that previously described for Qa/HK/G1/97 (6). In BALB/c mice and swine, virulence depended on the virus genotype. In swine, viruses of different genotypes induced different degrees of gross lung pathology and had different kinetics of growth. GF/HK/WF10/99 and GF/HK/NT184/03 caused the death of infected mice without prior adaptation, as H5N1 viruses and Qa/HK/G1/97 (H9N2) virus have been reported to do (8, 9, 15). Viruses of the other H9N2 genotypes did not kill infected mice, although loss of body weight was observed. The gene segments common to GF/HK/WF10/99 and GF/HK/NT184/03 are those from the PB2, PB1, NA, and M genes, suggesting that the mouse virulence factors reside within these genes. Conversely, the presence of the same lineage of PB2, PB1, NA, and M genes in Ck/HK/NT142/03, a virus not lethal to mice, argues against this hypothesis. Previous studies have shown that single-amino-acid changes can dramatically alter the virulence of influenza viruses (10, 24); therefore, hypotheses based on whole-gene homology may be misleading.
The H9N2 viruses isolated in 2003 have maintained the mammalian virus-like receptor HA signatures described by Matrosovich et al. in H9N2 viruses isolated in Hong Kong in 1997 and 1999 (17). Although the relative importance of this receptor trait and the potential for transmission are unknown, the viruses isolated in 2003 are able to replicate well in swine. H9N2 viruses have been reported to be found in swine in Asia, although their prevalence was apparently not high (19). Although the amount of virus shed from the H9N2-infected pigs was not as large as that previously reported for some adapted swine H3N2 viruses (22, 29), it was similar to titers obtained with other avian, human, and swine viruses upon experimental infection (12). Additional surveillance is needed to determine whether the prevalence of H9N2 viruses in swine in Asia has increased along with the diversity of circulating avian viruses. The importance of swine in the ecology of human influenza viruses clearly makes evaluation of H9N2 activity in swine in Asia a high priority.
The Qa/HK/G1/97-like viruses disappeared after the removal of quail from the Hong Kong bird markets and have been replaced by a genetically and biologically diverse group of viruses. One can only speculate on the factors that have induced this evolutionary burst. It is possible that Qa/HK/G1/97-like viruses were previously successful in competing with these reassortant viruses, but this possibility seems dubious considering that the Qa/HK/G1/97-like viruses were restricted primarily to quail, whereas the new viruses appear in chickens. It is also possible that the reintroduction of H9N2 viruses from domestic poultry into ducks (13) has promoted the emergence of the multiple reassortant viruses. Certainly the immense diversity of viruses within the wild aquatic bird reservoir would support the creation of multiple reassortants. Whatever may have caused the change in H9N2 viruses in the Hong Kong markets, the change is sure to be attributable to the intrinsic tendency of influenza viruses to constantly evolve in multiple and unpredictable directions.
Acknowledgments
These studies were supported by grants AI29680 and AI95357 from the National Institute of Allergy and Infectious Diseases, by Cancer Center Support (CORE) grant CA-21765 from the National Institutes of Health, and by the American Lebanese Syrian Associated Charities.
We thank Scott Krauss and Jennifer Humberd for technical assistance and Sharon Naron for editorial assistance.
REFERENCES
- 1.Bean, W. J., M. Schell, J. Katz, Y. Kawaoka, C. Naeve, O. Gorman, and R. G. Webster. 1992. Evolution of the H3 influenza virus hemagglutinin from human and nonhuman hosts. J. Virol. 66:1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cameron, K. R., V. Gregory, J. Banks, I. H. Brown, D. J. Alexander, A. J. Hay, and Y. P. Lin. 2000. H9N2 subtype influenza A viruses in poultry in Pakistan are closely related to the H9N2 viruses responsible for human infection in Hong Kong. Virology 278:36-41. [DOI] [PubMed] [Google Scholar]
- 3.Choi, Y. K., S. M. Goyal, M. W. Farnham, and H. S. Joo. 2002. Phylogenetic analysis of H1N2 isolates of influenza A virus from pigs in the United States. Virus Res. 87:173-179. [DOI] [PubMed] [Google Scholar]
- 4.Guan, Y., J. S. Peiris, A. S. Lipatov, T. M. Ellis, K. C. Dyrting, S. Krauss, L. J. Zhang, R. G. Webster, and K. F. Shortridge. 2002. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc. Natl. Acad. Sci. USA 99:8950-8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan, Y., M. Peiris, K. F. Kong, K. C. Dyrting, T. M. Ellis, T. Sit, L. J. Zhang, and K. F. Shortridge. 2002. H5N1 influenza viruses isolated from geese in southeastern China: evidence for genetic reassortment and interspecies transmission to ducks. Virology 292:16-23. [DOI] [PubMed] [Google Scholar]
- 6.Guan, Y., K. F. Shortridge, S. Krauss, P. S. Chin, K. C. Dyrting, T. M. Ellis, R. G. Webster, and M. Peiris. 2000. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J. Virol. 74:9372-9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan, Y., K. F. Shortridge, S. Krauss, and R. G. Webster. 1999. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. USA 96:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gubareva, L. V., J. A. McCullers, R. C. Bethell, and R. G. Webster. 1998. Characterization of influenza A/HongKong/156/97 (H5N1) virus in a mouse model and protective effect of zanamivir on H5N1 infection in mice. J. Infect. Dis. 178:1592-1596. [DOI] [PubMed] [Google Scholar]
- 9.Guo, Y. J., S. Krauss, D. A. Senne, I. P. Mo, K. S. Lo, X. P. Xiong, M. Norwood, K. F. Shortridge, R. G. Webster, and Y. Guan. 2000. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology 267:279-288. [DOI] [PubMed] [Google Scholar]
- 10.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 11.Kawaoka, Y., S. Krauss, and R. G. Webster. 1989. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 63:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kida, H., T. Ito, J. Yasuda, Y. Shimizu, C. Itakura, K. F. Shortridge, Y. Kawaoka, and R. G. Webster. 1994. Potential for transmission of avian influenza viruses to pigs. J. Gen. Virol. 75:2183-2188. [DOI] [PubMed] [Google Scholar]
- 13.Li, K. S., K. M. Xu, J. S. M. Peiris, L. L. M. Poon, K. Z. Yu, K. Y. Yuen, K. F. Shortridge, R. G. Webster, and Y. Guan. 2003. Characterization of H9 subtype influenza viruses from the ducks of southern China: a candidate for the next influenza pandemic in humans? J. Virol. 77:6988-6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, Y. P., M. Shaw, V. Gregory, K. Cameron, W. Lim, A. Klimov, K. Subbarao, Y. Guan, S. Krauss, K. Shortridge, R. Webster, N. Cox, and A. Hay. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. USA 97:9654-9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu, X., T. M. Tumpey, T. Morken, S. R. Zaki, N. J. Cox, and J. M. Katz. 1999. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J. Virol. 73:5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludwig, S., L. Stitz, O. Planz, H. Van, W. M. Fitch, and C. Scholtissek. 1995. European swine virus as a possible source for the next influenza pandemic? Virology 212:555-561. [DOI] [PubMed] [Google Scholar]
- 17.Matrosovich, M. N., S. Krauss, and R. G. Webster. 2001. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281:156-162. [DOI] [PubMed] [Google Scholar]
- 18.Palmer, D. F., W. R. Dowdle, M. T. Coleman, and G. C. Schild. 1975. Advanced laboratory techniques for influenza diagnosis. U.S. Department of Health, Education and Welfare Immunology Series, no. 6. Center for Disease Control, Atlanta, Ga.
- 19.Peiris, J. S. M., Y. Guan, D. Markwell, P. Ghose, R. G. Webster, and K. F. Shortridge. 2001. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 75:9679-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peiris, M., K. Y. Yuen, C. W. Leung, K. H. Chan, P. L. Ip, R. W. Lai, W. K. Orr, and K. F. Shortridge. 1999. Human infection with influenza H9N2. Lancet 354:916-917. [DOI] [PubMed] [Google Scholar]
- 21.Perez, D. R., W. Lim, J. P. Seiler, G. Yi, M. Peiris, K. F. Shortridge, and R. G. Webster. 2003. Role of quail in the interspecies transmission of H9 influenza A viruses: molecular changes on HA that correspond to adaptation from ducks to chickens. J. Virol. 77:3148-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richt, J. A., K. M. Lager, B. H. Janke, R. D. Woods, R. G. Webster, and R. J. Webby. 2003. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J. Clin. Microbiol. 41:3198-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rohm, C., N. Zhou, J. Suss, J. Mackenzie, and R. G. Webster. 1996. Characterization of a novel influenza hemagglutinin, H15: criteria for determination of influenza A subtypes. Virology 217:508-516. [DOI] [PubMed] [Google Scholar]
- 24.Seo, S. H., E. Hoffmann, and R. G. Webster. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 8:950-954. [DOI] [PubMed] [Google Scholar]
- 25.Subbarao, K., A. Klimov, J. Katz, H. Regnery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemphill, T. Rowe, M. Shaw, X. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393-396. [DOI] [PubMed] [Google Scholar]
- 26.Uyeki, T. M., Y. H. Chong, J. M. Katz, W. Lim, Y. Y. Ho, S. S. Wang, T. H. Tsang, W. W. Au, S. C. Chan, T. Rowe, J. Hu-Primmer, J. C. Bell, W. W. Thompson, C. B. Bridges, N. J. Cox, K. H. Mak, and K. Fukuda. 2002. Lack of evidence for human-to-human transmission of avian influenza A (H9N2) viruses in Hong Kong, China 1999. Emerg. Infect. Dis. 8:154-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuen, K. Y., P. K. Chan, M. Peiris, D. N. Tsang, T. L. Que, K. F. Shortridge, P. T. Cheung, W. K. To, E. T. Ho, R. Sung, and A. F. Cheng. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467-471. [DOI] [PubMed] [Google Scholar]
- 29.Zhou, N. N., D. A. Senne, J. S. Landgraf, S. L. Swenson, G. Erickson, K. Rossow, L. Liu, K. J. Yoon, S. Krauss, and R. G. Webster. 2000. Emergence of H3N2 reassortant influenza A viruses in North American pigs. Vet. Microbiol. 74:47-58. [DOI] [PubMed] [Google Scholar]