Abstract
The Sendai virus C protein acts to dismantle the interferon-induced cellular antiviral state in an MG132-sensitive manner, in part by inducing STAT1 instability. This activity of C maps to the first 23 amino acids (C1-23) of the 204-amino-acid (aa)-long protein (C1-204). C1-23 was found to act as an independent viral element that induces STAT1 instability, since this peptide fused to green fluorescent protein (C1-23/GFP) is at least as active as C1-204 in this respect. This peptide also induces the degradation of C1-23/GFP and other proteins to which it is fused. Most of C1-204, and particularly its amino-terminal half, is predicted to be structurally disordered. C1-23 as a peptide was found to be disordered by circular dichroism, and the first 11 aa have a strong potential to form an amphipathic α-helix in low concentrations of trifluoroethanol, which is thought to mimic protein-protein interaction. The critical degradation-determining sequence of C1-23 was mapped by mutation to eight residues near its N terminus: 4FLKKILKL11. All the large hydrophobic residues of 4FLKKILKL11, plus its ability to form an amphipathic α-helix, were found to be critical for STAT1 degradation. In contrast, C1-23/GFP self-degradation did not require 8ILKL11, nor the ability to form an α-helix throughout this region. Remarkably, C1-23/GFP also stimulated C1-204 degradation, and this degradation in trans required the same peptide determinants as for STAT1. Our results suggest that C1-204 coordinates its dual activities of regulating viral RNA synthesis and counteracting the host innate antiviral response by sensing both its own intracellular concentration and that of STAT1.
Protein degradation performs important roles in cellular regulation, including the disposal of defective proteins and control of levels of intrinsically and conditionally labile proteins that demand precise temporal adjustment (8). Cells have evolved complex mechanisms for destroying appropriate substrates and sparing other proteins. Three elements are commonly found (2): (i) proteolysis occurs within the interior of a proteolytic machine, the proteasome; (ii) authentic substrates are equipped with tags that license degradation (e,g., polyubiquitin); and (iii) another component of the proteolytic machine distinguishes marked proteins and admits them to its interior in an ATP-dependent manner, where they are processed to peptides. Viruses have also evolved mechanisms for destroying normally stable cellular substrates as part of their program to evade the innate cellular antiviral response, which includes the interferon (IFN)-induced antiviral state and programmed cell death. Signal transducer and activator of transcription 1 (STAT1) plays a key role in both these cellular antiviral responses and is normally very stable (22). However, STAT1 levels are strongly reduced via proteasomal degradation during infection with two groups of paramyxoviruses (reviewed in reference 18). In both cases, the Sendai virus C proteins and rubulavirus V proteins, the viral products responsible for STAT1 degradation, are encoded in their P genes.
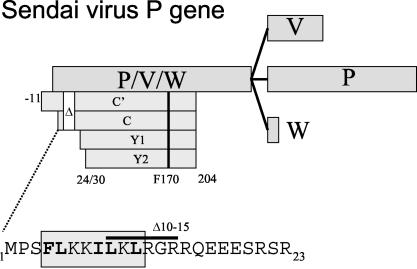
The Paramyxovirinae subfamily consists of three genera, Respirovirus (e.g., Sendai virus [SeV] and Parainfluenzavirus type 1 [PIV1]), Morbillivirus (e.g., Measles virus), and Rubulavirus (e.g., Mumps virus and Simian virus 5 [SV5]), which are classified largely by the organization of their complex P genes (30). P genes encode not only the P protein, which is an essential component of the viral RNA-dependent RNA polymerase, but also two to seven “accessory” proteins, including the C and V proteins, which play important roles in virus replication (Fig. 1). Paramyxovirus C and V proteins are referred to as accessory proteins because all members of this subfamily do not express them. P genes are divided in half by mRNA “editing” sites in the viral genome that induce pseudotemplated transcription by viral RNA-dependent RNA polymerase during mRNA synthesis (46, 50). The resulting nucleotide insertions in the mRNA fuse a common amino-terminal open reading frame ORF (P/V/W) to different carboxyl ORFs, yielding the P, V, and W proteins (Fig. 1). SeV, like all respiro- and morbilliviruses, contains two ORFs upstream of the editing site, a longer ORF that is fused to the different carboxyl segments, and a shorter C ORF that overlaps the beginning of the common P/V/W ORF. However, at least one (human parainfluenza virus type 1 [hPIV1]) (39) and probably two (hPIV3) (13) respiroviruses do not express V proteins. Rubulaviruses, in contrast, have only a single ORF upstream of the editing site, and rubulaviruses thus do not express C proteins. However, all paramyxoviruses express either a C protein or a V protein. The respirovirus C proteins and rubulavirus V proteins carry out different functions in viral RNA synthesis, and they also counteract host innate immunity, in part by inducing the proteasomal degradation of STAT1 (11). In cells that have established an IFN-induced antiviral (vesicular stomatitis virus [VSV]) state, the loss of STAT1 correlates with SeV-induced dismantlement of this state (14). The SeV C protein and the mumps virus V protein also appear to bind to the same surface of STAT1 in vitro (40).
FIG. 1.
SeV P gene organization. The various ORFs expressed as proteins are shown as horizontal boxes. The overlapping P/V/W and C ORFs are translated from the same mRNAs by ribosomal choice, whereas the overlapping V, P, and W ORFs are translated from differentially edited mRNAs. The four C proteins which initiate at different start codons and terminate on residue 204 are shown as crenelated boxes; the sites of the Δ10-15 and F170S mutations are indicated. The amino acid sequence of C1-23 is shown below in one-letter code; the boxed region shows that portion of the peptide with the strongest potential to form an α-helix. The location of the 10-15 deletion is also indicated.
Many paramyxoviruses express a single C protein, but SeV expresses a nested set of two longer C proteins (C′ and C) and two shorter proteins (Y1 and Y2) (33) (Fig. 1). Four C proteins are also expressed in hPIV1, which does not express a V protein (42). C′/C and Y1/Y2 carry out separate functions during virus replication (18, 32), and this difference appears to be due to the sequence that is uniquely present in the longer C proteins, i.e., residues 1 to 23 of C, or C1-23. For example, recombinant SeV (rSeV) that express only the Y proteins (because their C′ and C start codons are mutated) or those carrying a deletion of residues 10 to 15 of the C gene (rSeV-CΔ10-15) are unable to counteract the cellular antiviral response in some cells, even though these C proteins are abundantly expressed. Although rSeV-CΔ10-15 infection prevented IFN-induced pY701-STAT1 formation and IFN signaling, as with wild-type (wt) SeV (SeV-wt), this virus had simultaneously lost the ability to induce STAT1 degradation in mouse embryonic fibroblasts and to reverse their IFN-induced antiviral (VSV) state, i.e., as measured by the prevention of VSV replication. As might be expected, rSeV-CΔ10-15 was found to be 100-fold less virulent (by 50% lethal dose) than SeV-wt in mice (14).
We recently reported that C interacts with STAT1 in two different ways (18). The first interaction prevents IFN-induced pY701-STAT1 formation (i.e., that measured 45 min after addition of IFN to a culture) and IFN signaling to a reporter gene, and this aspect has been well documented (16, 17, 20). The other interaction paradoxically induces long-term pY701-STAT1 formation while decreasing bulk STAT1 levels in an IFN-independent manner (18). These two C-STAT1 interactions are distinguished genetically, since the IFN-independent increase in pY701-STAT1 and decrease in bulk STAT1 levels require a longer C protein but not phe170, whereas the IFN-dependent C-STAT1 interaction requires phe170 but not a longer C protein. This clean separation of phenotypes suggests that C is composed of two modules, albeit of unequal size: that of the Y proteins themselves (residues 24/30 to 204) and C1-23 present in C′ and C (Fig. 1). This paper reports that C1-23 does indeed act as an independent viral element that induces STAT1 instability, and it also induces the degradation of proteins to which it is fused.
MATERIALS AND METHODS
Cells.
2fTGH cells and their derived cell lines (e.g., U3A) were obtained from I. M. Kerr's lab (Imperial Cancer Research Fund, London, United Kingdom) and grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum in the presence of the relevant maintenance drug (hygromycin [250 μg/ml]).
Immunoprecipitation.
Cells were washed once with phosphate-buffered saline (PBS) and scraped into PBS. Cytoplasmic extracts were prepared with lysis buffer containing 0.5% NP-40, 50 mM Tris-Cl (pH 8), 150 mM NaCl, 10 mM EDTA, a 1:200 dilution of protease inhibitor cocktail (P8340; Sigma), and 1:200 phosphatase inhibitor cocktail (P2850; Sigma). The mixture was centrifuged at 13,000 × g for 2 min. The supernatant was used for immunoprecipitation with a rabbit antibody to green fluorescent protein (GFP) (anti-GFP; Clontech).
Immunoblotting.
Cytoplasmic extracts were prepared as described above. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon-P membranes by semidry transfer. The primary antibodies used included a rabbit antiserum to the SeV P, C, V, and W proteins, a mouse monoclonal antibody to the STAT1 C terminus (S21120; Transduction Laboratories), a rabbit antiserum to phospho-STAT (Y701) (06-657; Upstate Biotechnology), a rabbit antiserum to actin (provided by G. Gabbiani, Geneva, Switzerland), and anti-GFP (Clontech). Anti-STAT2 (A7) and anti-STAT3 (H-190) were obtained from Santa Cruz Biotechnology, Inc. The secondary antibodies used were alkaline phosphatase-conjugated goat antibodies specific for either rabbit or mouse immunoglobulin G (IgG) (Bio-Rad). The immobilized proteins were detected by light-enhanced chemiluminescence (Pierce), and the results were quantified in a Bio-Rad light detector with Quantity One software (Bio-Rad).
Plasmids and transient transfections.
Various SeV C and Stat1 genes were cloned into the episomal Epstein-Barr virus-based expression plasmid pEBS-PL (3). pEBS constructs expressing SV5 protein V and GFP were provided by Olivier Leupin and Michel Strubin (Geneva) (34, 35). All the C/GFP fusion proteins were constructed by PCR and cloned into pEBS-PL. All recombinant DNA work was carried out according to standard procedures. Details of the plasmid constructions are available upon request.
A plasmid expressing DDB1-specific small interfering RNA (siRNA) was the generous gift of Olivier Leupin and Michel Strubin (Geneva) (34).
A plasmid expressing GFP-specific siRNA used for silencing of GFP (Fig. 2, RNAi x GFP) was produced from the episomal EBP-SUP (34). A GFP targeting sequence directed against nucleotides 479 to 496 of the GFP coding region was inserted into the vector in the form of a double-stranded oligonucleotide (top strand, 5′-GATCCCCGAACGGCATCAAGGTGAACTTCAAGAGAGTTCACCTTGATGCCGTTCTTTTTGGAAA-3′; GFP sequences in boldface) according to the author's recommendations (5).
FIG. 2.
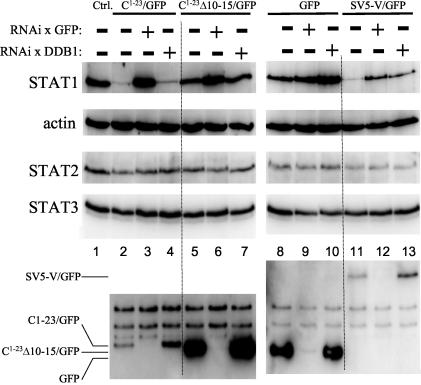
SeV C1-23-induced STAT1 degradation. Parallel cultures of U3A cells were transfected with plasmids expressing STAT1 and various GFPs as indicated above (and RFP as a transfection control). Some cultures were also transfected with plasmids expressing RNAi against GFP (RNAi x GFP) or DDB1 mRNA (RNAi x DDB1), also as indicated above. Cytoplasmic extracts were prepared at 48 h posttransfection, and samples containing equal amounts of total protein (Bradford) were separated by SDS-PAGE and then assayed by immunoblotting for their relative amounts of STAT1, STAT2, STAT3, actin, and the various GFPs.
The alpha/beta IFN (IFN-α/β)-responsive reporter plasmid p(9-27)4tk(239)lucter (28, 29), referred to here as pISRE-(f)luc, contains four tandem repeats of the IFN-inducible gene 9-27 IFN-stimulated response element fused to the firefly luciferase gene. pTK-(r)luc, used as a transfection standard, contains the herpes simplex virus thymidine kinase promoter region upstream of the Renilla luciferase gene (Promega).
For transfections, 100,000 cells were plated in six-well plates 20 h before transfection with 1 μg of pEBS expressing the various C, C/GFP, or GFP proteins, 0.5 μg of pDsRed (Clontech), 1 μg of pEBS-STAT1, and 7.5 μl of Fugene (Roche) according to the manufacturer's instructions. The transfected cells were systematically analyzed by fluorescence-activated cell sorting for green and red (control) fluorescence.
For luciferase assays, 2fTGH cells were transfected with 1 μg of pEBS-C, 1 μg of pISRE(f)luc, 0.3 μg of pTK-(r)luc, and 7.5 μl of Fugene (Roche). At 24 h posttransfection, the cells were treated with 1,000 IU of recombinant IFN-α2/α1 per ml (51) or left untreated. At 14 h post-IFN treatment, the cells were harvested and assayed for firefly and Renilla luciferase activity (dual-luciferase reporter assay system; Promega). Relative expression levels were calculated by dividing the firefly luciferase values by the Renilla luciferase values.
Metabolic labeling and chase experiments.
Transfected U3A cells were metabolically labeled 48 h posttransfection. The cells were starved for 45 min in methionine- and cysteine-free minimal essential medium (Sigma) and then pulse-labeled for 30 min at 37°C in fresh medium supplemented with a [35S]methionine-[35S]cysteine mix (Hartmann Analytic) at a final concentration of 0.10 mCi/ml. The cells were then rinsed twice in minimal essential medium containing 2% fetal calf serum, 10 mM methionine, and 10 mM cysteine (Sigma) and either harvested immediately (time zero) or further incubated in the same medium in the presence of 50 μg of cycloheximide/ml plus or minus 10 μM MG132 (Sigma). At each time point, cells were harvested and lysed as described previously. The lysates were immunoprecipitated with a rabbit anti-GFP antibody (Clontech).
RESULTS
C1-23 can act independently of C24-204.
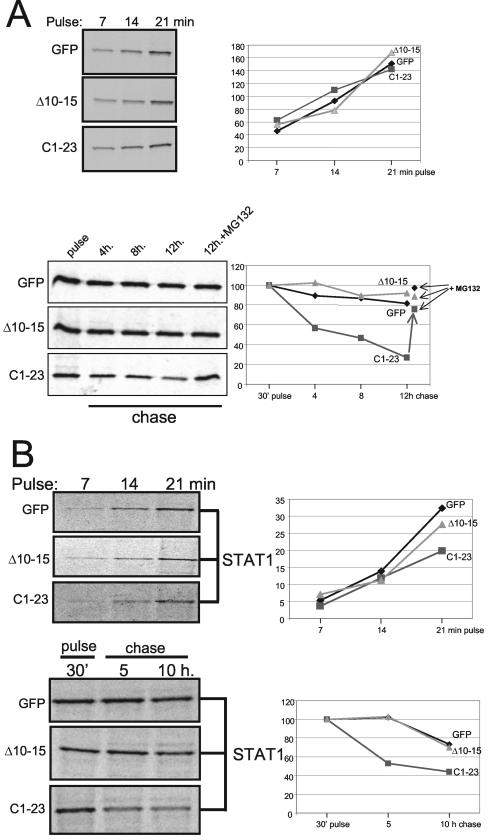
To examine whether C1-23 can act independently of the remainder of the Y1 polypeptide (C24-204) to which it is naturally fused, we constructed plasmids expressing C1-23 as an amino-terminal extension of GFP and examined their ability to reduce bulk STAT1 levels in (STAT1−) U3A cells cotransfected with plasmids expressing Myc-tagged STAT1. This approach minimizes the background of cells containing STAT1 without C1-23/GFP, since both proteins are expressed by transfection. We also examined C1-23Δ10-15/GFP, since this deletion in C1-204 specifically abolishes its effect on STAT1 levels. As shown in Fig. 2, C1-23/GFP and C1-23Δ10-15/GFP accumulate to very different levels in U3A cells (lanes 2 and 5, respectively), even though cotransfected red fluorescent protein (RFP) was expressed at similar levels in all cases (data not shown). Whereas C1-23Δ10-15/GFP levels were superior to those of unmodified GFP (lane 8), C1-23/GFP levels were reduced >10-fold. GFP has a compact structure and is normally very stable (7). Since C1-204 appears to be less stable than C24-204 (18), C1-23 might also act to destabilize other proteins to which it is fused. We therefore examined the half-lives of the various GFPs in [35S]Met pulse-chase experiments. In contrast to their very unequal levels of accumulation (Fig. 2), C1-23/GFP and C1-23Δ10-15/GFP were synthesized at similar rates during the pulse, namely that of unmodified GFP (Fig. 3A, top). Moreover, whereas GFP and C1-23Δ10-15/GFP were stable under the conditions of our chase, C1-23/GFP was clearly less stable, and this instability could be prevented by MG132 treatment (Fig. 3B, bottom). Thus, C1-23 as an amino-terminal extension of GFP appears to induce the proteasomal degradation of this fusion protein, and deleting residues 10 to 15 of C1-23 abolishes this effect.
FIG.3.
SeV C1-23 contains a degradation-determining element. Parallel cultures of U3A cells were transfected with plasmids expressing various GFPs as indicated and STAT1. The cultures were pulse-labeled with 35S-Translabel for 7, 14, and 21 min at 24 h posttransfection, and the amount of labeled protein was determined by SDS-PAGE after precipitation with anti-GFP (A) or anti-STAT1 (B). The kinetics of incorporation determined by autoradiography is plotted on the right. Other cultures were pulse-labeled for 30 min and chased for the various times in the presence of cycloheximide. Some cultures were also treated with MG132 after the pulse. Cell extracts were prepared at the times indicated, and samples containing equal amounts of total protein were immunoprecipitated with anti-GFP or anti-STAT1. The kinetics of protein loss is plotted on the bottom right.
We next examined the effects of C1-23/GFP and C1-23Δ10-15/GFP on STAT1, STAT2, and STAT3 levels relative to unmodified GFP as a negative control and to the SV5 V protein fused to GFP (GFP/SV5-V) as a positive control. The rubulavirus SV5 V protein induces the proteasomal degradation of STAT1, but SV5 V itself is relatively stable in this process (11). SV5 V-induced STAT1 degradation requires cellular DDB1 (1), and we also examined whether this was also the case for C1-23 (endogenous DDB1 was targeted by RNA interference (RNAi) (see Materials and Methods). The various GFP fusion proteins were also targeted by RNAi against GFP to ensure that their possible effects were due to the fusion proteins themselves. As shown in Fig. 2, all three STAT levels were unaltered by expression of unmodified GFP (lanes 8 versus the control cultures that received an empty plasmid [lanes 1]). Moreover, all three STAT levels were mostly unaltered by RNAi against GFP (lanes 9) or DDB1 (lanes 10), even though the RNAi against GFP appeared to operate effectively (lanes 3, 6, and 9, below). In contrast to unmodified GFP, expression of C1-23/GFP strongly and specifically reduced bulk STAT1 levels (lanes 2), whereas expression of C1-23Δ10-15/GFP had only a modest effect on decreasing STAT1 (lane 5 versus lane 1). RNAi directed against C1-23/GFP completely reversed the loss of STAT1 in these cultures (lanes 3), whereas RNAi directed against DDB1 was without effect (lane 4). RNAi directed against C1-23Δ10-15/GFP similarly reversed the modest loss of STAT1 in these cultures (lanes 6), whereas RNAi against DDB1 was again without effect (lane 7). Expression of GFP/SV5-V also led to the specific loss of STAT1 levels, as expected (lane 11), and in this case RNAi against either GFP (lane 12) or DDB1 (lane 13) reversed the loss of STAT1. The specificity of the RNAi confirms that the loss of STAT1 is due to expression of the C1-23 and V fusion proteins.
U3A cells contain endogenous STAT2 and STAT3, and thus, the extent to which these factors can be diminished by transfected C1-23/GFP in this experiment depends on the efficiency of transfection (60% in this case). However, in 2fTGH cells, endogenous STAT1 is clearly reduced by C expression, whereas STAT2 and STAT3 are not (Fig. 4; also data not shown). The effect of C1-23/GFP on STAT levels thus appears to be specific for STAT1. We have also directly examined the effect of C1-23/GFP on STAT1 stability in U3A cells in pulse-chase experiments. 35S-labeled STAT1 was found to be synthesized at similar rates during cotransfection with GFP, C1-23/GFP, or C1-23Δ10-15/GFP (Fig. 3B, top), but only C1-23/GFP clearly reduced the level of pulse-labeled STAT1 in the chase (Fig. 3B, bottom). Thus, C1-23 as an amino-terminal extension of GFP induces the instability of both itself and STAT1. In contrast to GFP/SV5-V, this induced instability does not require DDB1.
FIG. 4.
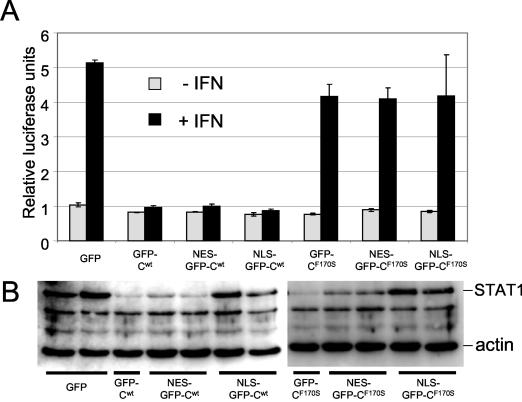
Restricting the intracellular location of C1-23/GFP. Parallel cultures of 2fTGH cells were transfected with plasmids expressing pISRE-(f)luc [and pTK-(r)luc as a transfection control] and plasmids expressing various GFP/C1-204 (wt) or GFP/C1-204 (F170S) constructs that either were not modified further or which contained a nuclear export signal (NES) or a nuclear localization signal (NLS) fused to the amino terminus of GFP, as indicated. Some of the cultures were treated with 1,000 IU of IFN-α at 24 h posttransfection for 16 h. Cell extracts were prepared at 40 h posttransfection, and samples containing equal amounts of total protein were used to determine relative firefly luciferase activity (normalized to Renilla luciferase activity) (A) and their relative amounts of STAT1 by immunoblotting (B). The double lanes in panel B represent duplicate transfections without IFN treatment. Only a single lane is shown for the two controls, GFP/Cwt and GFP/C(F170S).
Restricting the intracellular location of GFP/C1-204.
When GFP is fused to the amino or carboxyl ends of C1-204 (GFP/C1-204 and C1-204/GFP, respectively), the two GFP fusions have different effects on the activities of C1-204 that map to C1-23 and C24-204, i.e., to induce STAT1 instability and to stably bind STAT1 and prevent IFN signaling, respectively. Tagging the N terminus of C1-204 with GFP specifically decreases the first activity (and also decreases its own instability), whereas GFP fusion to the carboxyl end of C1-204 specifically decreases the second activity (15). Topological constraint of either end of C1-204 appears to specifically inhibit their respective activities. Although the activity of GFP/C1-204 in inducing STAT1 instability is reduced, this is largely compensated by its increased intracellular abundance due to its increased stability.
If C1-23 and C24/30-204 can act independently, they might also act in different cellular compartments. Transfected GFP/C1-204, like C1-204, is found throughout the cell but mainly in the cytoplasm. To determine whether the various activities of C operate when their cellular localization is restricted, either a nuclear export signal or a nuclear localization signal was appended to the amino-terminus of GFP/C and GFP/CF170S. Fluorescence microscopy of cells transfected with these constructs showed that their localization signals operated as expected (data not shown). We first examined these fusion proteins for their ability to prevent IFN signaling to a reporter gene in response to IFN-α treatment. The results (Fig. 4A) show that GFP/C is effective in preventing IFN signaling irrespective of its cellular localization, whereas GFP/CF170S was ineffective (as expected). Thus, GFP/C can apparently prevent IFN signaling by stably binding STAT1 in either the cytoplasm or nucleus. The ability of these fusion proteins to induce STAT1 degradation was also examined (Fig. 4B). In contrast to the above, both GFP/C and GFP/CF170S induce STAT1 instability, as expected. More importantly, both fusion proteins mostly lost their ability to reduce STAT1 levels when restricted to the nucleus (lanes NLS-GFP-Cwt/F170S) but appeared to be fully active when restricted to the cytoplasm (lanes NES-GFP-Cwt/F170S). C-induced STAT1 instability thus presumably occurs in the cytoplasm. We note that GFP/CF170S-transfected cells in which STAT1 levels have been strongly reduced nevertheless still efficiently respond to IFN-α (Fig. 4). The small amount of STAT1 that remains may be sufficient to account for this IFN-stimulated response element activation.
C1-23 contains a degradation determinant (degron) that may act via induced folding.
When U3A cells are cotransfected with plasmids expressing STAT1 and C1-23Δ10-15/GFP or C1-23/GFP, at least 10-fold more STAT1 and C/GFP accumulate during coexpression of C1-23Δ10-15/GFP than that of C1-23/GFP (see Fig. 6A, first 2 lanes, bottom). When C1-23 or C1-23Δ10-15 is added as an amino-terminal extension to STAT1 itself, the C1-23Δ10-15/STAT1 fusion protein again accumulates ∼10 times more abundantly than C1-23/STAT1 (not shown). C1-23 thus appears to contain a degradation-determining signal or degron (49), i.e., a short, often portable element that induces polypeptide degradation. Moreover, rSeV infections that express only CΔ10-15 or Y1/Y2 protein accumulate these deleted C proteins ca. fivefold more abundantly than infections that express C-wt or CF170S 18). The C1-23 degron thus also appears to be active within C1-204.
FIG. 6.
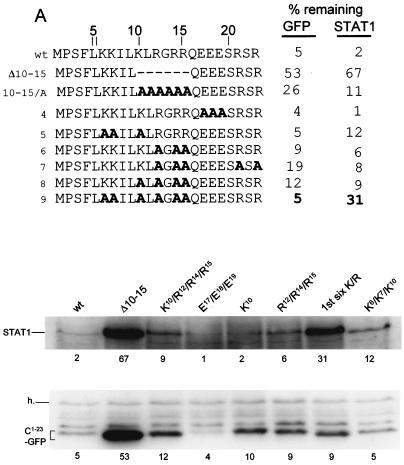
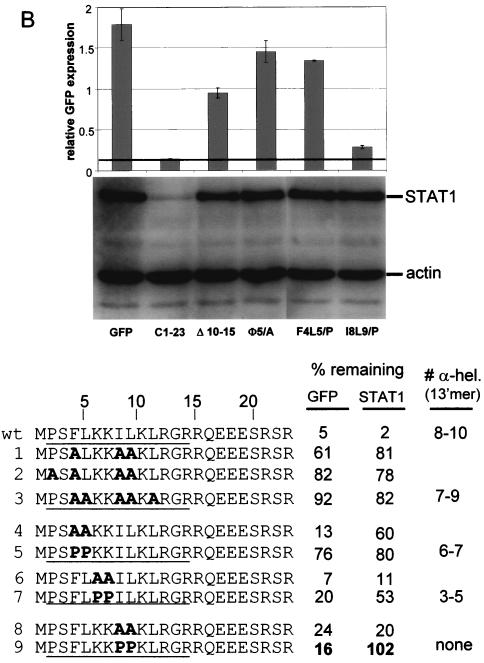
Mutational analysis of C1-23-induced GFP and STAT1 instability. Parallel cultures of U3A cells were transfected with plasmids expressing STAT1 and various GFPs as indicated. Those residues which were mutated to Ala in the various constructs are shown as A's in bold; those residues which were mutated to Pro (panel B) are shown as P in bold. Cytoplasmic extracts were prepared at 48 h posttransfection, and samples containing equal amounts of total protein were separated by SDS-PAGE. The samples were then assayed for their relative amounts of STAT1 by immunoblotting and were assayed for the various GFPs either by immunoblotting (A) or by measuring cellular fluorescence by fluorescence-activated cell sorting (B and C). In the latter cases, GFP fluorescence was normalized to that of RFP. Mutant C1-23/GFPs containing nested deletions from each end of C1-23 are reported in panel C. The critical region required for full activity determined from these deletions is boxed. The percentages of remaining GFP and STAT1 for each construct examined are relative to parallel cultures in which unmodified GFP was expressed. The average of at least two independent determinations is given; the duplicates in all cases varied by <15%.
The SeV C1-23 degron is not only transportable, it remarkably carries with it the ability to specifically induce STAT1 degradation, even without the remainder of the protein (C24-204) that binds stably to STAT1. We presume that C1-23 must interact with STAT1, at least transiently. However, unlike C1-204 or GFP/C1-204, C1-23/GFP did not form an immunoprecipitable complex with STAT1 in a cell-free binding assay in which GFP degradation should not occur (data not shown). The entire C protein, and especially its N-terminal half, is strongly predicted to be natively disordered (12). An attractive hypothesis that could explain the absence of stable C1-23-STAT1 interaction in vitro is that C1-23 is structurally disordered in aqueous solution and thus plastic in the unbound state but assumes a specific structure upon interaction with STAT1. Such induced folding interactions, although highly specific, are generally of limited affinity and may not survive repeated washes during immunoselection. Although most peptides as short as 23 residues can be expected to be unstructured in aqueous solutions, those that undergo induced-folding interactions often adopt structure in the presence of trifluoroethanol (TFE) (6, 23, 43). Moreover, the percentage of TFE at which the midpoint of the structural transition occurs can be used to estimate the likelihood that structure is adopted upon binding to its target protein.
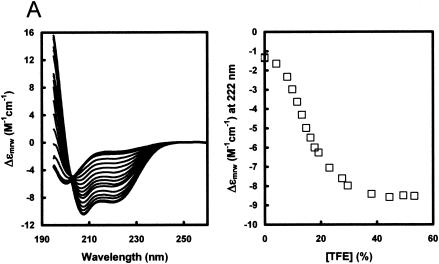
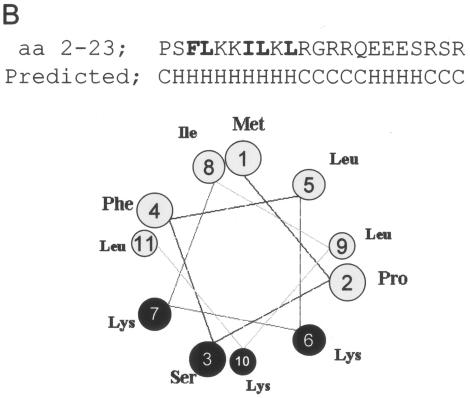
A peptide representing C residues 2 to 23 was examined by circular dichroism (CD) in increasing concentrations of TFE. As shown in Fig. 5A, the peptide in purely aqueous solution shows an intense negative band just below 200 nm and low intensity at longer wavelengths, characteristic for an unstructured peptide. The addition of TFE (at a constant peptide concentration) leads to the appearance of the characteristic α-helical spectrum, with intense negative bands at 207 and 222 nm and a strong positive band below 200 nm. The spectra are isodichroic at 203 nm, which is also characteristic of a random coil-to-helix transition. The midpoint of the TFE-induced transition was found to occur at unusually low TFE concentrations (14%); thus, this peptide has a strong tendency to be helical upon interaction with another protein. Analysis of the spectra at high TFE concentrations suggests a α-helical content of 74%, or 16 ± 2 peptide bonds in a α-helical conformation. The predicted structure of the peptide is shown in Fig. 5B. The first 11 residues are expected to form an amphipathic α-helix.
FIG. 5.
Circular dichroism of C1-23. A peptide representing C residues 2 to 23, whose N terminus was blocked by acetylation (the initiating Met may be removed in vivo) and whose C terminus was blocked by an amide, was dissolved in water at ∼2.2 mg/ml and then adjusted to a 25 mM concentration in Tris at pH 7.5. The concentration was determined from the absorption of the single Phe, using ɛ = 195 M−1 cm−1 at 257 nm. CD spectra were recorded in 1-mm cuvettes at 20oC in increasing concentrations of TFE at a constant peptide concentration. The midpoint of the structural transition is at 14% TFE (A) (right side). The predicted structure of the peptide, including the amphipathic nature of the N-terminal α-helix, is shown in panel B. Panel C shows the Δɛ at 222 nm as a function of the TFE concentration of wt and mutant peptides representing residues 2 to 14, as indicated (also see Fig. 6B).
Alanine scanning mutagenesis of C1-23.
To further characterize the degradative activities of C1-23 that act in cis and in trans, we turned to alanine scanning mutagenesis. Plasmids expressing various mutant forms of C1-23/GFP were transfected into U3A cells along with a plasmid expressing STAT1, and the percent mutant C1-23/GFP and STAT1 present, relative to that present when unmodified GFP is expressed, was determined (Fig. 6, % remaining). Since STAT1 is synthesized at similar rates here regardless of the various C1-23/GFPs coexpressed (Fig. 3B) and the various C1-23/GFPs are also synthesized at similar rates (Fig. 3A), the differences in steady-state levels of these two proteins presumably reflects their different stabilities.
One notable feature of C1-23 is its highly charged nature; the 18 residues from K6 to R23 contain three of Lys, five of Arg, and three of Glu. All 11 charged residues were mutated to alanine in various combinations (Fig. 6A). There are not many examples of degrons, but some, like the DSGXXS sequence of VpU, β-catenin, and IκB that act as docking sites for E3 ubitiquin ligases (Ub-ligases), are quite short (31). It is possible that C1-23 carries separate degrons that act in cis and in trans. Mutations that dissociate these activities would help clarify this point. We were unable to identify any individual positively charged residue of C1-23 that was important for activity. Moreover, the conversion of the three glutamates or three of the eight basic residues to Ala had minimal effects at best on the ability of C1-23/GFP to degrade either itself or STAT1 (Fig. 6A and data not shown). The conversion of four, five, or six of the eight positively charged residues to Ala had more noticeable effects. A mutant in which the first six positively charged residues were converted to Ala had the strongest effect, and in this case only STAT1 degradation was affected. Thus, although overall positive charge appears to play a role, the finding that the simultaneous mutation of six of these residues only partially diminished the degradative activity suggests that this role is not critical. For the majority of the mutants, there were no clear differences between the two degradative activities.
The DSGXXS degron acts as docking site for an E3 Ub-ligase only when both serines (in bold) are phosphorylated. Similarly, the degron of hypoxia-inducible factor 1α, a hypoxia-inducible regulator of angiogenesis, is activated through the oxygen-dependent hydroxylation of a single proline (24, 25). Amino acid modification as a result of signal transduction is not uncommon for cellular degrons, whereas human immunodeficiency virus type 1 (HIV-1) VpU is phosphorylated constitutively. We found that the simultaneous conversion of all three Ser residues or the single Pro residue of C1-23 to Ala did not affect the degradative activities of these mutants in any respect (data not shown). Furthermore, since none of the Lys or Arg residues is essential, it appears that amino acid modification of this degron is not important.
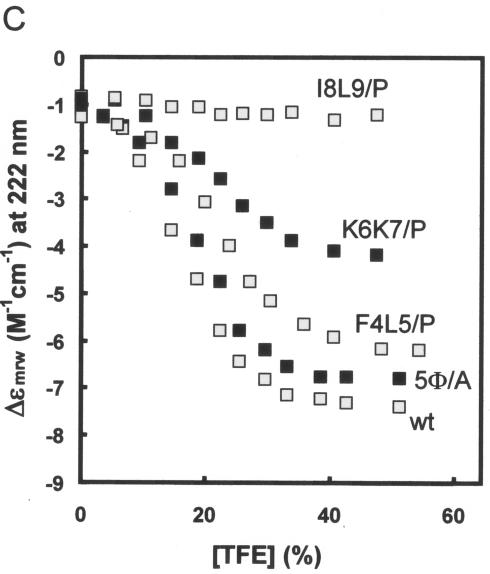
In contrast to short peptides with undefined structure that act in the proteasomal degradation of proteins like IκB, a prefolded surface appears to be a critical degradation determinant in the yeast mating type transcription factor α2. MATα2 is exceptionally short lived, and this property maps to a 19-residue element (Deg-1) that is critical for the rapid turnover of Deg1-containing substrates (26). This degradation determinant is part of a segment predicted to form an amphipathic helix, and inactivating mutations cluster on its hydrophobic face. C1-23 contains a cluster of five large hydrophobic residues (shown in bold) (Φ) near its amino terminus (4FLKKILKL11). The simultaneous conversion of three or more of these residues to Ala led to a strong loss of both activities (Fig. 6B, lines 1 to 3). 5Φ/A, in which all five large hydrophobic residues are converted to Ala, had the least activity towards itself or STAT1 (line 3). We have examined wt and 5Φ/A peptides containing residues 2 to 14 (underlined in Fig. 6B) by CD in increasing concentrations of TFE, and 5Φ/A was found to have much the same propensity to form an α-helix as the wt peptide, as predicted (Fig. 6B, lines wt and 3, and Fig. 5C), since Ala is not a helix-destabilizing residue. The large hydrophobic side chains themselves, independent of the amphipathic nature of the α-helix, are thus required for degradative activity towards both itself and STAT1. F4 and L5, K6 and K7, and I8 and L9 were also simultaneously converted to both Ala and Pro, and the optical properties in TFE of 13′mers containing the diproline substitutions were also examined as for 5Φ/A (Fig. 5C). The conversion of F4 and L5 to Ala led to a loss of more than half the activity towards STAT1 but only marginally affected its own degradation (Fig. 6B, line 4). The conversion of K6 and K7 or I8 and L9 to Ala had minimal effects on both activities, indicating that F4 and L5 are the most important of the large hydrophobic residues. The replacement of F4 and L5, K6 and K7, and I8 and L9 with diprolines progressively eliminates the ability of the peptides to form an α-helix in high concentrations of TFE (Fig. 5C). The number of α-helical residues determined from the CD spectra of each 13′mer peptide is listed in the right-hand column of Fig. 6B. Note that the I8 plus L9/P peptide had no detectable structure regardless of TFE concentration, again as predicted (45) (Fig. 5C). In contrast to C1-23 (I8+L9/A)/GFP, which had lost little of either degradative activity (Fig. 6B, line 8), C1-23 (I8+L9/P)/GFP had almost wt activity in self-degradation but had completely lost the ability to degrade STAT1 (Fig. 6B, line 9). STAT1 degradation thus requires both the large hydrophobic residues themselves and the potential to form an amphipathic α-helix, whereas self-degradation requires only the large hydrophobic residues. In all cases where the amino acid substitutions affected the two degradative activities differently, a more pronounced effect was found for STAT1. The large hydrophobic residues, 4FLKKILKL11 near the amino terminus of C1-23, which have a strong potential to form an amphipathic α-helix, thus appear to be the most important feature of this degron. In this respect, the C1-23 degron resembles Deg1 of Matα2, except that in this case the amphipathic α-helix is not part of a preformed structure.
The 11 amino-terminal residues of C1-23 are sufficient for both degradative activities.
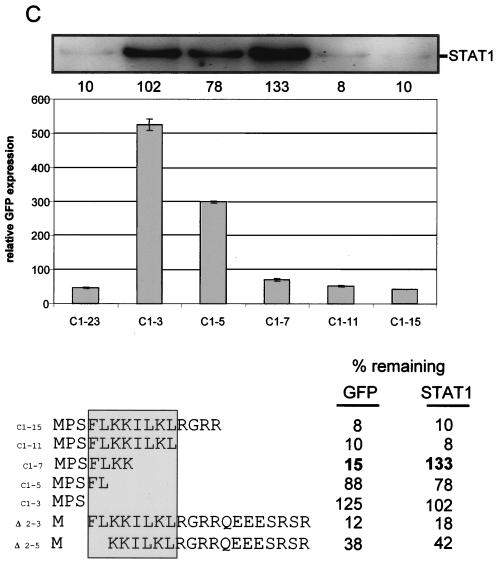
All the mutations that reduced the degradative activities of C1-23 mapped to the N terminus of the peptide. A series of C1-23/GFP mutants containing increasing deletions of the carboxyl segment of the peptide were examined to determine the minimal sequence that would act in cis and in trans. C1-15/GFP and C1-11/GFP were almost as active in both respects as C1-23/GFP (Fig. 6C). C1-7/GFP again appeared to be almost as unstable as C1-23/GFP; however, the first seven residues of C1-23 fused to GFP were unable to reduce STAT1 levels at all (Fig. 6C). C1-7/GFP is thus the second mutant (like I8L9/P; Fig. 6B, line 9) that has very specifically lost activity towards STAT1. Deletions from the N terminus showed that P2 plus S3 could be eliminated without much effect, whereas deletion of 2PSFL5 showed a clear but modest loss of both activities (Fig. 6C). In summary, our mutational analysis indicates that a short region (4FLKKILKL11) near the N terminus of C1-23/GFP is sufficient to induce the degradation of both itself and STAT1, whereas 8ILKL11 is specifically required to degrade STAT1. This differential requirement for 8ILKL11, as well as the different requirements for α-helical potential noted above, suggests that C1-23 could interact with different components of the degradation pathway to degrade itself and STAT1.
C1-23 also induces the degradation of C1-204 in trans.
SeV C1-23 presumably acts as a degradation signal for the protein to which it is fused and for STAT1 because it can direct both these proteins to cytoplasmic proteasomes. C1-23 is naturally found as the amino-terminal extension of the Y proteins that bind STAT1 with high affinity, permitting coimmunoprecipitation. The surprising result is that C1-23 induces STAT1 stability independently (i.e., when fused to GFP), even though it does not form a particularly stable complex with STAT1 in vitro. The available evidence suggests that STAT1 degradation is an important function of this SeV accessory protein. However, in contrast to SV5 V protein-induced STAT1 degradation or HIV-1 VpU-induced CD4 degradation, in which the viral proteins are not degraded in the process, SeV C1-23 is itself unstable, even in the absence of STAT1. This poses the question of how this short peptide can target both itself and STAT1 for degradation with seemingly similar efficiency when the degron is contained within C1-23 and its self-destruction does not require STAT1.
We know little about how SeV C1-23 connects either itself or STAT1 to the proteasomal pathway. It is possible that the activity that degrades C1-23-containing proteins in the absence of STAT1 requires another cellular partner. If C1-23 is indeed disordered when fused to either C24-204 or GFP, it may adopt specific structures under the influence of more than one cellular partner that mimic surfaces normally recognized as docking sites for the proteasome pathway. To gain insight into how C1-23 degrades itself, we examined whether the presence of C1-23/GFP affects the degradation of C1-204. Three forms of C1-23/GFP were expressed in increasing amounts (0, 1×, 3×, Fig. 7) in cells cotransfected with a constant amount of C1-204 and STAT1; namely, C1-11/GFP, which is fully active in both degradations, C1-7/GFP, which is active only towards itself, and (5Φ/α)/GFP, which is essentially inactive. If both C1-204 and C1-23/GFP degradation require the same putative cellular factor for proteasomal degradation, increasing C1-23/GFP should compete for this factor, sparing C1-204 degradation.
FIG. 7.
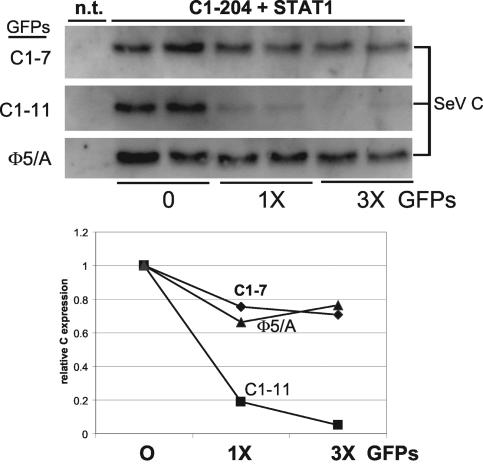
The effect of C1-23/GFP on C1-204 stability. Parallel cultures of U3A cells were transfected with constant amounts of plasmids expressing STAT1 and C1-204 and increasing amounts (0, 1×, 3×) of plasmids expressing various C1-23/GFPs as indicated. Some cultures were not transfected (n.t.) as controls. Cytoplasmic extracts were prepared at 48 h posttransfection, and samples containing equal amounts of total protein were separated by SDS-PAGE and then assayed for their relative amounts of STAT1, the various GFPs, and C1-204 by immunoblotting. Only the latter immunoblots are shown, and these results are plotted below.
Increasing expression of the null mutant 5Φ/α, or C1-7/GFP that cannot act on STAT1, had little if any effect on C1-204 levels. Increasing expression of C1-11/GFP, in contrast, strongly affected C1-204 levels. However, rather than sparing C1-204 degradation, C1-11/GFP clearly stimulated this degradation. The C1-11/GFP-stimulated degradation of C1-204 appears to be specific, since C1-11/GFP had no effect on cotransfected RFP expression (data not shown). Thus, C1-11/GFP apparently interacts with C1-204 in some manner to drive the latter's degradation. As expected, C1-11/GFP also drives C1-204 degradation in cells without STAT1 (data not shown). In summary, we have failed to find evidence that C1-204 instability itself requires a cellular factor such as STAT1. Rather, we have found evidence that C1-204 instability can also be driven by C1-23 in trans and that this activity requires the same peptide determinants that are required for C1-23 to induce STAT1 degradation.
DISCUSSION
Whereas all four SeV C proteins stably bind STAT1 and interdict IFN signaling, only SeV infections that express the longer C proteins can dismantle the IFN-induced cellular antiviral (VSV) state, and only the longer C proteins induce STAT1 instability. The latter activity has now been mapped to a short region at the NH2-terminal end of C1-23 that may be as short as 11 residues. Whereas STAT1 instability requires C1-23/GFP, C1-23/GFP instability occurs normally in cells devoid of STAT1. C1-23/GFP instability and its ability to induce that of STAT1 are thus, at least in part, separate processes, which can also be distinguished by mutation.
In contrast to human diploid fibroblasts (2fTGH) and mouse embryonic fibroblasts, SeV infection of HeLa cells does not lead to a reduction of endogenous STAT1 levels (27), and C1-23/GFP expression in these cells does not lead to the loss of transfected STAT1 (data not shown). The SeV C1-23-induced degradation of STAT1 is thus cell type specific. This situation may be similar to the recently discovered function of the HIV-1 Vif protein (44). Vif is required to prevent the cytosine deaminase APOBEC3G from being incorporated into HIV-1 virions (38) and to modify the newly synthesized minus-strand viral DNA upon reinfection (21, 36). Vif is essential for HIV-1 evasion of this aspect of host innate immunity, and it may act by targeting APOBEC3G for proteasomal degradation (53). Vif function is cell type specific, since APOBEC3G is not expressed in all cells, and cells that do not express APOBEC3G are permissive to infection with Vif-deficient HIV-1. It may be that HeLa cells lack a factor required for SeV C1-23-induced STAT1 degradation that is present in fibroblasts, either because the factor is cell type specific or because this factor is an aspect of innate immunity that has been lost in HeLa cells on continuous culture. We have little information about this putative factor, and our previous attempts here have led us to erroneously conclude that C1-23 induces the monoubiquitination of STAT1 (19). The cellular protein we so identified requires the expression of a SeV C protein in which C1-23 is intact, but this protein reacts with other unrelated mouse IgG monoclonal antibodies as well as antiubiquitin and anti-STAT1, and all we really know is that it is an IgG binding protein with the electrophoretic mobility expected for monoubiquitinated STAT1.
More is known about how the rubulavirus V proteins induce STAT instability. SV5, SV41, and mumps virus V proteins induce STAT1 instability, whereas hPIV2 V induces STAT2 instability. However, rubulavirus-induced STAT1 instability requires STAT2 and vice versa, as well as cellular DDB1 (41). DDB1 is known to be associated with Cullin 4A, and both are part of a larger SCF type Ub-ligase complex. The rubulavirus V, STAT1, STAT2, DDB1, and Cul4A proteins can be coimmunoprecipitated from infected cell extracts with specific antisera, and V can induce STAT1 ubiquitination in vitro (47, 48). Rubulavirus V is proposed to form a complex with STAT1 and STAT2, which then docks with an E3 Ub-ligase that includes DDB1 and Cul4A. The determinants of STAT instability do not map to a specific domain of V but are found throughout the protein (1), and different rubulavirus V proteins would presumably determine which STAT protein in the complex is marked for degradation. In keeping with the strictly essential function of the rubulavirus V proteins during genome replication (no V− rubulavirus has been reported), V itself is not destabilized in the process. SeV C-induced STAT1 instability, however, requires neither STAT2 (18) nor DDB1 (Fig. 2). Moreover, in contrast to the rubulavirus V protein (and HIV-1 VpU), SeV C itself is short-lived during virus infection. Mechanistically, rubulavirus V and SeV C protein-induced degradation of STAT1 appear to be unrelated.
HIV-1 VpU is another viral protein that targets a cellular partner (CD4) for degradation, by acting as an adaptor that connects CD4 to an E3 Ub-ligase (37). Unlike cellular IκB and β-catenin, whose DS GXXS motif is phosphorylated in response to specific signals, that of VpU appears to be constitutively phosphorylated. Thus, all three viral proteins constitutively induce protein degradation. VpU is anchored in the endoplasmic reticulum membrane, and its degron, which is unstructured, lies between two α-helices that stably bind the cytoplasmic tail of CD4 anchored in the same membrane (9). The crystal structure of a 26-aa segment of β-catenin bound to the βTrCp1-Skp1 complex has recently been reported, in which only the 11 residues centered on the DS*GXXS* motif are ordered in the crystal (52). This degron is thought to bind to its Ub-ligase without a preformed surface, and its final structure is determined by induced fit. HIV-1 VpU is thus an interesting model for SeV C1-23, since its degron is intrinsically unstructured and its activity depends on a structure that is induced upon association with its interacting partner.
SeV C, however, differs from HIV-1 VpU and SV5 V in important respects. The latter are relatively long-lived proteins that form stable complexes to connect their targets (CD4 and STAT1) to an ubiquitin ligase (4). SeV C1-23, in contrast, is itself unstable, and its activity does not appear to require the formation of such stable complexes. We note that one C1-204 protein does not form a stable complex with another C1-204 protein as judged by immunoprecipitation (data not shown), similar to C1-23/GFP and STAT1 interaction. Thus, the C1-23-C1-204 interaction that drives C1-204 degradation (Fig. 7), like that of C1-23 and STAT1, takes place without these stable interactions. Because of our inability to show that C1-23/GFP and STAT1 form a complex even in the presence of MG132, we have explored the notion that this complex is unstable due to the intrinsically disordered nature of this amino-terminal peptide. We have provided evidence that the C1-23 peptide is indeed disordered and forms an amphipathic α-helix in low concentrations of TFE, conditions that are thought to mimic protein-protein interaction. More importantly, mutational analysis of C1-23 has shown that the critical degradation region (4FLKKILKL11) is restricted to the amphipathic α-helix and that there is a good correlation between the ability of this peptide to form this helix in TFE and to function in vivo, i.e., to induce the loss of STAT1 in transfected cells when fused to GFP. This correlation supports the notion that C1-23 interacts with a cellular partner to degrade STAT1 via the induced formation of this amphipathic α-helix. Induced-fit interactions are of course possible with multiple partners, and this may help explain the many functions that map to C1-23, e.g., the formation of inactive pY701-STAT1 via JAK kinases (18; also unpublished data) and the induction of the curious protein that binds IgG and migrates like monoubiquitinated STAT1.
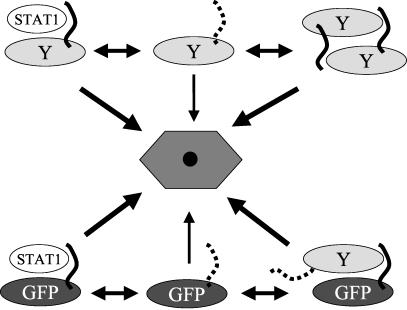
C1-23/GFP itself is unstable (at least as unstable as C1-204), and we presume that each C1-23/GFP connects itself to the degradation pathway independently of other C1-23/GFPs (Fig. 8, middle, bottom). Other things being equal, this direct pathway should predominate over that which requires STAT1, and STAT1 degradation should be relatively inefficient. However, just the opposite seems to be the case; STAT1 levels are in fact reduced efficiently by C1-23/GFP expression. To account for this efficient STAT1 degradation, we propose that although C1-23/GFP can independently connect to some component of the degradation pathway, this occurs infrequently due to the intrinsically disordered nature of this region and the apparent absence of any requirement to form an α-helical structure (Fig. 8). In contrast to self-degradation, STAT1 degradation requires the peptide's potential to form an amphipathic α-helix. If interaction of C1-23 with STAT1 (or C1-204) helps C1-23 form this N-terminal amphipathic α-helix (e.g., by limiting the number of conformations that the disordered peptide can adopt), this induced structure would presumably interact more efficiently with some component of the proteasome pathway, and this will lead to more efficient protein degradation (Fig. 8). During SeV infection, C1-204 protein degradation could occur either by direct interaction of C1-23 with the proteasomal pathway (Fig. 8, top middle,) or via interaction with another C1-204 protein (in trans; top right, Fig. 8). In the latter case, C would be able to sense both its own intracellular concentration and that of STAT1. STAT1 is an IFN-stimulated gene and plays an important role in both the IFN-induced antiviral state and apoptosis. The advantage to the virus in its battle with the innate immune system would be that STAT1 would be efficiently degraded when its intracellular levels are high, whereas C1-204 would mostly degrade itself when STAT1 levels are low, thus relieving its inhibition of viral RNA synthesis (10) when the threat of programmed cell death has been reduced.
FIG. 8.
A model for SeV C1-23-induced degradative activity. The SeV C1-204 protein is shown composed of two domains: the Y protein (C24-204, an oval) and C1-23 (as a wavy line). A dotted C1-23 line indicates that this peptide is disordered and rarely adopts a degradation-determining structure. A solid C1-23 line indicates that this peptide has adopted a degradation-determining structure. This degradation-determining structure can be induced by C1-23 interaction with either STAT1 or a separate C1-204. The various degradation pathways are represented by a single hexagonal structure in the center. The thicknesses of the arrows connecting the various substrates to the degradation pathways indicate the probability that the interaction will occur and that protein degradation will result.
.
REFERENCES
- 1.Andrejeva, J., E. Poole, D. F. Young, S. Goodbourn, and R. E. Randall. 2002. The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. J. Virol. 76:11379-11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumeister, W., J. Walz, F. Zuhl, and E. Seemuller. 1998. The proteasome: paradigm of a self-compartmentalizing protease. Cell 92:367-380. [DOI] [PubMed] [Google Scholar]
- 3.Bontron, S., C. Ucla, B. Mach, and V. Steimle. 1997. Efficient repression of endogenous major histocompatibility complex class II expression through dominant negative CIITA mutants isolated by a functional selection strategy. Mol. Cell. Biol. 17:4249-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bour, S., U. Schubert, and K. Strebel. 1995. The human immunodeficiency virus type 1 Vpu protein specifically binds to the cytoplasmic domain of CD4: implications for the mechanism of degradation. J. Virol. 69:1510-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 6.Buck, M. 1998. Trifluoroethanol and colleagues: cosolvents come of age. Recent studies with peptides and proteins. Q. Rev. Biophys. 31:297-355. [DOI] [PubMed] [Google Scholar]
- 7.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 8.Ciechanover, A., A. Orian, and A. L. Schwartz. 2000. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22:442-451. [DOI] [PubMed] [Google Scholar]
- 9.Coadou, G., N. Evrard-Todeschi, J. Gharbi-Benarous, R. Benarous, and J. P. Girault. 2001. Conformational analysis by NMR and molecular modelling of the 41-62 hydrophilic region of HIV-1 encoded virus protein U (Vpu). Effect of the phosphorylation on sites 52 and 56. C. R. Acad. Sci. Ser. II Fasc. C Chimie 4:751-758. [Google Scholar]
- 10.Curran, J., J.-B. Marq, and D. Kolakofsky. 1992. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology 189:647-656. [DOI] [PubMed] [Google Scholar]
- 11.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunker, A. K., C. J. Brown, and Z. Obradovic. 2002. Identification and functions of usefully disordered proteins. Adv. Protein Chem. 62:25-49. [DOI] [PubMed] [Google Scholar]
- 13.Galinski, M. S., R. M. Troy, and A. K. Banerjee. 1992. RNA editing in the phosphoprotein gene of the human parainfluenza virus type 3. Virology 186:543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcin, D., J. Curran, M. Itoh, and D. Kolakofsky. 2001. Longer and shorter forms of sendai virus C proteins play different roles in modulating the cellular antiviral response. J. Virol. 75:6800-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 74:8823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcin, D., M. Itoh, and D. Kolakofsky. 1997. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology 238:424-431. [DOI] [PubMed] [Google Scholar]
- 17.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcin, D., J. B. Marq, S. Goodbourn, and D. Kolakofsky. 2003. The amino-terminal extensions of the longer Sendai virus C proteins modulate pY701-Stat1 and bulk Stat1 levels independently of interferon signaling. J. Virol. 77:2321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcin, D., J. B. Marq, L. Strahle, P. Le Mercier, and D. Kolakofsky. 2002. All four Sendai virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology 295:256-265. [DOI] [PubMed] [Google Scholar]
- 20.Gotoh, B., K. Takeuchi, T. Komatsu, J. Yokoo, Y. Kimura, A. Kurotani, A. Kato, and Y. Nagai. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 459:205-210. [DOI] [PubMed] [Google Scholar]
- 21.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 22.Haspel, R. L., M. Salditt-Georgieff, and J. E. J. Darnell. 1996. The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends upon a protein tyrosine phosphatase. EMBO J. 15:6262-6268. [PMC free article] [PubMed] [Google Scholar]
- 23.Hua, Q. X., W. H. Jia, B. P. Bullock, J. F. Habener, and M. A. Weiss. 1998. Transcriptional activator-coactivator recognition: nascent folding of a kinase-inducible transactivation domain predicts its structure on coactivator binding. Biochemistry 37:5858-5866. [DOI] [PubMed] [Google Scholar]
- 24.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 25.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, P. R., R. Swanson, L. Rakhilina, and M. Hochstrasser. 1998. Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell 94:217-227. [DOI] [PubMed] [Google Scholar]
- 27.Kato, A., Y. Ohnishi, M. Kohase, S. Saito, M. Tashiro, and Y. Nagai. 2001. Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J. Virol. 75:3802-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King, P., and S. Goodbourn. 1994. The beta-interferon promoter responds to priming through multiple independent regulatory elements. J. Biol. Chem. 269:30609-30615. [PubMed] [Google Scholar]
- 29.King, P., and S. Goodbourn. 1998. STAT1 is inactivated by a caspase. J. Biol. Chem. 273:8699-8704. [DOI] [PubMed] [Google Scholar]
- 30.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 31.Laney, J. D., and M. Hochstrasser. 1999. Substrate targeting in the ubiquitin system. Cell 97:427-430. [DOI] [PubMed] [Google Scholar]
- 32.Latorre, P., T. Cadd, M. Itoh, J. Curran, and D. Kolakofsky. 1998. The various Sendai virus C proteins are not functionally equivalent and exert both positive and negative effects on viral RNA accumulation during the course of infection. J. Virol. 72:5984-5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Latorre, P., D. Kolakofsky, and J. Curran. 1998. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol. Cell. Biol. 18:5021-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leupin, O., S. Bontron, and M. Strubin. 2003. Hepatitis B virus X protein and simian virus 5 V protein exhibit similar UV-DDB1 binding properties to mediate distinct activities. J. Virol. 77:6274-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin-Marq, N., S. Bontron, O. Leupin, and M. Strubin. 2001. Hepatitis B virus X protein interferes with cell viability through interaction with the p127-kDa UV-damaged DNA-binding protein. Virology 287:266-274. [DOI] [PubMed] [Google Scholar]
- 36.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 37.Margottin, F., S. P. Bour, H. Durand, L. Selig, S. Benichou, V. Richard, D. Thomas, K. Strebel, and R. Benarous. 1998. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1:565-574. [DOI] [PubMed] [Google Scholar]
- 38.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 39.Matsuoka, Y., J. Curran, T. Pelet, D. Kolakofsky, R. Ray, and R. W. Compans. 1991. The P gene of human parainfluenza virus type 1 encodes P and C proteins but not a cysteine-rich V protein. J. Virol. 65:3406-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishio, M., D. Garcin, V. Simonet, and D. Kolakofsky. 2002. The carboxyl segment of the mumps virus V protein associates with Stat proteins in vitro via a tryptophan-rich motif. Virology 300:92-99. [DOI] [PubMed] [Google Scholar]
- 41.Parisien, J. P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Power, U. F., K. W. Ryan, and A. Portner. 1992. The P-genes of human parainfluenza virus type-1 clinical isolates are polycistronic and microheterogeneous. Virology 189:340-343. [DOI] [PubMed] [Google Scholar]
- 43.Reymond, M. T., G. Merutka, H. J. Dyson, and P. E. Wright. 1997. Folding propensities of peptide fragments of myoglobin. Protein Sci. 6:706-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 45.Sreerama, N., and R. W. Woody. 2000. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287:252-260. [DOI] [PubMed] [Google Scholar]
- 46.Thomas, S. M., R. A. Lamb, and R. G. Paterson. 1988. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell 54:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160-166. [DOI] [PubMed] [Google Scholar]
- 48.Ulane, C. M., J. J. Rodriguez, J. P. Parisien, and C. M. Horvath. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 77:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varshavsky, A. 1991. Naming a targeting signal. Cell 64:13-15. [DOI] [PubMed] [Google Scholar]
- 50.Vidal, S., J. Curran, and D. Kolakofsky. 1990. A stuttering model for paramyxovirus P mRNA editing. EMBO J. 9:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber, H., D. Valenzuela, G. Lujber, M. Gubler, and C. Weissmann. 1987. Single amino acid changes that render human IFN-alpha 2 biologically active on mouse cells. EMBO J. 6:591-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, G., G. Xu, B. A. Schulman, P. D. Jeffrey, J. W. Harper, and N. P. Pavletich. 2003. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol. Cell 11:1445-1456. [DOI] [PubMed] [Google Scholar]
- 53.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]