Abstract
Nuclear run-on (NRO) is a method that measures transcriptional activity via the quantification of biochemically labelled nascent RNA molecules derived from nuclear isolates. Widespread usage of this technique has been limited due to its technical difficulty relative to steady-state total mRNA analyses. Here we describe in detail a highly optimised and validated protocol for the quantification of transcriptional activity in human cell cultures using a modified bromouridine immunocapture NRO method and Reverse Transcription-quantitative Polymerase Chain Reaction (RT-qPCR). This method is rapid (the protocol can be completed in two days), cost-effective, exhibits negligible detection of background noise from unlabelled transcripts, requires no radioactive materials, and can be performed from as little as 500,000 nuclei. It also takes advantage of the high sensitivity, specificity and dynamic range of RT-qPCR. Protocol-specific issues such as appropriate qPCR assay design, data normalisation, changes in nuclear RNA content between experimental conditions, unequal label incorporation, and background signal determination are also discussed.
Keywords: Nuclear run-on, Bromouridine, RT-qPCR, transcription
INTRODUCTION
Transcription is the process by which a transient RNA transcript is generated through the copying of a complementary genomic DNA template. As such, transcription constitutes the first stage of gene expression and is therefore a fundamental cellular function. The vast majority of gene expression studies utilise steady-state total mRNA measurements (e.g. by semi-quantitative RT-PCR or RT-qPCR). These methods provide a snapshot of transcript abundance in the cell. However, steady-state transcript levels are a function of both transcriptional synthesis and post-transcriptional degradation. With conventional gene expression analysis it is therefore impossible to distinguish transcriptional from post-transcriptional gene regulatory effects.
Nuclear run-on (NRO) analysis is a biochemical method for quantifying changes in transcriptional activity in cultured cells. Cell nuclei are first harvested by gentle lysis of the plasma membrane and low-speed centrifugation. Maintaining nuclear preparations at low temperature ensures that the nuclear membrane remains intact and that active RNA polymerase II (RNAPII), and other associated proteins, remain paused on the genome. Subsequently, transcription is allowed to continue by warming the nuclear samples and allowing RNAPII activity to resume. This in vitro NRO transcription reaction is performed in the presence of labelled ribonucleotide pre-cursors. Incorporation of labelled nucleotides into nascent transcripts (NRO-RNAs) enables their specific quantification against a background of unlabelled nuclear RNAs. Changes in NRO-RNA abundance (determined by blotting, nucleic amplification, microarray or high-throughput sequencing techniques) are therefore a direct reflection of transcriptional activity.
Classically, NRO has been performed using 32P-containing UTP to label nascent RNA. Post-NRO samples are blotted onto membranes and nascent transcription visualised by autoradiography 1–4. Non-radioactive variants which utilise biotin-16-aminoallyluridine or bromouridine have since been reported 5, with nascent transcripts quantified by RT-qPCR 6 or semi-quantitative RT-PCR with fluorescent primers 7.
The NRO assay is technically more difficult and labour intensive than conventional steady-state (total mRNA) gene expression analysis, which has likely limited its widespread usage. Typically NRO has required large number of cells (e.g. at least 6 million cells in 15 cm2 plates8) meaning that analysis of rare samples or small subsets of sorted cells may not be possible. Furthermore, a number of specific technical hurdles can potentially confound NRO analysis including appropriate qPCR primer design, qPCR data normalisation, changes in nuclear RNA content between experimental groups, high background signal, and unequal label incorporation between experimental groups.
The quantification of unspliced pre-mRNA has been proposed as a simple alternative, and indirect, method for determining transcriptional activity 9. This method however, while facile, is subject to its own problems. Specifically, multiple studies have demonstrated that post-transcriptional gene silencing events can occur in the nucleus 10–12 meaning that changes in pre-mRNA levels may not necessarily reflect changes in transcriptional activity. Furthermore, such an approach is also limited to intron-containing genes.
A major development in the study of transcription is the combination of the nuclear run-on method with deep sequencing methodologies such as GRO-seq (Global Run-On followed by high-throughput sequencing of RNA) 8,13 and more recently PRO-seq (Precision Run-On followed by sequencing) 14. These techniques have enabled fundamental new insights into the process of transcription, such as the detection of widespread divergent transcription from human promoters and high resolution mapping of polymerase pause sites. However, in many cases genome-wide analysis is unnecessary or uneconomical.
In some cases, it may be useful to use NRO-RT-qPCR as a quality control tool during the preparation of GRO-seq libraries. Similarly, GRO-seq results may require validation on a gene-by-gene basis. In these cases, a complementary method for analysing a small number of genes is desirable. RT-qPCR is an ideal method for analysing NRO-RNA levels on account of its widespread adoption in research laboratories, low cost, ease of use, high specificity, and wide dynamic range (typically >6 logs). There is therefore a need for RT-qPCR-based NRO protocols which are suitable for addressing single gene questions, and that are not technically or economically cumbersome or prohibitive.
An alternative approach to nuclear run-on is metabolic labelling. This strategy consists of adding modified ribonucleotide precursors to the media of cells in culture. Labelled RNA precursors are taken up by cells and incorporated into nascent transcripts. Labelled transcripts are then purified and quantified. A disadvantage of metabolic labelling is that incorporation of the labelled nucleotide may have adverse effects on cell physiology. For example, culturing cells for extended period of time in the presence of 4-thiouridine, or 5-ethynyluridne results in inhibition of cell growth 15. Furthermore, 4-thiouridine can bind to guanine which introduces base changes in label-containing nascent transcripts. In the case of both 4-thiourinde and 5-ethynyluridine, additional chemical reaction steps are required for both metabolic labelling methods (thiol-biotinylation 16 and azide-biotin cycloaddition click chemistry17 respectively).
In contrast, metabolic labelling with 5-bromouridine does not affect cell growth, introduce base changes and does not require chemical addition of an additional moiety for affinity purification 15. This approach has also been combined with high-throughput sequencing as in the cases of Bru-seq (bromouridine-sequencing), BruChase-seq (bromouridine-chase sequencing)18,19 and BRIC-seq (5′-bromouridine (BrU) immunoprecipitation followed by sequencing) 15,20. An advantage of these approaches is that, in addition to labelling nascent RNA, global transcript stability can also be determined using a pulse-chase strategy (chasing with unlabelled uridine). A disadvantage of bromouridine metabolic labelling is that it requires relatively long (24 hours) treatments for cellular uptake and incorporation to occur 15. Alternatively, microinjection, permeabilisation, transfection and osmotic shock have all been used to facilitate bromouridine uptake. The issues surrounding bromouridine uptake add additional experimental complexity and potential confounding factors which are not applicable to NRO methodologies. In contrast, NRO also enables the analysis of short term changes in transcription and the investigation of polymerase pausing 21–23 for which metabolic labelling techniques are unsuitable.
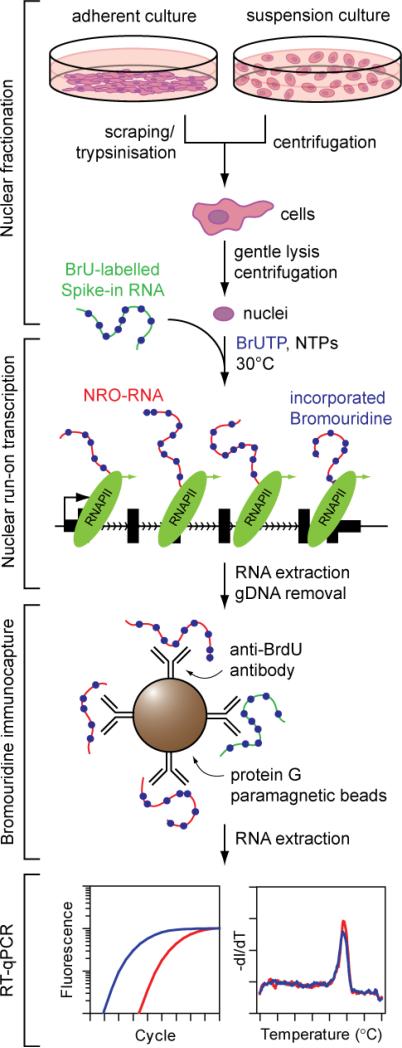
Updated protocols for studying transcriptional changes at specific genes of interest using magnetic bead-based separation and RT-qPCR are currently lacking. Here we describe a highly optimised and validated NRO protocol based on bromouridine immunocapture and RT-qPCR that enables facile, rapid, reproducible, cheap and readily automatable detection of nascent transcription in human cell cultures (Fig. 1).
Figure 1.
Complete workflow for nuclear run-on, bromouridine immunocapture and RT-qPCR.
Applications of the method
NRO-RT-qPCR is suitable for analysing any change in transcriptional activity (e.g. regulation of specific promoters by transcription factors, non-coding RNAs or small molecule interactions) and the reliable determination of promoter elements. Moreover, NRO also enables the investigation of phenomena such as promoter bidirectional transcription 8, enhancer transcription 24, and the differential regulation of transcription at the initiation and elongation stages (e.g. by polymerase pausing) 21–23. NRO is particularly useful for distinguishing transcriptional from post-transcriptional gene regulatory mechanisms. For example, a plethora of studies have shown that small RNA molecules can induce transcriptional gene silencing (TGS) via a process analogous to RNA interference (or post-transcriptional gene silencing, PTGS) 25–28. NRO analysis has been used to distinguish the novel TGS phenomena from the much more commonly reported PTGS effects. Similarly, long non-coding RNAs (lncRNAs) have been implicated in the epigenetic control of gene expression 29. NRO can therefore be used to demonstrate the gene regulatory effects of specific lncRNAs at the level of transcription 30.
Experimental design
Correct qPCR assay design is critical to success of the NRO technique. As with conventional qPCR, primers should be 18-22 nucleotides in length, have GC content of 40-60%, have Tm values ~60 °C, and produce a short (50-150 base pairs) amplicon. Of critical importance is the observation that nascent NRO-RNAs produced by run-on transcription are ectopic transcripts lacking splicing or polyadenylation 8 (Supplementary Figure 1a). As a result, NRO reverse transcription should be performed using random primers (or strand-specific primers for distinguishing between overlapping sense and antisense transcripts) whereas the use of oligo-dT primers should be avoided. Similarly, several conventional qPCR assay configurations for measuring steady-state mature mRNA transcript levels (frequently employed in commercially available assays) are unsuitable for detecting nascent transcripts. Inappropriate designs include: (i) the use of primers placed in different exons that are separated by large distances (kilobases) in genomic space but only short distances (~100 bp) in the spliced transcriptome, and (ii) primers or probe complementary to exon-exon junctions (Supplementary Figure 1b). Suitable designs for NRO-RT-qPCR therefore include: (i) primers within the same exon, (ii) primers within the same intron, or (iii) primer sequences spanning an intron-exon exon boundary (Supplementary Figure 1b). In fact, primers that span intron-exon boundaries may be preferable for NRO-RT-qPCR in many cases as this design configuration excludes the amplification of processed mRNA contamination. Importantly, all NRO-RT-qPCR assays have the potential to amplify gDNA, and so removal of DNA background during nuclear RNA preparation is therefore essential.
In order to investigate transcriptional pausing, NRO-RT-qPCR assays can be designed to amplify distinct regions within the same gene (specifically the promoter/5′ terminus, and the gene body respectively). Changes in transcriptional initiation would be expected to affect the signal from both assays equivalently. Conversely, changes in transcriptional elongation (e.g. following the release of paused polymerase) would be expected to result in changes in NRO signal from the gene body but not at the promoter/5′ terminus. To perform these analyses it is important that the length of NRO-RNAs be restricted to ~200-300 nucleotides. This can be accomplished by ensuring that the concentration of one ribonucleotide in the NRO reaction (typically CTP) is limiting. (CTP restriction is also commonly employed in GRO-seq methodologies 8).
Typically one or more experimental conditions are compared to untreated cultures. Three replicate cultures are sufficient in most cases, although a power analysis may be required to determine the sample size required to achieve statistical significance.
A number of controls can be used to determine the performance of the PCR reaction. qPCR reactions lacking template cDNA (No template controls, NTCs) should be included to monitor possible reagent contamination, typically a result of amplicon carry-over. Similarly, performing the reverse transcription reaction without enzyme (RT- control) can be used to monitor gDNA contamination. We have routinely observed no amplification in RT- control samples consistent with complete gDNA removal (Supplementary Figure 2).
Nascent gene-of-interest transcription is normalised to a reference (housekeeping) gene to control for equal sample loading between reactions as with steady-state total mRNA analysis. Normalisation to the geometric mean of multiple reference genes is recommended in order to minimise biases associated with the variability of any one gene 31. Notably, we have observed that reference genes that are suitable for total mRNA analysis are not necessarily suitable for NRO-RT-qPCR. For example, the RPL10 mRNA is highly abundant in total RNA extracts but is detected at low levels by NRO (data not shown). As a result the low rate of transcription and high transcript stability make RPL10 an excellent reference gene for steady-state total mRNA expression studies and a poor reference gene for NRO-RT-qPCR.
In 2009 the MIQE (Minimum Information for publication of Quantitative real-time PCR experiments) guidelines were established to promote consistency and transparency in gene expression studies 32,33. To date, these guidelines have not been applied to NRO experiments. In order to conform to community standards, and to help facilitate research in this area, we provide details of twelve validated and MIQE-compliant RT-qPCR assays for human NRO-RNA reference genes: ACTB, B2M, GAPDH, GUSB, HPRT1, PPIA, RPL10, RPLP0, TBP, TUBB, UBC, and YWHAZ. Primer sequences are shown in Table 2 and additional validation data are shown in Supplementary Figure 3 and Supplementary File 1.
Table 2.
Validated NRO-qPCR assays for commonly used human reference genes.
| Forward | Reverse | |
|---|---|---|
| ACTB | AGCTCATTGTAGAAGGTGTGG | GGCATGGGTCAGAAGGATTC |
| B2M | ACACCAAGTTAGCCCCAAG | ACCCAGACACATAGCAATTCAG |
| GAPDH | AATCCCATCACCATCTTCCAG | GAGCCACACCATCCTAGTTG |
| GUSB | CGACTTCTCTGACAACCGAC | GATCGCACCTCCCACAG |
| HPRT1 | CATTTACCACTTCTAGGCCCC | TCAGTCCATAACAAGCACCC |
| PPIA | GCAGATTTGGCACACTTCATG | GTTCTCATCTTCAAATTTCTCCCC |
| RPL10 | TTATCATGTCCATCCGCACC | GCATTGAACTTGGTGAAGCC |
| RPLP0 | CGCAGCCAATAGACAGGAG | GCGCGTGCCTTTTATAATGC |
| TBP | GCTGTTTAACTTCGCTTCCG | CCGAACCGTCAGAAAAGAGTAC |
| TUBB | ATATGTTCCTCGTGCCATCC | GTCTGGTCTAAAGATCTGGCC |
| UBC | GCCTTAGAACCCCAGTATCAG | AAGAAAACCAGTGCCCTAGAG |
| YWHAZ | CGGTACAAGAGGAAGTGGATAAG | AGGTAAGGGTTAAATGGGTGC |
Development of the protocol
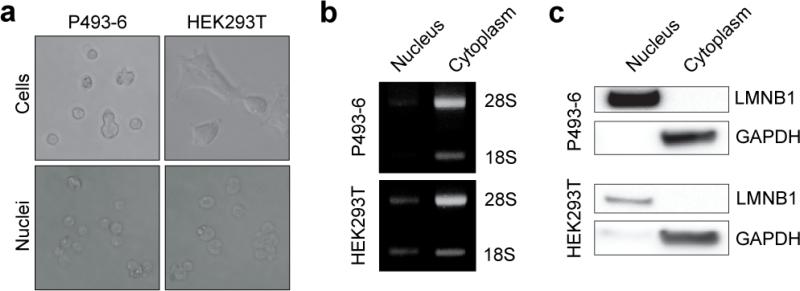
The first stage of the NRO protocol is the preparation of intact nuclei using standard methodologies. To test the fidelity of our nuclei harvesting protocol we prepared nuclei and cytoplasmic fractions from HEK293T embryonic kidney cells (adherent), and P493-6 B-cell (suspension) cultures 34. Cells were subjected to gentle lysis using a buffer containing the non-ionic detergent NP-40 (also called IGEPAL CA-630). NP-40 (at low concentrations) lyses the plasma membrane but is too weak to dissolve the nuclear envelope. Following low speed centrifugation, intact nuclei were visualised by light microscopy (Fig. 2a). RNA samples from the nuclear and cytoplasmic fractions were separated by agarose gel electrophoresis and the 18S and 28S ribosomal RNAs were found to be predominantly cytoplasmic (Fig. 2b). Analysis of protein expression by western blot also showed a clear distinction between the two fractions, with LMNB1 (Lamin B) restricted to the nucleus and GAPDH exclusively cytoplasmic (Fig. 2c). These results demonstrate successful isolation of intact nuclei from both adherent and suspension cells.
Figure 2. Preparation of nuclear extracts.
P493-6 and HEK293T nuclei were harvested by gentle lysis and low speed centrifugation. (a) Whole cells and nuclear extracts were visualised by light microscopy. (b) RNA was extracted from each fraction and 28S and 18S ribosomal RNAs visualised by agarose gel electrophoresis. (c) Protein was harvested from both fractions and LMNB1 and GAPDH expression measured by western blot.
Previously, we had terminated the NRO reaction by incubating with DNase I followed by a second incubation with Proteinase K 6. However, strong association between proteins and genomic DNA inhibits complete digestion, and proteolysis to remove these proteins results in loss of DNase I activity. We therefore chose to simplify this stage of the protocol by directly extracting RNA. The use of extraction buffers containing chaotropic salts such as guanidinium isothiocyanate allows the disruption of protein-DNA interactions while simultaneously ensuring rapid and complete termination of the NRO reaction. To stop the NRO reaction and harvest NRO-RNA, we have utilised the Maxwell 16 RNA extraction system from Promega. This method is rapid (~50 minutes), consistent between samples, and includes a rigorous DNA removal step. Alternatively, other automated or manual RNA extraction methodologies can be used such as MEGA clear/TRIzol/RNeasy followed by a separate DNase I step. The preparation of pure RNA samples allows for clean immunoprecipitation pull-downs with low background binding. It is important to measure the concentration of nuclear RNA at this stage as this value may vary between experimental groups (discussed below). Measuring the concentration in each sample of nuclear RNA also allows the inter- and intra-experimental standardisation of input RNA at the immunoprecipitation stage.
Agarose beads conjugated to anti-BrdU antibodies have been used in multiple studies reporting NRO or GRO-seq data 8,13. The GRO-seq protocol utilises a triple selection strategy to massively enrich for labelled NRO-RNAs 8. However, for NRO-RT-qPCR, the requirement for NRO-RNA purity is less stringent. In this protocol we have utilised superparamagnetic beads derivatised with Protein G in order to precipitate bromouridylated RNA-antibody complexes. Magnetic handling offers a number of advantages over agarose slurry-based methods: (i) complete wash buffer removal with minimal sample loss, (ii) low background binding, (iii) fast diffusion kinetics and therefore reduced wash times as a result of defined bead sizes, and (iv) magnetic separation steps are readily automatable. If necessary, additional rounds of immunoprecipitation can be used to enrich for labelled transcripts and reduce background.
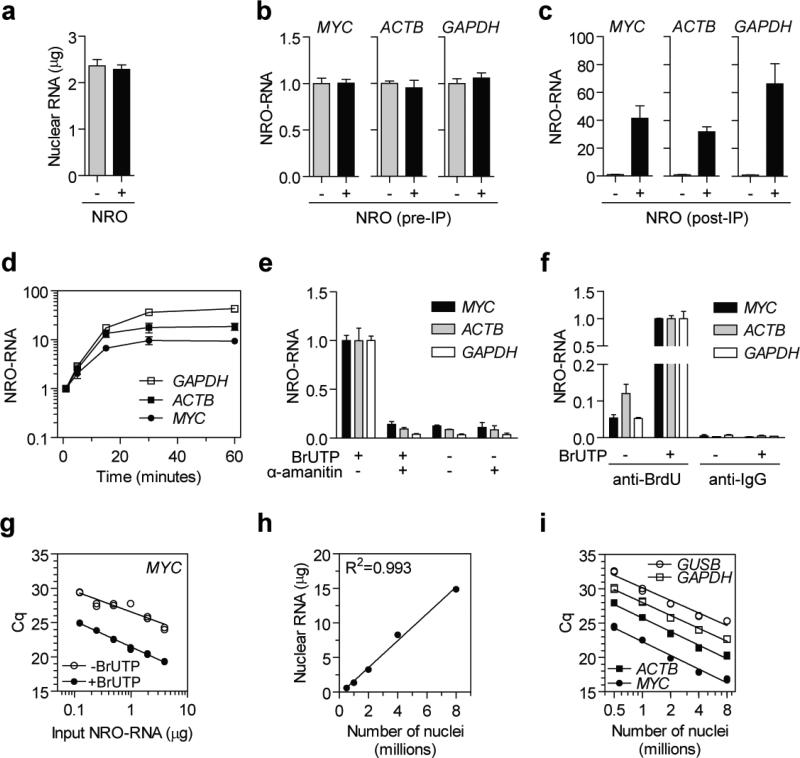
A number of experiments were performed in order to determine if the protocol specifically detects nascent transcription. Firstly, P493-6 nuclear isolates were either incubated at 30 °C for 30 minutes with BrUTP and unlabelled ribonucleotides before RNA extraction (NRO+) or harvested immediately without incubation (NRO−). Nuclear RNA concentration was not significantly affected by the nuclear run-on reaction (Fig. 3a) and NRO-RNA levels of MYC, ACTB and GAPDH in nuclear RNA samples (before immunoprecipitation) were similarly unchanged (Fig. 3b). Labelled and unlabelled nuclear RNA samples were immunoprecipitated with anti-BrdU (IIB5) antibodies and transcript levels measured by RT-qPCR. Labelled transcripts were detected at levels 35-70 fold above the unlabelled transcript background indicative of nascent transcript detection (Fig. 3c). Together these observations show that the nascent transcripts produced by the NRO reaction represent a tiny minority of the nuclear transcript pool.
Figure 3. Development of NRO-RT-qPCR protocol.
P493-6 nuclei were harvested and RNA extracted either immediately (NRO−) or following NRO transcription (NRO+). (a) Nuclear RNA content was determined by Nanodrop spectrophotometry and (b) pre-immunoprecipitation (pre-IP) NRO-RNAs were quantified by RT-qPCR for MYC, ACTB and GAPDH. (c) Nuclear RNA samples were immunoprecipitated using anti-BrdU antibodies and (post-IP) NRO-RNAs quantified by RT-qPCR. (d) P493-6 nuclei were harvested and NRO transcription performed. NRO reactions were terminated by extracting RNA at a series of time points and NRO-RNAs measured by RT-PCR. (e) NRO was performed in the presence and absence of the RNA polymerase II inhibitor α-amanitin (1 μM) and NRO-RNAs quantified by RT-qPCR. (f) Specificity of anti-BrdU immunoprecipitation was confirmed by comparison to a non-specific anti-IgG control antibody. (g) Immunoprecipitation was performed with a range of input nuclear RNA amounts. (h) NRO was performed using a range of different numbers of nuclei and nuclear RNA content was determined by Nanodrop. (i) Labelled NRO-RNAs derived from different numbers of starting nuclei were quantified for MYC, ACTB, GAPDH and GUSB. Values are mean +/− SEM. Cq, quantification cycle. NRO-RNA quantification was performed using the ΔCq method (i.e. not normalised to a reference gene) and the value of one experimental group returned to a value of one for each assay.
Secondly, NRO reactions were performed on P493-6 nuclear extracts and the reaction stopped after 1, 5, 15, 30 and 60 minutes by snap freezing the samples. Bromouridine-containing NRO-RNAs were immunoprecipitated and MYC, ACTB and GAPDH levels determined by RT-qPCR. (Fig. 3d). Levels of labelled NRO-RNAs increased over time and reached a plateau by 30 minutes, consistent with the detection of nascent transcription.
Thirdly, NRO reactions were performed in the presence or absence of the RNAPII inhibitor α-amanitin 35 (additional information can be found in Supplementary Methods). NRO-RNA transcripts were only detected at background levels (similar to mock NRO samples lacking BrUTP) in the α-amanitin-treated samples thereby confirming that the protocol specifically measures nascent transcription (Fig. 3e).
Specificity of the bromouridine immunocapture step was confirmed by performing the immunoprecipitation with either anti-BrdU (specific) or anti-IgG (non-specific) antibodies on labelled or unlabelled nuclear RNA samples (Fig. 3f). The highest signal was obtained from the specific pull-down, which was 200-450 fold higher than the background signal from non-specific binding to the IgG antibody. Importantly, unlabelled NRO-RNAs were also precipitated by the anti-BrdU antibodies although at levels 7-41 fold higher than the signal from the IgG negative control antibody. This finding suggests that the anti-BrdU antibody has a weak affinity for unlabelled RNA (further discussed below). Two additional anti-BrdU antibodies (Bu20A and ICR1) were tested for their ability to precipitate Bromouridine-containing NRO-RNAs (Figure 4a). ICR1 performed similarly to IIB5, whereas Bu20A was much less efficient at recovering labelled NRO-RNAs (Supplementary Figure 4b).
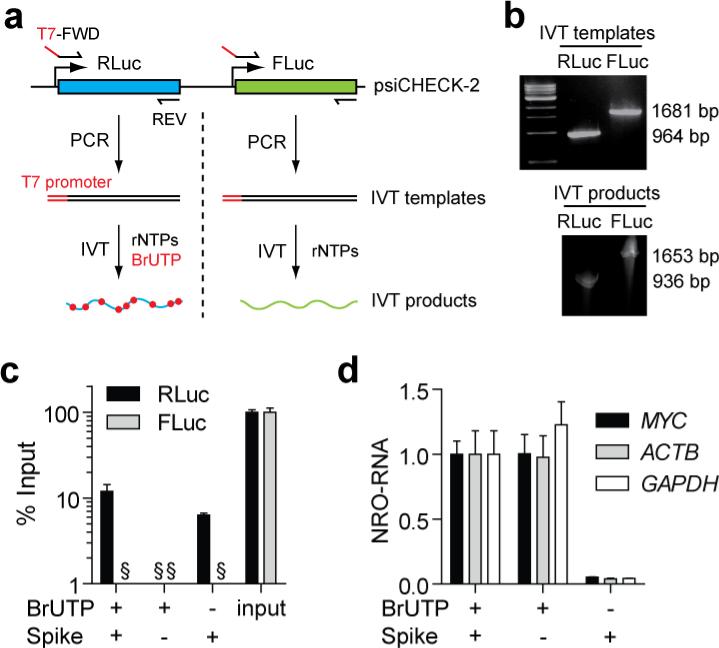
Figure 4. Monitoring background signal using exogenous spike-in RNA controls.
(a) Approach for generation of positive and negative spike-in controls. RLuc and FLuc sequences were amplified from the psiCHECK-2 plasmid in separate PCR reactions to generate in vitro transcription (IVT) templates. IVT products were generated using T7 polymerase in the presence or absence of BrUTP. (b) Generation of IVT templates and IVT products were confirmed by agarose and polyacrylamide gel electrophoresis respectively. (c) NRO was performed in the presence or absence of the exogenous spike-in controls. NRO-RNAs were precipitated and quantified by RT-qPCR. Data were compared relative to input samples that were not precipitated. (d) Addition of the exogenous spike-in controls did not significantly affect the levels of NRO-RNAs for MYC, ACTB or GAPDH. Values are mean + SEM, § not detected. NRO-RNA quantification was performed using the ΔCq method (i.e. not normalised to a reference gene) and the value of one experimental group returned to a value of one for each assay.
To assess the binding capacity of the bead-antibody complexes a two-fold dilution series of nuclear RNA was immunoprecipitated and MYC NRO-RNA levels measured by RT-qPCR (Fig. 3g). A one-to-one linear relationship was observed between input nuclear RNA and qPCR signal over a range of 0.125 μg to 8 μg (R2=0.993). Background binding in the BrUTP- control was also linear although with weaker goodness of fit (R2=0.846). Similar results were observed for ACTB and GAPDH (data not shown). Taken together, these experiments (Figs. 3h,i) suggest that the anti-BrdU antibodies have a low affinity for unlabelled RNAs in the absence of BrUTP.
To determine the optimal number of input nuclei, the NRO protocol was performed using a range of 0.5 to 8 million P493-6 starting cells. (It was difficult to visualise P493-6 nuclear pellets when using less than 0.5 million cells). A one-to-one linear relationship was observed between number of nuclei and nuclear RNA concentration (R2=0.993) (Fig. 3h). Nuclear RNA samples were immunoprecipitated and MYC, ACTB, GAPDH and GUSB NRO-RNAs quantified by RT-qPCR (Fig. 3i). (Primer binding locations are shown in Supplementary Figure 5 for the RT-qPCR assays used here). All four transcripts could be detected from as little as 0.5 million nuclei. The optimal number of input nuclei was 1-2 million as all NRO-RNAs tested could be detected with Cq values between 20 and 30 in this range. A linear relationship was observed between number of nuclei and qPCR signal. However, the relationship was not one-to-one. Instead a doubling in nuclei numbers resulted in ~4 fold increase in qPCR signal. This observation highlights the importance of standardising the number of nuclei between experimental samples to avoid unequal label incorporation between experimental groups. This is especially important if cell viability or proliferative activity varies between experimental groups (e.g. in the case of anticancer therapeutics).
To further quantify background binding we utilised a dual spike-in strategy. Two non-mammalian luciferase RNAs (RLuc and FLuc) were generated by in vitro transcription (IVT) with T7 polymerase. BrUTP was incorporated into the RLuc transcript (‘signal’ control spike) but not the FLuc transcript (‘noise’ control spike) (Fig. 4a). Generation of IVT templates and products was confirmed by agarose and polyacrylamide gel electrophoresis respectively (Fig. 4b). The two spike-in controls were added to P493-6 nuclei, NRO performed in the presence and absence of BrUTP, and the two exogenous spikes measured by RT-qPCR compared to non-precipitated input samples. RLuc was measured at levels ~12% of input (representing the capture rate of BrU-containing NRO-RNAs), whereas FLuc RNA was not detected in any of the NRO samples (Fig. 4c) suggesting that detection of unlabelled background RNA is negligible in the presence of BrUTP. Furthermore, detection of endogenous NRO-RNAs was unaffected by the addition of the exogenous spikes (Fig. 4d). Exogenous spike-in controls can therefore be utilised for three purposes (i) to monitor the efficiency of bromouridylated NRO-RNA recovery after immunoprecipitation, (ii) to determine the signal-to-noise (i.e. RLuc:FLuc) ratio in experimental samples, or (iii) as external controls for RT-qPCR normalisation in situations where suitable reference genes are lacking or unknown.
MATERIALS
REAGENTS
5-Bromouridine 5′-Triphosphate (catalogue #: B7166 [Sigma-Aldrich, Carlsbad, CA, USA]) ! CAUTION Always wear gloves and a face-mask when handling Bromouridine powder. Bromouridine is a mutagen.
Ribonucleoside Triphosphate Set (catalogue #: 11277057001 [Roche, Pleasanton, CA, USA])
RNaseOUT Recombinant Ribonuclease Inhibitor (catalogue #: 10777-019 [Life Technologies, Carlsbad, CA, USA]).
TRIzol Reagent (catalogue #: 15596-018 [Life Technologies]). ! CAUTION Always work with TRIzol reagent in a fume hood. Always wear a lab coat, gloves and safety glasses.
Chloroform (molecular biology grade). ! CAUTION Always work with Chloroform in a fume hood. Always wear a lab coat, gloves and safety glasses.
Isopropanol (molecular biology grade).
75% Ethanol wash (molecular biology grade)
Glycogen for molecular biology, 20 mg/ml (catalogue #: 10901393001 [Roche])
Dulbecco's Phosphate Buffered Saline (PBS) (catalogue #: 21-031-CV [Corning, Union City, CA, USA])
Trypsin-EDTA (catalogue #: 25-053-CI [Corning])
Nuclease-free water (catalogue #: AM9932 [Life Technologies]).
Mouse monoclonal IgG1 anti-BrdU antibody IIB5 (sc-32323) (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Tris-HCl (catalogue #: T5941 [Sigma-Aldrich])
NaCl (catalogue #: BP358-1 [Fisher Scientific, San Diego, CA, USA])
MgCl2 (catalogue #: M33-500 [Fisher Scientific])
KCl (catalogue #: BO366-500 [Fisher Scientific])
0.5 M EDTA (catalogue #: BDH8730-1 [VWR, Visalia, CA, USA])
NP-40 (catalogue #: P-877 [Boston BioProducts, Ashland, MA, USA])
Glycerol (catalogue #: BP229-1 [Fisher Scientific])
DTT (catalogue #: V3151[Fisher Scientific])
Tween-20 (catalogue #: BP337-100 [Fisher Scientific])
UltraPure BSA (catalogue #: AM2616 [Life Technologies])
Polyvinylpyrrolidone (PVP) (360 kDa) (catalogue #: P5288 [Sigma-Aldrich])
Protein G Dynabeads (catalogue #: 10003D [Life Technologies])
High-Capacity cDNA Reverse Transcription Kit (catalogue #: 4368814 [Life Technologies])
qPCR Mastermix (e.g. KAPA SYBR FAST qPCR Kit catalogue #: KK4600 [Kapa Biosystems, Wilmington, MA, USA])
Oligonucleotide primers (Integrated DNA Technologies, Boulder, Coralville, IA, USA)
psiCHECK-2 (catalogue #: C8021 [Promega, San Luis Obispo, CA, USA])
KAPA 2G HS PCR ReadyMix (catalogue #: KK5601 ReadyMix [Kapa Biosystems])
AmpliScribe T7-Flash Biotin-RNA Transcription Kit (catalogue #: ASB71110 [Epicentre, Madison, WI, USA])
MEGAclear Transcription Clean-Up Kit (catalogue #: AM1908 [Life Technologies])
QIAquick PCR Purification Kit (catalogue #: 28104 [Qiagen, Valencia, CA, USA])
TURBO DNA-free Kit (catalogue #: AM1907 [Life Technologies])
EQUIPMENT
Refrigerated microcentrifuge
Bench top microcentrifuge
Nanodrop spectrophotometer (or equivalent method of nucleic acid quantification)
Rotating platform
Vortex mixer
Magnet for bead separation (e.g. MagneSphere magnetic Separation Stand, catalogue #: Z5332 [Promega])
OPTIONAL: Method for counting cells hemocytometer or automated cell counter (e.g. Countess [Life Technologies])
OPTIONAL: Automated RNA extraction robot (e.g. Maxwell 16 and simplyRNA Cells Kit [Promega])
Real-time PCR instrument
Conventional PCR thermocycler
RNase-free plasticware for RT-qPCR (PCR plates, optical seals, plate sealer)
1.7 ml RNase-free microcentrifuge tubes
0.2 ml PCR strip tubes
REAGENT SETUP
Prepare all solutions using nuclease-free water.
NP-40 Lysis Buffer: 10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40
Nuclei Storage Buffer: 50 mM Tris-HCl pH 8.3, 0.1 mM EDTA, 5 mM MgCl2, 40% glycerol
2X Transcription Buffer: 20 mM Tris-HCl pH 8.3, 5 mM MgCl2, 300 mM KCl, 4 mM DTT
PBST: 0.1% TWEEN-20 in PBS
PBSTR: add 2 μl RNase OUT per 10 ml of PBST (Prepare immediately before use)
Blocking Buffer: 0.1% PVP, 0.1% UltraPure BSA in PBST
PROCEDURE
Harvesting Cell Nuclei ● TIMING 1-2 hours
▲ CRITICAL STEP Ensure that NP-40 Lysis Buffer and PBS are cold (4°C) before commencing protocol. Keep samples on ice throughout harvesting procedure.
-
1)
Culture cells as required for experiments. The optimal quantity of nuclei for NRO is between 1 and 4 million.
-
2)Harvest cells by one of the following methods. For adherent cells either mechanical scraping or cold trypsinsation can be used. We have found that trypsinisation is generally the most convenient method as fewer cells remain attached to the plate than with scraping. Cell scraping can also damage cells and so may be unsuitable in some instances. However, treatment with trypsin may have an effect on cell metabolism 36, in which case scraping would be desirable.
- Adherent cells (scraping)
- Remove cell culture media.
- Wash cells with ice-cold PBS.
- Add 1.5 ml of ice-cold PBS to each cell culture dish and scrape cells using a disposable cell scraper.
- Transfer scraped cells to a 1.5 ml microcentrifuge tube and place on ice. Mix by pipetting to ensure a uniform cell suspension.
- RECOMMENDED STEP: Count cells using a hemocytometer/automate cell counter. Standardise the number of cells between experimental groups and replicates.
- Pellet cells by centrifugation at 400 g for 4 minutes at 4°C.
- Discard supernatant.
- Adherent cells (trypsinisation)
- Remove cell culture media.
- Wash cells with ice-cold PBS.
- Add ice-cold Trypsin-EDTA to cell culture wells/dishes (volume as appropriate).
- Incubate cultures at room temperature for 1-3 minutes.
- Pipette cells up and down to detach cells from the cell culture well/dish.
- Neutralise the trypsin by adding 1 volume of growth medium.
- Pellet cells by centrifugation at 400 g for 4 minutes at 4°C.
- Resuspend the cells in ice-cold PBS.
- RECOMMENDED STEP: Count cells using a hemocytometer/automated cell counter. Standardise the number of cells between experimental groups and replicates.
- Pellet cells by centrifugation at 400 g for 4 minutes at 4°C.
- Discard supernatant.
- Suspension cells
-
xii.RECOMMENDED STEP: Count cells using a hemocytometer/automated cell counter. Standardise the number of cells between experimental groups and replicates.
-
xiii.Pellet cells by centrifugation at 400 g for 4 minutes at 4°C.
-
xiv.Discard supernatant.
-
xv.Resuspend the cells in ice-cold PBS.
-
xvi.Pellet cells by centrifugation at 400 g for 4 minutes at 4°C.
-
xvii.Discard supernatant
-
xii.
-
3)
OPTIONAL: Aliquot 5-10% of the cells into separate tubes for steady-state total mRNA and/or protein analysis.
-
4)
Resuspend pellet in 1 ml NP-40 Lysis Buffer and incubate on ice for 5 minutes.
-
5)
Pellet nuclei by centrifugation at 300 g for 4 minutes at 4°C.
-
6)
Discard supernatant.
-
7)
Resuspend nuclear pellet in a further 1 ml of NP-40 Lysis Buffer.
-
8)
Immediately pellet nuclei by centrifugation at 300 g for 4 minutes at 4°C.
-
9)
Discard supernatant.
-
10)
Resuspend the nuclear pellet in 40 μl of Nuclei Storage Buffer. Mix by pipetting or gentle vortexing and store on ice.
-
11)
Proceed immediately to the next step.
Nuclear Run-On Transcription ● TIMING 45 minutes
-
12)Prepare the following Transcription Buffer reaction cocktail for each sample:
final 1X (μl) 2X Transcription Buffer 1X 50 RNase OUT (40 U/μl) 100 U 2 BrUTP (100 mM) 0.5 mM 1 ATP (100 mM) 1 mM 2 GTP (100 mM) 1 mM 2 CTP (100 mM) 1 mM 2 UTP (100 mM) 0.5 mM 1 -
13)
Add 60 μl Transcription Buffer reaction buffer cocktail to each nuclei suspension sample.
-
14)
OPTIONAL: Add 10 μl of spike-in control oligonucleotide(s) (1×107 copies/μl stock solution) to each sample (described below).
-
15)
Ensure nuclei are fully resuspended by gentle pipetting.
-
16)
Incubate at 30°C for 30 minutes.
-
17)
Stop the nuclear run-on reaction by proceeding immediately to the next step.
Extraction of Nuclear RNA ● TIMING 1.5 hours
-
18)Extract nuclear RNA by one of the following methods:
- Automated RNA extraction (e.g. Maxwell 16 system)
- Assemble Maxwell 16 simplyRNA LEV cartridge in the cartridge rack by adding 5 μl of DNase to well #4 of the cartridge and a plunger to well #8.
- Add 200 μl of chilled Homogenization Solution (containing 1-Thioglycerol) and 200 μl Lysis Buffer to the NRO sample and mix by vortexting for 15 seconds.
- Transfer the mixture into well #1 of the LEV cartridge.
- Place a 0.6 ml collection tube in the cartridge rack for each sample. Add 50 μl of nuclease-free water to each tube and leave the tubes uncapped.
- Place the cartridge rack in the Maxwell 16 Instrument and initiate the simplyRNA LEV program.
- Column-based RNA extraction (MEGAclear Transcription Clean-Up Kit)
- Add 350 μl of Binding Solution Concentrate to the sample and mix by pipetting.
- Add 250 μl of 100% Ethanol to the sample and mix by pipetting.
- Assemble a filter cartridge in a collection tube.
- Add RNA mixture to membrane.
- Centrifuge columns for 30 seconds at 10,000 g.
- Discard flow-through and re-assemble cartridge.
- Wash each column twice with 500 μl Wash Solution.
- After second wash spin the column for an additional 30s to remove residual liquid.
- Elute RNA by adding 50 μl of nuclease-free water (pre-heated to 95 °C) to the filter and spin for 1 minute at 10,000 g.
- Remove genomic DNA contamination using the TURBO DNA-free Kit. Add 6 μl of 10X TURBO Buffer, 2 μl TURBO DNase and 2 μl nuclease-free water to each sample.
- Incubate samples at 37 °C for 30 minutes.
- Add 3 μl of DNase Inactivation Reagent.
- Incubate samples at room temperature (24 °C) for 5 minutes with regular vortexing.
- Centrifuge samples for 5 minutes at 10,000 g.
- Carefully transfer the supernatant to a fresh microcentrifuge tube.
-
19)
Quantify nuclear RNA by Nanodrop or equivalent method.
■ PAUSE POINT Nuclear RNA samples can be stored indefinitely at −70°C.
Immunoprecipitation of Bromouridylated NRO-RNAs ● TIMING 3 hours
-
20)
Resuspend Protein G Dynabeads by repetitive pipetting.
-
21)
Transfer enough beads (30 μl per run-on sample) to a fresh 1.7 ml microcentrifuge tube.
-
22)
Collect the beads by placing the tube on the magnet and incubate for 3 minutes at room temperature.
-
23)
Discard the bead storage buffer.
-
24)
Wash the beads twice with PBST. To perform the wash step resuspend the beads in 0.5 ml PBST and then place the tubes on the magnet. Incubate for three minutes and then discard the supernatant.
-
25)
Resuspend the beads in PBST (volume = 30 μl × number of samples) and aliquot 30 μl of beads into separate tubes for each NRO reaction.
-
26)
Add 2 μg of anti-BrdU monoclonal antibody to each tube and incubate on a rotating platform for 10 minutes at room temperature.
-
27)
Add 150 μl of Blocking Buffer to each tube and incubate for 30 minutes at room temperature.
-
28)
Capture the beads using the magnet and discard buffer.
-
29)
Wash the beads twice with 300 μl of PBSTR.
-
30)
Resuspend the beads for each reaction in 100 μl PBSTR.
-
31)
Incubate NRO-RNA samples at 65°C for 5 minutes in a thermal cycler in order to denature any RNA secondary structure which may reduce the efficiency of antibody binding.
-
32)
Add an equal volume of NRO-RNA to each tube. The amount of RNA added should be within the range of 0.5 – 8 μg per immunoprecipitation.
-
33)
Incubate the beads and NRO-RNA samples for 30 minutes at room temperature on a rotating platform.
-
34)
Capture the beads by placing the tubes on the magnet for 3 minutes. (Beads may require a brief pulse spin to collect liquid at the bottom of the tube before magnetic separation).
-
35)
Discard the buffer.
-
36)
Wash the beads three times with PBSTR.
-
37)
After discarding the final wash proceed directly to RNA extraction.
Extraction of NRO RNAs ● TIMING 2.5 hours
-
38)
Add 0.5 ml (1 volume) of TRIzol reagent directly to the beads and mix by pipetting.
■ PAUSE POINT NRO-RNA samples in TRIzol can be stored indefinitely at −70°C.
-
39)
Incubate the samples for 5 minutes at room temperature.
-
40)
Add 100 μl (0.2X volume) of chloroform to each sample.
-
41)
Vortex for 15 seconds and then incubate the samples for 10 minutes at room temperature.
-
42)
Centrifuge the samples at 13,000 g for 10 minutes at 4°C.
-
43)
The mixture separates into two phases. The top clear layer is the aqueous phase which contains the RNA. Carefully aspirate this layer and transfer to a clean microcentrifuge tube. Take care not to touch the phenolic (red) phase.
-
44)
Precipitate the nucleic acids by adding 250 μl (0.5X volume) of Isopropanol and 20 μg of RNase-free glycogen (1 μl of 20 mg/ml stock) to each sample.
▲ CRITICAL STEP The amount of labelled RNA that is precipitated at this stage is very small and so the addition of glycogen (which acts as an inert carrier for nucleic acids) maximises yield and ensures that a pellet is visible after isopropanol precipitation.
-
45)
Vortex each sample for 10 seconds and then incubate for 10 minutes at room temperature.
▲ CRITICAL STEP If the samples are not adequately mixed the RNA will not precipitate and the yield will be very low.
-
46)
Centrifuge the samples at 13,000 g for 10 minutes at 4°C.
-
47)
The nucleic acids form a white pellet. Carefully remove and discard the supernatant.
-
48)
Wash the pellet with 1 ml (2X volumes) of 70% ethanol.
-
49)
Centrifuge the samples at 5,500 g for 5 minutes at 4°C.
-
50)
Carefully remove and discard the wash.
-
51)
Pulse spin the tubes to collect any residual wash solution. Carefully remove and discard the supernatant.
-
52)
Un-cap all the tubes and incubate for 10 minutes at room temperature to dry the pellets.
-
53)
Resuspend each precipitated NRO-RNA pellet in 30 μl of nuclease-free water. (Note: At this stage the RNA concentration will be too low to measure by Nanodrop. We have also found that the RNA concentration is typically below the lower limit of detection of the Qubit fluorometer. As a result sample concentrations are expected to be <0.4 ng/μl and total RNA yields <12 ng).
■ PAUSE POINT Precipitated NRO-RNA samples can be stored indefinitely at −70°C.
Reverse Transcription ● TIMING 1 hour
-
54)Prepare the following reaction cocktail in 0.2 ml PCR tubes.
1X (μl) 10X RT Buffer 2 10X Random Primers 2 dNTP mix (100 mM) 0.8 MultiScribe RT 1 Nuclease-free water 4.2 Precipitated NRO-RNA 10 -
55)
Prepare the following reaction cocktail in 0.2 ml PCR tubes.
-
56)
Mix by gentle pipetting and pulse spin to collect liquid at the bottom of the tubes.
-
57)Incubate sample tubes in a PCR thermal cycler as follows:
Time Temperature (°C) 10 25 30 37 5 85 ∞ 4
■ PAUSE POINT cDNA samples can be stored indefinitely at −20°C.
Quantitative Polymerase Chain Reaction ● TIMING 3 hours
! CAUTION Prevent PCR amplicon carryover during preparation of qPCR by using clean gloves, barrier pipette tips, assign dedicated pre- and post-PCR areas and avoid opening plates after run completion.
-
58)Prepare the following qPCR cocktail for each sample. Ensure that at least 10% excess cocktail is prepared to account for losses during pipetting.
1X (μl) 2X qPCR SYBR Mastermix 10 Forward primer (10 μM) 0.5 Reverse primer (10 μM) 0.5 cDNA from RT reaction 2 Nuclease-free water 7 -
59)
Pipette qPCR reaction cocktails onto the qPCR plate.
-
60)
Seal the plate using an optical seal.
-
61)
Spin the plate at 1,000 g for 1 minute to collect the liquid at the bottom of the plate.
-
62)Place the sealed plate in the real-time instrument and perform the following cycling profile. (Reaction conditions may vary for qPCR mastermixes from different manufacturers).
Time Temperature (°C) 3 minutes 95 then 40 cycles of: 3 seconds 95 30 seconds 60* *Set the software to take a fluorescence measurement during this stage.
■ PAUSE POINT Data analysis can be performed at any time following acquisition of Cq data following completion of the qPCR run.
Generation of Spike-in Control Oligonucleotides (Optional) ● TIMING 6 hours
Spike-in oligonucleotides can be used to control for changes in nuclear RNA content and for determination of signal:nosie ratios in experimental samples. The following steps describe how to generate synthetic transcripts with non-mammalian sequences by in vitro transcription using a T7 polymerase capable of incorporating Bromouridine into its RNA products.
-
63)Templates for each spike-in oligonucleotide are generated in two separate PCR reactions. Prepare T7 transcription templates in two separate PCR amplifications using the following reaction cocktail:
1X (μl) 2X KAPA 2G ReadyMix 10 Template psiCHECK-2 (1 ng/μl) 1 Forward Primer (10 μM) 1 Reverse Primer (10 μM) 1 Water 7
And the following PCR primer sets:
| RLuc T7-FWD | ATGCATGCTAATACGACTCACTATAGGGATGGCTTCCAAGGTGTACGA |
| RLuc T7-REV | TTACTGCTCGTTCTTCAGCAC |
| FLuc T7-FWD | ATGCATGCTAATACGACTCACTATAGGGATGGCCGATGCTAAGAACAT |
| FLuc T7-REV | TTACACGGCGATCTTGCC |
-
64)Perform the PCR reaction to amplify the templates by incubating the reactions in a thermal cycler according to the following cycling protocol:
Time Temperature (°C) 2 minutes 95 then 30 cycles of: 15 seconds 95 15 seconds 55 90 seconds 72 Then final extension: 3 minutes 72 -
65)
Successful PCR amplification can be confirmed by separating the reaction products on a 1% agarose gel stained with Ethidium Bromide and observing a single amplicon products.
-
66)
Clean-up the PCR products using a QIAquick PCR Purification Kit as according to manufacturer's instructions and quantify DNA by Nanodrop (or equivalent method).
-
67)Prepare the following T7 reaction cocktails:
1X (μl) AmpliScribe T7-Flash 10X Reaction Buffer 2 100 mM DTT 2 RiboGuard RNase Inhibitor 0.5 T7 transcription template DNA (500 ng/μl) 1 AmpliScribe T7-Flash Enzyme Solution 2 ATP 2 CTP 2 GTP 2 UTP 1 / 2* BrUTP 1 / 0* Nuclease-free water 4.5 *Adjust according to whether BrU-labelled IVT products are required or not. -
68)
Incubate the reactions in a thermal cycler at 37 °C for 3 hours.
-
69)
Add 1 μl of RNase-free DNase I to each reaction and incubate at 37 °C for 30 minutes.
-
70)
Clean-up T7 transcription products using the MEGAclear Transcription Clean-Up Kit as according to manufacturer's instructions and quantify IVT-RNA by Namedrop (or equivalent).
-
71)
Success of the T7 reaction can be confirmed by observing a single product on a 4% Urea denaturing PAGE gel stained with Ethidium Bromide.
-
72)
Spike-in oligonucleotides should be diluted to 1×107 and divided into single-use 100 μl aliquots for storage at −80 °C.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting Advice
| Step | Problem | Solution |
|---|---|---|
| 1-11 | Nuclei form aggregates | Ensure nuclei are prepared at 4° C. Ensure NP-40 lysis buffer is prepared correctly. |
| Insufficient lysis | Increase NP-40 concentration in lysis buffer. Consider dounce homogenization to harvest nuclei. | |
| No nuclear pellet visible | Increase number of cells used. Pellets may be difficult to see when using less than 1 million cells. | |
| Nuclear pellets are different sizes between experimental groups | Count cells and standardise input cell numbers between samples before performing lysis. | |
| 18-19 | Low nuclear RNA yield | Expected yield is ~1-2 μg/million nuclei. RNA content may vary between nuclei derived from different cell lines and under different experimental groups. Increase the number of input nuclei in order to increase RNA yield. |
| Significant difference in nuclear RNA content between experimental groups | Utilise an external spike control to normalise NRO-RT-qPCR data between experimental groups with differing nuclear RNA content. | |
| 20-37 | High background binding | We have found that background binding is negligible using this protocol. Positive and negative spike-in controls can be used to monitor the signa:noise ratio in experimental samples. Background can also be measured in IgG immunoprecipitated samples and then subtracted from the specific NRO signal. |
| 38-53 | Low NRO-RNA yield | At this stage RNA concentration is expected to be below the lower limit of detection for methods like Nanodrop. Reducing elution volume can increase signal. |
| 54-72 | Low/no amplification | May be a real result (low transcriptional activity) if other primer assays amplify successfully. Check that qPCR primer designs appropriate. Consider parallel steady-state total mRNA analysis to confirm expression of the gene of interest. |
| Amplification in RT- control | Indicates gDNA contamination. Ensure gDNA removal by rigorous DNase I treatment. | |
| Amplification in no template control (NTC) | Indicates reagent contamination. Switch to new reagents and clean work surfaces. | |
| Atypical melt profile (if applicable) | Exclude wells. Melting artefacts may occurs at low copy numbers or be caused by sub-optimal assay design. Switch to a probe-based detection technology or re-design assay. | |
| High variability in reference normaliser gene | Utilise multiple reference genes and normalise to the geometric mean. Alternatively, use an external spike-in control to normalise data when no appropriate reference gene can be identified. | |
● TIMING
Steps 1-11, Harvesting Cell Nuclei: 1-2 hours
Steps 12-17, Nuclear Run-On Transcription: 45 minutes
Steps 18-19, Extraction of Nuclear RNA: 1.5 hours
Steps 20-37 Immunoprecipitation of Bromouridylated NRO-RNAs: 3 hours
Steps 38-53, Extraction of NRO RNAs: 2.5 hours
Steps 54-57, Reverse Transcription: 1 hour
Steps 58-62 Quantitative Polymerase Chain Reaction: 3 hours
Steps 63-72 Generation of Spike-in Control Oligonucleotides (Optional): 6 hours
ANTICIPATED RESULTS
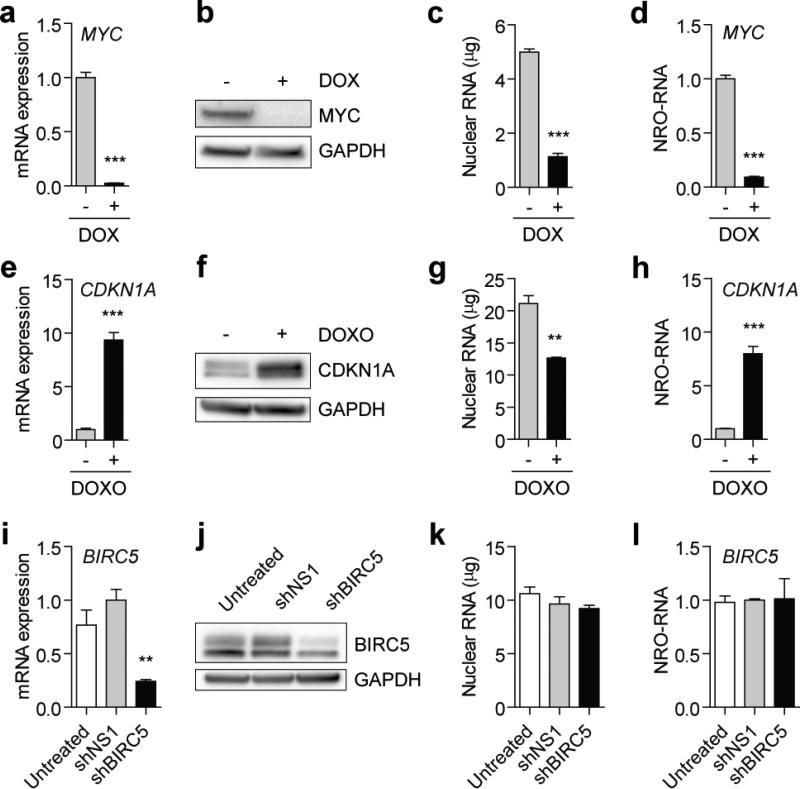
To validate the protocol in an experimental setting we performed NRO analysis on three simple model systems where the transcriptional activity of specific genes was known: (i) P493-6 cells have been engineered such that transcription from the MYC promoter can be silenced with the addition of doxycycline (DOX) 37. Treatment with DOX for 24 hours reduced steady-state MYC mRNA (Fig. 5a) and protein (Fig. 5b) expression. NRO was performed on cells cultured in parallel. Total nuclear RNA content was reduced in the DOX-treated cultures (Fig. 5c) consistent with findings reported by Lin and co-workers 38. In line with steady-state total mRNA observations, nascent transcription was significantly down-regulated following treatment with DOX (Fig. 5d). (ii) To examine a positive change in transcription HCT116 colorectal carcinoma cells were treated with Doxorubicin (DOXO). DOXO treatment is known to induce the DNA damage response which leads to TP53 localising to the CDKN1A (p21) promoter and activating its transcription 39,40. Treatment with DOXO for 24 hours increased CDKN1A mRNA (Fig. 5e) and protein (Fig. 5f) expression by ~8 fold. NRO was performed on cells cultured in parallel. Total nuclear RNA was reduced by ~40% in DOXO-treated cultures (Fig. 5g). NRO analysis demonstrated a statistically significant increase in CDKN1A nascent transcription (Fig. 5h). (iii) In the final example we targeted BIRC5 (Survivin) by RNA interference (RNAi) using a short hairpin RNA (shRNA). RNAi is an endogenous mechanism of post-transcriptional silencing whereby target mRNAs are selectively degraded while transcription is unaffected. Transfection of HE293T cells with an shRNA targeting BIRC5 3′ untranslated region (shBIRC5) resulted in significant down-regulation of steady-state mRNA (Fig. 5i) and protein (Fig. 5j) expression relative to a non-specific shRNA (shNS1). NRO analysis performed on cells cultured in parallel showed no significant change in total nuclear RNA (Fig. 5k) or nascent BIRC5 transcription (Fig. 5l).
Figure 5. Experimental validation of NRO-RT-qPCR protocol.
The NRO-RT-qPCR protocol was validated using three models of gene regulation. Firstly, P493-6 cells were treated with 0.1 μg/ml doxycycline (DOX) for 24 hours and MYC expression determined by (a) RT-qPCR and (b) western blot. P493-6 nuclei were harvested and (c) nuclear RNA content determined by Nanodrop spectrophotometry. (d) MYC expression was down-regulated as determined by NRO-RT-qPCR. In the second model, HCT116 cells were treated with 1 μM doxorubicin (DOXO) for 24 hours and CDKN1A expression determined by (e) RT-qPCR and (f) western blot. HCT116 nuclei were harvested and (g) nuclear RNA content determined by Nanodrop. (h) CDKN1A was up-regulated as determined by NRO-RT-qPCR. In the final model, HEK293T cells were transfected with plasmids encoding shRNAs targeting BIRC5 (shBIRC5) or a non-targeting control (shNS1) and BIRC5 expression determined by (i) RT-qPCR and (j) western blot 72 hours post transfection. (k) HEK293T nuclei were harvested and (k) nuclear RNA content was quantified by Nanodrop from nuclear extracts. (l) No change in BIRC5 expression was measured by NRO-RT-qPCR. All values are mean + SEM, **P<0.01, ***P<0.0001. All qPCR data were analysed using the ΔΔCq method and internally normalised to GAPDH expression.
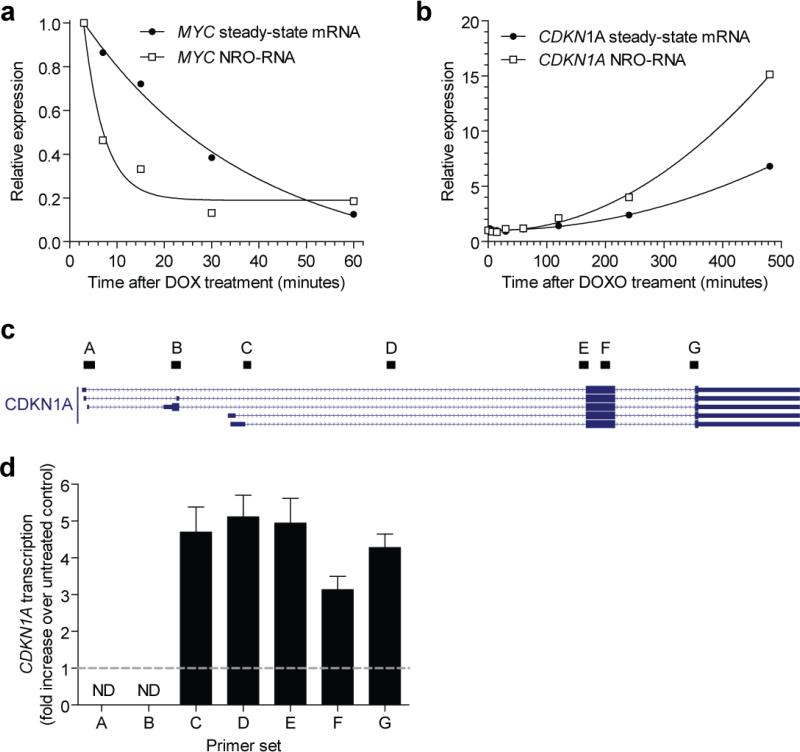
An advantage of the NRO method is that it can be used to detect changes in transcriptional activity that occur on the order of minutes to hours 22,23. In these cases, a change in transcriptional activity may be apparent before the cellular pool of total mRNA has had time to adapt. Consequently, NRO has the power to detect rapid and/or transient changes in transcriptional activity that are inaccessible to steady-state total mRNA analyses. To demonstrate the potential of the present NRO-RT-qPCR protocol to investigate these phenomena we repressed MYC expression in P493-6 cells (Fig. 6a) and induced CDKN1A expression in HCT116 cells (Fig. 6b) as described above. Cells were harvested at a series of time points after repression/induction and transcriptional activity analysed by NRO-RT-qPCR and steady-state total mRNA levels by conventional RT-qPCR respectively. In both model systems a change in transcription was observed prior to a change in steady-state mRNA expression (Fig. 6a,b). Repression of MYC transcription by DOX happened rapidly (within minutes). In contrast, induction of CDKN1A was much slower (on the order of hours) as DOXO acts as an indirect inducer.
Figure 6. Detection of short-term changes in transcriptional activity by NRO-RT-qPCR.
(a) P493-6 cells were treated with 0.1 μg/ml doxycycline (DOX) and harvested after 3, 7, 15, 30 and 60 minutes. NRO-RT-qPCR and steady-state total mRNA analyses were performed for MYC transcripts at each time point. (b) HCT116 cells were treated with 1 μM doxorubicin (DOXO) and harvested after 3, 7, 15, 30, 60, 120, 240 and 480 minutes. NRO-RT-qPCR and steady-state total mRNA analyses were performed for CDKN1A transcripts at each point. In both cases, changes in transcriptional activity precede changes in steady-state transcript levels. (c) Seven qPCR primer sets (labelled A-G) were designed targeting different parts of the CDKN1A locus. (d) HCT116 cells were treated with DOXO for 4 hours and CDKN1A transcriptional activity analysed by NRO-RT-qPCR using primer sets A-G. The dotted grey line indicates the value of the untreated control. No transcription was detected at the alternatively spliced exons 1 and 2. Approximately equal transcriptional activation was observed across the remainder of the CDKN1A gene. All values are mean + SEM. All qPCR data were analysed using the ΔΔCq method and internally normalised to GAPDH expression. ND, not detected.
NRO-RT-qPCR analysis can also be used to distinguish between changes in transcription initiation and elongation (i.e. the release of paused polymerases). To demonstrate this HCT116 cultures treated with DOXO for four hours, such that transcriptional activation had not reached a maximum value. NRO was performed with a limiting concentration of CTP (10 μM, 100 fold less than the other nucleotides) in order to restrict the length of NRO-RNA transcripts. Seven qPCR assays were designed to amplify different regions of the CDKN1A NRO-RNA. (Fig. 6c). After NRO, no specific amplification was observed at the first two exons (primer sets A and B) indicating that it is primarily the short isoforms of CDKN1A that are expressed in HCT116 cells (Fig. 6d). Furthermore, the level of transcriptional activation was comparable across the body of the gene (primer sets C to G) which is indicative of an increase in transcriptional initiation as opposed to transcriptional elongation. (An increase in elongation is typified by an increase in signal in the body of the gene relative to the 5′ promoter-proximal part of the gene). Together these data demonstrate that the NRO protocol described here is capable of detecting rapid or transient changes in the promoter transcriptional activity of specific genes, and can distinguish between transcriptional and post-transcriptional gene regulatory events.
Supplementary Material
ACKNOWLEDGEMENTS
T.C.R. is supported by a Medical Research Council UK Centenary Early Career Award. This is TSRI manuscript #29005.
Footnotes
AUTHOR CONTRIBUTIONS
Experimental work was performed by T.C.R. and J.R.H. All authors contributed to the protocol and analysed data. T.C.R. wrote the initial draft and all authors contributed to the final draft.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Greenberg ME, Bender TP. Identification of newly transcribed RNA. Curr Protoc Mol Biol. 2007 doi: 10.1002/0471142727.mb0410s78. Chapter 4, Unit 4.10. [DOI] [PubMed] [Google Scholar]
- 2.Folta KM, Kaufman LS. Isolation of Arabidopsis nuclei and measurement of gene transcription rates using nuclear run-on assays. Nat. Protocols. 2006;1:3094–3100. doi: 10.1038/nprot.2006.471. [DOI] [PubMed] [Google Scholar]
- 3.Khraiwesh B. Using nuclear run-on transcription assays in RNAi studies. Methods Mol. Biol. 2011;744:199–209. doi: 10.1007/978-1-61779-123-9_14. [DOI] [PubMed] [Google Scholar]
- 4.Smale ST. Nuclear Run-On Assay. 2009. Cold Spring Harb Protoc. 2009:pdb.prot5329. doi: 10.1101/pdb.prot5329. [DOI] [PubMed] [Google Scholar]
- 5.Ohtsu M, et al. Novel DNA microarray system for analysis of nascent mRNAs. DNA Res. 2008;15:241–251. doi: 10.1093/dnares/dsn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner A-MW, De La Cruz J, Morris KV. Mobilization-competent Lentiviral Vector-mediated Sustained Transcriptional Modulation of HIV-1 Expression. Mol. Ther. 2009;17:360–368. doi: 10.1038/mt.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patrone G, et al. Nuclear run-on assay using biotin labeling, magnetic bead capture and analysis by fluorescence-based RT-PCR. BioTechniques. 2000;29:1012–1014. 1016–1017. doi: 10.2144/00295st02. [DOI] [PubMed] [Google Scholar]
- 8.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elferink CJ, Reiners JJ., Jr. Quantitative RT-PCR on CYP1A1 heterogeneous nuclear RNA: a surrogate for the in vitro transcription run-on assay. BioTechniques. 1996;20:470–477. doi: 10.2144/19962003470. [DOI] [PubMed] [Google Scholar]
- 10.Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 11.Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi Factors Are Present and Active in Human Cell Nuclei. Cell Rep. 2014;6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts TC. The MicroRNA Biology of the Mammalian Nucleus. Mol Ther Nucleic Acids. 2014;3:e188. doi: 10.1038/mtna.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tani H, et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dölken L, et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008;14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulsen MT, et al. Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA. Methods. 2014;67:45–54. doi: 10.1016/j.ymeth.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulsen MT, et al. Coordinated regulation of synthesis and stability of RNA during the acute TNF-induced proinflammatory response. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2240–2245. doi: 10.1073/pnas.1219192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imamachi N, et al. BRIC-seq: A genome-wide approach for determining RNA stability in mammalian cells. Methods. 2014;67:55–63. doi: 10.1016/j.ymeth.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Min IM, et al. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danko CG, et al. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol. Cell. 2013;50:212–222. doi: 10.1016/j.molcel.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hah N, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hah N, et al. Inflammation-sensitive super enhancers form domains of coordinately regulated enhancer RNAs. Proc Natl Acad Sci U S A. 2015;112:E297–E302. doi: 10.1073/pnas.1424028112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts TC, Andaloussi SE, Morris KV, McClorey G, Wood MJ. Small RNA-Mediated Epigenetic Myostatin Silencing. Mol Ther Nucleic Acids. 2012;1:e23. doi: 10.1038/mtna.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris KV, Chan SW-L, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–92. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 28.Janowski BA, Corey DR. Inhibiting transcription of chromosomal DNA using antigene RNAs. Nucleic Acids Symp Ser (Oxf) 2005;367–368 doi: 10.1093/nass/49.1.367. doi:10.1093/nass/49.1.367. [DOI] [PubMed] [Google Scholar]
- 29.Roberts TC, Morris KV, Weinberg MS. Perspectives on the mechanism of transcriptional regulation by long non-coding RNAs. Epigenetics. 2013;9 doi: 10.4161/epi.26700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hart JR, Roberts TC, Weinberg MS, Morris KV, Vogt PK. MYC regulates the non-coding transcriptome. Oncotarget. 2015;5:12543–12554. doi: 10.18632/oncotarget.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3:research0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 33.Taylor S, Wakem M, Dijkman G, Alsarraj M, Nguyen M. A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods. 2010;50:S1–5. doi: 10.1016/j.ymeth.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Pajic A, et al. Cell cycle activation by c-myc in a burkitt lymphoma model cell line. Int. J. Cancer. 2000;87:787–793. doi: 10.1002/1097-0215(20000915)87:6<787::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Bensaude O. Inhibiting eukaryotic transcription. Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Batista U, et al. Effects of different detachment procedures on viability, nitroxide reduction kinetics and plasma membrane heterogeneity of V-79 cells. Cell Biol. Int. 2010;34:663–668. doi: 10.1042/CBI20090276. [DOI] [PubMed] [Google Scholar]
- 37.Schuhmacher M, et al. Control of cell growth by c-Myc in the absence of cell division. Current Biology. 1999;9:1255–1258. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- 38.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang JJ, Shen C, Lu YJ. Requirement for pre-existing of p21 to prevent doxorubicin-induced apoptosis through inhibition of caspase-3 activation. Mol. Cell. Biochem. 2006;291:139–144. doi: 10.1007/s11010-006-9206-7. [DOI] [PubMed] [Google Scholar]
- 40.Gomes NP, et al. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.