Abstract
Template-switching events during reverse transcription are necessary for completion of retroviral replication and recombination. Structural determinants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) that influence its template-switching frequency are not known. To identify determinants of HIV-1 RT that affect the frequency of template switching, we developed an in vivo assay in which RT template-switching events during viral replication resulted in functional reconstitution of the green fluorescent protein gene. A survey of single amino acid substitutions near the polymerase active site or deoxynucleoside triphosphate-binding site of HIV-1 RT indicated that several substitutions increased the rate of RT template switching. Several mutations associated with resistance to antiviral nucleoside analogs (K65R, L74V, E89G, Q151N, and M184I) dramatically increased RT template-switching frequencies by two- to sixfold in a single replication cycle. In contrast, substitutions in the RNase H domain (H539N, D549N) decreased the frequency of RT template switching by twofold. Depletion of intracellular nucleotide pools by hydroxyurea treatment of cells used as targets for infection resulted in a 1.8-fold increase in the frequency of RT template switching. These results indicate that the dynamic steady state between polymerase and RNase H activities is an important determinant of HIV-1 RT template switching and establish that HIV-1 recombination occurs by the previously described dynamic copy choice mechanism. These results also indicate that mutations conferring resistance to antiviral drugs can increase the frequency of RT template switching and may influence the rate of retroviral recombination and viral evolution.
One of the most important mechanisms contributing to genetic variation and evolution of retroviral populations is recombination during viral replication (9, 74). As a result of recombination and selection pressure, a significant proportion of circulating human immunodeficiency virus type 1 (HIV-1) strains are recombinants (25, 42, 53, 61, 63, 68). It was shown that recombination leads to the rapid emergence of viruses that are resistant to multiple antiviral drugs, ensuring their continued propagation (21, 35, 45, 81). The HIV-1 recombination rate has been shown to be significantly higher than that of gammaretroviruses (30, 60, 84). In a single replication cycle, the HIV-1 recombination rate was reported to be 10-fold higher than that of spleen necrosis virus and murine leukemia virus (MLV) (4, 26, 30). The reasons for this difference in the rates of recombination are not well defined. Recombination occurs during reverse transcription as a result of reverse transcriptase (RT) switching templates between copackaged RNA molecules during viral DNA synthesis (26). Therefore, the frequency with which viral RNAs from different proviruses are copackaged as well as the template-switching properties of RT could influence the rate of recombination.
It is not known whether viral RNAs derived from two different proviruses are copackaged randomly or whether RNAs derived from the same provirus are preferentially copackaged. It was previously observed that during spleen necrosis virus replication, multiple crossover events occur much more frequently than predicted by the frequency of single crossover events (5, 27). This phenomenon is genetically defined as high negative interference, for which two possible mechanisms were proposed (24). First, it was postulated that a subset of virions is capable of carrying out template switching between copackaged RNA molecules and that multiple template-switching events occur during their replication. Second, it was postulated that viral RNAs derived from different proviruses are not copackaged randomly; thus, the frequency of heterozygote formation may be less than expected and defines the subpopulation of virions that undergo recombination. In transient transfection assays, it was shown that RNAs derived from different HIV-1 vectors could copackage randomly (51). These results suggest that differences in the ability of HIV-1 and MLV to copackage RNAs randomly could account for the observed differences in recombination rates (51).
Another possible explanation for the difference in the rate of recombination between HIV-1 and MLV is that they undergo template switching at different rates or by different mechanisms. Recently, it was shown that the direct-repeat deletion frequency for a portion of the bacterial β-galactosidase gene (lacZ) is similar for HIV-1 and MLV in single replication cycle assays (1, 51). These results suggest that the template-switching rates for MLV and HIV-1 RT are similar; however, it is unclear whether these results can be generalized to other template sequences. It is conceivable that templates with different RNA secondary structures might lead to different template-switching frequencies during MLV and HIV-1 replication.
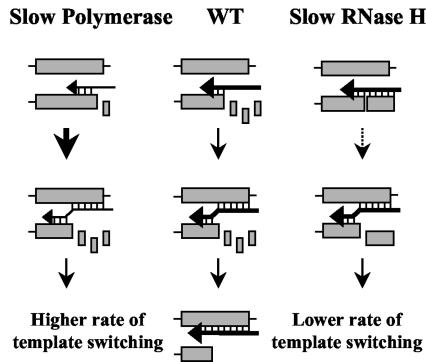
To date, the mechanism of HIV-1 RT template switching has not been fully elucidated. Several in vitro studies using purified nucleic acids and proteins have provided important insights into the process of RT template switching and the components of the reverse transcription complex that influence its frequency (3, 6, 12-15, 48-50, 62, 64). Another approach to studying the mechanism of template switching is in vivo direct-repeat deletion assays, which analyze template-switching events in directly repeated sequences derived from a reporter gene; a template switch within the repeats results in functional reconstitution of the reporter gene (1, 11, 29, 41, 56, 73). Based on results obtained using MLV vectors and direct-repeat deletion assays, we have proposed a mechanism that is referred to as dynamic copy choice (73). The dynamic copy choice model states that a steady state between the rate of DNA polymerization and the rate of RNA template degradation 3′ to the RT is an important determinant of RT template switching (Fig. 1). Experimental manipulations that influenced the steady state between DNA synthesis and RNA degradation altered the rate of template switching and supported the dynamic copy choice model in the MLV system. These experimental manipulations included mutations in the polymerase and RNase H domains of MLV RT, depletion of intracellular nucleotide pools (29, 73), introduction of secondary structures within the template RNA, and mutations in the MLV nucleocapsid protein that reduced its ability to melt RNA secondary structure and enhance the rate of DNA synthesis (83). Although these studies provide strong evidence in support of the dynamic copy choice mechanism for MLV RT template switching, it is unclear whether template switching occurs by this mechanism during replication of HIV-1 or other retroviruses. In addition, structural determinants of HIV-1 RT that influence the frequency of template switching have not been identified.
FIG. 1.
Dynamic copy choice model for RT template switching. Shaded boxes represent homologous sequences in two copackaged RNAs or in the same RNA. Horizontal arrows represent nascent DNA. The thickness of these arrows indicates the relative polymerization rate. When mutations are expected to reduce the rate of DNA synthesis (slow polymerase), the horizontal arrows are thinner relative to the wild-type RT (WT). Small boxes represent RNA degraded by the RNase H domain. When mutations are expected to reduce the rate of RNase H degradation (slow RNase H), the degraded RNA fragments are shown as larger boxes. Hydrogen bonds between the RNA template and nascent DNA are designated by vertical lines. Vertical arrows of various thicknesses indicate the relative efficiency of template switching.
We have now developed an HIV-1-based in vivo direct-repeat deletion assay to elucidate the mechanism of HIV-1 RT template switching and to identify structural determinants of HIV-1 RT that influence template switching. The results indicate that HIV-1 RT template switching occurs by the dynamic copy choice mechanism and that mutations in the HIV-1 RT polymerase active site that confer resistance to antiretroviral agents can significantly alter the frequency of RT template switching.
MATERIALS AND METHODS
Plasmids and cells.
All plasmid names are preceded by “p.” pKD-HIV-GFFP-IHy is a derivative of pHR′-CMVlacZ (47) in which lacZ was replaced by the overlapping GF and FP fragments of the green fluorescent protein gene (GFP) from pES-GFFP (73). This vector also contains the hygromycin phosphotransferase B gene (hygro) expressed from an internal ribosomal entry site (IRES) of encephalomyocarditis virus from plasmid pLW1 (32). pCMVΔR8.2 expresses all HIV-1 proteins, except envelope, under the control of the human cytomegalovirus (hCMV) immediate early promoter and lacks the packaging signal (Ψ) and adjacent sequences (46). pHCMV-G (80) and pCB6 WTAΔ513 (38) encode the G glycoprotein of vesicular stomatitis virus (VSV-G) and the envelope of avian sarcoma-leukosis virus (ASLV-A) truncated beyond the membrane-spanning region at residue 513, respectively.
pES-HIVApa-Sal is a derivative of pBluescript II SK (70) that has an insertion of the ApaI-SalI fragment from pCMVΔR8.2 containing the HIV-1 pol gene. This plasmid was used for the generation of RT mutations, which were introduced by using a Quick Change mutagenesis kit (Stratagene). Most of the oligonucleotide primers were designed to introduce additional silent mutations and generate new restriction sites for identification of mutated plasmids by restriction digestion analysis. A detailed description of the mutagenic oligonucleotides and the strategies to generate individual mutants is available upon request. The fragments containing RT mutations were subcloned from pES-HIVApa-Sal into pCMVΔR8.2, and the presence of desired mutations as well as the absence of undesired mutations were verified by DNA sequencing (Laboratory of Molecular Technology, Science Applications International Corporation-Frederick).
The human embryonal kidney cell line 293T was obtained from the American Type Culture Collection. The 293-TVA cell line, which expresses the receptor for ASLV-A, was a kind gift from Brian Lewis (38). The 293T-based cell line GN-HIV-GFFP, which contains an HIV-1 provirus derived from the vector pKD-HIV-GFFP-IHy, was created as follows: briefly, 293T cells were cotransfected with vector pKD-HIV-GFFP-IHy, helper construct pCMVΔR8.2, and envelope construct pHCMV-G; the resulting pseudotyped virus was used to infect 293T cells. After hygromycin selection, several nonfluorescent cell clones were isolated, expanded, and characterized by Southern analysis and infection assays. A cell clone containing one full-length provirus and producing the best virus titer was used as a producer cell line.
Cell culture, transfection, and infection.
Human 293T and 293-TVA cells were maintained at 37°C in Dulbecco's modified Eagle's medium supplemented with penicillin (50 U/ml; Gibco), streptomycin (50 μg/ml; Gibco), and fetal calf serum (10%; HyClone Laboratories). Hygromycin (Sigma) was used at a final concentration of 270 μg/ml.
All transfections of GN-HIV-GFFP cells were performed by calcium phosphate precipitation (MBS transfection kit; Stratagene). Cells were plated at a density of 2 × 106 per 100-mm-diameter dish and transfected with 20 μg of total plasmid DNA consisting of equivalent molar quantities of pHCMV-G and pCMVΔR8.2 (or variants containing mutated RT); after calcium phosphate exposure for 3 to 5 h, the culture medium was replaced. Virus-containing supernatants were harvested 24 to 48 h later and filtered through Millex-GS 0.45-μm-pore-size filters (Millipore). The amount of p24 capsid (CA) protein in the supernatants was determined using an HIV-1 p24 enzyme-linked immunosorbent assay (ELISA) kit (Perkin Elmer Life Sciences). Serial dilutions of virus-containing media were used to infect 293T target cells that were plated at a density of 1 × 105 cells per 60-mm-diameter dish the day before infection. The infected cells were subjected to hygromycin selection 24 h after infection, and hygromycin-resistant colonies were quantified to determine viral titers.
Determination of RT template switching in vivo.
Approximately 500 to 2,000 hygromycin-resistant colonies were pooled and expanded. Subsequently, the cells were harvested and resuspended in 0.5 to 2 ml of phosphate-buffered saline supplemented with 1% fetal calf serum and analyzed by flow cytometry to determine the percentage of GFP-expressing cells (FACScan; Becton Dickinson).
Virus preparation and RT assay.
To isolate and concentrate viruses, virus-containing medium was collected and centrifuged at 25,000 rpm for 90 min in an SW 28 rotor (Beckman) at 4°C. The virus pellet was resuspended in phosphate-buffered saline and stored at −80°C. The concentrated virus was thawed on ice, and RT activities were determined by modification of a previously described method (22). The reaction mixtures contained 100 mM Tris-HCl (pH 8.0), 100 mM KCl, 5 mM MgCl2, 10 mM dithiothreitol, 0.4% Igepal CA-630, 40 U of RNase inhibitor, 50 μM dTTP, 50 μg of 20-mer oligo(dT)/ml, 100 μg of poly(rA)/ml, 10 mCi of [3H]dTTP (specific activity, 72 Ci/mmol; ICN), and concentrated virus (100 to 200 ng of p24 CA equivalent) in a total volume of 60 μl. The reactions were carried out at 37°C for 1 h and were terminated by the addition of ice-cold 10% trichloroacetic acid. The reaction mixtures were precipitated and filtered through 0.45-μm-pore-size Metricel membranes (Gelman Sciences, Inc.), and the incorporated [3H]dTTP was determined using a scintillation counter.
Statistical analysis.
The statistical significance of differences in template-switching frequencies and RT activities between mutant and wild-type HIV-1 RTs were calculated by using a two-sample t test (SigmaPlot 8.0 software). Linear regression analysis of RT template-switching frequencies and viral titers or forward mutation frequencies was performed using SigmaPlot 8.0 software. The significance of the negative correlation was determined by using a t test, where t = r√[(n − 2)/(1 − r2)], and n is the number of data pairs.
RESULTS
HIV-1 direct-repeat deletion assay.
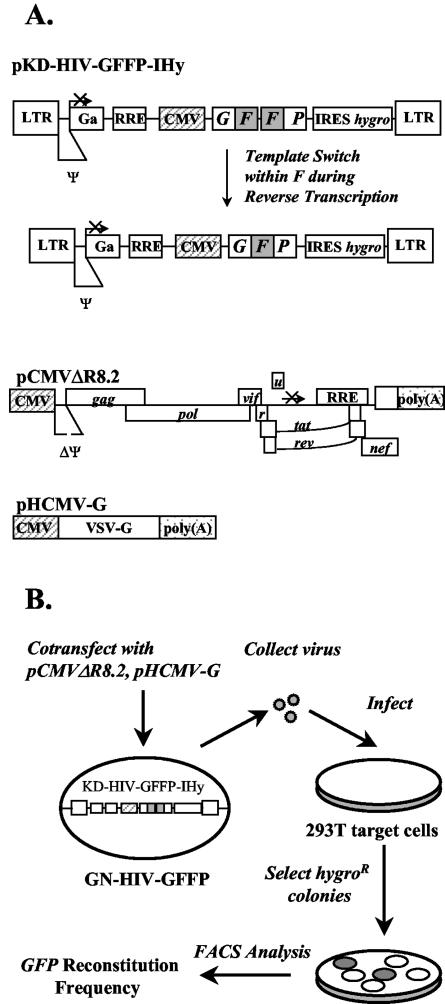
We have developed an in vivo direct-repeat deletion assay for HIV-1 RT template switching based on a previously described assay for MLV RT (73). In this assay, we used directly repeated overlapping fragments of GFP for identification of template-switching events that lead to functional reconstitution of GFP during one round of replication. We first constructed pKD-HIV-GFFP-IHy, an HIV-1-based retroviral vector shown in Fig. 2A, which contains all cis-acting elements required for HIV-1 reverse transcription, integration, and packaging. The selectable marker hygro is expressed from the vector with an encephalomyocarditis virus-derived IRES. The vector also contains the overlapping GF and FP fragments of GFP; the directly repeated F portion (250 bp in length) is separated by a 25-bp linker sequence. The vector does not express a functional GFP protein; however, a homologous template switch within the directly repeated F portion during reverse transcription results in the reconstitution of a functional GFP.
FIG. 2.
Schematic representation of HIV-1-based constructs and direct-repeat deletion assay for identification of structural determinants of HIV-1 RT that influence template switching. (A) Structures of HIV-1-based vectors pKD-HIV-GFFP-IHy, pCMVΔR8.2, and pHCMV-G. pKD-HIV-GFFP-IHy contains long terminal repeats (LTR) and all cis-acting elements of HIV-1. GFFP and hygro are transcribed from the LTR promoter or the hCMV promoter (hatched box). An IRES of encephalomyocarditis virus is used to express hygro. The directly repeated F portion of GFP is shaded. During reverse transcription, the repeated F portion may be deleted to reconstitute a functional GFP. The Rev-responsive element (RRE) and packaging signal (Ψ) are shown. The pCMVΔR8.2 helper construct (46) expresses all HIV-1 proteins except the envelope from the hCMV promoter (hatched box). The coding regions of viral proteins and a polyadenylation site [stippled box labeled poly(A)] from the insulin gene at the end of the nef reading frame are shown. The packaging signal and adjacent sequences, except the 5′ splice donor, were deleted from the 5′ untranslated region. The pHCMV-G construct expresses VSV-G from the hCMV promoter. (B) Experimental protocol. GN-HIV- GFFP, a 293T-based cell line expressing pKD-HIV-GFFP, is shown. The wild type or mutated pCMVΔR8.2 constructs were separately cotransfected with pHCMV-G into GN-HIV-GFFP cells, and the virus produced was used to infect 293T cells. The infected cell clones resistant to hygromycin were analyzed by flow cytometry to determine frequencies of direct-repeat deletion and GFP reconstitution. FACS, fluorescence-activated cell sorter.
Next, we created a cell line (GN-HIV-GFFP) containing a single provirus derived from the vector pKD-HIV-GFFP-IHy (shown in Fig. 2B). First, 293T cells were transfected with vector pKD-HIV-GFFP-Ihy, helper construct pCMVΔR8.2, and VSV-G expression construct pHCMV-G, and the resulting virus was used to infect fresh 293T cells. After hygromycin selection, several nonfluorescent cell clones were isolated and expanded. The absence of GFP expression was verified by flow cytometry analysis (data not shown). To identify a cell clone that exhibited a high virus titer, the cell clones were transfected with pCMVΔR8.2 and pHCMV-G, and the virus produced was used to infect 293T cells. A cell clone that exhibited the highest titer (>104 CFU/ml) and contained only one full-length provirus (confirmed by Southern blot analysis; data not shown) was named GN-HIV-GFFP and used in all subsequent experiments. Flow cytometry analysis of pools of infected hygromycin-resistant cells exhibited the presence of a high proportion of fluorescent cells, indicating functional reconstitution of GFP during one retroviral replication cycle.
The protocol used to identify protein determinants important for HIV-1 RT template switching during reverse transcription is outlined in Fig. 2B. First, pCMVΔR8.2-derived constructs containing mutations in HIV-1 RT were transiently cotransfected with pHCMV-G into GN-HIV-GFFP cells. Virus produced from the transfected cells was then used to infect 293T target cells. The infected 293T cells were selected for resistance to hygromycin, and the resulting colonies were pooled and analyzed by flow cytometry to determine the frequency of direct-repeat deletion. In general, the multiplicity of infection for the experiments analyzed by flow cytometry was <0.005; therefore, the probability of double infection was very low. In this assay, the virus titers and frequencies of RT template switching were examined during a single round of viral replication.
Viruses pseudotyped with VSV-G could potentially reinfect producer cells; possible reinfection of the producer cells would result in multiple rounds of viral replication during the course of the experiment. To rule out the possibility that multiple rounds of replication occurred during viral production, we performed a control experiment by using ASLV-A to pseudotype HIV-GFFP virus. Because the ASLV-A receptor was not expressed in the virus producer cells, the ASLV-A-pseudotyped virus could not reinfect the producer cells, ensuring that an only a single cycle of replication occurred during the experiment. 293-TVA cells, which expressed the ASLV-A receptor, were infected with HIV-GFFP virus pseudotyped with ASLV-A and VSV-G. The infected cells were selected for resistance to hygromycin and analyzed by flow cytometry to determine the direct-repeat deletion frequency. The results of the control experiment are presented in Table 1. The frequencies of template switching for wild-type RT were 17.6 and 17.8% for VSV-G- and ASLV-A-pseudotyped viruses, respectively, which were not statistically different (P > 0.9). The results indicated that under the conditions used for virus production, reinfection of the producer cells by VSV-G-pseudotyped HIV-GFFP virus did not occur with a sufficiently high frequency to affect the observed frequency of template switching.
TABLE 1.
Comparison of direct-repeat deletion frequencies for VSV-G- and ASLV-A-pseudotyped HIV-GFFP viruses
| Envelope | Deletion frequency (mean % ± SE)a | Virus titer (mean CFU/25 pg of p24 ± SE)b |
|---|---|---|
| VSV-G | 17.6 ± 0.8 | 10.6 ± 0.9 |
| ASLV-A | 17.8 ± 0.2 | 2.5 ± 0.2 |
Deletion frequency was determined for two independent experiments as the percentage of infected 293T target cells that exhibited fluorescence after hygromycin selection. SEs were determined by SigmaPlot 8.0 software.
Virus titer was determined as the number of CFU adjusted to p24 CA concentration as determined by ELISA. The average p24 CA production was 38.5 ± 1.5 ng/ml.
Effects of mutations in the HIV-1 RT polymerase domain on the frequency of template switching.
HIV-1 RT is a multifunctional enzyme that exhibits RNA- and DNA-dependent DNA polymerase as well as RNase H activities. Several substitution mutations in HIV-1 RT are associated with resistance to antiretroviral nucleoside analogs. To determine whether mutations associated with resistance to antiretroviral drugs influence the frequency of RT template switching, we introduced a number of mutations in HIV-1 RT and analyzed their effect on the rate of template switching during one cycle of replication.
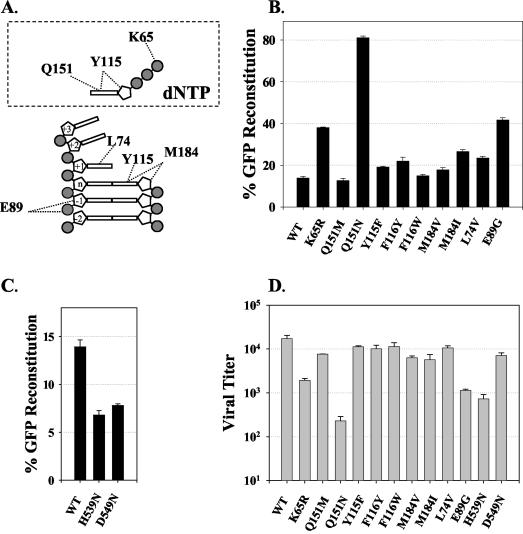
The first group of mutations in RT (outlined in Fig. 3A) targeted the polymerase domain and introduced substitutions in the deoxynucleoside triphosphate (dNTP)-binding site residues or residues adjacent to them. Amino acid K65, which donates hydrogen bonds to the γ-phosphate of the incoming dNTP substrate (28), was mutated to arginine because the K65R substitution is associated with resistance to 2′,3′-dideoxyinosine (ddI) (79), abacavir (44, 75), 2′,3′-dideoxy-3′-thiacytidine (3TC) (55), tenofovir (76), and 2′,3′-didehydro-3′-deoxythymidine (d4T) (19). The effect of the K65R substitution on RT template switching was determined in a single cycle of replication. The results indicated that the K65R substitution increased the frequency of GFP reconstitution nearly threefold (37.8 versus 13.5%; P < 0.0001).
FIG. 3.
HIV-1 RT mutations and their effects on RT template switching and viral titer. (A) Schematic representation of interactions between RT and its dNTP substrate in the polymerase active site and dNTP-binding site (modified from reference 28). Only the amino acids analyzed in this study are shown. (B) Direct-repeat deletion and GFP reconstitution frequencies of the polymerase domain RT mutants. WT, wild type. (C) Direct-repeat deletion and GFP reconstitution frequencies of the RNase H domain RT mutants. The percentage of GFP reconstitution represents the proportion of infected 293T target cells that exhibited fluorescence after hygromycin selection. Bar graphs and error bars represent the means and standard errors of the percentages of GFP reconstitution, respectively, for two to six independent experiments. (D) Viral titers represent the number of CFU per 50 ng of p24 CA as determined by ELISA. The average p24 CA concentration was 37.7 ± 2.8 ng/ml.
Amino acid residue Q151 of HIV-1 RT participates in stacking interactions with the base as well as other interactions with the 3′ OH of the deoxyribose sugar of the incoming dNTP substrate (28, 78). The Q151M substitution is associated with resistance to several nucleoside analogs, including ddI, 3′-azido-3′-deoxythymidine (AZT), and d4T (54, 66, 69). In addition, the Q151N substitution is associated with resistance to AZT, 2′,3′-dideoxycytidine, and other dideoxynucleosides (34, 78). Therefore, we generated RT mutants Q151M and Q151N and determined the effects of these mutations on template switching. The results showed that the Q151M substitution did not affect the template-switching frequency (P > 0.4). In contrast, the Q151N mutation increased the template-switching frequency to 81.1% relative to wild-type RT (13.5%; P < 0.0001).
Amino acids Y115 and F116 (data not shown) form the binding pocket for the 3′ OH of the dNTP substrate (28). The Y115F substitution occurs predominantly in patients treated with abacavir (44). In addition, the F116Y substitution arises in association with the Q151M substitution and is associated with a high level of resistance to several nucleoside RT inhibitors (NRTI) (43, 52). We generated RT mutants Y115F, F116Y, and F116W and determined the effects of these mutations on RT template switching. Mutations Y115F and F116Y increased the template-switching frequency to 19 and 21%, respectively, indicating a small but statistically significant increase (P < 0.05). In contrast, the F116W mutation did not reveal a significant difference in the template-switching frequency relative to wild-type RT (15.0 versus 13.5%; P > 0.1).
Amino acid residue M184 lies in the vicinity of the primer terminus, near the incoming dNTP substrate, and belongs to the highly conserved YXDD motif, which is found in all RTs; amino acids D185 and D186 of this motif, along with D110, form a catalytic triad (28). The M184I mutation appears transiently in response to 3TC treatment and is replaced with the M184V substitution, which is associated with a high level of resistance to 3TC (18, 36, 65). The RT mutations M184I and M184V increased the frequency of GFP reconstitution to 26.6% (P < 0.0001) and 17.8% (P < 0.002), respectively.
We also analyzed the effects of substitution mutations at the L74 and E89 positions, both of which directly contact the template strand. Amino acid substitution at position L74 could also potentially influence dNTP binding through interactions with the side chains of R72 and Q151 (28). The L74V substitution is associated with resistance to ddI and abacavir (44, 66, 79). E89 interacts with the template strand two to three nucleotides downstream of the polymerization site (28). The E89G substitution is associated with resistance to AZT, foscarnet, and several other nonnucleoside RT inhibitors in cell culture (37). The results of RT template-switching analysis indicated that the L74V and E89G substitution mutations increased the frequency of GFP reconstitution to 23.4% (P < 0.0001) and 41.7% (P < 0.0001), respectively; thus, the L74V and E89G mutations increased the frequency of template switching by 1.7- and 3.1-fold, respectively, relative to that for wild-type RT.
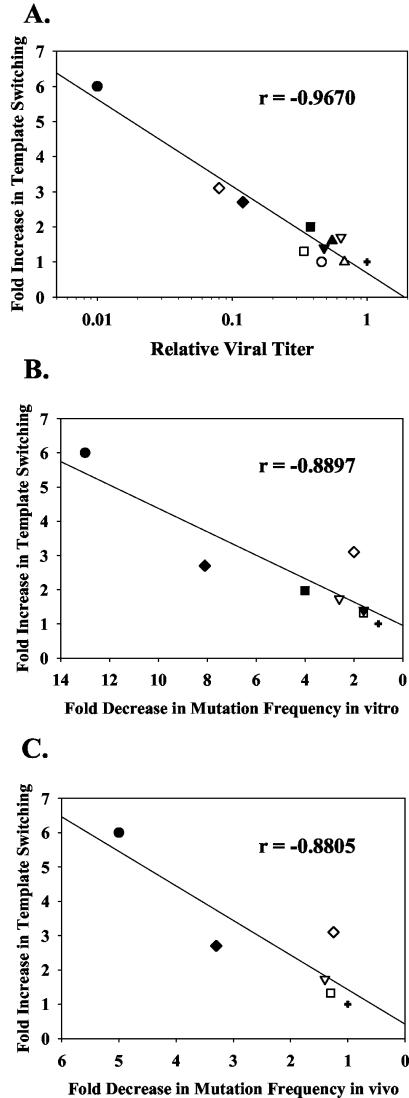
We compared the effects of mutations in the polymerase domain of HIV-1 RT on viral titers in a single cycle of replication (Fig. 3D). Interestingly, we observed an inverse correlation between RT template-switching frequencies and viral titers; for example, the K65R, E89G, and Q151N mutants, which demonstrated the highest increases in template-switching frequencies (Fig. 3B), exhibited the lowest viral titers, which were 12.8, 8, and 1.4% of the wild-type viral titer, respectively. This inverse correlation is presented in more detail later in Fig. 5A and in Discussion.
FIG. 5.
Linear regression analysis of template-switching frequencies, viral titers, and reverse transcription fidelity of HIV-1 RT polymerase domain mutants. The relative template-switching frequencies were correlated with relative viral titers (A) or mutation frequencies during reverse transcription in vitro (B) and in vivo (C). The correlation coefficients (r) were determined by using SigmaPlot 8.0 software. The relative template switching represents a ratio of the template-switching frequency of mutant HIV-1 RT obtained in each experiment divided by the template-switching frequency obtained for the wild-type HIV-1 RT. The relative viral titers represent ratios of the viral titer of mutant HIV-1 RT obtained in each experiment divided by the viral titer of wild-type HIV-1 RT in a parallel experiment. The average viral titer obtained with wild-type HIV-1 RT was 1.7 × 104 CFU/50 ng of p24 CA. The n-fold decrease in mutation frequency represents an average change in mutation frequency for mutants obtained in in vitro or in vivo forward mutation assays using the lacZ alphapeptide reporter gene (8, 16, 17, 31, 39, 40, 59, 67, 77; reviewed in reference 72). The following symbols for RT genotypes were used: +, wild type; •, Q151N; ○, Q151M; ▪, M184I; □, M184V; ▴, F116Y; ▵, F116W; ▾, Y115F; ▿, L74V; ♦, K65R; ⋄, E89G.
Effects of mutations in the HIV-1 RNase H domain on the frequency of template switching.
Based on results obtained in the MLV system, we hypothesized that mutations in the RNase H domain would reduce the frequency of RT template switching (73). Eight amino acid residues are conserved in all known bacterial and retroviral RNase H enzymes (D443, G444, E478, D498, S499, H539, Q545, and D549); four acidic residues play an important role in forming two metal-binding sites (D443, E478, D498, and D549) (10). We generated five RNase H substitution mutants: D443N, E478Q, D498N, H539N, and D549N. The first three mutants exhibited viral titers that were reduced by at least 1,000-fold; as a result, their effect on RT template switching could not be determined. Mutations H539N and D549N permitted viral replication and had sufficiently high viral titers for determination of the template-switching frequency (Fig. 3C and D). The RNase H mutations H539N and D549N decreased the frequency of GFP reconstitution by approximately twofold in comparison to that for wild-type HIV-1 RT (6.8 and 7.6%, respectively; P < 0.0001). The viral titers for the H539N and D549N mutants were 4 and 40%, respectively, relative to the wild-type viral titer.
RT activities of mutant HIV-1 RTs.
Viruses containing either wild-type or mutant RT were concentrated by ultracentrifugation and analyzed for RT activity by using poly(rA) as a template for reverse transcription (Table 2). The amounts of CA protein present in viral preparations were quantified using ELISA, and RT activities were determined in equivalent amounts of viral proteins. One set of HIV-1 RT mutants exhibited RT activities that ranged from 72 to 164% relative to wild-type HIV-1 RT and were not statistically different from the wild-type RT activity (P > 0.05); on the other hand, the RT activities for mutants M184I, M184V, D443N, D498N, H539N, and D549N (P < 0.05) ranged from 50 to 79% of the wild-type RT activity. No correlations were found between RT activities and viral titers or RT template-switching frequencies.
TABLE 2.
RT activity for virion-associated HIV-1 RTs
| HIV-1 RT | RT activity (mean% ± SE)a |
|---|---|
| Wild type | 100 ± 8 |
| K65R | 90 ± 11 |
| L74V | 72 ± 10 |
| E89G | 140 ± 19 |
| Y115F | 143 ± 24 |
| F116Y | 164 ± 30 |
| Q151N | 132 ± 12 |
| M184Ib | 47 ± 6 |
| M184Vb | 79 ± 2 |
| D443Nb | 54 ± 5 |
| E478Q | 127 ± 19 |
| D498Nb | 50 ± 8 |
| H539Nb | 52 ± 7 |
| D549Nb | 56 ± 8 |
Virion-associated RT activities were determined from three independent experiments. RT activities shown are relative to the wild-type HIV-1 RT activity (set to 100%). The amounts of p24 CA were determined by ELISA, and equivalent amounts of viral proteins were used to determine the RT activities. SEs were determined by SigmaPlot 8.0 software.
RT activities obtained for these mutant RTs were significantly different from the wild-type HIV-1 RT activity (P< < 0.05). The RT activities for the remaining mutants were not significantly different from the wild-type RT activity (P > 0.5).
Effect of HU treatment on the frequency of template switching.
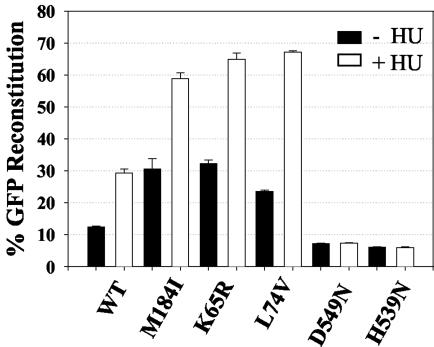
It has been shown that hydroxyurea (HU) treatment of infected cells resulted in a depletion of intracellular nucleotide pools and a significant reduction in the rate of reverse transcription (33). Based on the predictions of the dynamic copy choice mechanism of RT template switching, we hypothesized that treatment of target cells with HU would result in an increase in the frequency of RT template switching. To test this hypothesis, 293T target cells were infected with virus containing wild-type HIV-1 RT in the absence or presence of 0.2 mM HU as previously described (2, 73). Briefly, 293T cells were placed on culture medium containing 0.2 mM HU 4 h prior to infection, 4 h during infection, and 24 h postinfection. The frequency of direct-repeat deletion and GFP reconstitution in HU-treated target cells was determined. The results are shown in Fig. 4. In the presence of HU, the frequency of GFP reconstitution was increased by 2.3-fold for wild-type HIV-1 RT (P < 0.005), which was in agreement with previously published results for wild-type MLV RT (57, 73). HU treatment resulted in a twofold reduction in viral titers relative to untreated controls (data not shown). All template-switching experiments with MLV RT were performed using D17 target cells; to exclude any possible influence of the nature of the target cells and their response to HU treatment, we also performed infections with HIV-1 using D17 target cells. Similar to the previously obtained results for MLV RT (73), we observed a 1.8-fold increase (P < 0.005) in HIV-1 RT template switching when D17 target cells were treated with 1 mM HU (data not shown). These results indicated that slowing down the rate of HIV-1 reverse transcription increases the frequency of template switching and that MLV and HIV-1 RTs respond similarly to depletion of intracellular nucleotide pools by HU treatment.
FIG. 4.
Effects of HU treatment on the frequency of direct-repeat deletion and GFP reconstitution. Black bar graphs and error bars represent the means and standard errors of the frequency of direct-repeat deletion, respectively, for wild-type (WT) HIV-1 RT or mutant RTs for two to six independent experiments; white bar graphs and error bars represent the means and standard errors of the frequency of direct-repeat deletion observed for the same HIV-1 RTs in the presence of 0.2 mM HU.
The frequency of GFP reconstitution for the M184I, K65R, and L74V mutants in the presence of HU increased 2- to 2.8-fold, indicating that HU treatment could further increase the frequency of direct-repeat deletion for mutants that exhibited a higher deletion frequency in the absence of HU (P < 0.01). In contrast, HU treatment of the target cells and subsequent infection with RNase H mutants D549N and H539N did not affect template switching relative to the untreated control (P > 0.21) (Fig. 4).
DISCUSSION
The results of these studies show that mutations in the HIV-1 RT polymerase domain that are associated with antiretroviral drug resistance increase the frequency of template switching. The results also indicated that mutations in the RNase H domain decrease the frequency of RT template switching and that depletion of intracellular dNTP pools with HU treatment increases the frequency of RT template switching. These observations extend our previous studies with the MLV system and support the view that a steady state between the rates of DNA synthesis and RNA template degradation is an important determinant of HIV-1 RT template switching. Thus, the results indicate that the dynamic copy choice mechanism of RT template switching is likely to be responsible for recombination in both lentiviruses and gammaretroviruses.
A comparison of the HIV-1 structural determinants that influence RT template switching identified in this study to those previously identified for MLV RT has revealed interesting similarities and differences. Mutations at the M184 and Y115 positions affected RT template switching in a manner that was similar to the effects observed for the equivalent V223 and F155 positions, respectively, in MLV RT (73). In contrast, the F116W substitution had no effect on RT template switching relative to the 4.8-fold increase in template switching observed for the equivalent F156W substitution mutation in MLV RT. Similarly, the Q151M mutation in HIV-1 RT had no effect on RT template switching, whereas the equivalent Q190M substitution increased the MLV RT template-switching frequency by 1.8-fold. The reasons for these differences between substitutions in HIV-1 and MLV RTs are not known but likely reflect subtle structural differences in the dNTP-binding site and the effects of the substitutions on the kinetics and/or affinity of substrate dNTP binding.
The observation that substitution mutations of residues involved in dNTP binding as well as the dNTP pool depletion result in an increase in RT template switching indicates that the binding of dNTP substrate to RT is an important determinant of RT template switching. The interaction of RT with the incoming dNTP and proper base paring between the incoming dNTP and template nucleotide are likely to be important for ternary complex formation and the rate of DNA polymerization. Our results imply a direct relationship between the effects of dNTP-binding site mutations on reducing the affinity of substrate dNTP binding and an increase in RT template switching. The K65R and Q151N substitutions reduce the affinity of dNTP binding (20, 34, 77), while the Y115F substitution had little effect on dNTP binding (23). The reduced affinity for the incoming dNTP to RT would likely influence the rate of nucleotide incorporation or pyrophosphorolysis. This view is supported by previous observations that the K65R substitution also exhibited a selective 10-fold decrease in binding to ddCTP and ddATP, a moderate decrease in pyrophosphorolysis (71), and decreased processivity at limiting substrate concentrations in vitro (67). Similarly, the Q151N substitution diminished interactions with the 3′ OH of the dNTP substrate so that base pairing between the incoming dNTP and template nucleotide becomes the primary remaining determinant for nucleotide incorporation (78). Substitution of Q151 with asparagine diminished binding to both correct and incorrect dNTPs by up to 120-fold, resulting in greatly reduced processivity and efficiency of nucleotide incorporation (77).
Our observation that the K65R and Q151N substitutions substantially increased RT template-switching frequency while the Y115F substitution had a moderate effect on template switching suggests that reducing the affinity of dNTP binding is correlated with an increase in RT template switching.
Interestingly, we found a negative correlation between the effects of substitutions at the HIV-1 RT polymerase active site on RT template-switching frequency and viral titers (Fig. 5A). For example, Q151N, E89G, and K65R mutants, which exhibited the largest increases in template-switching frequencies (6-, 3.1-, and 2.7-fold, respectively), had the lowest viral titers (0.01, 0.08, and 0.12 of the wild-type titer, respectively). Linear regression analysis of the data obtained here for mutations at the polymerase active site of HIV-1 RT showed a high degree of negative correlation between template switching and viral titers (correlation coefficient r = −0.967; P < 0.001). This correlation suggests that the effects of these substitution mutations on the efficiency of nucleotide incorporation or pyrophosphorolysis, which are predicted to increase RT template switching, also result in a reduction in viral titers. Additionally, the observed correlation between titers and template-switching frequency is supported by the effect of HU treatment on viral titers (data not shown).
We also noted another intriguing negative correlation between RT template switching and fidelity of reverse transcription (Fig. 5B and C). Substitution mutations that were localized at the HIV-1 polymerase catalytic site and exhibited an increase in the template-switching frequency (Q151N, K65R, M184V, M184I, L74V, E89G, and Y115F) were previously observed to decrease the frequency of mutations introduced in the lacZ alphapeptide reporter gene in vitro (8, 16, 17, 31, 59, 67, 77) and in vivo (39, 40). For example, Q151N, K65R, and M184I, which in our experiments increased the template-switching frequency (6-, 2.7-, and 2-fold, respectively), exhibited a corresponding decrease in mutation frequency in vitro (13-, 8.1-, and 4-fold, respectively) (59, 67, 77). Linear regression analysis indicated a high degree of negative correlation between template switching and mutation frequency in vitro (correlation coefficient r = −0.8897; P < 0.01) and in vivo (r = −0.8805; P < 0.05). The explanation for the negative correlation between RT template-switching frequencies and fidelity of DNA synthesis is not known, but one hypothesis is that a reduction in the rate of DNA synthesis results in an increase in both the frequency of RT template switching and the accuracy of DNA synthesis.
The results of these studies also indicate that several mutations in RT that confer resistance to antiretroviral agents influence the rate of RT template switching. The Q151N substitution, which is associated with resistance to several NRTIs in vitro (34, 78), increased the frequency of RT template switching by sixfold. The K65R substitution, which confers resistance to several NRTIs (44, 75, 82), increased the rate of RT template switching by 2.7-fold. The M184V and M184I substitutions, which confer resistance to 3TC (7, 36), increased the rate of RT template switching by 1.3- and 2-fold, respectively. The L74V substitution, which confers resistance to didanosine and abacavir (44, 66, 79), increased the template-switching frequency by 1.7-fold. The E89G substitution, which is associated with resistance to ddNTPs, AZT-triphosphate and foscarnet in vitro (58) and to AZT, foscarnet, and several nonnucleoside RT inhibitors in cell culture (37), increased the template-switching frequency by threefold. These results indicate that selection of these resistance mutations during antiviral therapy could influence the rate of retroviral recombination and evolution. It is important to note that single drug resistance mutations generally arise at the beginning of antiretroviral therapy but most often exist in combination with other mutations that either enhance the drug resistance or improve viral fitness. Thus, the presence of additional mutations could either increase or decrease the viral recombination rate, depending on the effects of the additional mutations on overall viral titers and the rate of DNA polymerization. Additional studies are needed to assess the effects of multiple drug resistance mutations on RT template switching and recombination.
Acknowledgments
We are indebted to Rebekah Barr and Mollie Charon for technical assistance in generating RT mutants. We especially thank Wei-Shau Hu for intellectual input and valuable discussions throughout the project and Anne Arthur for expert editorial revisions. In addition, we thank Jean Mbisa and David Thomas for critical reading of the manuscript.
This work was supported by the HIV Drug Resistance Program, National Cancer Institute.
REFERENCES
- 1.An, W. F., and A. Telesnitsky. 2002. Effects of varying sequence similarity on the frequency of repeat deletion during reverse transcription of a human immunodeficiency virus type 1 vector. J. Virol. 76:7897-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, W. F., and A. Telesnitsky. 2001. Frequency of direct repeat deletion in a human immunodeficiency virus type 1 vector during reverse transcription in human cells. Virology 286:475-482. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, E. S., R. E. Jeeninga, C. K. Damgaard, B. Berkhout, and J. Kjems. 2003. Dimerization and template switching in the 5′ untranslated region between various subtypes of human immunodeficiency virus type 1. J. Virol. 77:3020-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, J. A., E. H. Bowman, and W. S. Hu. 1998. Retroviral recombination rates do not increase linearly with marker distance and are limited by the size of the recombining subpopulation. J. Virol. 72:1195-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson, J. A., R. J. Teufel, P. D. Yin, and W. S. Hu. 1998. Correlated template-switching events during minus-strand DNA synthesis: a mechanism for high negative interference during retroviral recombination. J. Virol. 72:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balakrishnan, M., B. P. Roques, P. J. Fay, and R. A. Bambara. 2003. Template dimerization promotes an acceptor invasion-induced transfer mechanism during human immunodeficiency virus type 1 minus-strand synthesis. J. Virol. 77:4710-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boucher, C. A. B., N. Cammack, P. Schipper, R. Schuurman, P. Rouse, M. A. Wainberg, and J. M. Cameron. 1993. High-level resistance to (−) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 37:2231-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer, P. L., and S. H. Hughes. 2000. Effects of amino acid substitutions at position 115 on the fidelity of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 74:6494-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burke, D. S. 1997. Recombination in HIV: an important viral evolutionary strategy. Emerg. Infect. Dis. 3:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, J. F., Z. Hostomska, Z. Hostomsky, S. R. Jordan, and D. A. Matthews. 1991. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science 252:88-95. [DOI] [PubMed] [Google Scholar]
- 11.Delviks, K. A., and V. K. Pathak. 1999. Effect of distance between homologous sequences and 3′ homology on the frequency of retroviral reverse transcriptase template switching. J. Virol. 73:7923-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derebail, S. S., M. J. Heath, and J. J. DeStefano. 2003. Evidence for the differential effects of nucleocapsid protein on strand transfer in various regions of the HIV genome. J. Biol. Chem. 278:15702-15712. [DOI] [PubMed] [Google Scholar]
- 13.Destefano, J. J. 1994. Kinetic analysis of the catalysis of strand transfer from internal regions of heteropolymeric RNA templates by human immunodeficiency virus reverse transcriptase. J. Mol. Biol. 243:558-567. [DOI] [PubMed] [Google Scholar]
- 14.Destefano, J. J., L. M. Mallaber, L. Rodriguez-Rodriguez, P. J. Fay, and R. A. Bambara. 1992. Requirements for strand transfer between internal regions of heteropolymer templates by human immunodeficiency virus reverse transcriptase. J. Virol. 66:6370-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeStefano, J. J., B. Roberts, and D. Shriner. 1997. The mechanism of retroviral recombination: the role of sequences proximal to the point of strand transfer. Arch. Virol. 142:1797-1812. [DOI] [PubMed] [Google Scholar]
- 16.Drosopoulos, W. C., and V. R. Prasad. 1998. Increased misincorporation fidelity observed for nucleoside analog resistance mutations M184V and E89G in human immunodeficiency virus type 1 reverse transcriptase does not correlate with the overall error rate measured in vitro. J. Virol. 72:4224-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Essink, B. B. O., and B. Berkhout. 1999. The fidelity of reverse transcription differs in reactions primed with RNA versus DNA primers. J. Biomed. Sci. 6:121-132. [DOI] [PubMed] [Google Scholar]
- 18.Gao, Q., Z. X. Gu, M. A. Parniak, J. Cameron, N. Cammack, C. Boucher, and M. A. Wainberg. 1993. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 37:1390-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Lerma, J. G., H. MacInnes, D. Bennett, P. Reid, S. Nidtha, H. Weinstock, J. E. Kaplan, and W. Heneine. 2003. A novel genetic pathway of human immunodeficiency virus type 1 resistance to stavudine mediated by the K65R mutation. J. Virol. 77:5685-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu, Z. X., R. S. Fletcher, E. J. Arts, M. A. Wainberg, and M. A. Parniak. 1994. The K65R mutant reverse transcriptase of HIV-1 cross resistant to 2′,3′-dideoxycytidine, 2′,3′-dideoxy-3′-thiacytidine, and 2′,3′-dideoxyinosine shows reduced sensitivity to specific dideoxynucleoside triphosphate inhibitors in vitro. J. Biol. Chem. 269:28118-28122. [PubMed] [Google Scholar]
- 21.Gu, Z. X., Q. Gao, E. A. Faust, and M. A. Wainberg. 1995. Possible involvement of cell fusion and viral recombination in generation of human immunodeficiency virus variants that display dual resistance to AZT and 3TC. J. Gen. Virol. 76:2601-2605. [DOI] [PubMed] [Google Scholar]
- 22.Halvas, E. K., E. S. Svarovskaia, and V. K. Pathak. 2000. Development of an in vivo assay to identify structural determinants in murine leukemia virus reverse transcriptase important for fidelity. J. Virol. 74:312-319. [PMC free article] [PubMed] [Google Scholar]
- 23.Harris, D., N. Kaushik, P. K. Pandey, P. N. S. Yadav, and V. N. Pandey. 1998. Functional analysis of amino acid residues constituting the dNTP binding pocket of HIV-1 reverse transcriptase. J. Biol. Chem. 273:33624-33634. [DOI] [PubMed] [Google Scholar]
- 24.Hu, W. S., E. H. Bowman, K. A. Delviks, and V. K. Pathak. 1997. Homologous recombination occurs in a distinct retroviral subpopulation and exhibits high negative interference. J. Virol. 71:6028-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu, W. S., T. Rhodes, Q. Dang, and V. Pathak. 2003. Retroviral recombination: review of genetic analyses. Front. Biosci. 8:D143-D155. [DOI] [PubMed] [Google Scholar]
- 26.Hu, W. S., and H. M. Temin. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. USA 87:1556-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu, W. S., and H. M. Temin. 1990. Retroviral recombination and reverse transcription. Science 250:1227-1233. [DOI] [PubMed] [Google Scholar]
- 28.Huang, H. F., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-I reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 29.Hwang, C. K., E. S. Svarovskaia, and V. K. Pathak. 2001. Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching. Proc. Natl. Acad. Sci. USA 98:12209-12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonckheere, H., E. De Clercq, and J. Anne. 2000. Fidelity analysis of HIV-1 reverse transcriptase mutants with an altered amino-acid sequence at residues Leu74, Glu89, Tyr115, Tyr183 and Met184. Eur. J. Biochem. 267:2658-2665. [DOI] [PubMed] [Google Scholar]
- 32.Julias, J. G., T. Kim, G. Arnold, and V. K. Pathak. 1997. The antiretrovirus drug 3′-azido-3′-deoxythymidine increases the retrovirus mutation rate. J. Virol. 71:4254-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Julias, J. G., and V. K. Pathak. 1998. Deoxyribonucleoside triphosphate pool imbalances in vivo are associated with an increased retroviral mutation rate. J. Virol. 72:7941-7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaushik, N., T. T. Talele, P. K. Pandey, D. Harris, P. N. S. Yadav, and V. N. Pandey. 2000. Role of glutamine 151 of human immunodeficiency virus type-1 reverse transcriptase in substrate selection as assessed by site-directed mutagenesis. Biochemistry (Moscow) 39:2912-2920. [DOI] [PubMed] [Google Scholar]
- 35.Kellam, P., and B. A. Larder. 1995. Retroviral recombination can lead to linkage of reverse transcriptase mutations that confer increased zidovudine resistance. J. Virol. 69:669-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keulen, W., N. K. T. Back, A. vanWijk, C. A. B. Boucher, and B. Berkhout. 1997. Initial appearance of the 184Ile variant in lamivudine-treated patients is caused by the mutational bias of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 71:3346-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kew, Y., H. Salomon, L. R. Olsen, M. A. Wainberg, and V. R. Prasad. 1996. The nucleoside analog-resistant E89G mutant of human immunodeficiency virus type 1 reverse transcriptase displays a broader cross-resistance that extends to nonnucleoside inhibitors. Antimicrob. Agents Chemother. 40:1711-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis, B. C., N. Chinnasamy, R. A. Morgan, and H. E. Varmus. 2001. Development of an avian leukosis-sarcoma virus subgroup A pseudotyped lentiviral vector. J. Virol. 75:9339-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mansky, L. M., and L. C. Bernard. 2000. 3′-azido-3′-deoxythymidine (AZT) and AZT-resistant reverse transcriptase can increase the in vivo mutation rate of human immunodeficiency virus type 1. J. Virol. 74:9532-9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mansky, L. M., E. Le Rouzic, S. Benichou, and L. C. Gajary. 2003. Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutant frequencies. J. Virol. 77:2071-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marr, S. F., and A. Telesnitsky. 2003. Mismatch extension during strong stop strand transfer and minimal homology requirements for replicative template switching during Moloney murine leukemia virus replication. J. Mol. Biol. 330:657-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCutchan, F. E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14:S31-S44. [PubMed] [Google Scholar]
- 43.Miller, M. D., N. A. Margot, K. Hertogs, B. Larder, and V. Miller. 2001. Antiviral activity of tenofovir (PMPA) against nucleoside-resistant clinical HIV samples. Nucleosides Nucleotides Nucleic Acids 20:1025-1028. [DOI] [PubMed] [Google Scholar]
- 44.Miller, V., M. Ait-Khaled, C. Stone, P. Griffin, D. Mesogiti, A. Cutrell, R. Harrigan, S. Staszewski, C. Katlama, G. Pearce, and M. Tisdale. 2000. HIV-1 reverse transcriptase (RT) genotype and susceptibility to RT inhibitors during abacavir monotherapy and combination therapy. AIDS 14:163-171. [DOI] [PubMed] [Google Scholar]
- 45.Moutouh, L., J. Corbeil, and D. D. Richman. 1996. Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc. Natl. Acad. Sci. USA 93:6106-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naldini, L., U. Blomer, F. H. Gage, D. Trono, and I. M. Verma. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA 93:11382-11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 48.Negroni, M., and H. Buc. 2000. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proc. Natl. Acad. Sci. USA 97:6385-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Negroni, M., and H. Buc. 2001. Mechanisms of retroviral recombination. Annu. Rev. Genet. 35:275-302. [DOI] [PubMed] [Google Scholar]
- 50.Negroni, M., and H. Buc. 1999. Recombination during reverse transcription: an evaluation of the role of the nucleocapsid protein. J. Mol. Biol. 286:15-31. [DOI] [PubMed] [Google Scholar]
- 51.Onafuwa, A., W. F. An, N. D. Robson, and A. Telesnitsky. 2003. Human immunodeficiency virus type 1 genetic recombination is more frequent than that of Moloney murine leukemia virus despite similar template switching rates. J. Virol. 77:4577-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palmer, S., R. W. Shafer, and T. C. Merigan. 1999. Highly drug-resistant HIV-1 clinical isolates are cross-resistant to many antiretroviral compounds in current clinical development. AIDS 13:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peeters, M., and P. M. Sharp. 2000. Genetic diversity of HIV-1: the moving target. AIDS 14:S129-S140. [PubMed] [Google Scholar]
- 54.Pellegrin, I., J. Izopet, J. Reynes, M. Denayrolles, B. Montes, J. L. Pellegrin, P. Massip, J. Puel, H. Fleury, and M. Segondy. 1999. Emergence of zidovudine and multidrug-resistance mutations in the HIV-1 reverse transcriptase gene in therapy-naive patients receiving stavudine plus didanosine combination therapy. AIDS 13:1705-1709. [DOI] [PubMed] [Google Scholar]
- 55.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pfeiffer, J. K., M. M. Georgiadis, and A. Telesnitsky. 2000. Structure-based Moloney murine leukemia virus reverse transcriptase mutants with altered intracellular direct-repeat deletion frequencies. J. Virol. 74:9629-9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfeiffer, J. K., R. S. Topping, N. H. Shin, and A. Telesnitsky. 1999. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J. Virol. 73:8441-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prasad, V. R., I. Lowy, T. Delossantos, L. Chiang, and S. P. Goff. 1991. Isolation and characterization of a dideoxyguanosine triphosphate resistant mutant of human immunodeficiency virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 88:11363-11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rezende, L. F., W. C. Drosopoulos, and V. R. Prasad. 1998. The influence of 3TC resistance mutation M184I on the fidelity and error specificity of human immunodeficiency virus type 1 reverse transcriptase. Nucleic Acids Res. 26:3066-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rhodes, T., H. Wargo, and W. S. Hu. 2003. High rates of human immunodeficiency virus type I recombination: near-random segregation of markers one kilobase apart in one round of viral replication. J. Virol. 77:11193-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robertson, D. L., P. M. Sharp, F. E. McCutchan, and B. H. Hahn. 1995. Recombination in HIV-1. Nature 374:124-126. [DOI] [PubMed] [Google Scholar]
- 62.Roda, R. H., M. Balakrishnan, J. K. Kim, B. P. Roques, P. J. Fay, and R. A. Bambara. 2002. Strand transfer occurs in retroviruses by a pause-initiated two-step mechanism. J. Biol. Chem. 277:46900-46911. [DOI] [PubMed] [Google Scholar]
- 63.Rodenburg, C. M., Y. Y. Li, S. A. Trask, Y. L. Chen, J. Decker, D. L. Robertson, M. L. Kalish, G. M. Shaw, S. Allen, B. H. Hahn, and F. Gao. 2001. Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents. AIDS Res. Hum. Retrovir. 17:161-168. [DOI] [PubMed] [Google Scholar]
- 64.Rodriguez-Rodriguez, L., Z. Tsuchihashi, G. M. Fuentes, R. A. Bambara, and P. J. Fay. 1995. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J. Biol. Chem. 270:15005-15011. [DOI] [PubMed] [Google Scholar]
- 65.Schinazi, R. F., R. M. Lloyd, M. H. Nguyen, D. L. Cannon, A. McMillan, N. Ilksoy, C. K. Chu, D. C. Liotta, H. Z. Bazmi, and J. W. Mellors. 1993. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob. Agents Chemother. 37:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shafer, R. W., M. J. Kozal, M. A. Winters, A. K. N. Iversen, D. A. Katzenstein, M. V. Ragni, W. A. Meyer, P. Gupta, S. Rasheed, R. Coombs, M. Katzman, S. Fiscus, and T. C. Merigan. 1994. Combination therapy with zidovudine and didanosine selects for drug-resistant human immunodeficiency virus type-1 strains with unique patterns of Pol gene mutations. J. Infect. Dis. 169:722-729. [DOI] [PubMed] [Google Scholar]
- 67.Shahs, F. S., K. A. Curr, M. E. Hamburgh, M. Parniak, H. Mitsuya, J. G. Arnez, and V. R. Prasad. 2000. Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 275:27037-27044. [DOI] [PubMed] [Google Scholar]
- 68.Sharp, P. M., E. Bailes, D. L. Robertson, F. Gao, and B. H. Hahn. 1999. Origins and evolution of AIDS viruses. Biol. Bull. 196:338-342. [DOI] [PubMed] [Google Scholar]
- 69.Shirasaka, T., M. F. Kavlick, T. Ueno, W. Y. Gao, E. Kojima, M. L. Alcaide, S. Chokekijchai, B. M. Roy, E. Arnold, R. Yarchoan, and H. Mitsuya. 1995. Emergence of human immunodeficiency virus type-1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc. Natl. Acad. Sci. USA 92:2398-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. D. Huse. 1988. Lambda-Zap: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sluis-Cremer, N., D. Arion, N. Kaushik, H. Lim, and M. A. Parniak. 2000. Mutational analysis of Lys65 of HIV-1 reverse transcriptase. Biochem. J. 348:77-82. [PMC free article] [PubMed] [Google Scholar]
- 72.Svarovskaia, E. S., S. R. Cheslock, W. H. Zhang, W. S. Hu, and V. K. Pathak. 2003. Retroviral mutation rates and reverse transcriptase fidelity. Front. Biosci. 8:D117-D134. [DOI] [PubMed] [Google Scholar]
- 73.Svarovskaia, E. S., K. A. Delviks, C. K. Hwang, and V. K. Pathak. 2000. Structural determinants of murine leukemia virus reverse transcriptase that affect the frequency of template switching. J. Virol. 74:7171-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Temin, H. M. 1991. Sex and recombination in retroviruses. Trends Genet. 7:71-74. [DOI] [PubMed] [Google Scholar]
- 75.Tisdale, M., T. Alnadaf, and D. Cousens. 1997. Combination of mutations in human immunodeficiency virus type 1 reverse transcriptase required for resistance to the carbocyclic nucleoside 1592U89. Antimicrob. Agents Chemother. 41:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wainberg, M. A., M. D. Miller, Y. D. Quan, H. Salomon, A. S. Mulato, P. D. Lamy, N. A. Margot, K. E. Anton, and J. M. Cherrington. 1999. In vitro selection and characterization of HIV-1 with reduced susceptibility to PMPA. Antivir. Ther. 4:87-94. [DOI] [PubMed] [Google Scholar]
- 77.Weiss, K. K., R. A. Bambara, and B. Kim. 2002. Mechanistic role of residue Gln151 in error prone DNA synthesis by human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT). J. Biol. Chem. 277:22662-22669. [DOI] [PubMed] [Google Scholar]
- 78.Weiss, K. K., S. J. Isaacs, N. H. Tran, E. T. Adman, and B. Kim. 2000. Molecular architecture of the mutagenic active site of human immunodeficiency virus type 1 reverse transcriptase: roles of the beta 8-alpha E loop in fidelity, processivity, and substrate interactions. Biochemistry (Moscow) 39:10684-10694. [DOI] [PubMed] [Google Scholar]
- 79.Winters, M. A., R. W. Shafer, R. A. Jellinger, G. Mamtora, T. Gingeras, and T. C. Merigan. 1997. Human immunodeficiency virus type 1 reverse transcriptase genotype and drug susceptibility changes in infected individuals receiving dideoxyinosine monotherapy for 1 to 2 years. Antimicrob. Agents Chemother. 41:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yee, J. K., A. Miyanohara, P. Laporte, K. Bouic, J. C. Burns, and T. Friedmann. 1994. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. USA 91:9564-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yusa, K., M. F. Kavlick, P. Kosalaraksa, and H. Mitsuya. 1997. HIV-1 acquires resistance to two classes of antiviral drugs through homologous recombination. Antiviral Res. 36:179-189. [DOI] [PubMed] [Google Scholar]
- 82.Zhang, D., A. M. Caliendo, J. J. Eron, K. M. Devore, J. C. Kaplan, M. S. Hirsch, and R. T. Daquila. 1994. Resistance to 2′,3′-dideoxycytidine conferred by a mutation in codon 65 of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 38:282-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang, W. H., C. K. Hwang, W. S. Hu, R. J. Gorelick, and V. K. Pathak. 2002. Zinc finger domain of murine leukemia virus nucleocapsid protein enhances the rate of viral DNA synthesis in vivo. J. Virol. 76:7473-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhuang, J. L., A. E. Jetzt, G. L. Sun, H. Yu, G. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2002. Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots. J. Virol. 76:11273-11282. [DOI] [PMC free article] [PubMed] [Google Scholar]