Abstract
Oxidative stress induces mitochondrial dysfunction and facilitates apoptosis, tissue damage or metabolic alterations following infection. We have previously discovered that the Pseudomonas aeruginosa (PA) quorum sensing (QS)-excreted small volatile molecule, 2-aminoacetophenone (2-AA), which is produced in infected human tissue, promotes bacterial phenotypes that favor chronic infection, while also compromising muscle function and dampens the pathogen-induced innate immune response, promoting host tolerance to infection. In this study, murine whole-genome expression data have demonstrated that 2-AA affects the expression of genes involved in reactive oxygen species (ROS) homeostasis, thus producing an oxidative stress signature in skeletal muscle. The results of the present study demonstrated that the expression levels of genes involved in apoptosis signaling pathways were upregulated in the skeletal muscle of 2-AA-treated mice. To confirm the results of our transcriptome analysis, we used a novel high-resolution magic-angle-spinning (HRMAS), proton (1H) nuclear magnetic resonance (NMR) method and observed increased levels of bisallylic methylene fatty acyl protons and vinyl protons, suggesting that 2-AA induces skeletal muscle cell apoptosis. This effect was corroborated by our results demonstrating the downregulation of mitochondrial membrane potential in vivo in response to 2-AA. The findings of the present study indicate that the bacterial infochemical, 2-AA, disrupts mitochondrial functions by inducing oxidative stress and apoptosis signaling and likely promotes skeletal muscle dysfunction, which may favor chronic/persistent infection.
Keywords: Pseudomonas aeruginosa, 2-aminoacetophenone, skeletal muscle, oxidative stress, apoptosis, mitochondria, nuclear magnetic resonance, microarrays, genomics
Introduction
Bacterial pathogens can manipulate the cell survival machinery of the host to establish infection by influencing the host signaling pathways that converge on the mitochondria (1). Mitochondria have been identified as the target of increasing numbers of bacterial products, which are transferred to the cell during infection, a process that often plays a crucial role in microbial pathogenesis (2). Bacterial small molecules have been previously shown to alter mitochondrial function (3,4) and induce apoptosis (5). In addition, the bacterial cell-wall component lipopolysaccharide (LPS) has been shown to promote apoptosis through oxidative stress in host cells (6).
Oxidative stress can be induced by pathogens either directly or indirectly through the induction of host inflammatory mediators (7,8). A major cause of oxidative stress is reactive oxygen species (ROS) generation in cells, which are a by-product of energy production in the mitochondria (9,10). The increased generation of ROS and changes in the redox homeostasis have been described in the context of a number of microbial infections (11–13), and the failure to maintain an appropriate redox balance contributes to microbial pathogenesis through alterations in mitochondrial functions and the induction of apoptosis (14,15).
Apoptosis plays a critical role in tissue homeostasis and regulates multiple physiological processes, including immune responses, infection and reduces inflammation-mediated tissue injury by removing damaged cells (16). Apoptosis is initiated through several processes: i) by the activation/stimulation of death receptors, including Fas, tumor necrosis factor (TNF)-α receptor I, and TNF-related apoptosis-inducing ligand receptors 1 and 2 (17); ii) by caspase activation that is independent of death receptors (18); iii) or by mitochondrial factors, such as apoptosis-inducing factor (AIF), which are independent of caspase activation (18). Increased apoptotic signals in the alveolar epithelium are related to the presence of chronic infections in patients with cystic fibrosis (CF) (19).
The opportunistic pathogen Pseudomonas aeruginosa (PA) thrives in patients with CF, burns and/or immunocompromised individuals (20,21) and utilizes cell-to-cell communication systems, termed quorum sensing (QS) (22) to establish acute and chronic infection (23–27). QS relies on the presence of small-excreted molecules termed herein 'infochemicals', to coordinate the concomitant expression of multiple virulence genes (28). It has been reported that PA QS-excreted infochemicals modulate the host innate immune response (29,30). The PA-excreted info-chemical, 2-aminoacetophenone (2-AA), promotes phenotypic changes in the pathogen that favor chronic infection (26,31) and dampens pathogen-induced inflammation that in turn favors host tolerance to infection (30). We have recently demonstrated that 2-AA reduces energy metabolism, promotes mitochondrial dysfunction and impairs muscle functions, which may further favor chronic and persistent infections (32) and potentially induces oxidative stress in skeletal muscle.
In this study, we used whole-genome expression and ex vivo nuclear magnetic resonance (NMR) to examine the effects of 2-AA on the oxidative response and apoptosis in skeletal muscle. NMR spectroscopy can be used to determine mitochondrial dysfunction and assess metabolic alterations in muscle (33,34). Furthermore, in vivo microscopy can be used to determine the reduction in mitochondrial membrane potential, which is a signature of mitochondrial dysfunction and induces apoptosis. The results of the present study demonstrated that the inhibition of genes involved in oxidative homeostasis resulted in the accumulation of ROS and increased apoptosis in skeletal muscle. Thus, 2-AA promotes mitochondrial dysfunction and compromises normal skeletal muscle functions (32) through the induction of oxidative stress and apoptosis, thus promoting pathogenicity.
Materials and methods
Ethics statement
This study was carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The study protocol was approved by the Committee on the Ethics of Animal Experiments at Massachusetts General Hospital (Boston, USA; permit nos. 2006N000093 and 2013N000034). All procedures were performed on mice which had been anaesthetized with a combination of xylazine (12.5 mg/kg) and ketamine (125 mg/kg), and every effort was made to minimize suffering.
Experimental animals
Six-week-old male CD1 mice weighing approximately 20–25 g were purchased from Charles River Laboratory (Boston, MA, USA). Twenty-eight mice were used in this study. The animals were kept in a controlled environment with a regular light-dark cycle (lights on from 8:00 to 20:00) with an ambient temperature of 22±1°C, and were allowed free access to food and water. The mice received an intraperitoneal (IP) injection of 2-AA (6.75 mg/kg), and mouse skeletal muscle was analyzed 4 days post-AA treatment. Ex vivo proton nuclear magnetic resonance (1H NMR) spectroscopy was performed on intact gastrocnemius muscle samples.
High-resolution magic-angle-spinning (HRMAS) 1H NMR spectroscopy of intact skeletal muscle tissue
At 4 days post-2-AA treatment, eight 2-AA-treated and eight untreated animals (that served as a control) were analyzed using HRMAS 1H NMR spectroscopy. The left gastrocnemius muscle was harvested, immediately frozen in liquid nitrogen, and stored at approximately −80°C. The same muscle from untreated animals served as a control. HRMAS 1H NMR spectroscopic analysis of muscle tissue was performed on a Bruker Bio-Spin Avance NMR spectrometer (proton frequency at 600.13 MHz, with an 89-mm vertical bore) using a 4-mm triple resonance (1H, 13C, 2H) HRMAS probe (Bruker, Billerica, MA, USA). The temperature was maintained at 4°C with a BTO-2000 thermocouple unit in combination with a magic angle spinning (MAS) pneumatic unit (Bruker). The MAS speed was stabilized at 4.0±0.001 kHz using a MAS speed controller. The 1H NMR spectra were acquired for all samples using a Carr-Purcell-Meiboom-Gill (CPMG) spin echo pulse sequence [90°−(τ−180°−τ−)n-acquisition], with an inter-pulse delay (τ−) of 250 µsec. Hard 90° (8 τ-s) and 180° (16 τ-s) were employed. As previously described (32), the relaxation delay was set to 2 sec, and spectra were collected both with and without water suppression. The transverse relaxation time (T2) was measured using the same CPMG pulse sequence by varying n from 0 to 520. Free induction decay (FID) signals were acquired with 8k points, a 600 msec acquisition time, 8 dummy scans and 128 scans. HRMAS 1H NMR spectra were analyzed using the MestRe-C NMR software package (Mestrelab Research, Santiago de Compostela, Spain; www.mestrec.com). FIDs were zero-filled to 16k points and apodized with exponential multiplication (1 Hz) prior to Fourier transformation. The spectra were then manually phased and corrected for baseline broad features (Whittaker smoother, smooth factor 10,000). The Levenberg-Marquardt algorithm was used to least-squares-fit a model of mixed Gaussian/Lorentzian functions to the data. The (CH2)n−2 peak at 1.32 ppm was selected for the quantification of intramyocellular lipids (IMCLs). As the sample was spun at a magic angle, and the sample was much smaller (25 μl) and more homogeneous (reduced bulk magnetic susceptibility effects) than the typical voxel size (1 ml) of in vivo 1H MRS, no chemical shift difference was observed between IMCL and extramyocellular lipids (EMCL). The small size of the muscle biopsies and the fact that the samples were collected from the most myocellular part of the muscle suggest that the main contribution to the (CH2)n−2 peak was made by IMCL lipids.
Absolute quantification of metabolites from 1-D CPMG spectra
Resonance intensities were measured for −CH3 protons of the trimethylsilyl-propionic-2,2,3,3-d4 acid (TSP) and compared to the resonance intensities measured for metabolites. The peak intensities of most of the metabolites, as well as of TSP, were calculated from the intensity of the respective resonance (X) measured from the T2-filtered HRMAS 1H MR spectrum. The calculated peak intensities were then corrected for T2 relaxation, using Ic(X) = Ir(X) * exp(TCPMG/T2(X))/n, where Ir(X) is the measured intensity, TCPMG is the CPMG echo time, and n is the number of protons in the functional group and corresponds to the resonance of the metabolite. In accordance with the 'external standard' technique employed (35), metabolite concentrations were quantified relative to the absolute concentration (μmol) of the respective metabolite [M] = Ic(M)/(IcTSP(M) * wt), where wt is the weight of the sample in grams.
Statistical analysis of HRMAS 1H NMR spectroscopy data
Data are reported as the means ± standard errors of the mean. Between-groups comparison was performed using analysis of variance with Bonferroni correction for multiple comparisons. A corrected p-value of <0.0125 was deemed to indicate a statistically significant difference. A comparison between measurements was performed in each group with a t-test (two-tailed, p<0.0125). All analyses were performed using SPSS software (SPSS version 12; SPSS Inc., Chicago, IL, USA).
Extraction of RNA samples
As previously described (32), the left gastrocnemius muscle was harvested at 4 days post 2-AA treatment (n=3), to determine the changes in gene expression in whole muscle. The muscle specimens of the anesthetized animals were excised and immediately immersed in 1 ml TRIzol (Gibco-BRL, Invitrogen, Carlsbad, CA, USA) for RNA extraction. Each muscle specimen was homogenized for 60 sec with a Brinkmann Polytron 3000 homogenizer prior to total RNA extraction. Chloroform (200 μl) was added to the homogenized muscle and mixed by inverting the tube for 15 sec. Following centrifugation at 12,000 × g for 15 min, the upper aqueous phase was collected and precipitated by the addition of 500 μl isopropanol. Further centrifugation at 12,000 × g for 10 min separated the RNA pellet, which was then washed with 500 μl of 70% ethanol and centrifuged at 7,500 × g for 5 min prior to air drying. The pellet was resuspended in 100 μl diethylpyrocarbonate (DEPC) water. An RNeasy kit (Qiagen, Germantown, MA, USA) was used to purify the RNA according to the manufacturer's instructions. The purified RNA was quantified by UV absorbance at 260 and 280 nm, and stored at −70°C for DNA microarray analysis.
Microarray hybridization
As previously described (32), biotinylated cRNA was generated with 10 μg of total cellular RNA according to the protocol outlined by Affymetrix Inc. (Santa Clara, CA, USA). cRNA was hybridized onto MOE430A oligonucleotide arrays (Affymetrix), stained, washed and scanned according to the Affymetrix protocol.
Genomic data analysis
As previously described (32), data of the scanned image files hybridized with probes from RNA extracted from the gastrocnemius muscle isolated at the specified times from the 2-AA-treated and untreated control mice (n=3) were converted to cell intensity files (CEL files) with the Microarray suite 5.0 (MAS; Affymetrix). The data were scaled to a target intensity of 500, and all possible pairwise array comparisons of the replicates to normal control mice were performed using a MAS 5.0 change call algorithm. Probe sets that had a signal value difference >100 and for which one of the two samples being compared was not termed 'absent', were scored as differentially modulated when i) the number of change calls in the same direction were at least 3, 4 and 6, when the number of comparisons were 4, 6 and 9, respectively; and ii) the other comparisons were unaltered. Based on the ratios of 100 genes determined to be invariant in most conditions tested (Affymetrix), an additional constraint of a minimum ratio of 1.65 was applied to control the known false positives at 5%. The microarray data are available at http://www.ncbi.nlm.nih.gov/geo/info/linking.html and the accession number is GSE43779. Gene Ontology (GO) analysis was performed using GeneSpring GX software (version 11) by Agilent Technologies (Santa Clara, CA, USA).
In vivo microscopic measurement of mitochondrial membrane potential
Following 4 days of treatment with 2-AA (6.75 mg/kg), the 2-AA treated mice and the untreated controls (n=3) were anesthetized and underwent in vivo microscopic observation as previously described (36,37) with modifications. Briefly, sternocleidomastoid muscles were exposed and stained with 40 nM 3,3′-dihexyloxacarbocyanine iodide (DiOC6) and 1 μM CellTracker Orange (Invitrogen) for 30 min. After washing, the fluorescence intensity of DiOC6, which reflects mitochondrial membrane potential, was recorded. The consistency of dye accessibility and image caption was confirmed by internal control staining with CellTracker Orange. The captured images were analyzed and their fluorescence intensity was analyzed by densitometry with ImageJ software (http://imagej.nih.gov/ij/).
Results
2-AA treatment affects ROS homeostasis and production and the response to oxidative stress
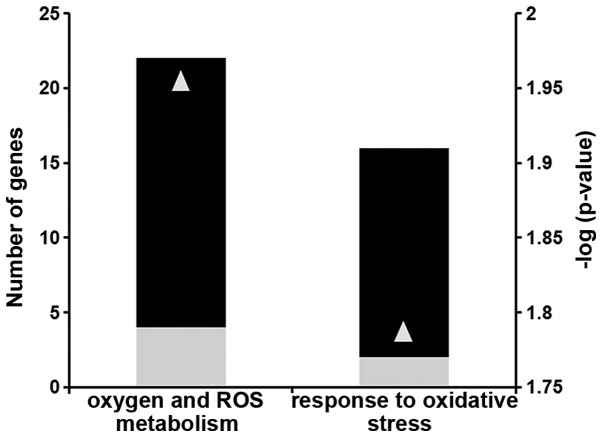
Using microarray analysis, we identified a subset of differentially expressed genes that have functional annotations of oxidative stress and mitochondrial function. Several genes involved in ROS homeostasis (group A) and in the response to oxidative stress (group B) were down-regulated (Fig. 1 and Table I).
Figure 1.
2-Aminoacetophenone (2-AA) affects reactive oxygen species (ROS) metabolism and the response to oxidative stress in murine skeletal muscle. Black bars indicate the number of downregulated genes; gray bars indicate the number of upregulated genes in the skeletal muscle of mice 4 days after 2-AA treatment versus the control mice (left vertical axis). The negative log10 of p-values represented by gray triangles are indicated in the right vertical axis.
Table I.
Differential expression of genes involved in ROS homeostasis and oxidative stress in mouse skeletal muscle 4 days following an injection of 2-AA.
| GenBank | Gene name | Fold change | p-value | GO Biological process |
|---|---|---|---|---|
| Group A | ||||
| NM_009127 | Stearoyl-coenzyme A desaturase 1 | (−)4.3 | 0.024 | Oxygen and ROS metabolism |
| AF173681 | Thioredoxin interacting protein | (−)4.4 | 0.038 | Oxygen and ROS metabolism |
| NM_009804 | Catalase | (−)8.6 | 0.013 | Oxygen and ROS metabolism |
| NM_020569 | Parkinson disease (autosomal recessive, early onset) 7 | (−)2.3 | 0.029 | Oxygen and ROS metabolism |
| NM_011435 | Superoxide dismutase 3, extracellular | (−)4.5 | 0.018 | Oxygen and ROS metabolism |
| NM_021356 | Growth factor receptor bound protein 2-associated protein 1 | (−)2.4 | 0.028 | Oxygen and ROS metabolism |
| NM_011563 | Peroxiredoxin 2 | (−)2.1 | 0.016 | Oxygen and ROS metabolism |
| NM_001111320 | Isocitrate dehydrogenase 1 (NADP+), soluble | (−)4.2 | 0.041 | Oxygen and ROS metabolism |
| NM_013603 | Metallothionein 3 | (+)2.8 | 0.023 | Oxygen and ROS metabolism |
| NM_010497 | Isocitrate dehydrogenase 1 (NADP+), soluble | (−)3.1 | 0.023 | Oxygen and ROS metabolism |
| NM_018881 | Flavin containing monooxygenase 2 | (−)4.6 | 0.022 | Oxygen and ROS metabolism |
| NM_023505 | Glutaredoxin 2 (thioltransferase) | (−)2 | 0.027 | Oxygen and ROS metabolism |
| BC019664 | Glutathione peroxidase 8 (putative) | (−)4 | 0.005 | Oxygen and ROS metabolism |
| NM_013711 | Thioredoxin reductase 2 | (−)3.5 | 0.025 | Oxygen and ROS metabolism |
| AF412308 | Thioredoxin reductase 2 | (−)2.4 | 0.009 | Oxygen and ROS metabolism |
| NM_027629 | Phosphoglucomutase 2-like 1 | (+)9.8 | 0.023 | Oxygen and ROS metabolism |
| NM_013671 | Superoxide dismutase 2, mitochondrial | (−)5.8 | 0.008 | Oxygen and ROS metabolism |
| M14222 | Cathepsin B | (−)2.5 | 0.028 | Oxygen and ROS metabolism |
| NM_008161 | Glutathione peroxidase 3 | (−)3.7 | 0.005 | Oxygen and ROS metabolism |
| AF274027 | Phospholipid hydroperoxide glutathione peroxidase | (−)3 | 0.020 | Oxygen and ROS metabolism |
| NM_018881 | Flavin containing monooxygenase 2 | (−)5.3 | 0.007 | Oxygen and ROS metabolism |
| XM_006508205 | Phosphoglucomutase 2-like 1 (predicted) | (+)5.1 | 0.013 | Oxygen and ROS metabolism |
| Group B | ||||
| AF173681 | Thioredoxin interacting protein | (−)4.4 | 0.038 | Response to oxidative stress |
| NM_009804 | Catalase | (−)8.6 | 0.013 | Response to oxidative stress |
| NM_020569 | Parkinson disease (autosomal recessive, early onset) 7 | (−)2.3 | 0.029 | Response to oxidative stress |
| NM_021356 | Growth factor receptor bound protein 2-associated protein 1 | (−)2.4 | 0.028 | Response to oxidative stress |
| NM_011563 | Peroxiredoxin 2 | (−)2.1 | 0.016 | Response to oxidative stress |
| NM_001111320 | Isocitrate dehydrogenase 1 (NADP+), soluble | (−)4.2 | 0.041 | Response to oxidative stress |
| NM_010497 | Isocitrate dehydrogenase 1 (NADP+), soluble | (−)3.1 | 0.023 | Response to oxidative stress |
| NM_023505 | Glutaredoxin 2 (thioltransferase) | (−)2 | 0.027 | Response to oxidative stress |
| BC019664 | Glutathione peroxidase 8 (putative) | (−)4 | 0.005 | Response to oxidative stress |
| NM_013711 | Thioredoxin reductase 2 | (−)3.5 | 0.025 | Response to oxidative stress |
| AF412308 | Thioredoxin reductase 2 | (−)2.4 | 0.009 | Response to oxidative stress |
| NM_027629 | Phosphoglucomutase 2-like 1 | (+)9.8 | 0.023 | Response to oxidative stress |
| NM_013671 | Superoxide dismutase 2, mitochondrial | (−)5.8 | 0.008 | Response to oxidative stress |
| M14222 | Cathepsin B | (−)2.5 | 0.028 | Response to oxidative stress |
| NM_008161 | Glutathione peroxidase 3 | (−)3.7 | 0.005 | Response to oxidative stress |
| AF274027 | Glutathione peroxidase 4 | (−)3 | 0.020 | Response to oxidative stress |
| XM_006508205 | Phosphoglucomutase 2-like 1 (predicted) | (+)5.1 | 0.013 | Response to oxidative stress |
Values represent the relative expression intensity of the 2-aminoacetophenone (2-AA)-treated versus the untreated control mice. Annotations for biological processes are from the Gene Ontology Consortium (http://geneontology.org/). +, Upregulation of genes compared with muscle from normal untreated mice; −, downregulation of genes compared with muscle from normal untreated mice. GenBank and gene names can be searched at http://www.ncbi.nlm.nih.gov/gene/. ROS, reactive oxygen species.
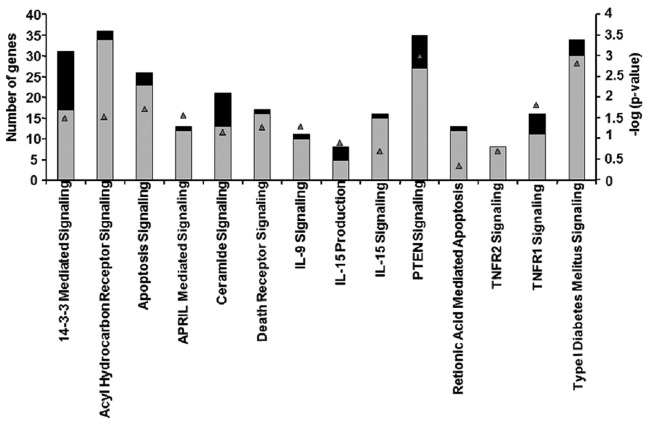
Differential expression of genes involved in the apoptosis signaling pathway in skeletal muscle of 2-AA-treated mouse
The microarray data presented in Table II and Fig. 2 demonstrated that certain genes, which had been identified as functioning in apoptosis via annotation by Gene Ontology Consortium and Ingenuity, were differentially expressed. Almost 97% of the genes involved in apoptosis signaling were upregulated in mouse skeletal muscle after 4 days of treatment with 2-AA (Table II and Fig. 2). For example, the TNF receptor superfamily member 1α gene exhibited an increased expression. This gene encodes a major TNF-α receptor that activates nuclear factor-κB (NF-κB), mediates apoptosis and regulates inflammation (17). In addition, genes encoding B-cell lymphoma (Bcl)-2, which function downstream of the TNF-α receptor in apoptosis signaling (38), exhibited an increased expression. Specifically, the upregulation of Bcl-2-like 13 (apoptosis facilitator), which is a member of Bcl-2 family of proteins, promotes apoptosis by blocking caspase inhibitors (39). The upregulation of PERP (p53 apoptosis effector), a p53 transcriptional target, promotes apoptosis (40). Fas apoptotic inhibitory molecules which confer resistance to Fas-induced apoptosis (41), were also downregulated following 2-AA treatment. The increased expression of growth arrest-specific 2 (Gas2) genes may activate apoptosis (42) in murine muscle of 2-AA-treated mice.
Table II.
Differential expression of genes involved in the apoptosis signaling pathway in mouse skeletal muscle samples following 4 days of treatment with 2-AA.
| GenBank | Gene name | Fold change | p-value |
|---|---|---|---|
| NM_030711 | Endoplasmic reticulum aminopeptidase 1 (Erap1) | 2.264 | 0.0158 |
| NM_134131 | Tumor necrosis factor, α-induced protein 8 | 3.863 | 0.0183 |
| NM_023517 | Tumor necrosis factor (ligand) superfamily, member 13 (Tnfsf13) | 3.293 | 0.0061 |
| NM_009396 | Tumor necrosis factor-α-induced protein | 5.796 | 0.00082 |
| NM_009425 | Tumor necrosis factor (ligand) superfamily, member 10 | 3.398 | 0.0226 |
| NM_011614 | Tumor necrosis factor (ligand) superfamily, member 12 (Tnfsf12) | 2.735 | 0.03 |
| NM_022310 | Heat shock 70 kDa protein 5 (glucose-regulated protein) | 4.146 | 0.0118 |
| AF250139 | Small stress protein-like protein (HSP22) | 6.137 | 0.0106 |
| NM_010481 | Heat shock protein 9A | 3.462 | 0.0107 |
| NM_013560 | Heat shock protein 1 | 3.248 | 0.0146 |
| NM_013559 | Heat shock protein 110 | −3.95 | 0.0367 |
| U03561 | Heat shock protein HSP27 internal deletion variant b | 4.008 | 0.025 |
| NM_010477 | Heat shock protein 1 (chaperonin) | 2.915 | 9.33E-05 |
| NM_010480 | Heat shock protein 90, α (cytosolic), class A member 1 (Hsp90aa1) | −2.63 | 0.0484 |
| NM_001163434 | Heat shock 70 kDa protein 5 (glucose-regulated protein) | 3.076 | 0.00638 |
| NM_001164708 | Heat shock protein 2 | 5.374 | 0.0412 |
| NM_013868 | Heat shock protein family, member 7 (cardiovascular) | 6.046 | 0.0427 |
| NM_011020 | Heat shock 70 kDa protein 4 like | −2.43 | 0.0368 |
| NM_008303 | Heat shock protein 1 (chaperonin 10) | 3.313 | 0.034 |
| M12573 | Heat shock protein 1B | 3.49 | 3.49 |
| XM_006500766 | Heat shock 70 kDa protein 4 like | −5.49 | 0.0266 |
| NM_028306 | Heat shock protein 12B | 8.802 | 0.0479 |
| NM_009883 | CCAAT/enhancer binding protein (C/EBP)β | 4.288 | 0.0215 |
| NM_009884 | CCAAT/enhancer binding protein (C/EBP)γ | 2.352 | 0.0479 |
| NM_010499 | Immediate early response 2 | 2.68 | 0.0161 |
| NM_008495 | Lectin, galactose binding, soluble 1 | 4.588 | 0.0481 |
| NM_001145953 | Lectin, galactose binding, soluble 3 | 2.464 | 0.0162 |
| NM_001199043 | Lectin, galactose binding, soluble 8 | 2.019 | 0.00659 |
| NM_010708 | Lectin, galactose binding, soluble 9 | 4.736 | 0.0464 |
| NM_019738 | Nuclear protein 1 | 2.299 | 0.0239 |
| NM_134141 | Cytokine induced apoptosis inhibitor 1 | 2.295 | 0.0222 |
| NM_022032 | PERP, TP53 apoptosis effector | 12.73 | 0.0252 |
| BC023121 | CASP8 and FADD-like apoptosis regulator | 2.91 | 0.0075 |
| NM_001177552 | Bifunctional apoptosis regulator | 2.006 | 0.0414 |
| NM_054056 | PRKC, apoptosis, WT1, regulator | 3.307 | 0.03 |
| NM_001038658 | Fas apoptotic inhibitory molecule 2 | −2.39 | 0.0493 |
| NM_153516 | BCL2-like 13 (apoptosis facilitator) | 2.796 | 0.0205 |
| NM_001039194 | Apoptosis-inducing factor (AIF)-like mitochondrion-associated inducer of death | 2.033 | 0.0445 |
| NM_007609 | Caspase 4, apoptosis-related cysteine peptidase (Casp4), mRNA | 3.446 | 0.00128 |
| NM_001042558 | Apoptotic peptidase activating factor 1 | 2.701 | 0.016 |
| NM_001165935 | Apoptosis, caspase activation inhibitor | 2.149 | 0.0206 |
| NM_001038658 | Fas apoptotic inhibitory molecule 2 | −22.32 | 0.00627 |
| BC003292 | Programmed cell death 8 | 3.129 | 0.0449 |
| BC026823 | Programmed cell death 6 interacting protein | 2.436 | 0.00988 |
| NM_001164677 | Programmed cell death 6 interacting protein | 4.606 | 0.000958 |
| NM_019746 | Programmed cell death 5 | 2.834 | 0.0486 |
| BC024876 | Death-associated protein | 3.706 | 0.0196 |
| NM_007566 | Baculoviral IAP repeat-containing 6 | 2.586 | 0.00413 |
| NM_001301639 | X-linked inhibitor of apoptosis (Xiap), transcript variant 1 | 2.44 | 0.0415 |
| NM_053207 | EGL nine homolog 1 (C. elegans) | 2.658 | 0.026 |
| AK017394 | Growth arrest specific 7 | −34.15 | 0.000771 |
| NM_026832 | Cell growth regulator with ring finger domain 1 | 3.016 | 0.0446 |
| NM_001109657 | Growth arrest-specific 7-cb protein (Gas7-cb) | −32.15 | 0.0113 |
| NM_008655 | Growth arrest and DNA-damage-inducible 45β | 3.297 | 0.0366 |
| NM_001033331 | Growth arrest-specific 2 like 3 | 4.312 | 0.0324 |
| NM_001277080 | Growth arrest-specific 7-cb protein (Gas7-cb) | −19.88 | 0.0303 |
| AF037370 | Cytochrome c oxidase, subunit VIIa 1 | 10.56 | 0.0445 |
| NM_007808 | Cytochrome c, somatic | 3.344 | 0.0391 |
| NM_007747 | Cytochrome c oxidase, subunit Va | 3.291 | 0.035 |
| NM_009941 | Cytochrome c oxidase subunit IV isoform 1 | 2.642 | 0.0202 |
| NM_007751 | Cytochrome c oxidase, subunit VIIIb | 5.591 | 0.0399 |
| AA190297 | Cytochrome c oxidase, subunit VIIc | 2.104 | 0.0296 |
| NM_025628 | Cytochrome c oxidase, subunit VIb polypeptide 1 | 2.015 | 0.0144 |
| NM_024226 | Reticulon 4 | −7.51 | 0.0255 |
| BF455257 | Reticulon 1 | −29.85 | 0.00175 |
| BM246564 | Phosphodiesterase 4B, cAMP specific | 2.161 | 0.0488 |
| NM_009811 | Caspase 6 | 3.503 | 0.0107 |
| NM_007611 | Caspase 7 | 3.837 | 0.00433 |
| NM_001163138 | Caspase recruitment domain family, member 6 | 3.344 | 0.00725 |
| NM_007611 | Caspase 7 | 4.957 | 0.0082 |
| BC008152 | Caspase 1 | 4.165 | 0.0361 |
| NM_001171007 | Nucleotide-binding oligomerization domain containing 1 (Nod1)/Caspase recruitment domain 4 | 2.35 | 0.0266 |
+, Upregulation of genes compared with normal untreated muscle; −, downregulation of genes compared with normal untreated muscle. 2-AA, 2-aminoacetophenone. GenBank and gene names can be found at http://www.ncbi.nlm.nih.gov/gene/.
Figure 2.
2-Aminoacetophenone (2-AA) affects genes involved in the apoptosis pathway in murine skeletal muscle. Black bars indicate the number of downregulated genes; gray bars indicate the number of upregulated genes in the skeletal muscle of mice 4 days after 2-AA treatment versus the control mice (left vertical axis). The negative log10 of p-values represented by gray triangles are indicated in the right vertical axis.
Several other genes were also upregulated, including heat shock proteins, lectins and AIF, the death associated protein. In addition, we noted that mitochondrial-dependent apoptosis genes which encode cytochrome c and caspases exhibited an increased expression.
1H NMR detects muscle lipid accumulation following 2-AA treatment
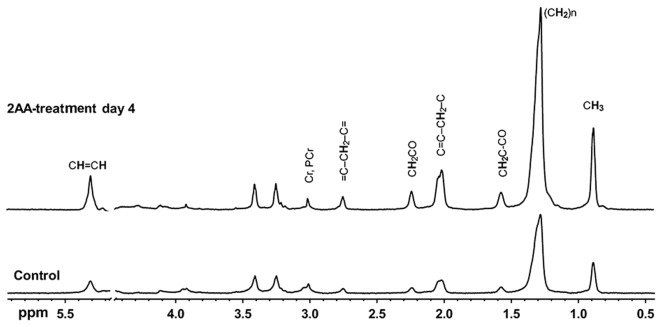
1H NMR spectroscopy has been used to explore metabolic changes (33,34,43). The accumulation of mobile lipids is associated with mitochondrial dysfunction, oxidative stress and apoptosis (44,45). As shown in Fig. 3 and Table III, the results of NMR spectroscopy demonstrated that most lipid peaks were consistently increased in the skeletal muscle samples from the 2-AA-treated mice compared to the control mice.
Figure 3.
Nuclear magnetic resonance (NMR) spectra from 1H-NMR high-resolution magic-angle-spinning (HRMAS) experiments performed on the gastrocnemius skeletal muscle specimens of mice. The spectra were acquired from normal and 2-aminoacetophenone (2-AA)-treated mice at 4 days post-2-AA treatment and scaled to the phosphocreatine and creatine peak (3.02 ppm). The lipid peak at 1.3 ppm is attributed to methylene protons of intra-myocellular triglyceride acyl chains, primarily due to intramyocellular lipids (IMCLs). Resonance signals are due to residual water (4.7–4.8 ppm); terminal methyl (0.8–1.0 ppm); acyl chain methylene (1.1–1.5 ppm); α- and β- methylene (2.0–2.5 ppm) and olefinic protons (5.4 ppm) of lipids; N-methyl protons of phosphocreatine and creatine (3.0 ppm); and N-trimethyl protons of betaines (3.2 ppm), which correspond to taurine and choline-containing compounds. Bisallylic methylene fatty acyl protons at 2.8 ppm correspond to polyunsaturated fatty acids (PUFAs), which accumulate due to apoptosis. Vinyl proton accumulation at 5.4 ppm, including protons from ceramide and possibly other sphingolipids suggests apoptosis.
Table III.
Results of 1H NMR HRMAS experiments performed on gastrocnemius muscle specimens from 2-AA-treated mice versus control mice.
| Chemical shift PPM | Chemical group | Control | 4 days post-2-AA | Percent change | p-value |
|---|---|---|---|---|---|
| 2.8 | =CH-CH2-CH= | 0.012±0.004a | 0.040±0.007 | +233 | 0.0075b |
| 5.4 | CH=CH- | 0.041±0.015 | 0.226±0.058 | +451 | 0.0233 |
Values (μmol/g muscle) are represented as the means ± standard error of the means from 8 samples/group;
p-value for comparisons between 2-aminoacetophenone (2-AA)-treated and normal mice obtained with the Student's t-test; +, indicates increase. 1H NMR, proton nuclear magnetic resonance; HRMAS, high-resolution magic-angle-spinning.
The stack plot presented in Fig. 3 shows typical examples of proton HRMAS 1H MRS CPMG spectra from skeletal muscle samples. This plot contains spectra of the control and 2-AA-treated animals sacrificed 4 days post-2-AA-treatment, with the spectra scaled to the phosphocreatine and creatine 3.02-ppm peak. Resonance signals are due to residual water (4.7–4.8 ppm); terminal methyl (0.8–1.0 ppm); acyl chain methylene (1.1–1.5 ppm); α- and β-methylene (2.0–2.5 ppm) and olefinic protons (5.4 ppm) of lipids; N-methyl protons of phosphocreatine and creatine (3.0 ppm); and N-trimethyl protons of betaines (3.2 ppm), which correspond to taurine and choline-containing compounds. The phospholipids at 3.2 ppm appear unaffected by 2-AA treatment and remain stable, along with other peaks between 3 and 4 ppm. The results from quantitative analysis of these high-resolution ex vivo HRMAS 1H MRS measurements are shown in Table III. Bisallylic methylene fatty acyl protons at 2.8 ppm correspond to polyunsaturated fatty acids (PUFAs), which accumulate due to apoptosis. Vinyl proton accumulation at 5.4 ppm, including protons from ceramide and possibly other sphingolipids suggests apoptosis.
Downregulation of mitochondrial membrane potential in skeletal muscle from 2-AA-treated mice
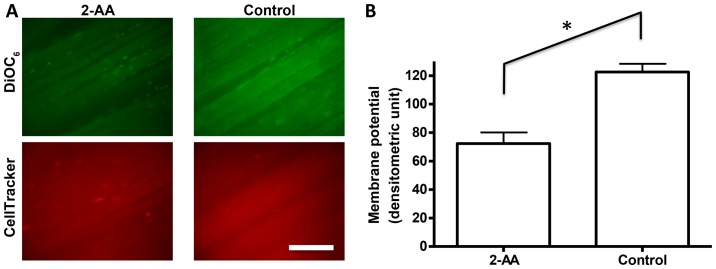
Mitochondrial membrane potential is considered a biomarker of oxidative stress and apoptosis (46,47). In vivo fluorescence microscopy with DiOC6 indicated that 2-AA treatment reduced skeletal muscle mitochondrial membrane potential by 41% compared to the untreated control group (p<0.05; Fig. 4). The fact that internal control staining with CellTracker Orange revealed equivalent staining between the two groups confirmed that the difference observed in membrane potential was not due to staining issues (i.e., dye accessibility or fluctuation in image capturing).
Figure 4.
2-Aminoacetophenone (2-AA) reduces mitochondrial membrane potential in murine skeletal muscle. Mitochondrial membrane potential was analyzed by in vivo microscopy. (A) In vivo fluorescence microscopic images are shown both for 2-AA-treated (left column) and untreated (right column) groups. Green fluorescence (top panel) from 3,3′-dihexyloxacarbocyanine iodide (DiOC6) staining represents mitochondrial membrane potential, and red signal (bottom panel) from CellTracker Orange staining is for internal control staining. The white scale bar at the bottom represents 200 μm. (B) The average fluorescence signal was quantified by densitometry and shown as a bar graph with the standard error of the means. 2-AA-treated group showed a significantly decreased signal as compared to the controls. *p<0.05, according to Student's t-test.
Discussion
In the present study, we demonstrate that 2-AA, a QS-regulated requisite for PA pathogenesis and a diagnostically important bacterial molecule, induces oxidative stress in skeletal muscle, disrupts the defense against oxidative stress, reduces mitochondrial membrane potential and enhances apoptotic signaling (Fig. 5). The reduced expression of genes involved in ROS metabolism and in the oxidative stress response may be responsible for the accumulation of ROS in skeletal muscle. ROS toxicity may lead to mitochondrial dysfunction, possibly via damage to mitochondrial DNA (mtDNA) and reduced mitochondrial potential. Our findings also suggest a reduction in the oxidative stress response as a potential mechanism through which 2-AA modulates the apoptotic pathway, and suggest candidate targets through which to limit oxidative stress and apoptosis in patients with infection.
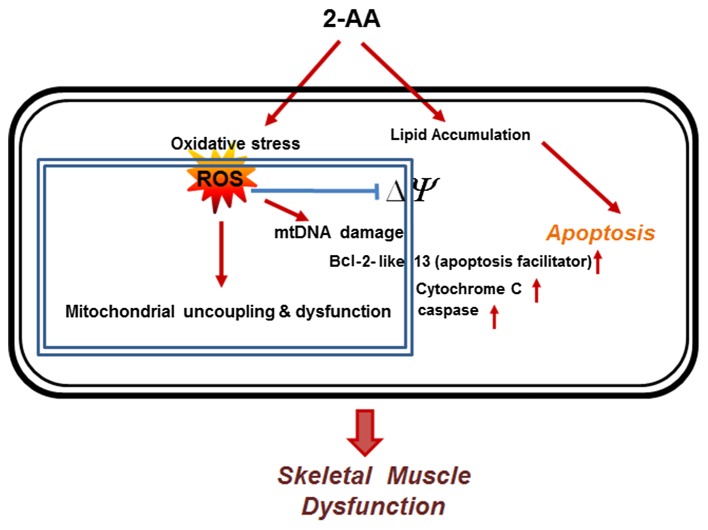
Figure 5.
Representative schematic diagram showing that 2-aminoacetophenone (2-AA) induces oxidative stress and apoptosis in skeletal muscle. 2-AA induces oxidative stress by generating reactive oxygen species (ROS). The oxidative damage and ROS reduce mitochondrial membrane potential (ΔΨ), and release Bcl-2 and cytochrome c, which promotes apoptosis. Lipid accumulation following 2-AA treatment potentially generates apoptotic signals in the cells (58,59). The red arrows denote the induction of the cellular components.
The principal finding of the present study was the effect of 2-AA on ROS production and the antioxidant status homeostasis in skeletal muscle, which correlates with the induction of apoptosis. Normally, increased ROS induce antioxidant defenses and prevent damage to mitochondrial macromolecules; in the absence of such defenses, oxidative damage ensues. In addition, the oxidative stress caused by increased ROS production results in apoptosis (8). Based on the findings of this study, we suggest that 2-AA interferes with ROS detoxification resulting in oxidative damage to mitochondrial macromolecules, possibly including mtDNA. ROS initiate damage to nucleic acids, proteins and lipids (10). Since mitochondrial DNA (mtDNA) is located closer to the site of ROS generation, lacks protective histones, and has more limited base excision repair mechanisms than the nucleus, it is more vulnerable to oxidative damage than nuclear DNA (nDNA) (48,49).
Another major finding of this study concurs with oxidative stress and is accompanied by mitochondrial dysfunction. In a recent study, we demonstrated that peroxisomal proliferator activator receptor (PPAR)-γ and PPAR-γ co-activator (PGC)1 expression was downregulated by 2-AA (32). PGC1 is hypothesized to play a central role in regulating energy homeostasis and metabolism (50). Therefore, the observed downregulation of genes involved in oxidative phosphorylation (32) and also in ROS detoxification by 2-AA may be explained by the effect of 2-AA on PGC1 (32). As a result of this dysregulation, the balance of detoxification and ROS generation may be shifted towards ROS accumulation. To this end, this study complies with the notion that certain components of the ROS scavenging pathway are linked by PGC1 to mitochondrial oxidative phosphorylation, apparently preventing cells from maintaining the normal redox status in response to a change in ROS production (51).
In this study, the expression of genes which encode proteins with pro- or anti-apoptotic functions was altered in response to 2-AA treatment (Fig. 2 and Table II); however, their exact regulatory role remains unclear. These data may reflect a heterogeneous mRNA population which is due to a direct effect of 2-AA on the underlying superficial layers of the hind limb muscle, and the asynchronous state of apoptotic muscle cells. Nonetheless, our data demonstrated that apoptotic pathways are activated in muscle from 2-AA-treated mice, which further demonstrates a link between the dysregulation of fatty acid oxidation (32), lipid accumulation (32) and apoptosis. Furthermore, the mitochondrial release of pro-apoptotic proteins, such as cytochrome c is promoted by the formation of specific channels in the outer membrane of the mitochondria by pro-apoptotic Bcl-2 family members (39). Once released, cytochrome c triggers the activation of caspases, which in turn regulates the apoptotic process (18) and has been suggested to induce contractile dysfunction (52). The orderly process of apoptosis is energy dependent and, consequently, damage to the mitochondria to the point that they can no longer produce ATP can easily shift to apoptosis (53). The core apoptotic pathway and cellular energy metabolism maintain an important inter-relationship between apoptosis and mitochondrial function (54), which is modulated by 2-AA.
In the present study, we noted that lipids accumulated in muscle tissue following 2-AA treatment, as assessed by 1H NMR. The lipid peak at 1.3 ppm in Fig. 3 has been attributed to methylene protons of intramyocellular triglyceride acyl chains, primarily due to IMCLs (55), which suggests that the increase in NMR-visible lipids at 1.4 ppm after 2-AA treatment is primarily due to increased quantifiable IMCL. This conclusion is further supported by the results of previous studies on humans (56,57). Conversely, EMCLs which may contribute to the 1.4 ppm peak are relatively metabolically inert and serve as a long-term energy storage depot with slow turnover. EMCLs are therefore unlikely to correspond to the lipids observed in this study, which rapidly accumulated post-2-AA treatment.
The accumulation of lipids is associated with apoptosis (58,59); and the present study demonstrates that 2-AA induced apoptosis-related gene expression (Table II) and increased the lipid profile (Fig. 3) in skeletal muscle, which could be linked. The increase in bisallylic methylene fatty acyl protons at 2.8 ppm is in accordance with the data of Hakumäki et al (60), which suggested that these protons correspond to PUFAs, and PUFA accumulation follows apoptosis. In addition, our data demonstrate vinyl proton accumulation at 5.4 ppm, including protons from ceramide and possibly other sphingolipids, such as sphingosine. Indeed, ceramide and sphingolipids are known to be involved in apoptosis (61). These increased signals suggest that ceramide, a product of sphingolipid metabolism, contributes to 2-AA-mediated apoptosis. Ceramide and sphingolipids, in particular, have been implicated as second messengers for apoptotic stimuli, including TNF-α (62), Fas ligand (63), ionizing radiation (64), heat shock (65) and oxidative stress (66).
The loss of mitochondrial membrane potential leads to the activation of apoptotic signals in several tissues, including skeletal muscle (67). When apoptotic signals reach the mitochondria, membrane permeability transition occurs, involving voltage-dependent anion channels (VDAC), adenine nucleotide translocase (ANT) and translocator protein (TSPO) (68). Damaged mitochondria are known to release apoptosis-inducing molecules, including cytochrome c, endonuclease G, AIF and Smac/Diablo and HtrA2/Omi (36,67). Other research has also demonstrated that uncoupled mitochondria become a major source for cellular ROS production (69). The data from our in vivo microscopic analysis support the molecular biological observation that the increase in apoptosis and ROS accumulation occurred in skeletal muscle of mice treated with 2-AA.
PA and 2-AA in the lungs of chronically infected patients with CF are considered pathogenomic (70,71). Patients with CF often develop skeletal muscle wasting due to inadequate antioxidant defenses which cannot cope with the elevated oxidative stress (72,73), and increased apoptosis is observed in airway cells of patients with CF (19,74). The characteristic loss of muscle mass, coupled with a decrease in strength, force output and alteration in oxidative stress, has previously been shown to be associated with apoptotic pathways and is an underlying mechanism of the pathogenesis of chronic disease (75,76). Oxidative stress and ROS accumulation can alter muscle gene expression, causing protein loss that consequently diminishes muscle mass or function (75). Nuclear receptor subfamily 2, group F, member 2 (NR2F2), a member of the steroid thyroid hormone superfamily of nuclear receptors, which is required for skeletal muscle development (77) and appears to be involved in the regulation of oxidative stress (78), was also downregulated (1.65-fold, p<0.0094) in the skeletal muscle of 2-AA-treated mice (data not shown). This suggests that 2-AA-mediated muscle dysfunction (32) may be the result of increased oxidative stress and apoptosis.
Mitochondrial dysfunction may interact with oxidative stress (14), lipid accumulation (59), apoptosis (79) and innate immune response (80). 2-AA abridges energy production and mitochondrial function in skeletal muscle, which may favor chronic diseases (32,81) and chronic infection (30). Recently, we demonstrated that 2-AA acts as an immunomodulatory signal (30) and likely affects insulin resistance associated with a molecular signature of mitochondrial dysfunction (32) that increases the ability of the host to live with the pathogen, enabling tolerance to infection and a long-term bacterial presence leading to bacterial persistence (30). Therefore, 2-AA-mediated oxidative stress and the activation of apoptotic signals may serve to eliminate key immune cells or evade host defenses (1) and further contribute to a long-term bacterial presence (30). The results of the present study are consistent with previous data of other chronic infections, including Escherichia coli (82), tuberculosis (83), Helicobacter pylori (84) and patients chronically infected with CF (85).
In conclusion, our results from transcriptome analysis and NMR spectroscopy provide strong evidence that the altered NMR-visible lipid profile is related to apoptosis which is induced by oxidative stress in response to the bacterial infochemical, 2-AA. 2-AA-mediated oxidative stress leads to mitochondrial dysfunction, which results in host metabolic dysfunction and apoptosis, and such a dysregulation in host metabolism can promote chronic/persistent disease state (30). Lipid accumulation may reflect apoptosis, rather than providing a direct measure of mitochondrial dysfunction. Since apoptosis and cellular energy metabolism are the two major determinants of cell survival, NMR-visible lipids may serve as biomarkers to monitor therapies for muscle wasting following bacterial infection, as well as other disease states. To this end, our results provide insight into the response of skeletal muscle to the PA-excreted small molecule, 2-AA, and point to novel therapeutic possibilities in targeting antioxidant pathways to reduce oxidative stress and apoptosis in skeletal muscle.
Acknowledgments
This study was supported in part by Shriner's Hospital for Children research grants (nos. 87100 and 85200), and Basic Research Award, W81XWH-10-DMRDP-BRA from US Army Medical Research Acquisition Act of US Department of Defense, Congressionally Directed Medical Research Programs (CDMRP), Defense Medical Research and Development Program (DMRDP) to Laurence G. Rahme, and by a Center grant of the National Institutes of Health (NIH) to the Stanford Genome Technology Cente, as well as by NIH grant AI105902. We acknowledge Dr Damien Maura at the Massachusetts General Hospital and Harvard Medical School for his insightful comments on the manuscript.
References
- 1.Rudel T, Kepp O, Kozjak-Pavlovic V. Interactions between bacterial pathogens and mitochondrial cell death pathways. Nat Rev Microbiol. 2010;8:693–705. doi: 10.1038/nrmicro2421. [DOI] [PubMed] [Google Scholar]
- 2.Jiang JH, Tong J, Gabriel K. Hijacking mitochondria: bacterial toxins that modulate mitochondrial function. IUBMB Life. 2012;64:397–401. doi: 10.1002/iub.1021. [DOI] [PubMed] [Google Scholar]
- 3.Trumpower BL. Cytochrome bc1 complexes of microorganisms. Microbiol Rev. 1990;54:101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Ark G, Berden JA. Binding of HQNO to beef-heart sub-mitochondrial particles. Biochim Biophys Acta. 1977;459:119–127. doi: 10.1016/0005-2728(77)90014-7. [DOI] [PubMed] [Google Scholar]
- 5.Schwarzer C, Fu Z, Shuai S, Babbar S, Zhao G, Li C, Machen TE. Pseudomonas aeruginosa homoserine lactone triggers apoptosis and Bak/Bax-independent release of mitochondrial cytochrome C in fibroblasts. Cell Microbiol. 2014;16:1094–1104. doi: 10.1111/cmi.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raza H, John A, Shafarin J. NAC attenuates LPS-induced toxicity in aspirin-sensitized mouse macrophages via suppression of oxidative stress and mitochondrial dysfunction. PLoS One. 2014;9:e103379. doi: 10.1371/journal.pone.0103379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valyi-Nagy T, Dermody TS. Role of oxidative damage in the pathogenesis of viral infections of the nervous system. Histol Histopathol. 2005;20:957–967. doi: 10.14670/HH-20.957. [DOI] [PubMed] [Google Scholar]
- 8.Pohanka M. Role of oxidative stress in infectious diseases. A review. Folia Microbiol (Praha) 2013;58:503–513. doi: 10.1007/s12223-013-0239-5. [DOI] [PubMed] [Google Scholar]
- 9.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding SZ, Minohara Y, Fan XJ, Wang J, Reyes VE, Patel J, Dirden-Kramer B, Boldogh I, Ernst PB, Crowe SE. Helicobacter pylori infection induces oxidative stress and programmed cell death in human gastric epithelial cells. Infect Immun. 2007;75:4030–4039. doi: 10.1128/IAI.00172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strengert M, Jennings R, Davanture S, Hayes P, Gabriel G, Knaus UG. Mucosal reactive oxygen species are required for antiviral response: role of Duox in influenza a virus infection. Antioxid Redox Signal. 2014;20:2695–2709. doi: 10.1089/ars.2013.5353. [DOI] [PubMed] [Google Scholar]
- 13.Paracha UZ, Fatima K, Alqahtani M, Chaudhary A, Abuzenadah A, Damanhouri G, Qadri I. Oxidative stress and hepatitis C virus. Virol J. 2013;10:251. doi: 10.1186/1743-422X-10-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pieczenik SR, Neustadt J. Mitochondrial dysfunction and molecular pathways of disease. Exp Mol Pathol. 2007;83:84–92. doi: 10.1016/j.yexmp.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Garofalo RP, Kolli D, Casola A. Respiratory syncytial virus infection: mechanisms of redox control and novel therapeutic opportunities. Antioxid Redox Signal. 2013;18:186–217. doi: 10.1089/ars.2011.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashida H, Mimuro H, Ogawa M, Kobayashi T, Sanada T, Kim M, Sasakawa C. Cell death and infection: a double-edged sword for host and pathogen survival. J Cell Biol. 2011;195:931–942. doi: 10.1083/jcb.201108081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guicciardi ME, Gores GJ. Life and death by death receptors. FASEB J. 2009;23:1625–1637. doi: 10.1096/fj.08-111005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madeo F, Carmona-Gutierrez D, Ring J, Büttner S, Eisenberg T, Kroemer G. Caspase-dependent and caspase-independent cell death pathways in yeast. Biochem Biophys Res Commun. 2009;382:227–231. doi: 10.1016/j.bbrc.2009.02.117. [DOI] [PubMed] [Google Scholar]
- 19.Galli F, Battistoni A, Gambari R, Pompella A, Bragonzi A, Pilolli F, Iuliano L, Piroddi M, Dechecchi MC, Cabrini G. Working Group on Inflammation in Cystic Fibrosis: oxidative stress and antioxidant therapy in cystic fibrosis. Biochim Biophys Acta. 2012;1822:690–713. doi: 10.1016/j.bbadis.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Navon-Venezia S, Ben-Ami R, Carmeli Y. Update on Pseudomonas aeruginosa and Acinetobacter baumannii infections in the healthcare setting. Curr Opin Infect Dis. 2005;18:306–313. doi: 10.1097/01.qco.0000171920.44809.f0. [DOI] [PubMed] [Google Scholar]
- 21.Kerr KG, Snelling AM. Pseudomonas aeruginosa: a formidable and ever-present adversary. J Hosp Infect. 2009;73:338–344. doi: 10.1016/j.jhin.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 22.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2012;76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Déziel E, Gopalan S, Tampakaki AP, Lépine F, Padfield KE, Saucier M, Xiao G, Rahme LG. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol Microbiol. 2005;55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- 24.Xiao G, Déziel E, He J, Lépine F, Lesic B, Castonguay MH, Milot S, Tampakaki AP, Stachel SE, Rahme LG. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol. 2006;62:1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- 25.Parker CT, Sperandio V. Cell-to-cell signalling during pathogenesis. Cell Microbiol. 2009;11:363–369. doi: 10.1111/j.1462-5822.2008.01272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kesarwani M, Hazan R, He J, Que YA, Apidianakis Y, Lesic B, Xiao G, Dekimpe V, Milot S, Deziel E, et al. A quorum sensing regulated small volatile molecule reduces acute virulence and promotes chronic infection phenotypes. PLoS Pathog. 2011;7:e1002192. doi: 10.1371/journal.ppat.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou L, Slamti L, Nielsen-LeRoux C, Lereclus D, Raymond B. The social biology of quorum sensing in a naturalistic host pathogen system. Curr Biol. 2014;24:2417–2422. doi: 10.1016/j.cub.2014.08.049. [DOI] [PubMed] [Google Scholar]
- 28.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rumbaugh KP, Kaufmann GF. Exploitation of host signaling pathways by microbial quorum sensing signals. Curr Opin Microbiol. 2012;15:162–168. doi: 10.1016/j.mib.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Bandyopadhaya A, Kesarwani M, Que YA, He J, Padfield K, Tompkins R, Rahme LG. The quorum sensing volatile molecule 2-amino acetophenon modulates host immune responses in a manner that promotes life with unwanted guests. PLoS Pathog. 2012;8:e1003024. doi: 10.1371/journal.ppat.1003024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Que YA, Hazan R, Strobel B, Maura D, He J, Kesarwani M, Panopoulos P, Tsurumi A, Giddey M, Wilhelmy J, et al. A quorum sensing small volatile molecule promotes antibiotic tolerance in bacteria. PLoS One. 2013;8:e80140. doi: 10.1371/journal.pone.0080140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzika AA, Constantinou C, Bandyopadhaya A, Psychogios N, Lee S, Mindrinos M, Martyn JA, Tompkins RG, Rahme LG. A small volatile bacterial molecule triggers mitochondrial dysfunction in murine skeletal muscle. PLoS One. 2013;8:e74528. doi: 10.1371/journal.pone.0074528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Astrakas LG, Goljer I, Yasuhara S, Padfield KE, Zhang Q, Gopalan S, Mindrinos MN, Dai G, Yu YM, Martyn JA, et al. Proton NMR spectroscopy shows lipids accumulate in skeletal muscle in response to burn trauma-induced apoptosis. FASEB J. 2005;19:1431–1440. doi: 10.1096/fj.04-2005com. [DOI] [PubMed] [Google Scholar]
- 34.Padfield KE, Astrakas LG, Zhang Q, Gopalan S, Dai G, Mindrinos MN, Tompkins RG, Rahme LG, Tzika AA. Burn injury causes mitochondrial dysfunction in skeletal muscle. Proc Natl Acad Sci USA. 2005;102:5368–5373. doi: 10.1073/pnas.0501211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morvan D, Demidem A, Papon J, Madelmont JC. Quantitative HRMAS proton total correlation spectroscopy applied to cultured melanoma cells treated by chloroethyl nitrosourea: demonstration of phospholipid metabolism alterations. Magn Reson Med. 2003;49:241–248. doi: 10.1002/mrm.10368. [DOI] [PubMed] [Google Scholar]
- 36.Asai A, Sahani N, Kaneki M, Ouchi Y, Martyn JA, Yasuhara SE. Primary role of functional ischemia, quantitative evidence for the two-hit mechanism, and phosphodiesterase-5 inhibitor therapy in mouse muscular dystrophy. PLoS One. 2007;2:e806. doi: 10.1371/journal.pone.0000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosokawa S, Koseki H, Nagashima M, Maeyama Y, Yomogida K, Mehr C, Rutledge M, Greenfeld H, Kaneki M, Tompkins RG, et al. Title efficacy of phosphodiesterase 5 inhibitor on distant burn-induced muscle autophagy, microcirculation, and survival rate. Am J Physiol Endocrinol Metab. 2013;304:E922–E933. doi: 10.1152/ajpendo.00078.2013. [DOI] [PubMed] [Google Scholar]
- 38.Burgmaier G, Schönrock LM, Kuhlmann T, Richter-Landsberg C, Brück W. Association of increased bcl-2 expression with rescue from tumor necrosis factor-alpha-induced cell death in the oligodendrocyte cell line OLN-93. J Neurochem. 2000;75:2270–2276. doi: 10.1046/j.1471-4159.2000.0752270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tzifi F, Economopoulou C, Gourgiotis D, Ardavanis A, Papageorgiou S, Scorilas A. The role of BCL2 family of apoptosis regulator proteins in acute and chronic leukemias. Adv Hematol. 2012;2012:524308. doi: 10.1155/2012/524308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davies L, Spiller D, White MR, Grierson I, Paraoan L. PERP expression stabilizes active p53 via modulation of p53-MDM2 interaction in uveal melanoma cells. Cell Death Dis. 2011;2:e136. doi: 10.1038/cddis.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huo J, Xu S, Lam KP. Fas apoptosis inhibitory molecule regulates T cell receptor-mediated apoptosis of thymocytes by modulating Akt activation and Nur77 expression. J Biol Chem. 2010;285:11827–11835. doi: 10.1074/jbc.M109.072744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou H, Ge Y, Sun L, Ma W, Wu J, Zhang X, Hu X, Eaves CJ, Wu D, Zhao Y. Growth arrest specific 2 is up-regulated in chronic myeloid leukemia cells and required for their growth. PLoS One. 2014;9:e86195. doi: 10.1371/journal.pone.0086195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tzika AA, Astrakas LG, Cao H, Mintzopoulos D, Zhang Q, Padfield K, Yu H, Mindrinos MN, Rahme LG, Tompkins RG. Murine intramyocellular lipids quantified by NMR act as metabolic biomarkers in burn trauma. Int J Mol Med. 2008;21:825–832. [PubMed] [Google Scholar]
- 44.Yuzefovych LV, Musiyenko SI, Wilson GL, Rachek LI. Mitochondrial DNA damage and dysfunction, and oxidative stress are associated with endoplasmic reticulum stress, protein degradation and apoptosis in high fat diet-induced insulin resistance mice. PLoS One. 2013;8:e54059. doi: 10.1371/journal.pone.0054059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blankenberg FG. In vivo detection of apoptosis. J Nucl Med. 2008;49(Suppl 2):81S–95S. doi: 10.2967/jnumed.107.045898. [DOI] [PubMed] [Google Scholar]
- 46.Vayssier-Taussat M, Kreps SE, Adrie C, Dall'Ava J, Christiani D, Polla BS. Mitochondrial membrane potential: a novel biomarker of oxidative environmental stress. Environ Health Perspect. 2002;110:301–305. doi: 10.1289/ehp.02110301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchi S, Giorgi C, Suski JM, Agnoletto C, Bononi A, Bonora M, De Marchi E, Missiroli S, Patergnani S, Poletti F, et al. Mitochondria-ros crosstalk in the control of cell death and aging. J Signal Transduct. 2012;2012:329635. doi: 10.1155/2012/329635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37:2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blasiak J, Glowacki S, Kauppinen A, Kaarniranta K. Mitochondrial and nuclear DNA damage and repair in age-related macular degeneration. Int J Mol Sci. 2013;14:2996–3010. doi: 10.3390/ijms14022996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 51.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 52.Supinski GS, Callahan LA. Caspase activation contributes to endotoxin-induced diaphragm weakness. J Appl Physiol (1985) 2006;100:1770–1777. doi: 10.1152/japplphysiol.01288.2005. [DOI] [PubMed] [Google Scholar]
- 53.Tatsumi T, Shiraishi J, Keira N, Akashi K, Mano A, Yamanaka S, Matoba S, Fushiki S, Fliss H, Nakagawa M. Intracellular ATP is required for mitochondrial apoptotic pathways in isolated hypoxic rat cardiac myocytes. Cardiovasc Res. 2003;59:428–440. doi: 10.1016/S0008-6363(03)00391-2. [DOI] [PubMed] [Google Scholar]
- 54.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/S0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 55.Szczepaniak LS, Babcock EE, Schick F, Dobbins RL, Garg A, Burns DK, McGarry JD, Stein DT. Measurement of intracellular triglyceride stores by H spectroscopy: validation in vivo. Am J Physiol. 1999;276:E977–E989. doi: 10.1152/ajpendo.1999.276.5.E977. [DOI] [PubMed] [Google Scholar]
- 56.Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300:1140–1142. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004;350:664–671. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blankenberg FG, Katsikis PD, Storrs RW, Beaulieu C, Spielman D, Chen JY, Naumovski L, Tait JF. Quantitative analysis of apoptotic cell death using proton nuclear magnetic resonance spectroscopy. Blood. 1997;89:3778–3786. [PubMed] [Google Scholar]
- 59.Boren J, Brindle KM. Apoptosis-induced mitochondrial dysfunction causes cytoplasmic lipid droplet formation. Cell Death Differ. 2012;19:1561–1570. doi: 10.1038/cdd.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hakumäki JM, Poptani H, Sandmair AM, Ylä-Herttuala S, Kauppinen RA. 1H MRS detects polyunsaturated fatty acid accumulation during gene therapy of glioma: implications for the in vivo detection of apoptosis. Nat Med. 1999;5:1323–1327. doi: 10.1038/15279. [DOI] [PubMed] [Google Scholar]
- 61.Mullen TD, Obeid LM. Ceramide and apoptosis: Exploring the enigmatic connections between sphingolipid metabolism and programmed cell death. Anticancer Agents Med Chem. 2012;12:340–363. doi: 10.2174/187152012800228661. [DOI] [PubMed] [Google Scholar]
- 62.Martinez TN, Chen X, Bandyopadhyay S, Merrill AH, Tansey MG. Ceramide sphingolipid signaling mediates Tumor Necrosis Factor (TNF)-dependent toxicity via caspase signaling in dopaminergic neurons. Mol Neurodegener. 2012;7:45. doi: 10.1186/1750-1326-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giussani P, Tringali C, Riboni L, Viani P, Venerando B. Sphingolipids: key regulators of apoptosis and pivotal players in cancer drug resistance. Int J Mol Sci. 2014;15:4356–4392. doi: 10.3390/ijms15034356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aureli M, Murdica V, Loberto N, Samarani M, Prinetti A, Bassi R, Sonnino S. Exploring the link between ceramide and ionizing radiation. Glycoconj J. 2014;31:449–459. doi: 10.1007/s10719-014-9541-y. [DOI] [PubMed] [Google Scholar]
- 65.Jenkins GM. The emerging role for sphingolipids in the eukaryotic heat shock response. Cell Mol Life Sci. 2003;60:701–710. doi: 10.1007/s00018-003-2239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Brocklyn JR, Williams JB. The control of the balance between ceramide and sphingosine-1-phosphate by sphingosine kinase: oxidative stress and the seesaw of cell survival and death. Comp Biochem Physiol B Biochem Mol Biol. 2012;163:26–36. doi: 10.1016/j.cbpb.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 67.Yasuhara S, Asai A, Sahani ND, Martyn JA. Mitochondria, endoplasmic reticulum, and alternative pathways of cell death in critical illness. Crit Care Med. 2007;35(Suppl):S488–S495. doi: 10.1097/01.CCM.0000278045.91575.30. [DOI] [PubMed] [Google Scholar]
- 68.Bernardi P, Di Lisa F. The mitochondrial permeability transition pore: molecular nature and role as a target in cardioprotection. J Mol Cell Cardiol. 2015;78:100–106. doi: 10.1016/j.yjmcc.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nabben M, Shabalina IG, Moonen-Kornips E, van Beurden D, Cannon B, Schrauwen P, Nedergaard J, Hoeks J. Uncoupled respiration, ROS production, acute lipotoxicity and oxidative damage in isolated skeletal muscle mitochondria from UCP3-ablated mice. Biochim Biophys Acta. 2011;1807:1095–1105. doi: 10.1016/j.bbabio.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 70.Reynolds HY, Di Sant'Agnese PA, Zierdt CH. Mucoid Pseudomonas aeruginosa. A sign of cystic fibrosis in young adults with chronic pulmonary disease? JAMA. 1976;236:2190–2192. doi: 10.1001/jama.1976.03270200028024. [DOI] [PubMed] [Google Scholar]
- 71.Scott-Thomas AJ, Syhre M, Pattemore PK, Epton M, Laing R, Pearson J, Chambers ST. 2-Aminoacetophenone as a potential breath biomarker for Pseudomonas aeruginosa in the cystic fibrosis lung. BMC Pulm Med. 2010;10:56. doi: 10.1186/1471-2466-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Velsor LW, Kariya C, Kachadourian R, Day BJ. Mitochondrial oxidative stress in the lungs of cystic fibrosis transmembrane conductance regulator protein mutant mice. Am J Respir Cell Mol Biol. 2006;35:579–586. doi: 10.1165/rcmb.2005-0473OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Divangahi M, Balghi H, Danialou G, Comtois AS, Demoule A, Ernest S, Haston C, Robert R, Hanrahan JW, Radzioch D, Petrof BJ. Lack of CFTR in skeletal muscle predisposes to muscle wasting and diaphragm muscle pump failure in cystic fibrosis mice. PLoS Genet. 2009;5:e1000586. doi: 10.1371/journal.pgen.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rottner M, Tual-Chalot S, Mostefai HA, Andriantsitohaina R, Freyssinet JM, Martínez MC. Increased oxidative stress induces apoptosis in human cystic fibrosis cells. PLoS One. 2011;6:e24880. doi: 10.1371/journal.pone.0024880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moylan JS, Reid MB. Oxidative stress, chronic disease, and muscle wasting. Muscle Nerve. 2007;35:411–429. doi: 10.1002/mus.20743. [DOI] [PubMed] [Google Scholar]
- 76.Schwartz LM. Atrophy and programmed cell death of skeletal muscle. Cell Death Differ. 2008;15:1163–1169. doi: 10.1038/cdd.2008.68. [DOI] [PubMed] [Google Scholar]
- 77.Aare S, Radell P, Eriksson LI, Akkad H, Chen YW, Hoffman EP, Larsson L. Effects of corticosteroids in the development of limb muscle weakness in a porcine intensive care unit model. Physiol Genomics. 2013;45:312–320. doi: 10.1152/physiolgenomics.00123.2012. [DOI] [PubMed] [Google Scholar]
- 78.Xu H, Lam SH, Shen Y, Gong Z. Genome-wide identification of molecular pathways and biomarkers in response to arsenic exposure in zebrafish liver. PLoS One. 2013;8:e68737. doi: 10.1371/journal.pone.0068737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maestre I, Jordán J, Calvo S, Reig JA, Ceña V, Soria B, Prentki M, Roche E. Mitochondrial dysfunction is involved in apoptosis induced by serum withdrawal and fatty acids in the beta-cell line INS-1. Endocrinology. 2003;144:335–345. doi: 10.1210/en.2001-211282. [DOI] [PubMed] [Google Scholar]
- 80.Lartigue L, Faustin B. Mitochondria: metabolic regulators of innate immune responses to pathogens and cell stress. Int J Biochem Cell Biol. 2013;45:2052–2056. doi: 10.1016/j.biocel.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 81.Nicolson GL. Mitochondrial dysfunction and chronic disease: treatment with natural supplements. Altern Ther Health Med. 2013:at5027. [PubMed] [Google Scholar]
- 82.Frick CG, Fink H, Gordan ML, Eckel B, Martyn JA, Blobner M. Chronic Escherichia coli infection induces muscle wasting without changing acetylcholine receptor numbers. Intensive Care Med. 2008;34:561–567. doi: 10.1007/s00134-007-0852-3. [DOI] [PubMed] [Google Scholar]
- 83.Macallan DC, McNurlan MA, Kurpad AV, de Souza G, Shetty PS, Calder AG, Griffin GE. Whole body protein metabolism in human pulmonary tuberculosis and undernutrition: evidence for anabolic block in tuberculosis. Clin Sci (Lond) 1998;94:321–331. doi: 10.1042/cs0940321. [DOI] [PubMed] [Google Scholar]
- 84.Machado AM, Desler C, Bøggild S, Strickertsson JA, Friis-Hansen L, Figueiredo C, Seruca R, Rasmussen LJ. Helicobacter pylori infection affects mitochondrial function and DNA repair, thus, mediating genetic instability in gastric cells. Mech Ageing Dev. 2013;134:460–466. doi: 10.1016/j.mad.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 85.Morton RE, Hutchings J, Halliday D, Rennie MJ, Wolman SL. Protein metabolism during treatment of chest infection in patients with cystic fibrosis. Am J Clin Nutr. 1988;47:214–219. doi: 10.1093/ajcn/47.2.214. [DOI] [PubMed] [Google Scholar]