Abstract
Human papillomavirus (HPV), particularly type 16 (HPV-16), is present in more than 99% of cervical cancers. The HPV oncoproteins E6 and E7 are constantly expressed and therefore represent ideal targets for HPV vaccine development. We previously developed DNA vaccines encoding calreticulin (CRT) linked to HPV-16 E7 and generated potent E7-specific CD8+ T-cell immune responses and antitumor effects against an E7-expressing tumor. Since vaccines targeting E6 also represent an important strategy for controlling HPV-associated lesions, we developed a DNA vaccine encoding CRT linked to E6 (CRT/E6). Our results indicated that the CRT/E6 DNA vaccine, but not a wild-type E6 DNA vaccine, generated significant E6-specific CD8+ T-cell immune responses in vaccinated mice. Mapping of the immunodominant epitope of E6 revealed that an E6 peptide comprising amino acids (aa) 48 to 57 (E6 aa48-57), presented by H-2Kb, is the optimal peptide and that the region of E6 comprising aa 50 to 57 represents the minimal core sequence required for activating E6-specific CD8+ T lymphocytes. We also demonstrated that E6 aa48-57 contains cytotoxic T-lymphocyte epitopes naturally presented by E6-expressing TC-1 cells. Vaccination with a CRT/E6 but not a CRT/mtE6 (lacking aa 50 to 57 of E6) DNA vaccine could protect vaccinated mice from challenge with E6-expressing TC-1 tumors. Thus, our data indicate that E6 aa48-57 contains the immunodominant epitope and that a CRT/E6 DNA vaccine may be useful for control of HPV infection and HPV-associated lesions.
Cervical cancer is the second leading cause of cancer death among women worldwide, and more than 99% of cervical cancers contain human papillomavirus (HPV), particularly the high-risk HPV type 16 (HPV-16) (26). Two HPV oncoproteins, E6 and E7, are consistently expressed in HPV-associated cancer cells and are responsible for their malignant transformation. These oncogenic proteins therefore represent ideal target antigens for developing vaccines and immunotherapeutic strategies against HPV-associated neoplasms. Numerous preclinical studies and some clinical studies have targeted the HPV oncogenic proteins E6 and E7 for the development of vaccines to control HPV-associated lesions (for a review, see reference 16).
DNA vaccines have become an appealing approach to generating antigen-specific immunotherapy because of their simplicity, stability, safety, and capacity for repeated administration (for reviews, see references 7, 17, 19, 22, and 24). Intradermal administration of DNA vaccines by means of a gene gun represents an efficient means of targeting dendritic cells, the most potent professional antigen-presenting cells, which are specialized to prime helper and killer T cells in vivo (5, 21). Using intradermally administered DNA vaccines, we have tested several intracellular targeting strategies to modify the properties of dendritic cells in order to enhance antigen presentation through the major histocompatibility complex (MHC) class I and class II pathways (for a review, see reference 11). Kim et al. have recently performed a head-to-head comparison of DNA vaccines encoding various fusion proteins and have determined that calreticulin (CRT) is the most effective protein for enhancing antigen-specific CD8+ T-cell immune responses (12).
CRT, an abundant 46-kDa Ca2+-binding protein located in the endoplasmic reticulum (18), is considered a relative of the family of heat shock proteins (1, 6). CRT has been shown to aid in antigen presentation by associating with peptides delivered to the endoplasmic reticulum by transporters associated with antigen processing (TAP-1 and TAP-2) (25) and by associating with MHC class I β2-microglobulin molecules (23). Previous studies have shown that CRT can be complexed with peptides in vitro to elicit peptide-specific CD8+ T-cell responses through exogenous administration (1). In addition, peptide-bound CRT purified from tumor extracts has been shown to elicit an antitumor effect specific to the source tumor (1). It was recently demonstrated that a DNA vaccine encoding CRT linked to the model antigen HPV-16 E7 generated potent E7-specific CD8+ T-cell responses and antitumor effects as a result of enhanced MHC class I presentation of the linked antigen (4). Thus, DNA vaccines encoding the HPV-16 E6 antigen linked to CRT may also elicit enhanced E6-specific CD8+ T-cell immune responses and antitumor effects in vaccinated mice.
In the past, most HPV researchers focused on E7, and therefore an E7 immunodominant epitope and the associated immune responses have been well characterized (8). Since E6 represents another important target for potential vaccines to control HPV-associated lesions, it is crucial to develop vaccines targeting E6. Therefore, we developed a DNA vaccine encoding CRT linked to E6. We found that the linkage of CRT to HPV-16 E6 can generate a significantly enhanced E6-specific CD8+ T-cell response in vaccinated mice. In addition, we have demonstrated that mice vaccinated with DNA encoding chimeric CRT/E6 are able to control E6-expressing tumors and that the immunodominant epitope of E6 is located within amino acids (aa) 48 to 57. These findings show that DNA vaccines encoding E6 have potential for future clinical application in the control of HPV.
MATERIALS AND METHODS
Mice.
C57BL/6 mice (6 to 8 weeks old) were purchased from the National Cancer Institute (Frederick, Md.). All animals were maintained under specific-pathogen-free conditions at the Johns Hopkins Hospital (Baltimore, Md.), and all procedures were performed according to approved protocols and in accordance with recommendations for the proper care of laboratory animals.
Peptides.
A panel of 10 peptides from the HPV-16 E6 protein (E6 aa1-45, E6 aa36-80, E6 aa71-115, E6 aa106-138, E6 aa134-158, E6 aa48-56, E6 aa48-57, E6 aa49-57, E6 aa50-57, and E6 aa51-58) and an H2-Kb-restricted ovalbumin-specific cytotoxic T-lymphocyte (CTL) peptide, SIINFEKL, were synthesized by Macromolecular Resources (Denver, Colo.) at a purity of ≥70%.
Cells.
Briefly, TC-1 cells were obtained by cotransformation of primary C57BL/6 mouse lung epithelial cells with HPV-16 E6 and E7 and an activated ras oncogene as described previously (15). The expression of E6 in TC-1 cells has also been characterized previously by He et al. (10). The lymphoma cell line EL-4 (ATCC TIB-39) was obtained from the American Type Culture Collection (Manassas, Va.). The C1R cell line is an Epstein-Barr virus-transformed B-cell line that has lost most of its HLA class I alleles, expressing only Cw0401 and trace amounts of B3503 (28). The C1R murine MHC class I transfectants C1R/Db and C1R/Kb were kindly provided by Michael Edidin (Johns Hopkins University, Baltimore, Md.). The T2-Kb cell line, a murine MHC class I transfectant of T2 (174 × CEM.T2) cells that are deficient in TAPs, was kindly provided by Jonathan Schneck (Johns Hopkins University). Cells were maintained in RPMI medium supplemented with 2 mM glutamine, 1 mM sodium pyruvate, 20 mM HEPES, 5 × 10−5 M β-mercaptoethanol, 100 IU of penicillin/ml, 100 μg of streptomycin/ml, and 10% fetal bovine serum.
Plasmid DNA constructs and preparation.
For the generation of pcDNA3-E6, HPV-16 E6 DNA was amplified by PCR using the primer set GGCCGAATTCATGCACCAAAAGAGAACTG and GGCCGGATCCCAGCTGGGTTTCTCTACGT with pCMVneoE6 as the template. pCMVneoE6 is a plasmid containing the E6 gene and was kindly provided by Kathleen Cho (University of Michigan, Ann Arbor). The amplified E6 DNA was then cloned into the EcoRI/BamHI site of the pcDNA3 vector (Invitrogen, Carlsbad, Calif.). The generation of pcDNA3-CRT has been described previously (4). For the generation of pcDNA3-CRT/E6, the E6 DNA was isolated from pcDNA3-E6 and further cloned into the EcoRI/BamHI sites of pcDNA3-CRT. To generate pcDNA3-CRT/mtE6, we first used PCR to generate mutant E6 with nucleotides 148 to 171 deleted (resulting in deletion of aa 50 to 57). The DNA fragment containing nucleotides 1 to 147 of E6 was amplified by using the primer pair 5′-CCCGAATTCATGCACCAAAAGAGAACTGCA-3′ and 5′-CCCATCTCTATATACTATGCATACCTCACGTCGCAGTAACTG-3′ with pcDNA3-E6 as the template. The DNA fragment containing nucleotides 172 to 474 of E6 was amplified by using the primer pair 5′-CAGTTACTGCGACGTGAGGTATGCATAGTATATAGAGATGGG-3′ and 5′-GGGAAGCTTTTACAGCTGGGTTTCTCTACG-3′ with pcDNA3-E6 as the template. The mtE6 DNA fragment with nucleotides 148 to 171 of E6 deleted was generated by using E6 DNA fragments 1-147 and 172-474 as templates and the primer pair 5′-CCCGAATTCATGCACCAAAAGAGAACTGCA-3′ and 5′-GGGAAGCTTTTACAGCTGGGTTTCTCTACG-3′. The final PCR product was cut with EcoRI and HindIII and cloned into pcDNA3-CRT to generate pcDNA3-CRT/mtE6. All plasmid constructs were confirmed by DNA sequencing. Expression of encoded gene products in cells transfected with the DNA plasmid constructs was confirmed by Western blot analysis (data not shown). DNA for vaccination was prepared by using an endotoxin-free kit (QIAGEN, Valencia, Calif.).
DNA vaccination.
DNA-coated gold particles were prepared, and gene gun particle-mediated DNA vaccination was performed, according to a protocol described previously (3). Gold particles coated with pcDNA3-E6, pcDNA3-CRT, pcDNA3-CRT/E6, or pcDNA3-CRT/mtE6 DNA were delivered to the shaved abdominal regions of mice by using a helium-driven gene gun (Bio-Rad Laboratories Inc., Hercules, Calif.) with a discharge pressure of 400 lb/in2. Mice were immunized with 2 μg of the DNA vaccine and received two boosts with the same regimen 1 and 2 weeks later. Splenocytes were harvested 1 week after the last vaccination.
Intracellular cytokine staining and flow cytometry analysis.
Before intracellular cytokine staining, 3.5 × 105 pooled splenocytes from each vaccination group were incubated overnight with 1 μg of the E6 peptide/ml. GolgiPlug (BD Pharmingen, San Diego, Calif.) was added to the cell culture at a ratio of 1 μl/ml based on the vendor's manual. Cells were subjected to intracellular cytokine staining by using the Cytofix/Cytoperm kit according to the manufacturer's instructions (BD Pharmingen). Intracellular gamma interferon (IFN-γ) was stained with fluorescein isothiocyanate-conjugated rat anti-mouse IFN-γ. Intracellular CD8 was stained with phycoerythrin-conjugated monoclonal rat anti-mouse CD8a (clone 53.6.7). All antibodies were purchased from BD Pharmingen. After staining, flow cytometry analysis was performed using FACScalibur with CellQuest software (BD Biosciences, Mountain View, Calif.).
For characterization of the optimal E6 peptide for activating E6-specific T-cell lines, T2-Kb, C1R, C1R/Db, and C1R/Kb cells were first pulsed with E6 or a control peptide at a concentration of 10−6 M at 37°C for 2 h. Peptide-pulsed cells were then washed extensively. A total of 104 peptide-pulsed cells together with 105 E6-specific T cells were seeded into each well of 96-well round-bottom plates in the presence of GolgiPlug for 5 h. Cells were then washed once with FACScan buffer. Intracellular staining for IFN-γ and CD8 was performed as described above.
CTL assay.
Mice were immunized as described above. One week after the last boost, splenocytes from immunized mice were harvested and stimulated in vitro with the HPV-16 E6 peptide aa48-57 in the presence of interleukin-2 (IL-2; 20 U/ml) for 5 days. The lytic activity of T cells was assessed by a standard chromium release assay similar to that described previously (27). TC-1 and EL-4 cells were labeled with 100 μCi of 51Cr per 106 cells at 37°C for 90 min. Labeled EL-4 cells pulsed with E6 aa48-57 were used as a positive control. After extensive washing, the labeled target cells were incubated with CTLs at various effector-to-target cell (E:T) ratios in triplicate in 96-well U-bottom plates. One hundred microliters of supernatants was harvested after a 4-h incubation, and 51Cr radioactivity was quantitated by using a gamma counter. Results are expressed as percent cytotoxicity ± standard error of the mean and were calculated by the following formula: percent specific lysis = [(experimental release value − spontaneous release value)/(maximum release value − spontaneous release value)] × 100, where the spontaneous release value is the counts in supernatants from wells containing target cells in medium only and the maximum release value is the counts in supernatants from wells containing target cells in medium supplemented with 10% sodium dodecyl sulfate.
In vitro generation of HPV-16 E6-specific CTL lines.
Splenocytes from pcDNA3-CRT/E6-immunized C57BL/6 mice were harvested 1 week after the last vaccination. These splenocytes were stimulated with irradiated TC-1 tumor cells in the presence of murine IL-2 (20 U/ml). The cells were restimulated every week for 8 weeks. The specificity of the CTL lines was characterized by staining for CD8 and by determining the ability of the T cells to kill HPV-16 E6 (aa 48 to 57) peptide-pulsed EL-4 cells.
In vivo tumor protection experiment.
For the tumor protection experiment, C57BL/6 mice (five per group) were immunized via gene gun with 2 μg of pcDNA3-E6, pcDNA3-CRT, pcDNA3-CRT/E6, or pcDNA3-CRT/mtE6. Mice were boosted twice with the same regimen as that used in the first vaccination. One week after the last vaccination, mice were each challenged with 5 × 104 TC-1 tumor cells subcutaneously in the right leg; then they were monitored twice a week by inspection and palpation.
In vivo antibody depletion experiment.
In vivo antibody depletions have been described previously (15). Briefly, C57BL/6 mice (five per group) were vaccinated with 2 μg of pcDNA3-CRT/E6 via gene gun and were boosted twice with the same regimen, 1 week and 2 weeks later. The mice were challenged subcutaneously with 5 × 104 TC-1 tumor cells/mouse 1 week after the last vaccination. Depletions were started 1 week before tumor challenge. Monoclonal antibody (MAb) GK1.5 was used for CD4 depletion, MAb 2.43 was used for CD8 depletion, and MAb PK136 was used for NK1.1 depletion as described previously (15). Depletion was ended on day 35 after tumor challenge. Lin et al. have previously characterized the efficacy of depletion of the antibodies used for the in vivo antibody depletion experiment, confirming more than 90% depletion of NK cells and more than 95% depletion of CD8+ and CD4+ T cells (14).
Statistical analysis.
All data expressed as means ± standard deviations (SD) are representative of at least two different experiments. Comparisons between individual data points were made by the Student t test. For statistical analysis of the tumor protection and antibody depletion experiments, we used Kaplan-Meier analysis.
RESULTS
DNA encoding CRT linked to E6 significantly enhances the E6-specific CD8+ T-cell immune response in vaccinated mice.
It has been shown previously that vaccination with DNA encoding HPV-16 E7 linked to CRT facilitated antigen presentation through the MHC class I pathway and significantly enhanced E7-specific CD8+ T-cell immune responses in vaccinated mice (4). We therefore hypothesized that DNA vaccines encoding E6 linked to CRT could also generate potent E6-specific CD8+ T-cell immune responses. pcDNA3-E6 and pcDNA3-CRT/E6 were generated and compared for their abilities to generate an E6-specific CD8+ T-cell response in vaccinated mice.
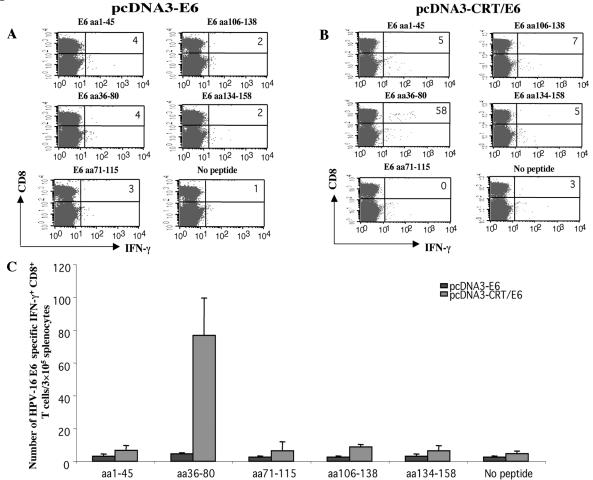
To determine if CRT/E6 or E6 DNA could generate E6-specific T-cell-mediated immune responses in C57BL/6 mice, we performed intracellular cytokine staining with flow cytometry analysis to characterize E6-specific CD8+ T-cell precursors by using overlapping E6 peptides as stimulants. We harvested splenocytes from mice vaccinated with pcDNA3-E6 or pcDNA3-CRT/E6 and stimulated them overnight with each of the overlapping E6 peptides (aa1-45, aa36-80, aa71-115, aa106-138, or aa134-158). As shown in Fig. 1, mice vaccinated with E6 DNA did not generate numbers of E6-specific CD8+ T cells above background levels when stimulated with any of the E6 peptides. In comparison, mice vaccinated with CRT/E6 DNA generated a significantly greater number of E6-specific IFN-γ-expressing CD8+ T-cell precursors (76.67 ± 22.7 per 3.5 × 105 splenocytes) than mice vaccinated with E6 DNA alone (4.50 ± 0.71 per 3.5 × 105 splenocytes) (P < 0.01) when stimulated with the E6 aa36-80 peptide but not when stimulated with any of the other E6 peptides. Thus, our data show that linkage of CRT to E6 in a DNA vaccine can generate E6-specific CD8+ T-cell responses in vaccinated mice. Our data also suggest that this E6-specific CD8+ T-cell response is specific for a peptide within aa 36 to 80 of E6.
FIG. 1.
Intracellular cytokine staining with flow cytometry analysis to determine the proportion of IFN-γ-expressing, E6-specific CD8+ T-cell precursors. Mice were immunized, and splenocytes were collected and cultured, as described in Materials and Methods. Pooled splenocytes from vaccinated mice (5 mice/group) were cultured in vitro with various overlapping E6 peptides overnight and were stained for both CD8 and intracellular IFN-γ. (A) Representative flow cytometry data for splenocytes harvested from mice vaccinated with pcDNA3-E6 and either left unstimulated or stimulated overnight with the E6 peptide aa1-45, aa36-80, aa71-115, aa106-138, or aa134-158. (B) Representative flow cytometry data for splenocytes harvested from mice vaccinated with pcDNA3-CRT/E6 and stimulated overnight with one of the E6 peptides listed above. (C) Bar graph depicting the number of E6-specific IFN-γ-expressing CD8+ T-cell precursors/3 × 105 splenocytes (mean ± SD) from mice vaccinated with pcDNA3-E6 or pcDNA3-CRT/E6. Data presented in this figure are from one experiment representative of two performed. Note that the E6 peptide aa36-80 was the only peptide to activate E6-specific CD8+ T cells.
The E6 peptide aa48-57 likely contains the E6 CTL immunodominant epitope.
Using the BioInformatics & Molecular Analysis Section (BIMAS) for HLA peptide binding predictions (20) and the SYFPEITHI database of MHC ligands and peptide motifs (http://syfpeithi.bmi-heidelberg.com/), we analyzed various peptides of 8, 9, or 10 residues between E6 aa 36 and 80 and determined their sequences, positions, and scores. As shown in Table 1, this analysis indicated that an 8-mer positioned from aa 50 to 57 (YDFAFRDL) and a 10-mer positioned from aa 48 to 57 (EVYDFAFRDL) were the candidates for H-2b-restricted CTL epitopes with the highest calculated MHC class I binding affinity in HPV-16 E6 (with restriction element Kb). We therefore synthesized these two peptides for further characterization of the E6-specific CD8+ T-cell immune response by intracellular cytokine staining with flow cytometry analysis.
TABLE 1.
Candidate H-2b-restricted CTL epitopes in HPV-16 E6 aa 36 to 80a
| Restriction element | Peptide length (aa) | BIMAS
|
SYFPEITHI
|
||||
|---|---|---|---|---|---|---|---|
| Peptide sequence | Position (aa) | Affinity score | Peptide sequence | Position (aa) | Affinity score | ||
| Db | 8 | NA | NA | ||||
| 9 | CVYCKQQLL | 37-45 | 2.7 | YRDGNPYAV | 61-69 | 16 | |
| 10 | EVYDFAFRDL | 48-57 | 0.3 | ECVYCKQLL | 36-45 | 13 | |
| Kb | 8 | YDFAFRDL | 50-57 | 20 | YDFAFRDL | 50-57 | 21 |
| 9 | KCLKFYSKI | 72-80 | 7.2 | NA | |||
| 10 | EVYDFAFRDL | 48-57 | 60 | NA | |||
Boldfaced peptides were selected for further testing. The sequence of the HPV-16 E6 protein (with aa 36 to 80 boldfaced) is as follows: NH2-MHQKRTAMFQDPQERPRKLPQLCTELQTTIHDIILECVYCKQQLLRREVYDFAFRDLCIVYRDGNPYAVCDKCLKFYSKISEYRHYCYSLYGTTLEQQYNKPLCDLLIRCINCQKPLCPEEKQRHLDKKQRFHNIRGRWTGRCMSCCRSSRTRRETQLZ-COOH. NA, not applicable.
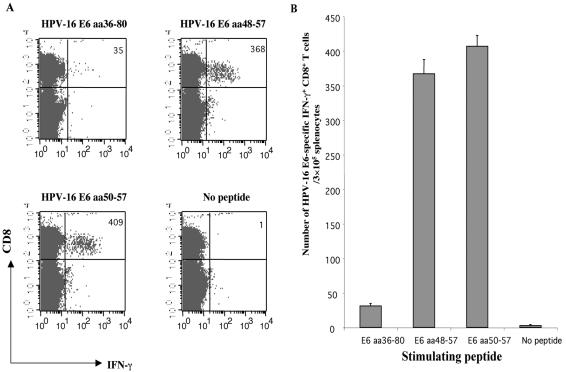
We either stimulated splenocytes from pcDNA3-CRT/E6-vaccinated mice with E6 peptide aa36-80, aa48-57, or aa50-57, or left them unstimulated. As shown in Fig. 2, splenocytes stimulated with the E6 aa48-57 peptide or the E6 aa50-57 peptide generated significantly greater numbers of E6-specific CTLs (367 ± 20.521 or 406.67 ± 15.63 per 3 × 105 splenocytes, respectively) than splenocytes stimulated with the E6 aa36-80 peptide (31 ± 3.61 per 3 × 105 splenocytes) (P < 0.001) or unstimulated splenocytes (2.67 ± 1.53 per 3 × 105 splenocytes) (P < 0.001). These data indicate that both the E6 peptides aa48-57 and aa50-57 can be recognized by CD8+ T cells generated by pcDNA3-CRT/E6-vaccinated mice, and they suggest that the immunodominant epitope for HPV-16 E6 may be located within the region between aa 48 and 57.
FIG. 2.
Intracellular cytokine staining with flow cytometry analysis of IFN-γ-expressing, E6-specific CD8+ T-cell precursors generated by splenocytes stimulated with various E6 peptides. Mice were immunized with pcDNA3-CRT/E6, and splenocytes were collected and cultured. (A) Representative flow cytometry data for splenocytes harvested from mice and either left unstimulated or stimulated overnight with the E6 peptide aa36-80, aa48-57, or aa50-57. (B) Bar graph showing the number of E6-specific IFN-γ-expressing CD8+ T-cell precursors per 3 × 105 splenocytes (mean ± SD) generated by in vitro stimulation. Data presented in this figure are from one experiment representative of two performed.
The MHC class I immunodominant epitope of E6 (aa 48 to 57) is H-2Kb restricted.
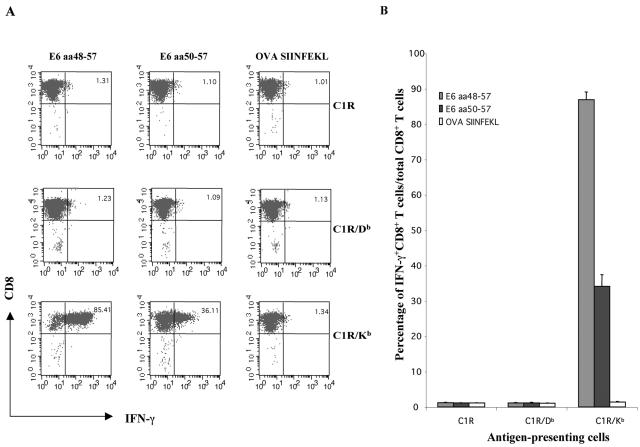
We sought to determine which H-2b MHC class I molecule presents the E6 immunodominant epitope (aa 48 to 57) to an E6-specific CD8+ T-cell line. We created an E6-specific CD8+ T-cell line by using splenocytes from pcDNA3-CRT/E6-immunized mice, stimulating them with irradiated TC-1 tumor cells in the presence of murine IL-2 (20 U/ml) for 8 weeks. We pulsed C1R, C1R/Db, and C1R/Kb stimulator cells with the E6 peptide aa48-57 or aa50-57 or with the ovalbumin control peptide SIINFEKL and coincubated them with the E6-specific CTL cell line. The C1R cell line is an Epstein-Barr virus-transformed B-cell line that has lost most of its HLA class I alleles, expressing only Cw0401 and trace amounts of B3503 (28). As shown in Fig. 3, only C1R/Kb cells can effectively present the aa48-57 and aa50-57 epitopes to the E6-specific CD8+ T-cell line; C1R and C1R/Db cells were unable to elicit appreciable responses in the E6-specific CTLs. In contrast, the C1R/Kb cell line pulsed with the ovalbumin control peptide failed to activate E6-specific CD8+ T cells. These data indicate that HPV-16 E6 peptides aa48-57 and aa50-57 are restricted by H-2Kb. Furthermore, 86.93% ± 2.15% of the total E6-specific CD8+ T cells expressed IFN-γ when coincubated with C1R/Kb cells pulsed with E6 aa48-57, but only 34.07% ± 3.35% expressed IFN-γ when coincubated with C1R/Kb cells pulsed with E6 aa50-57. These results suggest that the E6 aa48-57 epitope is more readily presented by H2-Kb molecules to activate an E6-specific CD8+ T-cell line than the E6 aa50-57 epitope.
FIG. 3.
Intracellular cytokine staining and flow cytometry analysis to determine the MHC class I H-2b binding restriction of the E6 aa50-57 epitope. An E6-specific CD8+ T-cell line was used to determine the MHC class I binding restriction. Two E6 peptides (aa48-57 and aa50-57) were used. A peptide from ovalbumin (OVA) was used as a negative control. (A) Representative flow cytometry data showing percentages of IFN-γ-expressing E6-specific CD8+ T cells in cultures of E6-specific CD8+ T cells coincubated with peptide-pulsed C1R, C1R/Db, or C1R/Kb cells. (B) Bar graph showing percentages of IFN-γ+ CD8+ T cells per total CD8+ T cells in cultures described above. Note that the data show that the E6 aa48-57 and aa50-57 epitopes are Kb restricted.
The 10-mer HPV-16 E6 peptide aa48-57 is the optimal peptide, and the 8-mer E6 peptide aa50-57 is the minimal epitope, recognized by an E6-specific CD8+ T-cell line.
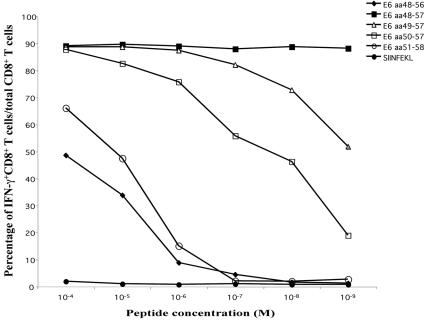
We have determined that the immunodominant epitope of E6 is located within aa 48 to 57. To determine the length and position of the immunodominant epitope recognized by an E6-specific CD8+ T-cell line, we pulsed T2-Kb cells with the originally identified 10-mer E6 peptide aa48-57, the 8-mer E6 peptide aa50-57, or other peptides generated by the addition or deletion of single amino acids on the 5′ and 3′ ends of the aa48-57 or aa50-57 sequence. The T2-Kb cell line is a murine MHC class I transfectant of T2 (174 × CEM.T2) cells deficient in TAP. The peptide-pulsed T2-Kb cells were then coincubated with an E6-specific CTL line at an E:T ratio of 10:1. Figure 4 represents the change in the percentage of IFN-γ-expressing E6-specific CD8+ T cells among the total number of E6-specific CD8+ T cells at different concentrations of E6 peptides. The data indicate that the E6 peptide aa50-57 is the minimal sequence necessary for activating E6-specific CD8+ T cells. The peptides lacking a residue on either end of the aa50-57 peptide (aa51-58 and aa49-56) failed to activate the CTL line at peptide concentrations lower than 10−6 M (Fig. 4). In contrast, the 10-mer E6 peptide aa48-57 elicited the secretion of IFN-γ in close to 90% of total E6-specific CD8+ T cells even at a very low (10−9 M) concentration. These results suggest that E6 aa50-57 is the essential core sequence and E6 aa48-57 is the optimal epitope of HPV-16 E6 for activating E6-specific CD8+ T cells.
FIG. 4.
Determination of the percentage of activated E6-specific CD8+ T cells within the total number of E6-specific CD8+ T cells at decreasing peptide concentrations. An E6-specific CD8+ T-cell line was used to determine the optimal and essential components of the E6 immunodominant epitope. T2-Kb cells were pulsed with various E6 peptides (aa48-56, aa48-57, aa49-57, aa50-57, or aa51-58) or a negative-control peptide from ovalbumin (SIINFEKL) at sequentially decreasing concentrations from 10−4 to 10−9 M. The peptide-pulsed T2-Kb cells were then coincubated with an HPV-16 E6-specific CTL line at an E:T ratio of 1:10. Intracellular cytokine staining followed by flow cytometry analysis was used to determine the percentage of IFN-γ-expressing E6-specific CD8+ T cells within the total E6-specific CD8+ T-cell population.
DNA encoding CRT linked to mtE6 (aa 50 to 57 deleted) fails to generate E6-specific CD8+ T cells in vaccinated mice, confirming E6 aa48-57 to be the immunodominant epitope.
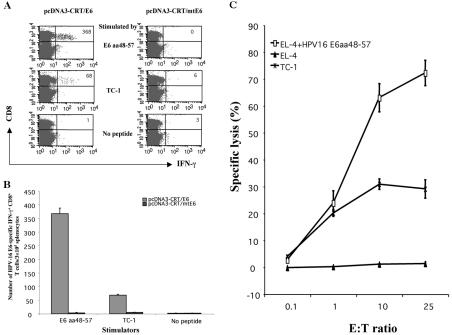
In order to confirm that the E6 sequence characterized (E6 aa48-57) contains the immunodominant epitope for HPV-16 E6, we created a DNA vaccine encoding CRT linked to mutant E6 (mtE6) DNA lacking the essential (aa 50 to 57) core (pcDNA3-CRT/mtE6). We harvested splenocytes from pcDNA3-CRT/E6- or pcDNA3-CRT/mtE6-vaccinated mice, stimulated them overnight with the E6 aa48-57 peptide or with E6-expressing TC-1 cells, and analyzed the number of IFN-γ-expressing CD8+ T cells present. As shown in Fig. 5A and B, the E6 aa48-57 peptide elicited the generation of a significantly larger number of IFN-γ-expressing CD8+ T cells in mice vaccinated with pcDNA3-CRT/E6 (367 ± 20.52 per 3 × 105 splenocytes) than in mice vaccinated with the mtE6 DNA (3.33 ± 1.53 per 3 × 105 splenocytes) (P < 0.001). More importantly, TC-1 cells are able to activate significantly more CD8+ T cells from splenocytes harvested from pcDNA3-CRT/E6-vaccinated mice (68.33 ± 3.51 per 3 × 105 splenocytes) than from pcDNA3-CRT/mtE6-vaccinated mice (5 ± 1 per 3 × 105 splenocytes) (P < 0.001) (Fig. 5A and B). Furthermore, E6-specific CTLs from mice vaccinated with pcDNA3-CRT/E6 can specifically lyse tumor cells that express HPV-16 E6 endogenously (Fig. 5C). When overlapping E6 peptides were used to stimulate splenocytes from pcDNA3-CRT/mtE6-immunized mice, we failed to observe an E6 aa36-80-specific CD8+ T-cell response. In addition, we did not detect any E6-specific CD8+ T-cell response against other E6 regions, even when stimulating with aa48-57 or with a previously published HPV-16 E6 H-2Kb epitope (E6 aa137-144) (data not shown). Taken together, our data confirm that E6 aa48-57 contains the immunodominant epitope of E6 and that the E6 epitope containing the essential component of the immunodominant epitope (aa50-57) may be processed and presented by TC-1 cells from endogenously expressed HPV-16 E6 protein.
FIG. 5.
Intracellular cytokine staining with flow cytometry analysis and chromium release assay to demonstrate that TC-1 cells naturally process the E6 epitope containing aa 50 to 57. (A) Mice were immunized with pcDNA3-CRT/E6 or pcDNA3-CRT/mtE6. Flow cytometry data indicate the number of IFN-γ-expressing CD8+ T-cell precursors generated from splenocytes harvested from vaccinated mice and pulsed with either E6 aa48-57 or E6-expressing TC-1 cells. (B) Bar graph shows the number of E6-specific IFN-γ+ CD8+ T cells/3 × 105 splenocytes (mean ± SD). (C) The lytic activity of T cells was assessed by using a standard chromium release assay. TC-1 and EL-4 cells were used as target cells. EL-4 cells pulsed with the E6 peptide aa48-57 were used as a positive control. Splenocytes stimulated in vitro with E6 peptide served as effector cells. Percent cytotoxicity (specific lysis) was calculated. Data were collected from cultures measured at E:T ratios of 1:10, 1:1, 10:1, and 25:1. Results are expressed as percent cytotoxicity ± SD.
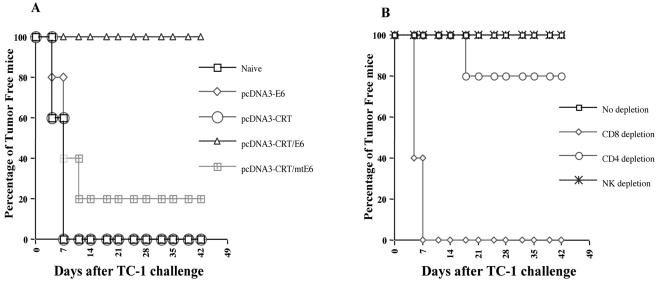
Vaccination with pcDNA3-CRT/E6 provides protection against challenge with an E6-expressing tumor cell line, TC-1.
We performed tumor protection experiments to test whether vaccination with pcDNA3-CRT/E6 can provide protection against tumor challenge with an E6-expressing tumor cell line, TC-1. As shown in Fig. 6A, 100% of mice vaccinated with pcDNA3-CRT/E6 remained tumor free 6 weeks after TC-1 challenge. In contrast, only 20% of mice vaccinated with pcDNA3-CRT/mtE6 remained tumor free after 6 weeks. Furthermore, all naïve mice and all mice vaccinated with pcDNA3-CRT or pcDNA3-E6 exhibited tumor growth within 1 week of tumor challenge. These data suggest that pcDNA3-CRT/E6 can generate significant protective effects against E6-expressing tumors and that the aa48-57 epitope of E6 is necessary for eliciting potent tumor protection.
FIG. 6.
In vivo tumor protection experiment to demonstrate the antitumor effect generated by pcDNA3-CRT/E6 against E6-expressing TC-1 tumors and the effects of lymphocyte subsets on tumor protection. (A) Mice were immunized with pcDNA3-CRT, pcDNA3-E6, pcDNA3-CRT/E6, or pcDNA3-CRT/mtE6. One week after vaccination, mice were challenged subcutaneously with 5 × 104 TC-1 cells/mouse; they were then monitored for evidence of tumor growth by palpation and inspection twice a week. (B) In vivo antibody depletion experiments to determine the effects of lymphocyte subsets on the tumor protection of the CRT/E6 DNA vaccine. CD4, CD8, and NK1.1 depletions were initiated 1 week before tumor challenge. Data shown in this figure are from one experiment representative of two performed.
CD8+ T cells are essential for the protective antitumor effect generated by pcDNA3-CRT/E6 vaccination.
To determine the subsets of lymphocytes that are important for protection against E6-expressing tumor cells, we performed in vivo antibody depletion experiments. As shown in Fig. 6B, all of the mice depleted of CD8+ T cells grew tumors within 1 week of tumor cell challenge. In contrast, all of the nondepleted and NK1.1-depleted mice, and 80% of CD4+ T-cell-depleted mice, remained tumor free for 6 weeks after tumor challenge. These results indicate that CD8+ T cells are essential for the protective antitumor immunity generated by the CRT/E6 DNA vaccine.
DISCUSSION
In this study, we investigated the feasibility of the HPV-16 oncoprotein E6 as a target for vaccine development to control HPV-associated neoplasms. We demonstrated that the linkage of CRT to E6 elicits an E6-specific CD8+ T-cell immune response in C57BL/6 mice. Further characterization of this immune response determined that the MHC class I CTL immunodominant epitope for E6 is located within aa 48 to 57 of E6 and is H2-Kb restricted. Furthermore, vaccination with DNA encoding CRT linked to wild-type E6, but not to mtE6 (aa 50 to 57 deleted), can protect mice against challenge with an E6-expressing tumor cell line. This suggests that the aa50-57 sequence of E6 is important for antitumor effects and that the epitope containing aa 50 to 57 is likely presented on the surface of the TC-1 tumor.
Thus, in the present study we have pioneered the characterization of an E6 immunodominant epitope that is responsible for a protective antitumor effect mediated by an effective E6-encoding DNA vaccine. Our results indicate that the E6-expressing TC-1 cell line processes and presents the epitope containing aa 50 to 57. This was confirmed by a tumor protection experiment, showing that mice vaccinated with pcDNA3-CRT/E6, but not pcDNA3-CRT/mtE6, exhibit 100% protection against the growth of E6-expressing tumor cells. While we cannot exclude the contribution of E6 subdominant epitopes to the observed 20% protective antitumor effect of vaccination with CRT/mtE6, a second set of experiments found that vaccination with CRT/mtE6 DNA resulted in 0% protection of mice against TC-1 challenge (data not shown). Thus, the contribution of subdominant epitopes to the antitumor effect is likely trivial.
We found that CRT can successfully elicit an E6-specific CD8+ T-cell response when linked to HPV-16 E6. Previous studies found that the linkage of CRT to HPV-16 E7 (4) and to the nucleocapsid protein of the severe acute respiratory syndrome coronavirus (13) in the context of a DNA vaccine results in enhancement of antigen-specific immune responses. The ability of CRT to enhance immune responses when linked to three different antigens in DNA vaccines suggests that CRT may be broadly applicable as a strategy to enhance DNA vaccines encoding a variety of antigens. This possibility will be explored further in future DNA vaccination studies, in which CRT will be linked to other viral or tumor-specific antigens.
Our study has shown that the aa48-57 region contains the H2-Kb-restricted immunodominant CTL epitope of E6. Previous studies have taken different approaches to identifying E6 CTL epitopes. Gao et al. used recombinant vaccinia virus to deduce H2-Kb-restricted E6 epitopes and found that an epitope from aa 131 to 140 exhibited some immunogenicity but bound only weakly to Kb (9). Bauer et al. demonstrated the ability of the E6 aa50-57 epitope to bind and stabilize Kb but concluded that the aa50-57 epitope was not able to prime mice in vivo (2). In comparison, our data indicate that mice vaccinated with CRT/E6 DNA are able to generate E6 peptide (aa48-57)-specific CD8+ T cells capable of killing E6-expressing tumors. In contrast, mice vaccinated with mtE6 DNA lacking the aa50-57 region cannot generate CTL activity against an E6-expressing tumor, suggesting that this region is essential for E6-specific CTL activity and indicating that aa 48 to 57 contain the immunodominant epitope for HPV-16 E6.
Identification of the H2-Kb-restricted E6 immunodominant epitope is important because its further characterization will allow for the development of E6-specific T-cell immunological assays that may aid in the future development of HPV vaccines incorporating E6 as a target antigen. Researchers have been able to study different vectors and vaccine enhancement strategies for HPV-16 E7 vaccines because thorough characterization of the E7-specific immune response and identification of the E7 CTL immunodominant epitope have led to the development of basic immunological assays specific for E7-specific CD8+ T cells. Similarly, the characterization of the immunodominant epitope for E6 (aa48-57) may facilitate further studies of E6 vaccines by providing immunological assays such as peptide-specific ELISPOT, intracellular cytokine staining, or tetramer assays that are specific for E6. Such assays are critical for HPV vaccine development, because reliable assays will be necessary to determine the efficacy of future E6 vaccines. Furthermore, this identified epitope will be potentially useful for the design of other E6 vaccine strategies, such as peptide vaccines or DNA vaccines. Thus, the present study represents an important step toward the development of quantitative assays to characterize E6-specific immune responses and will provide a foundation for future development of vaccines targeting the HPV E6 protein.
Since E6 and E7 are both important for the malignant transformation of HPV-associated neoplasia and are coexpressed in a majority of HPV-associated tumors, both antigens represent ideal targets for HPV vaccine development. We have found that linkage of CRT to either antigen in a DNA vaccine is capable of enhancing antigen-specific immune responses and antitumor effects in vivo. Because TC-1 tumor cells express both E6 and E7, protection against TC-1 challenge provides a means for comparing the relative efficacies of vaccines encoding E6 and/or E7. This study and prior studies have shown that both CRT/E6 and CRT/E7 DNA vaccines are capable of providing 100% protection to C57BL/6 mice against challenge with identical doses of TC-1 tumor cells (4). Therefore, to distinguish between the protective antitumor effects of the CRT/E7 and CRT/E6 DNA vaccines, it will be necessary to increase the dose of TC-1 cells, reduce the vaccine dosages, or compare the therapeutic efficacies of the two vaccines. It will be of interest in future studies to develop DNA vaccines encoding both E6 and E7 linked to CRT and compare their efficacies with those of CRT/E6 and CRT/E7 DNA vaccines in vivo.
In summary, the observation that DNA vaccines encoding CRT/E6 can significantly enhance E6-specific CD8+ T-cell responses and elicit protection against E6-expressing tumors carries with it important clinical implications. The majority of cervical carcinomas are associated with HPV-16, and E6 and E7 are the principal oncoproteins responsible for the malignant transformation of HPV-containing cancer cells. We have shown that DNA vaccines encoding E6 can generate strong E6-specific CD8+ T-cell immunity and can control the growth of E6-expressing tumor cells. Therefore, E6 vaccines, and perhaps E6 and E7 vaccines in combination, may represent an important approach to controlling HPV-associated cancers.
Acknowledgments
We thank Richard Roden and Ken-Yu Lin for helpful discussions. We also thank Michael Edidin, Jonathan Schneck, and Kathleen Cho for generously providing us with reagents necessary for our studies.
This work was supported by a NIAID grant (1PO1AI48203-01).
REFERENCES
- 1.Basu, S., and P. K. Srivastava. 1999. Calreticulin, a peptide-binding chaperone of the endoplasmic reticulum, elicits tumor- and peptide-specific immunity. J. Exp. Med. 189:797-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, S., K. Heeg, H. Wagner, and G. B. Lipford. 1995. Identification of H-2Kb binding and immunogenic peptides from human papilloma virus tumour antigens E6 and E7. Scand. J. Immunol. 42:317-323. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C. H., T. L. Wang, C. F. Hung, Y. Yang, R. A. Young, D. M. Pardoll, and T. C. Wu. 2000. Enhancement of DNA vaccine potency by linkage of antigen gene to an HSP70 gene. Cancer Res. 60:1035-1042. [PubMed] [Google Scholar]
- 4.Cheng, W. F., C. F. Hung, C. Y. Chai, K. F. Hsu, L. He, M. Ling, and T. C. Wu. 2001. Tumor-specific immunity and antiangiogenesis generated by a DNA vaccine encoding calreticulin linked to a tumor antigen. J. Clin. Investig. 108:669-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condon, C., S. C. Watkins, C. M. Celluzzi, K. Thompson, and L. D. Falo, Jr. 1996. DNA-based immunization by in vivo transfection of dendritic cells. Nat. Med. 2:1122-1128. [DOI] [PubMed] [Google Scholar]
- 6.Conway, E. M., L. Liu, B. Nowakowski, M. Steiner-Mosonyi, S. P. Ribeiro, and M. Michalak. 1995. Heat shock-sensitive expression of calreticulin. In vitro and in vivo up-regulation. J. Biol. Chem. 270:17011-17016. [DOI] [PubMed] [Google Scholar]
- 7.Donnelly, J. J., J. B. Ulmer, J. W. Shiver, and M. A. Liu. 1997. DNA vaccines. Annu. Rev. Immunol. 15:617-648. [DOI] [PubMed] [Google Scholar]
- 8.Feltkamp, M. C., H. L. Smits, M. P. Vierboom, R. P. Minnaar, B. M. de Jongh, J. W. Drijfhout, J. ter Schegget, C. J. Melief, and W. M. Kast. 1993. Vaccination with cytotoxic T lymphocyte epitope-containing peptide protects against a tumor induced by human papillomavirus type 16-transformed cells. Eur. J. Immunol. 23:2242-2249. [DOI] [PubMed] [Google Scholar]
- 9.Gao, L., J. Walter, P. Travers, H. Stauss, and B. M. Chain. 1995. Tumor-associated E6 protein of human papillomavirus type 16 contains an unusual H-2Kb-restricted cytotoxic T cell epitope. J. Immunol. 155:5519-5526. [PubMed] [Google Scholar]
- 10.He, Z., A. P. Wlazlo, D. W. Kowalczyk, J. Cheng, Z. Q. Xiang, W. Giles-Davis, and H. C. Ertl. 2000. Viral recombinant vaccines to the E6 and E7 antigens of HPV-16. Virology 270:146-161. [DOI] [PubMed] [Google Scholar]
- 11.Hung, C. F., and T. C. Wu. 2003. Improving DNA vaccine potency via modification of professional antigen presenting cells. Curr. Opin. Mol. Ther. 5:20-24. [PubMed] [Google Scholar]
- 12.Kim, J. W., C. F. Hung, J. Juang, L. He, T. W. Kim, D. K. Armstrong, S. I. Pai, P. J. Chen, C. T. Lin, and T. C. Wu. 2004. Comparison of HPV DNA vaccines employing intracellular targeting strategies. Gene Ther. 11:1011-1018. [DOI] [PubMed] [Google Scholar]
- 13.Kim, T. W., J. H. Lee, C. F. Hung, S. Peng, R. Roden, M. C. Wang, R. Viscidi, Y. C. Tsai, L. He, P. J. Chen, D. A. K. Boyd, and T. C. Wu. 2004. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 78:4638-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, C. T., C. F. Hung, J. Juang, L. He, K. Y. Lin, T. W. Kim, and T. C. Wu. 2003. Boosting with recombinant vaccinia increases HPV-16 E7-specific T cell precursor frequencies and antitumor effects of HPV-16 E7-expressing Sindbis virus replicon particles. Mol. Ther. 8:559-566. [DOI] [PubMed] [Google Scholar]
- 15.Lin, K.-Y., F. G. Guarnieri, K. F. Staveley-O'Carroll, H. I. Levitsky, T. August, D. M. Pardoll, and T.-C. Wu. 1996. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 56:21-26. [PubMed] [Google Scholar]
- 16.Ling, M., M. Kanayama, R. Roden, and T. C. Wu. 2000. Preventive and therapeutic vaccines for human papillomavirus-associated cervical cancers. J. Biomed. Sci. 7:341-356. [DOI] [PubMed] [Google Scholar]
- 17.Moniz, M., M. Ling, C. F. Hung, and T. C. Wu. 2003. HPV DNA vaccines. Front. Biosci. 8:D55-D68. [DOI] [PubMed] [Google Scholar]
- 18.Nash, P. D., M. Opas, and M. Michalak. 1994. Calreticulin: not just another calcium-binding protein. Mol. Cell. Biochem. 135:71-78. [DOI] [PubMed] [Google Scholar]
- 19.Pardoll, D. M., and A. M. Beckerleg. 1995. Exposing the immunology of naked DNA vaccines. Immunity 3:165-169. [DOI] [PubMed] [Google Scholar]
- 20.Parker, K. C., M. A. Bednarek, and J. E. Coligan. 1994. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 152:163-175. [PubMed] [Google Scholar]
- 21.Porgador, A., K. R. Irvine, A. Iwasaki, B. H. Barber, N. P. Restifo, and R. N. Germain. 1998. Predominant role for directly transfected dendritic cells in antigen presentation to CD8+ T cells after gene gun immunization. J. Exp. Med. 188:1075-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson, H. L., and C. A. Torres. 1997. DNA vaccines. Semin. Immunol. 9:271-283. [DOI] [PubMed] [Google Scholar]
- 23.Sadasivan, B., P. J. Lehner, B. Ortmann, T. Spies, and P. Cresswell. 1996. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity 5:103-114. [DOI] [PubMed] [Google Scholar]
- 24.Shedlock, D. J., and D. B. Weiner. 2000. DNA vaccination: antigen presentation and the induction of immunity. J. Leukoc. Biol. 68:793-806. [PubMed] [Google Scholar]
- 25.Spee, P., and J. Neefjes. 1997. TAP-translocated peptides specifically bind proteins in the endoplasmic reticulum, including gp96, protein disulfide isomerase and calreticulin. Eur. J. Immunol. 27:2441-2449. [DOI] [PubMed] [Google Scholar]
- 26.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 27.Wu, T.-C., F. G. Guarnieri, K. F. Staveley-O'Carroll, R. P. Viscidi, H. I. Levitsky, L. Hedrick, K. R. Cho, T. August, and D. M. Pardoll. 1995. Engineering an intracellular pathway for MHC class II presentation of HPV-16 E7. Proc. Natl. Acad. Sci. USA 92:11671-11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zemmour, J., A. M. Little, D. J. Schendel, and P. Parham. 1992. The HLA-A,B “negative” mutant cell line C1R expresses a novel HLA-B35 allele, which also has a point mutation in the translation initiation codon. J. Immunol. 148:1941-1948. [PubMed] [Google Scholar]