Abstract
We recently reported that retroviral pseudotypes bearing the hepatitis C virus (HCV) strain H and Con1 glycoproteins, genotype 1a and 1b, respectively, require CD81 as a coreceptor for virus-cell entry and infection. Soluble truncated E2 cloned from a number of diverse HCV genotypes fail to interact with CD81, suggesting that viruses of diverse origin may utilize different receptors and display altered cell tropism. We have used the pseudotyping system to study the tropism of viruses bearing diverse HCV glycoproteins. Viruses bearing these glycoproteins showed a 150-fold range in infectivity for hepatoma cells and failed to infect lymphoid cells. The level of glycoprotein incorporation into particles varied considerably between strains, generally reflecting the E2 expression level within transfected cells. However, differences in glycoprotein incorporation were not associated with virus infectivity, suggesting that infectivity is not limited by the absolute level of glycoprotein. All HCV pseudotypes failed to infect HepG2 cells and yet infected the same cells after transduction to express human CD81, confirming the critical role of CD81 in HCV infection. Interestingly, these HCV pseudotypes differed in their ability to infect HepG2 cells expressing a panel of CD81 variants, suggesting subtle differences in the interaction of CD81 residues with diverse viral glycoproteins. Our current model of HCV infection suggests that CD81, together with additional unknown liver specific receptor(s), mediate the virus-cell entry process.
Hepatitis C virus (HCV) is an enveloped virus classified in the Hepacivirus genus of the family Flaviviridae (32). An estimated 170 million individuals are infected with HCV worldwide. Infection is associated with the development of chronic hepatitis, cirrhosis, and hepatocellular carcinoma. B-cell abnormalities, including cryoglobulinemia and an increased risk of non-Hodgkins B-cell lymphoma, have also been reported (11, 13, 38). The principal site of virus replication is thought to be the liver; however, several reports suggest that HCV RNA or proteins associate with lymphoid cells, particularly B cells (8, 30, 49), a view consistent with the clinical abnormalities observed in B-lymphocyte growth and function.
HCV encodes two envelope glycoproteins (gp's) E1 and E2, which are believed to be type I integral transmembrane proteins. Our understanding of gp maturation and virus assembly is limited by the lack of a tissue culture system supporting particle assembly and release. In the absence of a cell culture system, surrogate assays have been developed to study HCV-cell attachment, including the expression of truncated version(s) of the E2 gp (19, 43), E1E2 gp-liposomes (28), and virus-like particles expressed in insect cell systems (6, 51). Truncated soluble versions of E2 bind specifically to human cells and were used to identify interactions with a number of cell surface molecules, including CD81 (19, 43), scavenger receptor class B type I (SR-BI) (47), and DC-specific ICAM-3 grabbing nonintegrin (DC-SIGN) (22, 35, 44). In addition, HCV purified from human plasma is associated with low-density lipoprotein, suggesting that the virus may use the low-density lipoprotein receptor to enter cells (2, 52).
The development of infectious retroviral pseudotypes bearing unmodified HCV gp's has provided a model system to study HCV cell entry (4, 12, 25). Pseudotypes bearing strain H and Con1 HCV gp's show a restricted tropism for human liver cell lines, infection is pH-dependent and can be neutralized by monoclonal antibodies (MAbs) specific for E2 and by HCV-positive human sera (3). We recently reported that the infectivity of pseudotypes harboring these gp strains is CD81 dependent (55). However, CD81 expression alone is not sufficient to allow pseudotype infection of a target cell, and additional liver specific molecule(s) are thought to be required.
HCV is grouped into six major genotypes (20 to 30% overall sequence difference) and more than 50 subtypes (10 to 20% difference) (39). Within an infected individual, HCV exists as a group of different but closely related variants referred to as a viral quasispecies, a characteristic shared by many RNA viruses. Although variability has been documented across the entire genome, the most variable proteins are the envelope gp's. Distinct gp variants have been reported between the liver and peripheral blood mononuclear cell (PBMC) fractions, supporting a model where HCV may replicate in extrahepatic sites (29, 30, 49). This tropism is most likely determined at the level of virus gp-receptor interaction(s). Several reports have suggested that soluble E2 cloned from diverse genotypes fail to interact with CD81, suggesting that viruses of diverse origin may demonstrate altered cell tropism and potentially utilize different receptors (45, 48). We have used the pseudotyping system to study the tropism of viruses bearing HCV gp strains of diverse origin. HCV pseudotypes bearing these gp's failed to infect lymphoid cells and demonstrated a range of infectivities for liver-derived cell lines, infection was CD81 dependent, and some viruses differed in their ability to infect HepG2 cells expressing variant CD81 molecules. These data suggest that CD81 is used as a coreceptor by a wide range of HCV strains.
MATERIALS AND METHODS
Cells and antibodies.
293T (obtained from the American Type Culture Collection [ATCC]), Huh-7.5 (7), Hep3B (ATCC), and HepG2 (gift of Y. Matsuura, Osaka University) cells were propagated in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS). HepG2 cells were cultured on collagen type I-coated tissue culture plastic. Human lymphoid MT-2, Daudi, and LAZ 221 cells (gifts of C. Cheng-Mayer, Aaron Diamond AIDS Research Center, and E. Meffre, The Rockefeller University) were propagated in RPMI-10% FBS.
PBMC were separated from whole blood on lymphocyte separation medium (ICN, Costa Mesa, Calif.) by centrifugation at 800 × g. PBMC were propagated as resting cultures in RPMI-10% FBS, with or without activation with phytohemagglutinin (PHA; 3 μg/ml) for 72 h, and subsequently maintained in RPMI-10% FBS-interleukin-2 (20 U/ml). CD19+ B lymphocytes were positively selected by using immunomagnetic separation according to the manufacturer's protocol (MACS; Miltenyi Biotech, Auburn, Calif.). B cells fractionated in this manner are routinely >90% pure as determined by flow cytometric analysis. B-cell-enriched and -depleted fractions were cultured overnight at 3 × 106 cells/ml in RPMI-10% FBS and 1 μg of pokeweed mitogen (Sigma, St. Louis, Mo.)/ml. All cells were grown at 37°C in 5% CO2.
Murine MAbs used in the present study include 1.3.3.22 anti-CD81 (Santa Cruz Biochemicals, Santa Cruz, Calif.), 1D6 anti-CD81 (Serotec, Ltd., Oxford, United Kingdom), CLA1 anti-SRBI (BD Biosciences), and AC-15 anti-actin (Sigma). Rat MAbs specific for HCV E1 and E2 were previously described (19). Anti-E1 MAb A4 was kindly provided by H. Greenberg (Stanford University) and anti-E1 MAb 725P was purchased from Maine Biotechnology.
PCR amplification, cloning, and sequence analysis of HCV E1E2.
The plasmids encoding strain H and Con1 E1E2 gp's (polyprotein residues 171 to 746), SF162 gp160 and murine leukemia virus (MLV) envelope (env) were described previously (25, 55). The E1E2 open reading frame (ORF) was amplified by a nested PCR protocol from plasmid DNA encoding the infectious molecular clones H77 (GenBank accession no. AF009606) (27), H (M67463) (26), Con1 (AJ238799) (33), HCJ4 (AF054250) (54), and HCJ6 (AF177036) (53) and from total RNA prepared from HCV-infected plasma (HCV viral RNA levels >105/ml) by using commercial kits (Qiagen) (23). Briefly, cDNA was synthesized in a reaction volume of 20 μl, containing 2 to 5 μl of template RNA, 2.5 U of “Multiscribe” Moloney MLV reverse transcriptase with 400 μM concentrations of each of the four deoxynucleoside triphosphates and a 200 nM concentration of the antisense primer p7-2710 (AGC AGG AGG AGN GGC CAY ATC CCR TAG A, where Y = C/T mixture, R = A/G, and N = A/G/C/T) in the manufacturer's recommended buffer (N808-0234; ABI, Foster City, Calif.) for 2 h at 42°C. This cDNA was used as the template for PCR amplification of the E1E2 region as previously described (17). Briefly, a 50-μl PCR was set up containing 2.5 μl of cDNA, 2.5 U of the proofreading Expand polymerase mixture (1 681 834; Roche, Mannheim, Germany) in 1× Expand Buffer 3 (2.25 mM Mg2+) with 400 μM concentrations of each of the four deoxynucleoside triphosphates and 200 nM concentrations of each of the primers core+813 (GAG GAC GGY RTR AAY TAY GCA ACA GG; sense) and p7-2710. The PCR consisted of 30 cycles of 92°C for 45 s, 45°C for 45 s, and 68°C for 300 s and was performed in an Eppendorf thermal cycler. Then, 2 μl of the completed reaction was used as a template for a second amplification, containing the same reaction components as described above with 200 nM concentrations of the primers core+843 (CACC ATG GGT TGC TCT TTC TCT ATC TT; sense) and E2-2580 (CTA CTA GGC CTC AGC CTG GGC TAT CAG CAG CAT CAT CCA; antisense). This second-round PCR comprised 25 cycles of 92°C for 35 s, 55°C for 35 s, and 68°C for 150 s. In most cases, full-length E1E2 (OH8, CH35, CH129, OH5, CH46, and CH54) was readily amplified by these two rounds of PCR. However, for genotype 6 (HK6), two separate but overlapping fragments covering the E1E2 region were first amplified with primers specific for a Hong Kong 6a sequence, EUHK2 (1), and then combined to generate full-length HK6 sequences. PCR products were cloned into pcDNA3.1-TA (Invitrogen), and the sense and antisense strands were sequenced (Big Dye 3.0 Terminator Kit; ABI). All sequences were deposited with GenBank and have the accession numbers AY545951 to AY545980.
Pseudotype production and infection.
Pseudotypes were generated by the transfection of 293T cells (plated at 8 × 105 cells per well of a six-well dish 24 h prior to transfection) with 2 μg of pNL4-3.Luc.R−E− plasmid containing the env-defective human immunodeficiency virus type 1 (HIV-1) proviral genome and 2 μg of expression plasmid encoding the HCV gp's, MLV env gp, vesicular stomatitis virus protein G (VSV-G), or an empty vector with Lipofectamine-2000 (Invitrogen, Carlsbad, Calif.) (9, 25). The supernatants were collected 48 to 72 h posttransfection. Transfected cells were collected, washed with phosphate-buffered saline (PBS), and resuspended in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, Complete protease inhibitors) on ice for 30 min. Lysates were clarified by centrifugation at 4°C for 15 min at maximum speed (20,000 × g) in a microcentrifuge; and the resulting supernatants were stored at −80°C prior to analysis.
Virus was pelleted through a sucrose cushion by layering 1 ml of cell culture medium onto 0.3 ml of 20% (wt/vol) sucrose-PBS before centrifugation at 20,000 × g for 4 h. The medium and cushion were discarded, and the virus pellet was resuspended in 100 μl of PBS on ice. Pelleted virus was inactivated by resuspension in 1% Empigen-PBS and incubated at 56°C for 30 min. Virus and cell lysates were tested for HIV-1 p24 antigen content by using a commercially available HIV-1-p24 antigen enzyme-linked immunosorbent assay (ELISA) protocol (Aalto Bio Reagents, Dublin, Ireland).
Hepatoma cell lines were seeded in 96-well plates (8 × 103 cells/well) 24 h before infection. PBMC and lymphoid cells were seeded in 96-well plates (2 × 105 cells/well) on the day of infection, and 100 μl of virus-associated p24 normalized viral supernatant, diluted in 3% FBS-DMEM plus 4 μg of Polybrene/ml, was added per well. Cells were centrifuged at 400 × g for 1 h (40) and incubated at 37°C overnight, and virus was removed, followed by incubation for a total of 72 h. Cells were lysed with 40 μl of cell lysis buffer (Promega, Madison, Wis.)/well, 35 μl of lysate was tested for luciferase activity by the addition of 50 μl of luciferase substrate, and light output was measured for 10 s in a luminometer (Lumat LB 9507). Luciferase values more than twofold the mean value obtained for the no-env control virus were considered infectious.
In neutralization experiments MAb 3/11, specific for E2, or an irrelevant MAb 10/76b (final concentration, 5 μg/ml) were incubated with pseudotype virus for 1 h at 37°C, and virus-ligand mixtures were tested for infectivity.
Quantitative ELISA.
Briefly, GNA lectin (Sigma) was used to coat Immulon II ELISA plates (Nunc) at 1 μg/ml for 4 h at 37°C. After a wash with PBS, the plates were blocked with 5% bovine serum albumin-PBS, and cell lysates or pelleted virus were allowed to bind overnight at 4°C. A preparation of truncated E2661 was used as an internal calibrant in all ELISAs, and this allowed the comparison of data between different assays. Bound antigen was visualized with MAbs specific for E2 or pooled HCV-positive human sera, an anti-species immunoglobulin G-horseradish peroxidase conjugate (Jackson Laboratories, West Grove, Pa.) and tetramethylbenzidene (BioFX Laboratories). Absorbance values were measured at 450 nm (Fusion Plate Reader; Perkin-Elmer, Boston, Mass.).
Transduction of cells to express CD81.
Wild type and human CD81 mutants (T163A, F186L, E188K, and D196E) were cloned into the BamHI/XhoI site of the lentiviral vector TRIP as previously reported (55). HepG2 cells were plated at 8 × 105 cells per well of a six-well dish and infected 24 h later with a packaged lentivirus expressing the wild type and CD81 mutants (TRIP-CD81) at an approximate multiplicity of infection of 1 to 3 in DMEM-3% FBS plus 4 μg of Polybrene/ml for 12 h. Cells were stained with the CD81-specific MAb 1D6 and phycoerythrin-conjugated secondary antibody, and positively stained cells were sorted using a FACSVantage sorter (Becton Dickinson). CD81-positive cell populations were seeded at 8 × 103 cells/well in a 96-well plate and infected 24 h later with pseudotype viruses.
Flow cytometric fluorescence-activated cell-sorting (FACS) analysis.
E2 expression was quantified as previously described (25). All cells were incubated with an irrelevant isotype-matched immunoglobulin G or anti-E2 MAb 6/1a, and the mean fluorescence signal(s) was determined. Expression of CD81 and SR-BI was quantified as previously described (55). All cells were incubated with an irrelevant isotype-matched immunoglobulin G or the antibody of interest, and the fluorescence signal(s) was used to establish threshold values of detection for the test MAbs. Analyses were performed by using a FACScalibur flow cytometer (Becton Dickinson) and FlowJo software (Tree Star, San Carlos, Calif.).
RESULTS
Infectivity of pseudotype viruses bearing diverse HCV gp's.
Recent studies have reported on the generation of infectious retroviral pseudotypes bearing HCV gp's from strains H (HIV-HCV H) and Con1 (HIV-HCV Con1), genotypes 1a and 1b, respectively. These viruses showed a restricted tropism for human liver cells, and infection was CD81 dependent (25, 55). To assess whether CD81 is utilized by viruses of different genotypes, pseudotype viruses harboring diverse strains of gp's were generated, and their infectivity and tropism was studied.
The E1E2 ORF was PCR amplified from RNA extracted from plasma obtained from HCV-infected individuals or chimpanzees infected with viruses of genotypes 1 to 6 and from plasmid DNA encoding infectious HCV molecular clones (H77, Con1, HCJ4, and HCJ6) (Table 1). The PCR products were cloned into the eukaryotic expression vector pcDNA3.1, and at least three clones were sequenced to confirm genotype and to assess intrasample sequence diversity. Genetic analysis of replicate clones derived from plasma showed a low level of intrapatient diversity (0.06 to 0.73%, Table 1); the frequencies of synonymous and nonsynonymous changes were similar in all samples studied (0 to 2%) and were consistent with previous reports (10, 14-16, 34, 36, 49). Phylogenetic reconstruction with referent sequences representative of the major subtypes confirmed the genotype designations (Table 1; Los Alamos HCV sequence database [http://hcv.lanl.gov]). Genetic distances between clones were considerable (up to 46%), reflecting the high diversity of the E1E2 region between different HCV genotypes.
TABLE 1.
Characterization of divergent HCV E1E2 sequences
| Clone | Genotype | No. of clones | Avg difference (avg distance [%])a | Genetic distance from strain H77 (%) | No. of potential N-linked glycosylation sites (E1/E2) | Sequence of epitope:
|
||
|---|---|---|---|---|---|---|---|---|
| 3/11 | 6/1a | 9/75 | ||||||
| H77 | 1a | 1 | NA | 0 | 15 (5/11) | QLINTNGSWHIN | DFAQGWGP | APTYSWGA |
| H | 1a | 1 | NA | 0.7 | 15 (5/10) | ------------ | -------- | -------- |
| OH8 | 1b | 3 | 0.7 (0.04) | 32.5 | 15 (5/10) | --V--------- | K-D----- | V---N--D |
| CH35 | 1b | 3 | 3.3 (0.52) | 31.1 | 17 (6/11) | --V--------- | K-S----- | V------E |
| Con1 | 1b | 1 | NA | 31.1 | 15 (5/10) | --V--------- | A------- | V------E |
| HCJ4 | 1b | 1 | NA | 34.5 | 15 (5/10) | --V--------- | W------- | V------E |
| CH129 | 2a | 3 | 1.3 (0.08) | 43.9 | 15 (4/11) | --V--------- | D-M----- | L---NF-G |
| HCJ6 | 2a | 1 | NA | 46.9 | 14 (4/10) | --V-S------- | A-RV---A | ----T--E |
| OH5 | 3a | 3 | 1.7 (0.08) | 42.6 | 14 (4/10) | --V--------- | SFN----- | V---T--G |
| CH46 | 3a | 4 | 2.0 (0.07) | 42.4 | 14 (4/10) | --V--------- | SFN----- | V---T--G |
| CH54 | 3b | 3 | 3.7 (0.27) | 43.9 | 14 (4/10) | --V--------- | SFN----- | V---T--G |
| C4a1 | 4 | 2 | 0 (0) | 42.6 | 15 (4/11) | ----S------- | SYG----- | V---T--E |
| C5a1 | 5 | 2 | 1 (0.05) | 43.5 | 15 (4/11) | ------------ | A-D----T | N---N--E |
| C6a1 | 6 | 5 | 7.1 (0.41) | 46.6 | 15 (5/10) | ------------ | --R----Q | I---N--E |
| HK6 | 6 | 3 | 2.7 (0.19) | 45.5 | 16 (5/11) | --V--------- | --R----Q | I---N--D |
Average number of nucleotides differing between clones. Distances were calculated by using the HKY85 method after the gaps were aligned to codon boundaries. NA, not applicable.
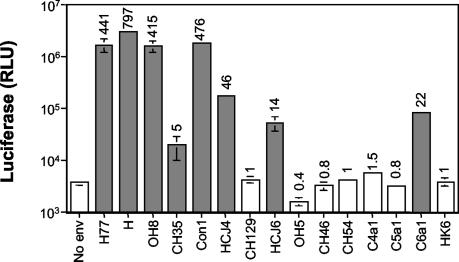
All plasmids encoding a complete E1E2 ORF were cotransfected with pNL4-3.Luc.R−E− into 293T cells, and pseudotype viruses were tested for their ability to infect the hepatoma cell line, Hep3B. Control pseudotype viruses were generated bearing MLV or no envelope gp (no env) and tested for infectivity. Previous studies have shown Hep3B to be the most permissive cell line for pseudotypes bearing strain H and Con1 gp's (25). All virus preparations were normalized for particle number by quantifying the HIV p24 antigen associated with virus after they were pelleted through a sucrose cushion before infection. HIV p24 can exist both as a soluble protein and as a virus-associated antigen. We consistently observed reduced particle/soluble p24 antigen ratios in the extracellular medium from 293T cells transfected to express HCV gp's compared to the no-env or MLV gp controls, suggesting that HCV gp's affect the secretion of HIV particles compared to soluble p24 antigen (data not shown). Pseudotypes bearing H77, H, OH8, CH35, Con1, HCJ4, HCJ6, and C6a1 gp's were infectious for Hep3B and Huh-7.5 cells compared to particles lacking an envelope gp (no env) (Fig. 1 and Table 2). All infections that showed an increase in luciferase signal relative to the no-env virus were blocked by the broadly cross-reactive anti-E2 MAb 3/11 but were unaffected by an irrelevant anti-HIV gp120 MAb, 10/76b (data not shown). Pseudotypes demonstrated a 150-fold range in luciferase activity, which we have previously shown to be proportional to the infectious viral titer (Fig. 1) (25).
FIG. 1.
Infectivity of pseudotypes bearing diverse HCV gp's. Pseudotype viruses were pelleted through a sucrose cushion, and the particle amounts were estimated by quantifying HIV p24 antigen. Hep3B cells were infected with pseudotype viruses bearing a panel of diverse HCV gp's or no envelope gp (No env) containing 10 ng of particulate HIV p24 antigen. At 72 h postinfection, cells were lysed and assayed for luciferase activity. Values are the means of triplicate wells with the standard deviations indicated and are represented as relative light units (RLU). The values above each bar represent the relative infectivity compared to the no-env control virus; values >2-fold the mean of no-env virus infection are considered infectious (shaded bars).
TABLE 2.
Infectivity of pseudotypes bearing diverse HCV gp's for hepatoma and lymphoid cells
| Viral gp | Infectiona of cells (RLU [104])
|
||||||
|---|---|---|---|---|---|---|---|
| Hep3B (hepatoma cell) | Huh-7.5 (hepatoma cell) | PBMC (donor 1) | PBMC (donor 2) | Daudi (B cell) | LAZ 221 (pre-B cell) | MT-2 (T cell) | |
| No env | 0.3 | 0.5 | 0.3 | 0.3 | 0.3 | 0.3 | 0.2 |
| MLV | 16000.0 | 9188.9 | 415.5 | 811.0 | 3,552.1 | 12.2 | 2,587.0 |
| SF162 | 0.3 | 0.5 | 21.8 | 32.7 | 0.4 | 0.4 | 0.3 |
| H77 | 984.9 | 137.8 | 0.4 | 0.4 | 0.5 | 0.3 | 0.4 |
| OH8 | 876.4 | 189.4 | 0.3 | 0.4 | 0.3 | 0.4 | 0.3 |
| CH35 | 15.5 | 4.8 | 0.2 | 0.3 | 0.3 | 0.3 | 0.2 |
| Con1 | 1,329.0 | 312.9 | 0.4 | 0.3 | 0.4 | 0.6 | 0.4 |
| HCJ4 | 108.8 | 9.7 | 0.3 | 0.3 | 0.5 | 0.4 | 0.4 |
| HCJ6 | 38.8 | 16.7 | 0.3 | 0.2 | 0.5 | 0.4 | 0.3 |
| CH46 | 0.4 | 0.5 | 0.4 | 0.3 | 0.5 | 0.4 | 0.3 |
| C6a1 | 42.3 | 12.6 | 0.2 | 0.2 | 0.2 | 0.3 | 0.2 |
The various cell types were infected with pseudotype viruses bearing control MLV gp (0.01 ng of HIV p24 antigen), no gp (no env), HIV SF162 gp, or a range of HCV gp's (10 ng of HIV p24 antigen). At 72 h postinfection, the cells were lysed and assayed for luciferase activity. Values are the means of triplicate wells. RLU, relative light units. PBMC from two donors with or without phytohemagglutinin-interleukin-2 stimulation were tested for their ability to support pseudotype infection; data are shown for the activated cells. Comparable data were obtained for resting PBMC, except that the HIV SF162 pseudotype failed to infect the resting cells.
Since many of the pseudotype viruses bearing non-genotype 1 gp's cloned from infected plasma were not infectious for Hep3B cells, we assessed their ability to infect PBMC, Daudi, LAZ 221, and MT-2 cells. Pseudotypes bearing diverse HCV gp's were tested alongside viruses bearing control MLV envelope, HIV SF162, or no gp. Viruses bearing the MLV envelope gp infected all cell types tested with various efficiencies, whereas pseudotypes bearing the HIV CCR-5-tropic gp160, SF162, only infected activated PBMC (Table 2). In contrast, none of the pseudotypes bearing HCV gp's infected lymphoid cells (Table 2). Since HCV RNA has been reported to preferentially associate with B cells, we tested these pseudotype viruses for their ability to infect purified resting and activated B-cell and non-B-cell fractions from two donors. All pseudotype viruses failed to infect the B cells, including viruses bearing MLV and VSV-G gp's (data not shown). In conclusion, pseudotypes bearing diverse HCV gp's infect human hepatoma liver cells with various efficiencies but fail to infect cells of lymphoid origin.
Expression and incorporation of HCV gp's.
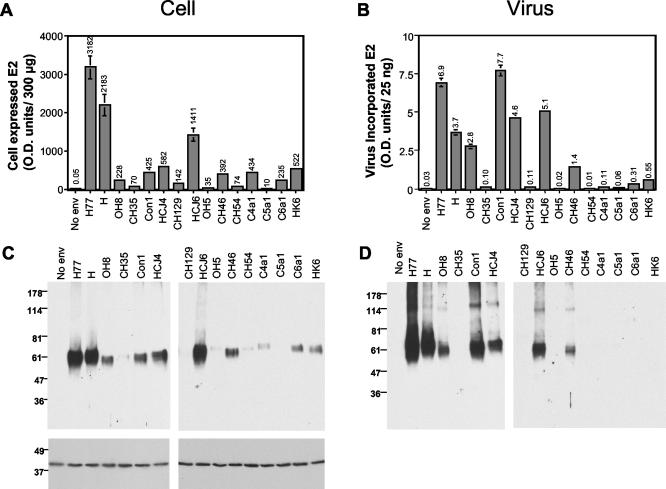
Since pseudotype virus infectivity is dependent upon HCV gp expression (25), we were interested in determining whether the range of infectivities was defined by the level of gp expression and incorporation into particles. 293T cells cotransfected with pNL4-3.Luc.R−E− and plasmids encoding the various HCV gp's were screened for E2 expression by quantitative ELISA and Western blotting with anti-E2 MAb 3/11 (Fig. 2). E2 expression was confirmed for all plasmids; however, the levels varied 3,000-fold, with strain H and H77 proteins being expressed at the highest level (Fig. 2). All clones from a single sample showed comparable levels of E2 expression (data not shown). The epitope recognized by MAb 3/11 was invariant within clones from a single sample and is well conserved between divergent clones, with only four alternative sequences observed. With the exception of HCJ6 (two changes); all sequences differed from H77 by a single conserved amino acid (Table 1). However, these single amino acid changes within the epitope could alter the binding affinity of the MAb for a particular strain, biasing our determination of E2 levels. To address this issue, we used both a cocktail of anti-E2 MAbs (3/11, 6/1a, and 9/75; Table 1) and a mixture of human HCV+ sera from individuals infected with HCV genotypes 1, 2, 3, and 5 to realize the E2 bound in the ELISA. Comparable data were obtained with respect to differences in expression levels between plasmids; however, the sensitivity of the ELISA was reduced ∼20-fold when polyclonal human sera was used as the detecting agent (data not shown). The anti-E1 MAbs A4, 3/8ow, and 725P were able to detect E1 in lysates from cells transfected with plasmids encoding strain H and H77 gp's; however, these MAbs failed to react with other gp strains (data not shown). Analysis of cell lysates by reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with MAb 3/11 showed an E2 species migrating at ∼60 kDa, with some minor changes in migration patterns observed between clones, a finding consistent with changes in the number of predicted N-linked glycosylation sites (Fig. 2 and Table 1). E2 expression levels observed by Western blotting of cell lysates were consistent with the ELISA data and varied between different HCV strains.
FIG. 2.
Expression and incorporation of diverse HCV gp's into pseudotype particles. 293T cells were cotransfected with pNL4-3.Luc.R−E− and plasmids expressing representative HCV gp's listed in Table 1 or an empty vector (No env). At 72 h posttransfection, the virus was pelleted through a sucrose cushion. The cells and virus were lysed, and both fractions were quantified for E2 expression by ELISA and Western blotting. The total E2 expressed per well of 293T cells (300 μg of cellular protein) (A) and that incorporated into pseudotype particles (B) (25 ng of particulate HIV p24 antigen) were measured by ELISA, and the data are expressed as optical density units (OD) at 450 nm and annotated above each bar. Virus preparations that gave optical density signals >2-fold the mean of the no-env virus control were considered to have incorporated gp's. Transfected cell lysate (10 μg of total protein) (C) and pelleted virus particles (D) (25 ng of particulate HIV p24 antigen) were separated by reducing SDS-PAGE and immunoblotted for E2 (MAb 3/11) and actin (MAb AC-15). The migration of molecular mass markers is indicated in kilodaltons.
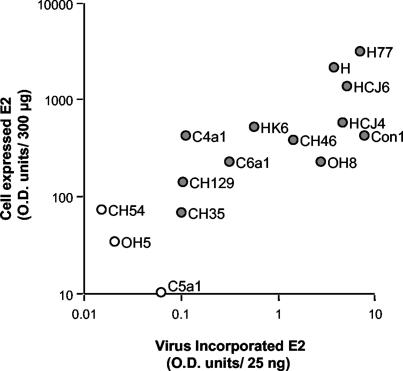
To quantify E2 incorporation into HIV pseudotypic particles, the virus present in the extracellular medium was pelleted by centrifugation through a 20% sucrose cushion. Virus particles were quantified by measuring HIV p24 antigen, and 25 ng of each preparation was evaluated by ELISA and immunoblotting with anti-E2 MAb 3/11. The level of E2 incorporated into particles varied between different gp strains, generally reflecting the differences in gp expression observed in the cell lysates (Fig. 2). An association was observed between E2 expression levels and incorporation into particles; however, the absolute level of incorporated gp was not a predictor of pseudotype virus infectivity (Fig. 3). In replicate experiments, the levels of E2 incorporated into particles varied by up to fivefold, depending on the transfection efficiency; however, the relative differences in incorporation noted between diverse gp's were maintained. Quantification of the levels of E2 antigen in both the cell and the virus preparations by ELISA demonstrated that between 0.7 and 4.6% of the E2 antigen within the cell was incorporated into particles. If we assume that incorporation occurs at the plasma membrane, the observation that only a minor fraction of the total cellular antigen is incorporated into particles is consistent with the low level of E2 detected at the surface of transfected 293T cells. Analysis of the incorporated gp's by SDS-PAGE demonstrated E2 species migrating at ca. 60 and 100 kDa; the latter band may represent an uncleaved E1E2 species or a form of E2 that is resistant to SDS treatment (Fig. 2) (18). In general, there was good agreement in the inferred levels of incorporated E2 protein between the two methodologies, with the ELISA being more sensitive.
FIG. 3.
Association between cellular HCV E2 expression levels and incorporation into pseudotype particles. Relationship between the level of E2 expressed in cells and that incorporated into pseudotype particles as determined by ELISA (r2 = 0.643, P = 0.0003). HCV gp strains OH5, CH54, and C5a1 failed to show any specific incorporation into particles and are indicated by open circles.
HCV gp's (OH5, CH54, and C5a1) failed to incorporate into particles or to yield infectious virus (Fig. 1 and 2), whereas viruses bearing CH46 gp's, which appeared to incorporate E2 at levels comparable to HCJ4 and HCJ6, were not infectious. In contrast, moderate levels of infectivity were noted for pseudotypes bearing low levels of CH35 and C6a1 gp's, suggesting that the absolute level of gp incorporated into particles is not associated with virus infectivity. To assess the relationship between the level of incorporated gp(s) and infectivity, pseudotype viruses were generated by transfection of 293T cells with pNL4-3.Luc.R−E− and different amounts of pH77 E1E2. Cells were monitored for both total and cell surface-expressed E2 by FACS and for E2 antigen levels within cell lysates and sucrose cushion-pelleted virus by ELISA. Decreasing the H77 plasmid concentration by 5- and 10-fold had a modest but detectable effect on the level of cellular E2, as measured by FACS and ELISA, suggesting that E2 expression levels were close to saturation (Fig. 4A and B). In contrast, significant differences were noted in the levels of E2 expressed at the cell surface and subsequently incorporated into particles, a finding consistent with a minor fraction of the total cellular antigen being incorporated into particles (Fig. 4C). However, all H77 virus preparations showed comparable levels of infectivity for Hep3B (Fig. 4D) and for Huh-7.5 cells (data not shown). These data suggest that incorporation of functional gp's is near saturation and that the infectivity of particles may not be limited by the apparent low levels at which some HCV gp's incorporate. Hence, the range of infectivities observed for viruses bearing diverse HCV gp's may not simply be to attributable differences in gp incorporation per se but may reflect differences in their ability to generate and transport a functional gp complex to the cell surface and in different receptor binding and/or fusion activities of the assembled gp complex.
FIG. 4.
gp incorporation and infectivity of pseudotypes bearing H77 E1E2. 293T cells were cotransfected with pNL4-3.Luc.R−E− and different amounts of pH77 E1E2 (2.0, 0.4, or 0.2 μg) or an empty vector (No env) to produce pseudotypes with different amounts of incorporated gp's. (A) Transfected cells were fixed with paraformaldehyde and stained for E2 expression with (dark gray shading) or without permeabilization (light gray shading) to detect total and cell surface E2 antigen, respectively. The data are expressed as mean fluorescence units (M.F.I.). The level of E2 in both the transfected cells (300 μg of cellular protein) (B) and pelleted pseudotype particles (C) (25 ng of particulate HIV p24 antigen) was measured by ELISA, and the data are expressed as optical density units (OD) at 450 nm. Hep3B cells were infected with pseudotype viruses bearing different amounts of H77 gp's or no envelope gp (No env) containing 10 (gray) and 2 ng (unshaded) of particulate HIV p24 antigen (D). At 72 h postinfection, cells were lysed and assayed for luciferase activity. Values are the mean of triplicate wells with the indicated standard deviations and are represented as relative light units (RLU).
HCV pseudotype infection is CD81 dependent.
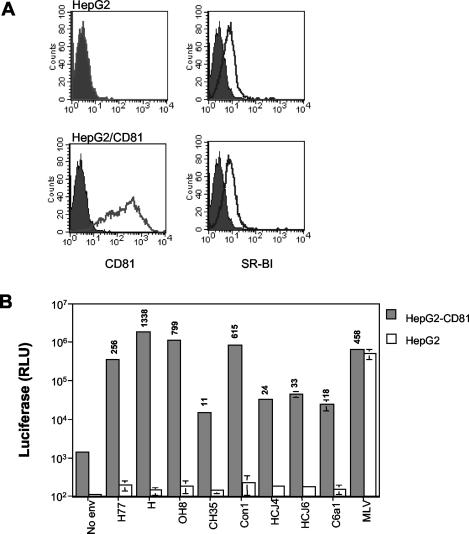
We previously reported that the infectivity of pseudotypes bearing strain H and Con1 E1E2 gp's was CD81 dependent. However, CD81 expression alone is not sufficient to confer susceptibility to infection by HCV pseudotypes, and additional liver specific factor(s) are thought to be required (55). A recent report by Bartosch et al. suggested that both SR-BI receptor and CD81, in addition to liver-specific factor(s), are required for HCV pseudotype infection (5). The HepG2 hepatoma cell line expresses SR-BI but not CD81 and allows one to assess the relative contribution(s) of these cell surface molecules for HCV pseudotype infection (Fig. 5). HCV pseudotypes were tested for their ability to infect parental HepG2 cells and those transduced to express human CD81 (HepG2-CD81). A control pseudotype virus bearing MLV envelope gp infected both HepG2 and HepG2-CD81, whereas HCV pseudotypes only infected HepG2 cells expressing CD81 (Fig. 5B). These data support a role for CD81 in HCV pseudotype infection that is conserved across diverse HCV strains.
FIG. 5.
Infectivity of pseudotype viruses bearing divergent HCV gp's is CD81 dependent. (A) Cell surface expression of CD81 and SR-BI on parental HepG2 cells and cells transduced to express human CD81. (B) HepG2 and HepG2-CD81cells were infected with pseudotype viruses bearing a panel of diverse HCV gps or no envelope gp (No env) containing 10 ng of particulate HIV p24 antigen. In order to give comparable relative light unit (RLU) signals, the MLV pseudotype virus was infected at a lower dose (0.01 ng of particulate p24 antigen). At 72 h postinfection, cells were lysed and assayed for luciferase activity. Values are the means of triplicate wells with the indicated standard deviation. Values above each bar represent the relative infectivity compared to the no-env control virus; values >2-fold the mean for the no-env virus infection were considered infectious.
The ability of CD81 to render HepG2 cells susceptible to HCV pseudotype infection provides an ideal system to test the determinants of CD81 required for infection. We previously reported that soluble truncated E2 failed to interact with African green monkey CD81, which differed from the human molecule at four amino acid residues (24). Mutation of the human CD81 sequence at each of these residues identified amino acid 186 to be critical for its interaction with soluble E2. However, expression of the CD81 variants T163A, F186L, E188K, and D196E in HepG2 cells conferred comparable levels of permissivity to HCV pseudotypes bearing strain H and Con1 gp's (55). Since pseudotype viruses bearing diverse HCV gp's display different infectivities for Hep3B and Huh-7.5 cells, we compared their infectivity for HepG2 cells expressing these variant CD81 molecules. All CD81 variants were expressed at comparable levels on the cell surface and in >95% of cells (data not shown). As previously reported, a control pseudovirus bearing MLV envelope gp and viruses bearing strain H or Con1 HCV gp's infected HepG2 expressing wild-type or variant CD81 molecules equally. In contrast, viruses bearing CH35, HCJ4, and C6a1 gp's infected HepG2-CD81 D196E cells with 70 to 96% reduced efficiency compared to cells expressing wild-type CD81 (Table 3). Since viruses bearing these gp's were generally less infectious than those bearing H, H77, and Con1 gp's, their reduced titers may contribute to the altered permissivity of the CD81 D196E variant line. However, when HepG2 cell lines were infected with a reduced inoculum of virus bearing strain H gp's, a comparable luciferase activity was observed in all clones, suggesting that the permissivity of the variant CD81 HepG2 cells lines was not dependent on the infecting viral dose (Table 3). These data confirm that all of the HCV pseudotypes require CD81 to infect HepG2 cells. However, some HCV gp's differ in their interaction(s) with CD81 and in particular amino acid 196 appears to be an important determinant of such differences.
TABLE 3.
Infectivity of pseudotypes for HepG2 cells expressing wild-type and variant human CD81 moleculesa
| Viral gp | Relative infection of HepG2 cells expressing CD81 (RLU [103])
|
||||
|---|---|---|---|---|---|
| Wild type | CD81 T163A | CD81 F186L | CD81 E188K | CD81 D196E | |
| No env | 0.3 | 0.3 | 0.4 | 0.2 | 0.3 |
| H77 | 352.0 | 205.7 | 253.8 | 346.8 | 220.1 |
| OH8 | 1099.5 | 657.1 | 684.5 | 879.4 | 609.5 |
| CH35 | 14.5 | 3.4 | 8.9 | 4.8 | 3.9 |
| Con1 | 846.7 | 697.5 | 844.6 | 824.8 | 567.6 |
| HCJ4 | 33.6 | 20.1 | 35.4 | 27.7 | 10.1 |
| HCJ6 | 45.3 | 29.1 | 39.1 | 54.1 | 21.6 |
| C6a1 | 22.4 | 28.0 | 31.3 | 21.2 | 3.1 |
| H77 (reduced inoculum) | 34.5 | 36.7 | 25.9 | 22.4 | 22.8 |
HepG2 cells transduced to express wild-type and variant CD81 molecules were infected with pseudotype viruses bearing no gp (No env) or a range of HCV gp's containing 10 ng of particulate HIV p24 antigen. Control MLV pseudotype infections were performed with a reduced inoculum of 0.01 ng of HIV p24 antigen. HIV-HCV H77 was also infected with a reduced inoculum of 1 ng of particulate HIV p24 antigen. At 72 h postinfection, the cells were lysed and assayed for luciferase activity. Luciferase values for HCV pseudotype infection of each cell line were normalized relative to the signal obtained from an MLV infection of the same target cell.
DISCUSSION
In the present study we show that retroviral pseudotypes bearing a diverse panel of HCV gp's are infectious for liver-derived cells and fail to infect lymphoid cells. These data are consistent with the liver being the primary reservoir for HCV replication in vivo and support a model where liver-specific receptor(s) may contribute to the tissue specificity of HCV infection. The observation that all HCV pseudotypes fail to infect HepG2 cells and yet infect the same cells after transduction to express human CD81 confirms the critical role of CD81 in HCV infection. The inability of HepG2 cells to support HCV pseudotype infection suggests that SR-BI expression alone is not sufficient to confer infection (5). Further clarification of the relative contribution of CD81 and SR-BI in HCV cell attachment and fusion awaits the identification of the liver specific factor(s).
Pseudotypes bearing gp's cloned from infectious molecular clones (H77, Con1, HCJ4, and HCJ6) were all infectious for Hep3B cells. In contrast, only three of ten virus preparations bearing gp's (OH8, CH35, and C6a1) cloned directly from infected plasma were infectious. There are several possible interpretations of these results. First, viruses in the plasma may infect cell types not tested in the present study; however, it should be noted that previous attempts to infect immature and mature blood-derived dendritic cells with pseudotype viruses bearing strain H and Con1 gp's failed (25). Second, viruses in the plasma may encode for a high frequency of defective gp's, as has been noted for HIV (31, 41, 46); however, this is unlikely, given the high infectivity to particle number noted for a number of related flavi- and pestiviruses (32). Third, PCR amplification of the E1E2 ORFs from HCV-infected plasma may have resulted in in vitro errors (37). However, in samples from which infectious virus was obtained, all replicate clones generated virus of comparable infectivity, suggesting that the in vitro error rate (which we previously estimated to be <1 base/clone for this method [23]) was not an adequate explanation for the lack of infectivity observed. Fourth, these gp's may be infectious in a native virion but defective in the retroviral pseudotyping system, which involves gp processing and particle assembly pathways distinct from those likely to be used by HCV.
Several of the plasma-derived E1E2 ORFs expressed low levels of gp compared to H77 and showed minimal incorporation into particles (Fig. 2). It is possible that immune detection and quantitation of these diverse gp's may misrepresent differences in gp expression between divergent clones. However, the epitope recognized by MAb 3/11 is of low variability, and the changes that are observed are generally conservative (Table 1). One method of overcoming this potential issue would be to epitope tag the E1E2 ORFs; however, Dubuisson and coworkers recently reported that introduction of a tag sequence into the carboxyl regions of E1 or E2 reduced the infectivity of pseudotypes bearing these gp's (42). The use of a polyclonal HCV-positive human sera mixture to detect the gp's by ELISA provided data comparable to that obtained with MAb 3/11, confirming the differences in expression levels noted between clones. An experiment to address the relationship between HCV gp incorporation and infectivity showed that a 10-fold reduction in the level of virion-associated E2 did not affect viral entry, suggesting that the level of gp incorporation is not rate limiting (Fig. 4). We have recently reported that HIV pseudotypes incorporate a heterogeneous mixture of HCV gp's with respect to their glycosylation pattern, disulfide linkages, and predicted molecular weight (18). Hence, one possible interpretation for the differences in infectivity of these pseudotypes is that some HCV strains express and incorporate a greater frequency of functionally active gp's than others. However, at the present time it is not possible to define the functionally active gp species and to evaluate this possibility.
Pseudotypes bearing gp's amplified from infectious molecular clones demonstrated a 30-fold range in infectivity (Fig. 2), which failed to associate with the level of incorporated gp(s). Similar differences in infectivity have been reported for HIV particles bearing envelope gp's cloned from diverse genotypes and has been attributed to variations in affinity for CD4 and chemokine coreceptors (20, 21, 23, 50). The soluble form of HCV strain H E2 has been used extensively as a model to study HCV-CD81 interactions, and several reports have shown that soluble E2 gp's cloned from other genotypes fail to interact with CD81 (45, 48). The minimal interaction of non-strain H E2 gp's with CD81 may simply reflect differences in gp folding and conformation of the truncated protein or may represent true differences in the affinity of E2 gp's for CD81. Pseudotype viruses bearing the Con1 gp's infect cells in a CD81-dependent manner, and yet a truncated form of Con1 E2 shows negligible interaction with CD81, suggesting that pseudotype virus interaction with CD81 is more complex than the interaction of soluble E2 with CD81 and is likely to involve other cellular molecules (55). It is interesting that viruses bearing HCJ4, CH35, and C6a1 gp's infected HepG2 cells expressing the D196E CD81 variant less efficiently than cells expressing wild-type CD81 (Table 3). These data suggest that there may be subtle differences in the CD81 residues interacting with diverse viral gp's; however, all viruses studied to date require CD81 to initiate infection of a target cell. Our current model of HCV infection suggests that CD81, together with additional unknown liver-specific receptor(s), mediates the virus-cell entry process, and it is likely that interaction of gp's with these factors will be a critical determinant in defining the permissivity of a cell to HCV infection.
Acknowledgments
We are grateful to Hernan Jaramillo, Merna Torres, Jack Hietpas, and Jennifer Alt for excellent technical support and to Cecilia Cheng-Mayer for kindly providing PBMC. We thank Jens Bukh, Harry Greenberg, Shoshana Levy, Eric Meffre, and Andy Talal for antibodies, cell lines, and HCV-infected plasma.
C.L., M.F., J.Z., L.B.D, D.B, C.M.R., and J.A.M are supported by the Greenberg Medical Research Institute and PHS grants CA57973 and AI40034.
REFERENCES
- 1.Adams, N. J., R. W. Chamberlain, L. A. Taylor, F. Davidson, C. K. Lin, R. M. Elliott, and P. Simmonds. 1997. Complete coding sequence of hepatitis C virus genotype 6a. Biochem. Biophys. Res. Commun. 234:393-396. [DOI] [PubMed] [Google Scholar]
- 2.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartosch, B., J. Bukh, J. C. Meunier, C. Granier, R. E. Engle, W. C. Blackwelder, S. U. Emerson, F. L. Cosset, and R. H. Purcell. 2003. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. USA 100:14199-14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of coreceptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 6.Baumert, T. F., S. Ito, D. T. Wong, and T. J. Liang. 1998. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 72:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boisvert, J., X. S. He, R. Cheung, E. B. Keeffe, T. Wright, and H. B. Greenberg. 2001. Quantitative analysis of hepatitis C virus in peripheral blood and liver: replication detected only in liver. J. Infect. Dis. 184:827-835. [DOI] [PubMed] [Google Scholar]
- 9.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 10.Curran, R., C. L. Jameson, J. K. Craggs, A. M. Grabowska, B. J. Thomson, A. Robins, W. L. Irving, and J. K. Ball. 2002. Evolutionary trends of the first hypervariable region of the hepatitis C virus E2 protein in individuals with differing liver disease severity. J. Gen. Virol. 83:11-23. [DOI] [PubMed] [Google Scholar]
- 11.Dammacco, F., P. Gatti, and D. Sansonno. 1998. Hepatitis C virus infection, mixed cryoglobulinemia, and non-Hodgkin's lymphoma: an emerging picture. Leuk. Lymphoma 31:463-476. [DOI] [PubMed] [Google Scholar]
- 12.Drummer, H. E., A. Maerz, and P. Poumbourios. 2003. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 546:385-390. [DOI] [PubMed] [Google Scholar]
- 13.El-Serag, H. B., H. Hampel, C. Yeh, and L. Rabeneck. 2002. Extrahepatic manifestations of hepatitis C among United States male veterans. Hepatology 36:1439-1445. [DOI] [PubMed] [Google Scholar]
- 14.Erickson, A. L., Y. Kimura, S. Igarashi, J. Eichelberger, M. Houghton, J. Sidney, D. McKinney, A. Sette, A. L. Hughes, and C. M. Walker. 2001. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity 15:883-895. [DOI] [PubMed] [Google Scholar]
- 15.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 16.Farci, P., R. Strazzera, H. J. Alter, S. Farci, D. Degioannis, A. Coiana, G. Peddis, F. Usai, G. Serra, L. Chessa, G. Diaz, A. Balestrieri, and R. H. Purcell. 2002. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc. Natl. Acad. Sci. USA 99:3081-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flint, M., J. Dubuisson, C. Maidens, R. Harrop, G. R. Guile, P. Borrow, and J. A. McKeating. 2000. Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J. Virol. 74:702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flint, M., C. Logvinoff, C. M. Rice, and J. A. McKeating. 2004. Characterization of infectious retroviral pseudotype particles bearing hepatitis C virus glycoproteins. J. Virol. 78:6875-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao, F., S. G. Morrison, D. L. Robertson, C. L. Thornton, S. Craig, G. Karlsson, J. Sodroski, M. Morgado, B. Galvao-Castro, H. von Briesen, et al. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J. Virol. 70:1651-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, F., L. Yue, S. Craig, C. L. Thornton, D. L. Robertson, F. E. McCutchan, J. A. Bradac, P. M. Sharp, B. H. Hahn, et al. 1994. Genetic variation of HIV type 1 in four World Health Organization-sponsored vaccine evaluation sites: generation of functional envelope (glycoprotein 160) clones representative of sequence subtypes A, B, C, and E. AIDS Res. Hum. Retrovir. 10:1359-1368. [DOI] [PubMed] [Google Scholar]
- 22.Gardner, J. P., R. J. Durso, R. R. Arrigale, G. P. Donovan, P. J. Maddon, T. Dragic, and W. C. Olson. 2003. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 100:4498-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond, A. L., J. Lewis, J. May, J. Albert, P. Balfe, and J. A. McKeating. 2001. Antigenic variation within the CD4 binding site of human immunodeficiency virus type 1 gp120: effects on chemokine receptor utilization. J. Virol. 75:5593-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higginbottom, A., E. R. Quinn, C. C. Kuo, M. Flint, L. H. Wilson, E. Bianchi, A. Nicosia, P. N. Monk, J. A. McKeating, and S. Levy. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inchauspe, G., S. Zebedee, D. H. Lee, M. Sugitani, M. Nasoff, and A. M. Prince. 1991. Genomic structure of the human prototype strain H of hepatitis C virus: comparison with American and Japanese isolates. Proc. Natl. Acad. Sci. USA 88:10292-10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 28.Lambot, M., S. Fretier, A. Op De Beeck, B. Quatannens, S. Lestavel, V. Clavey, and J. Dubuisson. 2002. Reconstitution of hepatitis C virus envelope glycoproteins into liposomes as a surrogate model to study virus attachment. J. Biol. Chem. 277:20625-20630. [DOI] [PubMed] [Google Scholar]
- 29.Laskus, T., M. Radkowski, L. F. Wang, M. Nowicki, and J. Rakela. 2000. Uneven distribution of hepatitis C virus quasispecies in tissues from subjects with end-stage liver disease: confounding effect of viral adsorption and mounting evidence for the presence of low-level extrahepatic replication. J. Virol. 74:1014-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lerat, H., S. Rumin, F. Habersetzer, F. Berby, M. A. Trabaud, C. Trepo, and G. Inchauspe. 1998. In vivo tropism of hepatitis C virus genomic sequences in hematopoietic cells: influence of viral load, viral genotype, and cell phenotype. Blood 91:3841-3849. [PubMed] [Google Scholar]
- 31.Li, Y., J. C. Kappes, J. A. Conway, R. W. Price, G. M. Shaw, and B. H. Hahn. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65:3973-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 33.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 34.Love, A., V. Molnegren, A.-S. Mansson, A. Smaradottir, S. B. Thorsteinsson, and A. Widell. 2004. Evolution of hepatitis C virus variants following blood transfusion from one infected donor to several recipients: a long-term follow-up. J. Gen. Virol. 85:441-450. [DOI] [PubMed] [Google Scholar]
- 35.Lozach, P. Y., H. Lortat-Jacob, A. De Lacroix De Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houles, F. Fieschi, Schwartz, J. L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high-affinity binding receptors for hepatitis C Virus glycoprotein E2. J. Biol. Chem. 278:20358-20366. [DOI] [PubMed] [Google Scholar]
- 36.Lu, M., J. Kruppenbacher, and M. Roggendorf. 2000. The importance of the quasispecies nature of hepatitis C virus (HCV) for the evolution of HCV populations in patients: study on a single source outbreak of HCV infection. Arch. Virol. 145:2201-2210. [DOI] [PubMed] [Google Scholar]
- 37.McAllister, J., C. Casino, F. Davidson, J. Power, E. Lawlor, P. L. Yap, P. Simmonds, and D. B. Smith. 1998. Long-term evolution of the hypervariable region of hepatitis C virus in a common-source-infected cohort. J. Virol. 72:4893-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mele, A., A. Pulsoni, E. Bianco, P. Musto, A. Szklo, M. G. Sanpaolo, E. Iannitto, A. De Renzo, B. Martino, V. Liso, C. Andrizzi, S. Pusterla, F. Dore, M. Maresca, M. Rapicetta, F. Marcucci, F. Mandelli, and S. Franceschi. 2003. Hepatitis C virus and B-cell non-Hodgkin lymphomas: an Italian multicenter case-control study. Blood 102:996-999. [DOI] [PubMed] [Google Scholar]
- 39.Mellor, J., E. C. Holmes, L. M. Jarvis, P. L. Yap, P. Simmonds, et al. 1995. Investigation of the pattern of hepatitis C virus sequence diversity in different geographical regions: implications for virus classification. J. Gen. Virol. 76:2493-2507. [DOI] [PubMed] [Google Scholar]
- 40.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohagen, A., A. Devitt, K. J. Kunstman, P. R. Gorry, P. P. Rose, B. Korber, J. Taylor, R. Levy, R. L. Murphy, S. M. Wolinsky, and D. Gabuzda. 2003. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J. Virol. 77:12336-12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Op De Beeck, A., C. Voisset, B. Bartosch, Y. Ciczora, L. Cocquerel, Z. Keck, S. Foung, F. L. Cosset, and J. Dubuisson. 2004. Characterization of functional hepatitis C virus envelope glycoproteins. J. Virol. 78:2994-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pileri, P., Y. Uematsu, S. Compagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 44.Pohlmann, S., J. Zhang, F. Baribaud, Z. Chen, G. J. Leslie, G. Lin, A. Granelli-Piperno, R. W. Doms, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 77:4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roccasecca, R., H. Ansuini, A. Vitelli, A. Meola, E. Scarselli, S. Acali, M. Pezzanera, B. B. Ercole, J. A. McKeating, A. Yagnik, A. Lahm, A. Tramontano, R. Cortese, and A. Nicosia. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain-specific and is modulated by a complex interplay between the hypervariable regions 1 and 2. J. Virol. 77:1856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez, G., X. Xu, J. C. Chermann, and I. Hirsch. 1997. Accumulation of defective viral genomes in peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected individuals. J. Virol. 71:2233-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaw, M. L., J. McLauchlan, P. R. Mills, A. H. Patel, and E. A. McCruden. 2003. Characterization of the differences between hepatitis C virus genotype 3 and 1 glycoproteins. J. Med. Virol. 70:361-372. [DOI] [PubMed] [Google Scholar]
- 49.Sung, V. M., S. Shimodaira, A. L. Doughty, G. R. Picchio, H. Can, T. S. Yen, K. L. Lindsay, A. M. Levine, and M. M. Lai. 2003. Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection. J. Virol. 77:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tscherning, C., A. Alaeus, R. Fredriksson, A. Bjorndal, H. Deng, D. R. Littman, E. M. Fenyo, and J. Albert. 1998. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology 241:181-188. [DOI] [PubMed] [Google Scholar]
- 51.Wellnitz, S., B. Klumpp, H. Barth, S. Ito, E. Depla, J. Dubuisson, H. E. Blum, and T. F. Baumert. 2002. Binding of hepatitis C virus-like particles derived from infectious clone H77C to defined human cell lines. J. Virol. 76:1181-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wunschmann, S., J. D. Medh, D. Klinzmann, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 74:10055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250-263. [DOI] [PubMed] [Google Scholar]
- 54.Yanagi, M., M. St. Claire, M. Shapiro, S. U. Emerson, R. H. Purcell, and J. Bukh. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244:161-172. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]