Abstract
The mK3 protein of gammaherpesvirus 68 and the kK5 protein of Kaposi's sarcoma-associated herpesvirus are members of a family of structurally related viral immune evasion molecules that all possess a RING-CH domain with ubiquitin ligase activity. These proteins modulate the expression of major histocompatibility complex class I molecules (mK3 and kK5) as well as other molecules like ICAM-1 and B7.2 (kK5). Previously, mK3 was shown to ubiquitinate nascent class I molecules, resulting in their rapid degradation, and this process was found to be dependent on TAP and tapasin, endoplasmic reticulum molecules involved in class I assembly. Here, we demonstrate that in murine cells, kK5 does not affect class I expression but does downregulate human B7.2 molecules in a TAP/tapasin-independent manner. These differences in substrate specificity and TAP/tapasin dependence between mK3 and kK5 permitted us, using chimeric molecules, to map the sites of mK3 interaction with TAP/tapasin and to determine the requirements for substrate recognition by mK3. Our findings indicate that mK3 interacts with TAP1 and -2 via their C-terminal domains and with class I molecules via their N-terminal domains. Furthermore, by orienting the RING-CH domain of mK3 appropriately with respect to class I, mK3 binding to TAP/tapasin, rather than the presence of unique sequences in class I, appears to be the primary determinant of substrate specificity.
Many viruses have developed elaborate mechanisms to evade immune detection (11, 26, 40, 46). These mechanisms are typically specific for a given type of virus and are highly host adapted (10). Thus, viruses have clearly evolved under the selective pressure of the host immune system to develop counter strategies to prevent their elimination. Given the importance of CD8+ T cells in immune surveillance against many viral infections, it is not surprising that viruses have evolved genes whose products function to block the expression of class I molecules. Recently, a novel family of viral and cellular proteins (termed here the K3 family) has been identified and found to possess E3 ubiquitin (Ub) ligase activity. Several members of this family have been shown to target class I molecules and/or T-cell costimulation molecules for Ub-dependent degradation (3, 15, 21). E3 Ub ligase activity is conferred to members of the K3 family by a consensus N-terminal sequence encoding a special type of RING (for “really interesting new gene”) finger motif, known as the RING-CH type of zinc finger (38), characterized by a cysteine residue in the fourth zinc-coordinating position and a histidine residue in the fifth. Alternatively, this motif has been classified as a subclass of the plant homeodomain (PHD)/leukemia-associated protein (LAP) finger (6). Although proteins in this family are structurally and functionally similar, their target specificities and sites of ubiquitination and degradation are distinct. Understanding how disparate members of the K3 family target different proteins at different subcellular sites is an area of intense investigation that promises to define the role of ubiquitination in regulating intracellular transport and endoplasmic reticulum (ER)-associated degradation pathways (9).
Belonging to this family, the mK3 protein encoded by gamma-2 herpesvirus 68 (γHV68) consists of a conserved RING-CH finger domain in its N terminus followed by two closely spaced transmembrane (TM) segments and a C-terminal tail. Investigation of the topology of mK3 showed that it is a type III ER protein, with both N- and C-terminal domains projecting into the cytoplasm and a short segment between the two TM regions in the lumen of the ER (3). As with other members of the K3 family, mK3's RING-CH domain is critical for cysteine-dependent E3 Ub ligase activity that mediates the rapid destruction of major histocompatibility complex (MHC) class I proteins (3). However, the mechanisms underlying the mK3-induced MHC class I degradation are different from those in even its closest homologs, the kK3 and kK5 proteins. The kK3 and kK5 proteins encoded by Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8, target surface class I molecules via accelerated endocytosis, resulting in their Ub-mediated degradation in the lysosome (15). In addition, kK5 is known to also target B7.2 and ICAM-1 molecules (36), and the TM regions of both kK5 and its substrates are critical for targeted degradation, presumably by mediating protein-protein interactions (7, 33). Recent studies by Stevenson et al. and Yu et al. have shown that, in contrast to kK3 and kK5, mK3 induces the rapid turnover of nascent, ER-resident class I molecules mainly through the ubiquitination-proteasome pathway (36, 47). Furthermore, Stevenson et al. demonstrated that mice infected with an mK3-deficient virus had a reduced number of latently infected spleen cells and an increased number of virus-specific CD8 T cells compared with mice infected with wild-type (wt) virus (37). These findings established the physiologic relevance of mK3 in immune evasion of CD8 T cells. More recently, Lybarger et al. reported that two class I assembly-specific proteins (TAP and tapasin) are required for mK3 stabilization and for mK3-mediated class I downregulation (20). In the presence of mK3, TAP/tapasin-associated class I heavy chains (H chains) were ubiquitinated while mutants of class I incapable of TAP/tapasin interaction were not ubiquitinated and, therefore, not rapidly degraded. The association of mK3 with TAP/tapasin was found to be class I independent, suggesting that mK3 can associate with TAP/tapasin in a substrate-independent manner. Thus, the interaction of mK3 with TAP/tapasin is class I independent but is essential for mK3 to Ub conjugate class I proteins and target them for rapid destruction. An important implication of these findings is that mK3 must have separate sites by which it interacts with TAP/tapasin versus class I proteins.
As for how mK3 interacts with class I, a basic cluster motif (KRRRNT) located in the membrane-proximal cytoplasmic tail of the class I protein Db was found to be required for mK3-class I association and for optimal downregulation of surface Db molecules by mK3. Thus, it was proposed that this membrane-proximal cluster may be a specific site of interaction between class I and mK3 (3). Interestingly, a similar basic cluster (KKKKRP) is found in an analogous region of the cytoplasmic tail of human T-cell costimulation protein B7.2, and this cluster was found to be necessary and sufficient for kK5-mediated downregulation of B7.2 (7). Alternatively, our data suggested that substrate specificity of mK3 might be conferred secondarily, by its obligate interaction with TAP and tapasin. These combined findings raised interesting questions about how mK3 interacts with class I molecules versus TAP/tapasin and, more importantly, which of these interactions contributes to the substrate specificity for mK3-mediated degradation. Here, we investigated these issues by exploiting the disparate substrate specificities and TAP/tapasin dependencies of mK3 versus kK5. These differences permitted us to use chimeric molecules of mK3 and kK5 as well as of class I and B7.2 to address several important issues regarding mK3 function.
MATERIALS AND METHODS
Cell lines.
B6/WT3 murine embryo fibroblasts (MEFs; H-2b), L-Ld fibroblasts (H-2k), β2m-deficient MEFs (β2m−/−; H-2b), and tapasin-deficient MEFs (Tpn−/−; H-2b) have been described previously (12, 19, 20, 30). RMA/S (H-2b) (41) TAP2-deficient cells were obtained from the American Type Culture Collection (ATCC; Manassas, Va). 293T cells (8) were used for production of ecotropic retrovirus (to transduce B6/WT3, Tpn−/−, and β2m−/− cells), and ProPak-A.6 cells (ATCC) were used for production of amphotropic retrovirus for transduction of L-Ld and RMA/S cells. All cells were maintained in complete RPMI 1640 (Invitrogen, Carlsbad, Calif.) supplemented with 10% fetal calf serum (HyClone, Logan, Utah) as described previously (20). Where indicated, cells were cultured for 24 h in the presence of 100 U of mouse gamma interferon (IFN-γ)/ml (Biosource, Sunnyvale, Calif.).
DNA constructs and retroviral transductions.
By using reverse transcription-PCR, mouse and human B7.2 cDNAs were obtained from total RNA of C57BL/10 mouse splenocytes and MOU cells, a human Epstein-Barr virus-transformed B cell line, respectively. wt mK3 and kK5 constructs were generated by PCR amplification of the K3 gene from a γHV68 subclone (43) and the kK5 gene from a KSHV subclone (kindly provided by H. W. Virgin IV, Washington University School of Medicine), respectively. All mK3 truncations were made by PCR amplification of the particular region of mK3. For the generation of point mutations of mK3, a QuikChange mutagenesis kit (Stratagene, La Jolla, Calif.) was used according to the manufacturer's instructions. Chimeric molecules of Ld and human or mouse B7.2 or of mK3 and kK5 were made by overlap PCR. The correct sequences for all of the constructs were confirmed by DNA sequence analysis. For retroviral expression, two retroviral vectors, pMSCV-IRES-GFP (pMIG) (42) and pMSCV-IRES-Neo (pMIN), were used. They permit the gene of interest and the gene for green fluorescent protein (GFP) or neomycin resistance to be expressed from a single bicistronic mRNA. pMIG was used to express all of the mK3 and kK5 constructs, while pMIN was used for expression of all of the Ld and B7.2 constructs. Retrovirus-containing supernatants were produced as described previously (20) using either (i) the Vpack vector system (Stratagene) to generate ecotropic virus in 293T cells for infection of B6/WT3, Tpn−/−, and β2m−/− cells or (ii) ProPakA.6 cells to generate amphotropic virus for infection of L-Ld and RMA/S cells. Cells transduced by pMIN-containing retrovirus were enriched by neomycin selection. When necessary, the GFP+ cells from pMIG-transduced lines were enriched by cell sorting.
Antibodies.
Rabbit antisera to C-terminal sequences of mK3 (residues 167 to 187; anti-mK3-c) or N-terminal sequences of mK3 (residues 1 to 25; anti-mK3-n) were generated by immunization with keyhole limpet hemocyanin-coupled peptides. A rabbit serum against C-terminal sequences of kK5 (residues 240 to 256; anti-kK5-c) was produced by similar means. The anti-mouse tapasin monoclonal antibody (MAb) 5D3 and anti-mouse TAP1 serum have been previously described (13). Anti-GFP MAb was obtained from Covance (Princeton, N.J.), and anti-Ub MAb (PD41) was obtained from Santa Cruz Biotech (Santa Cruz, Calif.). MAbs B8-24-3 (ATCC), 28-14-8 (24), and 15-5-5 (25) were used to detect Kb, Db, and Dk, respectively. MAb 64-3-7 is specific for the α1 domain of open forms (unassembled) of Ld (34, 48), and MAb 30-5-7 is specific for the α2 domain of folded forms of Ld (34). MAbs GL1 for mouse B7.2 and FUN-1 for human B7.2 were obtained from BD Pharmingen (San Diego, Calif.).
Flow cytometry.
All flow cytometric analyses were performed using a FACSCalibur (Becton Dickinson, San Jose, Calif.). Data were analyzed using CellQuest software (Becton Dickinson). Staining was performed as described previously (47). Phycoerythrin-conjugated goat anti-mouse immunoglobulin G (IgG; BD Pharmingen, San Diego, Calif.) was used to visualize class I and human B7.2 expression. Phycoerythrin-conjugated mouse anti-rat Ig, κ light-chain MAb (BD Pharmingen) was used to visualize mouse B7.2 detection.
Immunoprecipitations and immunoblotting.
Coimmunoprecipitations were performed essentially as described previously (20, 47) by using 1.0% digitonin (Wako, Richmond, Va.) in lysis buffer (Tris-buffered saline, pH 7.4). For immunoblotting of cell lysates, cells were lysed in Tris-buffered saline containing 1.0% NP-40 (Sigma). Postnuclear lysates were mixed with lithium dodecyl sulfate sample buffer, and 2-mercaptoethanol (Sigma) was added to a final concentration of 1%. Immunoblotting was performed following separation of precipitated proteins or cell lysates by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (47). Biotin-conjugated goat anti-mouse IgG, goat anti-hamster IgG (both from Caltag, San Francisco, Calif.), or donkey anti-rabbit IgG (Jackson Immunoresearch, West Grove, Pa.) was used in the second step, followed by streptavidin-horseradish peroxidase (Zymed Laboratories, San Francisco, Calif.). Specific proteins were visualized by chemiluminescence using the ECL system (Amersham Biosciences, Piscataway, N.J.).
Metabolic labeling and pulse-chase.
After ≈45 min of preincubation in Cys/Met-free medium (Dulbecco's modified Eagle medium with 5% dialyzed fetal calf serum), cells (107 cells/ml) were pulse-labeled with Express [35S]Cys/Met labeling mix (Perkin-Elmer Life Sciences, Boston, Mass.) at 300 μCi/ml for 10 to 15 min. Chase was initiated by the addition of an excess of unlabeled Cys/Met (5 mM each). Immunoprecipitation and endoglycosidase H (endo H; ICN Pharmaceuticals, Costa Mesa, Calif.) treatment were performed as described previously (20). Samples were subjected to SDS-PAGE, and gels were treated with Amplify (Amersham), dried, and exposed to BioMax-MR film (Kodak, New Haven, Conn.).
RESULTS
mK3 and kK5 have disparate substrate specificities and TAP/tapasin dependencies in mouse cells.
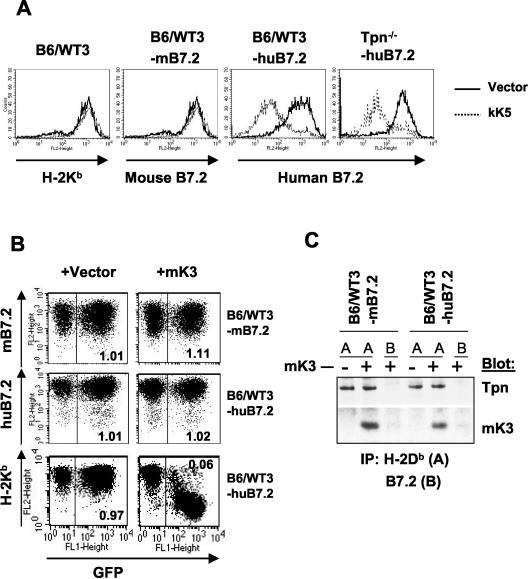
To define how mK3 interacts with class I molecules versus TAP/tapasin, we made chimeric molecules of mK3 and one of its homologs with distinct substrate specificity and TAP/tapasin dependency. kK5 is a good candidate for these comparisons because it does not downregulate mouse class I in either human or murine cells (36), yet it is similar to mK3 in sequence, domain organization, and topology. However, it was essential to first determine the substrate specificity and TAP/tapasin dependency of kK5 versus mK3 in murine cells. Flow cytometric analysis of pMIG-kK5-transduced mouse fibroblasts (B6/WT3; H-2b) indicated that kK5 downregulated human B7.2 but not mouse B7.2 and class I (Fig. 1A). Furthermore, in contrast to mK3, which failed to downregulate class I in a tapasin-deficient (Tpn−/−) cell line (12), kK5 downregulation of human B7.2 in the Tpn−/− cells was as efficient as it was in the wt cells (Fig. 1A). Thus, kK5 regulates human B7.2 in murine cells in a manner independent of substrate association with TAP/tapasin.
FIG. 1.
kK5 but not mK3 downregulates human B7.2 in mouse cells in a tapasin-independent manner. (A) wt (B6/WT3) and tapasin-deficient fibroblasts (Tpn−/−) (both H-2b) were first transduced with either human or mouse B7.2, followed by transduction with pMSCV-IRES-GFP (pMIG; vector) only or pMIG-kK5. Stable lines were analyzed for surface expression of human or mouse B7.2, or Kb, as indicated. The thick solid lines indicate staining of vector-only cells; dashed lines indicate staining of kK5-expressing cells. (B) wt cells expressing either mouse B7.2 (mB7.2) or human B7.2 (huB7.2) were transduced with pMIG (vector) only or pMIG-mK3 (mK3). After culture for 24 h with 100 U of IFN-γ/ml, surface B7.2 or Kb expression (y axis) versus GFP fluorescence (x axis) was monitored. The GFP+ and GFP− populations in each transductant represent the cells transduced or not transduced by pMIG-containing virus, respectively. The number in each plot represents the ratio of B7.2 or Kb staining between GFP+ and GFP− populations. (C) Digitonin cell lysates from the cells used in panel B were immunoprecipitated with MAb 28-14-8 for H-2Db (A) or GL1/FUN-1 for mouse/human B7.2 (B). Precipitates were then blotted with the antibody listed.
The ability of mK3 to target human and mouse B7.2 has not been reported but is particularly interesting since B7.2 is one of the specific targets of kK5. Furthermore, as shown in Fig. 2A, both mouse and human B7.2 possess multiple lysine residues in their cytoplasmic tails. More strikingly, in the cytoplasmic tail of human B7.2, the region proximal to the membrane contains a KKKKRP cluster that is required for maximal downregulation by kK5 (7), and this cluster is very similar to the KRRRNT cluster in the cytoplasmic tail of mouse class I molecules that is required for mK3 association (3). Based on sequence homologies in their cytoplasmic tails, mouse and human B7.2 were considered potential substrates for mK3-induced downregulation. The ability of mK3 to target B7.2 for degradation was tested in mouse or human B7.2-expressing B6/WT3 murine fibroblasts that were transduced with a bicistronic retroviral vector (pMSCV-IRES-GFP; pMIG) encoding mK3 plus GFP or GFP only (vector only). Flow cytometric analysis of these cells showed that the expressions of both mouse and human B7.2 were unaffected by mK3 expression, although class I expression on these cells was significantly reduced, as expected (Fig. 1B). Thus, mK3 had no effect on the surface expression of either mouse or human B7.2 molecules in spite of the presence in their tails of similar motifs that contain multiple lysine residues. Furthermore, consistent with its failure to downregulate B7.2 surface molecules, no physical association was observed between either mouse or human B7.2 and mK3 (Fig. 1C).
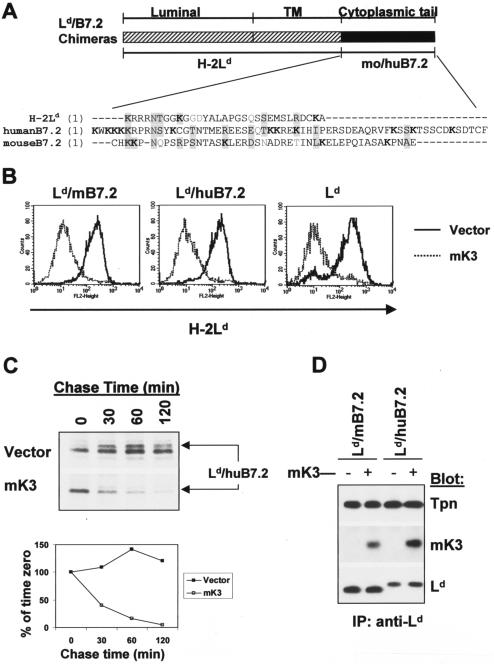
FIG. 2.
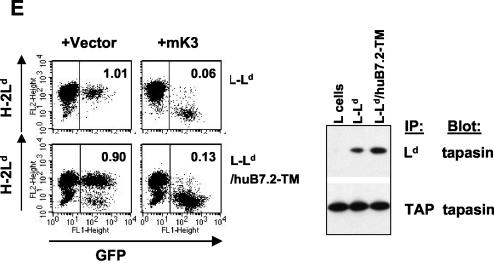
Ld/B7.2 chimeras are sensitive to mK3, and this correlates with their ability to associate with TAP/tapasin. (A) Schematic representation of Ld/B7.2 chimeras constructed with the luminal and TM regions of H-2Ld and cytoplasmic tail of either mouse B7.2 (Ld/mB7.2) or human B7.2 (Ld/huB7.2). A sequence alignment of the cytoplasmic tails of the indicated proteins is shown below the diagram. Lysine residues (K) are shown in bold. Identical or similar residues are shaded. (B) Constructs in panel A were introduced into B6/WT3 cells, followed by transduction with pMIG only (vector) or pMIG-mK3 (mK3). Sorting was used to enrich for GFP+ cells. Cells were incubated for 24 h with 100 U of IFN-γ/ml and then analyzed for expression of wt Ld or the Ld/B7.2 constructs by using MAb 30-5-7. Solid lines, staining of vector-only cells; dashed lines, staining of the mK3-expressing cells. (C) IFN-γ-treated cells were pulse-labeled with [35S]Cys/Met and chased for the indicated times with unlabeled Cys/Met. Ld/huB7.2 molecules were precipitated with MAb 64-3-7. Precipitates were resolved by SDS-PAGE and visualized by autoradiography. Ld/huB7.2 bands are indicated (upper panel). In the lower panel, relative band intensities from the gels are plotted as a percentage of the intensity at time zero for each cell line. (D) Digitonin cell lysates from cells were immunoprecipitated with anti-Ld MAb 64-3-7. Precipitates were then blotted with the antibodies listed. Loading of each sample was normalized on the basis of an equal level of Ld/B7.2 molecules. (E) The Ld/huB7.2-TM construct was introduced into L cells. Surface Ld expression was monitored (MAb 30-5-7) after transduction of the cells with pMIG (vector) or pMIG-mK3. Right panels show anti-tapasin immunoblotting of anti-TAP-1 or anti-Ld precipitates from the indicated cell lines. The Ld/huB7.2-TM construct consists of the luminal domain of Ld (residues 1 to 282) followed by the TM and cytoplasmic tail of human B7.2 (residues 225 to 306).
The cytoplasmic domain of B7.2 supports mK3-mediated degradation when appended to class I.
To determine whether the cytoplasmic tail or the luminal portion of B7.2 is responsible for its failure to be targeted by mK3, two chimeric molecules were made with the luminal and TM domains of the class I molecule Ld and the tail of either mouse or human B7.2 (designated Ld/mB7.2 or Ld/huB7.2) (schematized in Fig. 2A). These Ld/B7.2 chimeras or wt Ld were then coexpressed with or without mK3 in B6/WT3 cells. In contrast to wt B7.2 molecules, both chimeric molecules (Ld/mB7.2 and Ld/huB7.2) displayed a greater-than-fivefold surface reduction in the presence of mK3 (Fig. 2B). The extent of downregulation of these chimeras was comparable to those of intact Ld and endogenously expressed Kb proteins in the same cells (Fig. 2B and data not shown). To determine whether surface downregulation of the Ld/B7.2 chimeric molecules resulted from their rapid intracellular turnover, we next compared the half-lives of Ld/B7.2 molecules in the presence or absence of mK3 by pulse-chase analysis. As shown in Fig. 2C, Ld/huB7.2 molecules were turned over with a half-life of less than 30 min in the presence of mK3, whereas no detectable turnover was observed in the absence of mK3 within the 2 h tested. Similar findings were also observed with the other chimeric molecule, Ld/mB7.2 (data not shown). The presence of a cytoplasmic tail on Ld was required for rapid degradation, since Ld molecules that completely lacked a cytoplasmic tail were not rapidly degraded in the presence of mK3 although associated with TAP/tapasin (unpublished observation). In addition, primarily endo H-sensitive forms of both chimeric molecules were Ub conjugated in the presence of mK3 when examined by immunoblotting with anti-Ub antibody (data not shown). These data demonstrate that mK3 is capable of functionally interacting with the tails of mouse and human B7.2, since it induces the ubiquitination and rapid intracellular turnover of these chimeric molecules. Furthermore, these findings suggest that the mechanism by which mK3 regulates expression of the Ld/B7.2 chimeric molecules is analogous to that previously defined for mK3 regulation of class I proteins.
To extend these observations, we examined the ability of mK3 to regulate an additional chimeric molecule consisting of the luminal domain of Ld linked to the cytoplasmic and TM domains of human B7.2 (Ld/huB7.2-TM). It was important to consider a possible contribution of the Ld TM domain to mK3 targeting, since class I TM domains have been implicated in binding to kK5 and kK3 (7, 33). However, as shown in Fig. 2E, this chimeric molecule displayed sensitivity to mK3-mediated surface downregulation that was comparable to that of wt Ld. Therefore, mK3 does not require sequences unique to class I in the TM or cytoplasmic domain in order to target class I turnover.
Correlation between mK3 susceptibility and TAP/tapasin association of the Ld/B7.2 chimeric molecules.
Given that the class I luminal domain rendered the Ld/huB7.2 chimeras susceptible to mK3 regulation, we next examined whether this regulation correlated with TAP/tapasin interaction. It should be noted that class I directly associates with tapasin and not TAP (32), though tapasin and TAP are always detected as a complex in cells that express both (18, 32). Sites located in the luminal domains of class I profoundly affect its association with TAP/tapasin (45), although the cytoplasmic tail of class I may also influence association with TAP/tapasin (4). Therefore, we examined the TAP/tapasin association status of B7.2 and the Ld/B7.2 chimeras and observed that the various Ld/B7.2 constructs all associated with TAP/tapasin (Fig. 2D and E) but that intact B7.2 did not (Fig. 1C). These results demonstrate that TAP/tapasin association correlates with mK3 association for the Ld/B7.2 molecules, and this association leads to mK3-mediated ubiquitination and degradation. This indicates that human and mouse B7.2 are resistant to mK3 regulation due to a failure to interact with TAP/tapasin and not because they lack particular sequence motifs in their cytoplasmic tails that are necessary for recognition. Thus, the substrate specificity (class I versus B7.2) of mK3-targeted degradation is primarily determined by the ability of the substrate to bind the host proteins TAP and tapasin rather than the direct interaction of mK3 with the substrate. In addition, the fact that kK5 does target human B7.2 (and not mouse B7.2 or class I) in mouse cells in a TAP/tapasin-independent manner, validates the use of mK3/kK5 chimeras to study how mK3 interacts with class I versus TAP/tapasin.
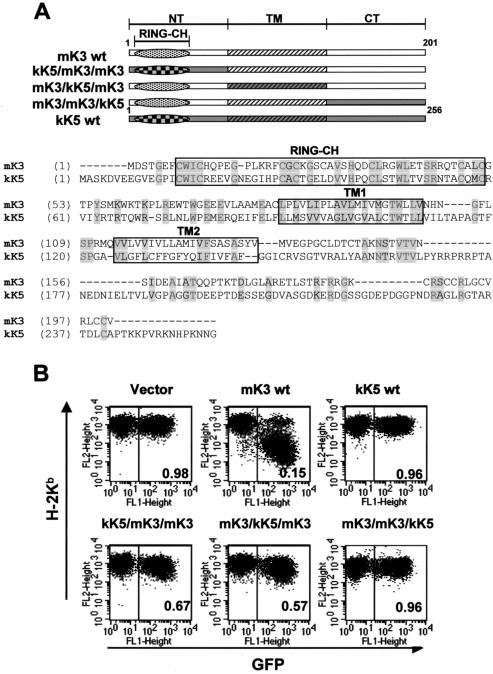
Replacement of the N-terminal, TM, or C-terminal domain of mK3 with analogous domains of kK5 impairs its ability to regulate class I. To define which domains of mK3 are responsible for TAP/tapasin interaction versus class I regulation, several chimeric constructs consisting of mK3 and kK5 were made. These chimeric constructs take advantage of the fact that the kK5 protein of KSHV does not downregulate mouse class I proteins and does not associate with TAP/tapasin yet is similar to mK3 in terms of domain organization and topology (33, 36). Therefore, analogous domains of kK5 can be used to replace the corresponding domains of mK3 without interfering with the overall topology, which could occur when an entire domain is deleted. All mK3/kK5 chimeras are shown schematically in Fig. 3A and were introduced into B6/WT3 cells by using the aforementioned pMIG retroviral vector. As a control, wt kK5 transductants were also generated. As expected, intact mK3 sharply reduced class I expression, whereas intact kK5 had no effect on class I expression. Using different mK3/kK5 chimeras, we observed that replacement of the C-terminal domain of mK3 with that of kK5 (mK3/mK3/kK5) profoundly abolished its ability to downregulate class I. Further, replacement of the TM domains of mK3 with analogous kK5 domains (mK3/kK5/mK3) also impaired class I regulation (Fig. 3B). Thus, mK3-specific information in the C-terminal and TM regions is required for mK3 function. Surprisingly, the N-terminal domain swap (kK5/mK3/mK3) also displayed an impaired ability to downregulate class I in spite of the fact that mK3 and kK5 share a highly conserved RING-CH motif in their N-terminal domains. Furthermore, the RING-CH domain of kK5 is clearly functional in murine cells, at least in terms of its ability to regulate human B7.2 molecules (Fig. 1). These findings demonstrate that additional information in the N-terminal domain of mK3 besides the RING-CH motif influences class I regulation and/or that the minor sequence differences between the RING-CHs of mK3 and kK5 are important. These possibilities were explored further by comparing the entire N-terminal domain swap with a construct in which only the RING-CH motif was swapped (see below). In any case, mK3-specific information in all three domains, N terminus, TM, and C terminus, is required for mK3 to effectively regulate the expression of class I molecules.
FIG. 3.
Replacement of the N-terminal (NT), TM, or C-terminal (CT) domain of mK3 with analogous domains of kK5 affects surface downregulation. (A) Schematic description and nomenclature of mK3 chimeras with KSHV kK5 (upper portion). An alignment of mK3 and kK5 sequences is shown in the lower portion. The numbers indicate the first and last amino acids for full-length mK3 or kK5. (B) Each mK3 chimera was introduced into B6/WT3 cells by using the pMIG vector. An analysis of surface Kb expression versus GFP fluorescence is shown. The number in each plot represents the ratio of Kb staining intensities between GFP+ and GFP− populations. Analysis of Db yielded very similar results (data not shown).
The C-terminal domain of mK3 is the major domain responsible for interaction with TAP/tapasin. We next examined which domain(s) of mK3 is primarily responsible for its interaction with TAP/tapasin. To do so, various mK3 mutants were tested for functional interaction with TAP/tapasin in β2m−/− MEF cells. Using these cells, Lybarger et al. previously described a highly sensitive assay to detect the functional interaction of mK3 with TAP/tapasin (20). The principle of this assay rests on the fact that the interaction with TAP/tapasin is required for stabilization of mK3, and the β2m−/− fibroblasts normally express barely detectable levels of TAP/tapasin. Consequently, the steady-state levels of mK3 are very low in these cells. However, treatment of these cells with IFN-γ dramatically increases TAP and tapasin expression (>10-fold), leading to a commensurate increase in mK3 protein levels (20). It should be noted that (i) the IFN-γ-induced increase in the steady-state level of mK3 is not seen in the absence of TAP1 or tapasin (20) and (ii) the expression levels of mK3 are not noticeably affected by IFN-γ treatment, as indicated by the expression of GFP, which is expressed from the same mRNA as mK3. Therefore, detection of IFN-γ-induced changes in mK3 protein levels is a sensitive means to assess TAP/tapasin interaction with mK3 mutants, including ones present at low steady-state levels.
To define regions of mK3 important for TAP/tapasin interaction, various mK3 deletion constructs or chimeras were introduced into β2m−/− MEF cells via transduction with the pMIG vector. Transduced cells were enriched by sorting the GFP+ population. The GFP levels of the corresponding cell lysates then serve as an equal loading control as well as an indicator of the effect of IFN-γ treatment on the expression levels of the introduced vector (which is minimal). As previously observed, the level of wt mK3 increased more than five times after IFN-γ treatment, demonstrating significant interaction between mK3 and TAP/tapasin (Fig. 4, left panel). Similarly, the chimeric molecule (kK5/mK3/mK3) generated by replacement of the N-terminal domain of mK3 with that of kK5 was highly inducible by IFN-γ treatment. A high level of induction of the kK5/mK3/mK3 chimera with IFN-γ was also observed in a Kb−/− Db−/− β2m−/− triple knockout MEF line (20), confirming the class I independence of this response (data not shown). Therefore, the entire mK3 N-terminal domain can be replaced without a loss of TAP/tapasin interaction, indicating that it does not contribute to this interaction. In addition, the lack of class I regulation by this mutant cannot be explained by a failed association with the peptide-loading complex (including class I).
FIG. 4.
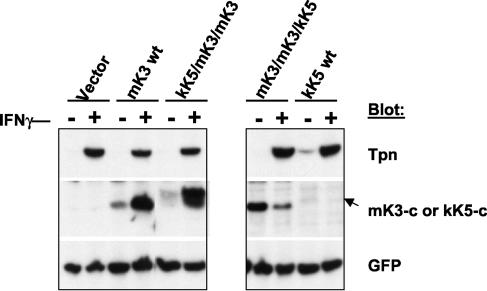
C-terminal domain of mK3 is required for functional interaction with TAP/tapasin. Each mK3 chimera in Fig. 3A was introduced into β2m−/− cells (H-2b) by using pMIG, followed by sorting to enrich the GFP+ fraction. Following culture with or without 100 U of IFN-γ/ml for 24 h, lysates from the cells were blotted with the indicated antibodies. The GFP signal intensity serves as a loading control. The arrow indicates the position of wt kK5.
The blots shown in the right panel of Fig. 4 were obtained using an antibody to the C-terminal domain of kK5 (anti-kK5-c). The wt kK5 was detected only at a low steady-state level by immunoblotting, although by metabolic labeling and immunoprecipitation, we confirmed its specific expression and migration in the gel. By contrast, the replacement of the C-terminal domain of mK3 with that of kK5 resulted in a very stable chimeric molecule (mK3/mK3/kK5). Although the significance of their stability difference remains unclear, neither wt kK5 nor mK3/mK3/kK5 was inducible by IFN-γ treatment. Thus, mK3-specific information in the C-terminal domain is required to confer functional TAP/tapasin interaction. Interestingly, expression of the mK3/mK3/kK5 chimera was reduced in the presence of IFN-γ, suggesting that TAP/tapasin association may protect against IFN-γ-induced degradation.
It has been reported that the TM regions of the KSHV proteins kK3 and kK5 mediate target selectivity (33). It is not known whether the TM domains of mK3 are likewise involved in TAP/tapasin interaction and, thus, substrate specificity. To examine this, we tested the mK3/kK5/mK3 chimera, in which the TM region of mK3 was replaced by kK5. In spite of comparable levels of translation as evidenced by GFP expression, the mK3/kK5/mK3 chimeric molecule was very unstable and displayed a more rapid turnover than wt mK3 (data not shown). In addition, the steady-state level of this chimeric molecule did not increase in the presence of IFN-γ, implying poor TAP/tapasin association (data not shown). As shown in Fig. 3B, the mK3/kK5/mK3 chimera also displayed poor class I regulation. Thus, the TM regions of mK3 are clearly important for the stability of the mK3 protein, which in turn may secondarily affect its ability to regulate class I proteins. Taken together, the above data demonstrate that the C-terminal domain of mK3 is critical for its specific association with TAP/tapasin, although an additional role for the TM domains cannot be excluded.
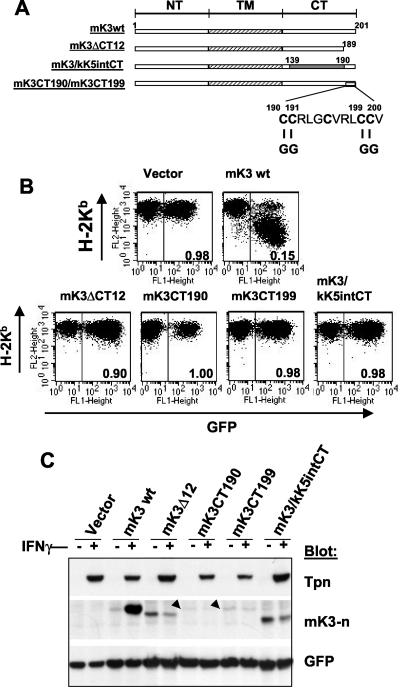
Information in the final 12 residues of the C-terminal domain of mK3 is essential for TAP/tapasin interaction.
To more precisely map the C-terminal region critical for TAP/tapasin association, 40-, 24-, and 12-amino-acid deletions of the C terminus of mK3 were made. Unlike the case with kK5 and kK3, where a large portion of their C termini can be deleted without significantly affecting their activity against class I (22, 33), the 40-, 24-, and 12-amino-acid deletions in the C terminus of mK3 all resulted in impaired regulation of class I as well as poor TAP/tapasin interaction (Fig. 5B for the 12-residue deletion; data not shown for the others). A unique characteristic of the C-terminal 12-amino-acid fragment is its high cysteine content (5 of 12 residues [Fig. 5A]), which is quite different from kK5 and kK3. To determine whether these cysteine residues play a role in the interaction with TAP/tapasin and mK3 function, two additional mutations, mK3 C→G at residues 190 and 191 (mK3CT190) and mK3 C→G at residues 199 and 200 (mK3CT199), were generated (Fig. 5A). Strikingly, neither of these double mutants displayed an increase in their steady-state levels in β2m−/− MEF cells treated with IFN-γ (Fig. 5C), indicating an inability to associate with TAP/tapasin. This correlated with an inability to regulate surface class I levels (Fig. 5B). To determine whether information in these 12 residues of mK3 is sufficient to confer TAP/tapasin interaction, a construct was engineered with these 12 residues attached to a spacer of the same length as the remaining portion (residues 1 to 139) of wt mK3. The spacer was derived from analogous sequence in kK5 (residues 147 to 194) (mK3/kK5intCT [Fig. 5A]). As shown in Fig. 5B and C, this mK3 construct did not regulate class I or display functional association with TAP/tapasin. Thus, we conclude that the cysteine-rich final 12 amino acid residues in the tail of mK3 are necessary but not sufficient to provide TAP/tapasin interaction. The importance of these cysteine residues is intriguing since a mutant with an arginine-to-isoleucine point mutation at residue 197 within the final 12 amino acids was still responsive to the IFN-γ induction though its steady-state level was relatively low compared with that of wt mK3 (data not shown). Consistent with the fact that the N- and C-terminal domains of mK3 protrude from the ER membrane into the cytosol, we found no evidence for the presence of cysteine-mediated intra- or interchain disulfide bonds involving mK3; mK3 mobilities in both reducing and nonreducing gels were essentially identical (data not shown). Interestingly, free cysteine residues can be important sites for posttranslational modifications that may influence protein-protein interaction (35).
FIG. 5.
Information in the final 12 amino acids of mK3 is critical for TAP/tapasin interaction. (A) Schematic depiction of the C-terminal domain (CT) mutants of mK3. NT, N-terminal domain. In the mK3/kK5intCT construct, mK3 residues 140 to 189 were replaced by kK5 residues 147 to 194. (B) Each mutant was introduced into B6/WT3 cells. Flow cytometric analysis of Kb versus GFP fluorescence is shown. The numbers in each plot represent the ratio of Kb staining intensities between GFP+ and GFP− fractions. Analysis of Db expression yielded very similar results (data not shown). (C) The GFP+ cells from each line shown in panel B were enriched by sorting. Lysates from these cells were blotted with the indicated antibodies following culture for 24 h with or without 100 U of IFN-γ/ml. GFP staining levels serve as a loading control as well as an indicator of the effect of IFN-γ on pMIG expression. Bands corresponding to mK3CT190 and mK3CT199 are indicated by arrowheads.
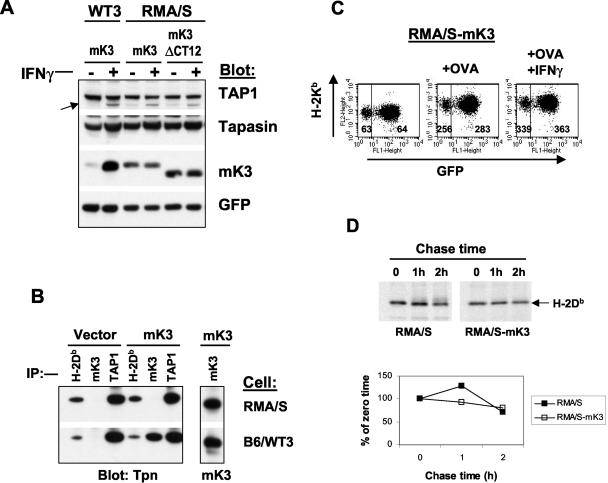
Both TAP1 and TAP2 are required for mK3 regulation of class I.
A previous study demonstrated that mK3 fails to induce the rapid degradation of class I in the absence of TAP1 or tapasin, and reintroduction of TAP1 or tapasin into these deficient cells results in marked increases in steady-state mK3 levels (20). To determine whether these findings can be extended to TAP2, we tested mK3 stabilization and class I regulation in TAP2-deficient RMA/S cells. Surprisingly, we detected significant levels of steady-state mK3 in RMA/S cells (Fig. 6A). However, the relatively high levels of mK3 in RMA/S were not due to mK3 interaction with TAP1 and tapasin, since comparable levels of expression of mK3ΔCT12 and wt mK3 were observed (Fig. 6A). Accordingly, mK3 levels were unchanged in RMA/S cells following IFN-γ induction, although tapasin and TAP1 levels were clearly inducible (Fig. 6A). Furthermore, coimmunoprecipitation failed to detect association between mK3 and TAP1/tapasin in RMA/S cells, though mK3-TAP/tapasin association was readily observed in wt cells; similar levels of mK3 were present in both lines (Fig. 6B). Thus mK3 is not efficiently brought into association with TAP1/tapasin in RMA/S cells, even though TAP1, tapasin, and class I molecules associate in the absence of TAP2 (1, 29, 31) (Fig. 6B). Failed association between mK3 and TAP1/tapasin in RMA/S cells correlated with the inability of mK3 to downregulate Kb expression on cells cultured with the Kb-binding peptide OVA (5) or OVA plus IFN-γ (Fig. 6C). This treatment of RMA/S cells resulted in a substantial surface induction of Kb (four- to fivefold) that was unaffected by mK3 (Fig. 6C). Further, mK3 did not induce the rapid turnover of nascent Db molecules in RMA/S cells (Fig. 6D). The lack of mK3 function in TAP2-deficient RMA/S cells is in accordance with the previously observed lack of class I regulation by mK3 in TAP1-deficient MEFs (20). Therefore, these combined findings reveal that mK3 function requires both TAP1 and TAP2. It should be noted that subsequent to submission of this manuscript, the Stevenson group reported on the function of mK3 in RMA-S cells (2). They, too, observed that mK3 was nonfunctional in the absence of TAP-1. However, their data indicate an association between mK3 and TAP-1 in the absence of TAP-2. We did not observe such an association as detected by coimmunoprecipitation or IFN-γ induction (Fig. 6). In fact, all of our available data indicate that mK3 association with the peptide-loading complex requires both TAP subunits for mK3 association and stabilization. The apparent discrepancy between the results of these studies remains unclear but may reflect a quantitative difference in the amount of mK3 associated with TAP-1 in the absence of TAP-2, consistent with a contributing role of TAP-2 in mK3 association with the peptide-loading complex. Although tapasin is clearly required for mK3 stabilization and function as well, our data suggest that it is not a direct binding partner for mK3, since TAP1- and TAP2-deficient cells both display very high tapasin levels following IFN-γ treatment, yet mK3 levels remain unchanged (Fig. 6A and data not shown). This dependency on TAP could reflect the fact that mK3 must bind to both TAP1 and TAP2 or that mK3 binding is dependent on a conformational change that occurs within TAP1 and/or TAP2 when they associate.
FIG. 6.
Both TAP1 and TAP2 are required for mK3 function. (A) B6/WT3 cells and TAP2-deficient cells (RMA/S) were transduced with mK3 or mK3ΔCT12 (see Fig. 5A) by using pMIG, followed by sorting to enrich for GFP-expressing cells. After culture with or without 100 U of IFN-γ/ml for 24 h, lysates from these cells were immunoblotted with the antibodies listed. GFP staining indicated equivalent expressions of the vector in both lines. The arrow indicates TAP1. (B) Lysates from RMA/S or B6/WT3 (with or without mK3) were immunoprecipitated with the indicated antibodies. Precipitates were blotted as indicated. (C) Kb expression on RMA/S cells with mK3 that were cultured in the presence or absence of the Kb binding peptide OVA (200 μM) (5) with or without 100 U of IFN-γ/ml for 24 h. The numbers represent the geometric mean fluorescence of Kb expression for the GFP+ or GFP− population. (D) RMA/S cells with or without mK3 were pulse-labeled with [35S]Cys/Met and chased for the indicated times. H-2Db immunoprecipitates (MAb 28-14-8) were resolved by SDS-PAGE, and labeled proteins were visualized by autoradiography (upper panel). The lower panel shows the relative band intensities from the gels as a percentage of the intensity at time zero for each cell line.
Unique information in the N terminus of mK3 both within and outside the RING-CH domain is important for class I ubiquitination.
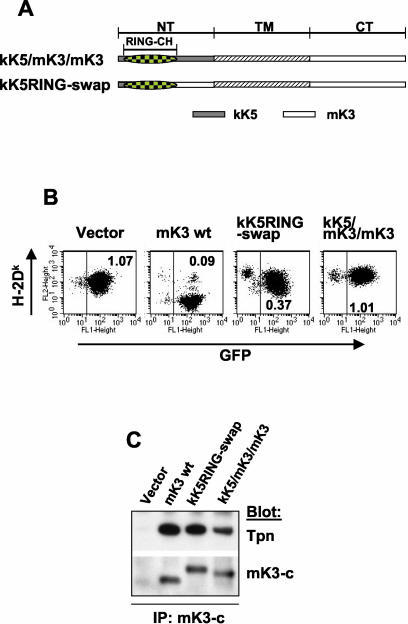
We showed above that the kK5/mK3/mK3 chimera, in which the entire N-terminal region of mK3 was replaced by kK5, had a similar capacity to interact with TAP/tapasin as mK3 (Fig. 4), yet it was incapable of downregulating surface class I expression (Fig. 3B). This indicates that unique information in the N-terminal domain of mK3 not shared with the N-terminal domain of kK5 must be important for Ub conjugation in mouse cells. Given that the identity between kK5 and mK3 in the RING-CH domain region is 40% and the rest of N-terminal region is only 20% identical, we generated another chimeric molecule with the RING-CH of kK5 (and a short upstream segment) replacing the homologous sequence in mK3 (kK5RING-swap) (Fig. 7A). Comparisons were made between wt mK3, kK5RING-swap, and the kK5/mK3/mK3 chimera to determine which regions of the N-terminal domain are important for class I-specific regulation in mouse cells (L-Ld). As shown in Fig. 7B, kK5RING-swap showed a moderate downregulation of surface class I compared with wt mK3, whereas the kK5/mK3/mK3 chimera had no effect on the surface expression of class I. A similar pattern was also observed in B6/WT3 cells, where the kK5/mK3/mK3 chimera displayed no obvious downregulation and the kK5RING-swap displayed intermediate downregulation compared to what was seen with wt mK3 (data not shown). The failure of these chimeric molecules to efficiently regulate class I did not result from their inability to interact with TAP/tapasin, as determined using (i) IFN-γ induction in β2m−/− cells or triple knockout (Kb−/− Db−/− β2m−/−) MEFs (Fig. 4 and data not shown) or (ii) coimmunoprecipitation of mK3 followed by anti-tapasin immunoblotting (Fig. 7C). However, consistent with the surface expression data, kinetic analysis showed that the half-life of newly synthesized Ld molecules in the presence of kK5RING-swap was much longer (t1/2 ≈ 2 h) than that in the presence of wt mK3 (t1/2 ≈ 30 min), while kK5/mK3/mK3 had no obvious effect on the stability of newly synthesized Ld proteins (Fig. 7D). Thus, sequences in the N-terminal domain of mK3 outside of its RING-CH domain are involved in its ability to regulate class I proteins without affecting its ability to associate with TAP/tapasin.
FIG. 7.
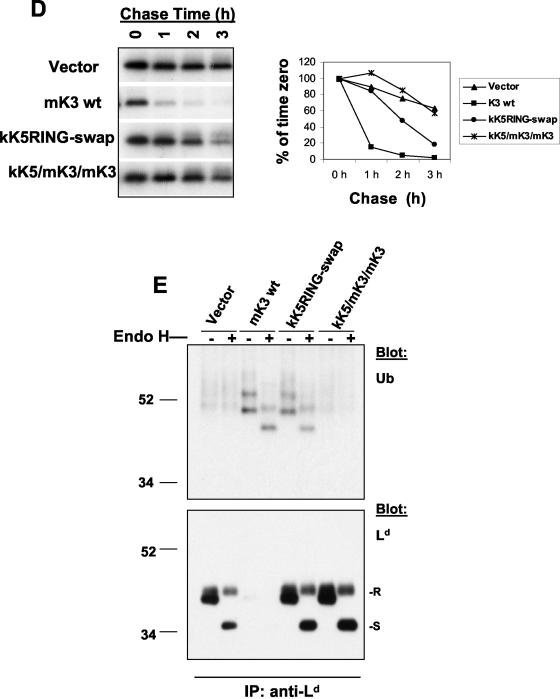
Unique information in the N-terminal domain of mK3 not shared with kK5 is involved in its E3 ligase activity. (A) Schematic depiction of two mK3/kK5 chimeras in which different portions of the N-terminal domain of mK3 were replaced by the corresponding portions of kK5. (B) L-Ld cells were transduced with constructs encoding the chimeras or wt mK3, and the GFP+ transductants were enriched by cell sorting. Dk expression (MAb 15-5-5) versus GFP intensity is shown. The numbers indicate the ratio of Dk staining intensities between GFP+ and GFP− populations in each line. Similar profiles were observed with Ld expression (data not shown). (C) Digitonin lysates from the cells in panel B were precipitated with rabbit anti mK3-c serum. Precipitates were then blotted with the antibodies listed. Note that the mK3 blot indicates comparable steady-state levels of both chimeras and wt mK3. (D) Cells were pulse-labeled with [35S]Cys/Met and chased for the indicated times. Lysates were precipitated with a mixture of anti-Ld MAbs 64-3-7 and 30-5-7. Following SDS-PAGE, labeled Ld molecules were visualized by autoradiography (left panel). Relative band intensities from the gels in the left panel are plotted as a percentage of the intensity at time zero for each cell line (right panel). (E) Digitonin lysates from the indicated cell lines were precipitated with MAb 64-3-7. After treatment with or without endo H, precipitates were blotted with either anti-Ub MAb PD41 (upper panel) or anti-Ld (MAb 64-3-7) (lower panel). In the lower panel, R and S indicate endo H-resistant and -sensitive forms of the heavy chain, respectively. Note that ubiquitinated Ld bands can only be detected in the presence of wt mK3 or kK5RING-swap (upper panel), and these bands shift down upon endo H digestion, confirming their endo H-sensitive nature.
To determine which N-terminal domain sequences of mK3 determine E3 ligase activity, we next examined whether the class I H chain is Ub conjugated in the cells expressing kK5RING-swap or kK5/mK3/mK3 chimeric molecules. Class I Ld H chains were immunoprecipitated from each cell line and then blotted with anti-Ld to detect class I H chains or with anti-Ub to monitor E3 ligase activity. As expected (47), there was a dramatic reduction of total class I H chains in the presence of wt mK3 (visible only with longer exposure) (Fig. 7E, lower panel). By contrast, high steady-state levels of H chains were detected in cells expressing both the kK5/mK3/mK3 and the kK5RING-swap chimeric constructs (Fig. 7E, lower panel). Furthermore, consistent with its complete failure to cause degradation of class I, ubiquitinated H chains were not observed in kK5/mK3/mK3-expressing cells (Fig. 7E, upper panel), indicating that the RING-CH of kK5 (within the native N-terminal domain) displays no detectable E3 ligase activity though it interacts with TAP/tapasin. Interestingly, class I H chains in the presence of the kK5RING-swap displayed a ubiquitination pattern very similar to that of H chains in the presence of wt mK3; this ubiquitination affected predominantly ER-resident (endo H-sensitive) H chains (Fig. 7E, upper panel). Thus, the RING-CH of kK5 can form detectable levels of Ub-conjugated class I when expressed in the context of the additional N-terminal domain sequences of mK3. This reveals that the N-terminal domain of mK3 outside of the RING-CH motif contains information that is required for ubiquitination of class I. It should be noted that the kK5RING-swap construct is clearly not as proficient at targeting the degradation of class I as wt mK3, as shown by the high steady-state levels of endo H-resistant H chains (Fig. 7E, lower panel). Comparison of the amount of class I H chain versus the amount of Ub-conjugated forms in the presence of kK5RING-swap or the wt mK3 suggests that the E3 ligase activity of the RING-CH swap is inefficient. These data indicate that the kK5 RING-CH domain can function as an E3 ligase in the context of mK3 in mouse cells but not as efficiently as the native RING-CH domain of mK3. Conversely, the mK3 RING-CH in the context of kK5 is completely unable to downregulate human B7.2 (or class I) in mouse cells compared to what was seen with wt kK5 (data not shown). Thus, while tethered in the ER to TAP/tapasin, the RING-CH of mK3 may only recruit ER-related cellular ubiquitination machinery, such as an ER-associated E2. In any case, both reciprocal RING-CH swaps between mK3 and kK5 have impaired functional activity, highlighting their unique host adaptation.
DISCUSSION
Ub-mediated signals are known to play a central role in many basic cellular processes, including quality control of cellular proteins as well as regulation of signal transduction, cell cycle, gene transcription, and intracellular trafficking. The nature of its role requires Ub-mediated degradation to be highly substrate specific. Where defined, substrate specificity appears to be mediated by specific ubiquitination signals, which can be sequence or structural features of the substrate recognized by E3 ligases (17, 28). The sequences or structural requirements for mK3-mediated Ub conjugation of MHC class I remain unclear. It is apparent that the cytoplasmic tail of class I is important for mK3 targeting (3). Indeed, we have observed that a tailless Ld molecule was not rapidly degraded by mK3 but was associated with TAP/tapasin and mK3 (X. Wang and L. Lybarger, unpublished data). However, molecules that are not substrates of mK3 (such as human B7.2) have similar cytoplasmic tails. Additional clues into mK3 substrate selection were provided by the previous study of Lybarger et al., which demonstrated that mK3 regulation of class I requires an interaction between mK3 and the host MHC class I-dedicated ER accessory proteins TAP and tapasin (20). Data presented here provide compelling evidence that substrate binding to TAP/tapasin is the primary determinant of susceptibility to mK3, as opposed to the presence of an mK3-specific recognition motif uniquely within the tail of class I. Indeed, B7.2 molecules have a tail that supports mK3-mediated degradation, but only when transferred to Ld, which provides the requisite interaction with TAP/tapasin.
Our results point to promiscuity in terms of the cytoplasmic tail sequences that can support mK3-mediated degradation of substrate molecules that enter the peptide-loading complex. It has been reported that lysine residues in the cytoplasmic tails of substrates are important for mK3 and its homologs to mediate the Ub conjugation which links the substrates to their degradation (3, 7). It is noteworthy that tapasin also possesses multiple lysine residues in its cytoplasmic tail and is capable of associating with mK3 yet is not rapidly degraded in the presence of mK3 (47). These observations permit us to propose a model wherein mK3-mediated ubiquitination is restricted by the physical proximity and orientation of its RING-CH domain in relation to the lysine residues in the tail of the substrate. Therefore, it is attractive to speculate that mK3 binding to TAP/tapasin provides the proper proximity and/or orientation for the RING-CH domain of mK3 to interact primarily with the cytoplasmic tail of class I molecules (Fig. 8).
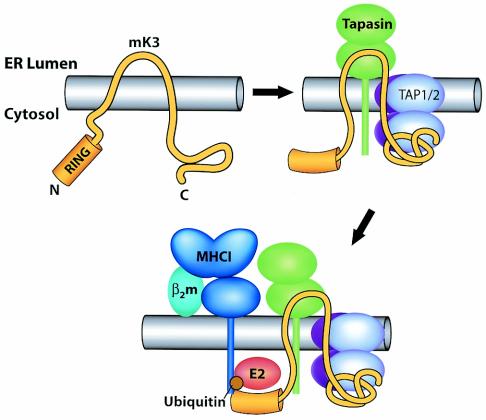
FIG. 8.
Proposed proximity model depicting how mK3 interacts with host proteins to specifically target the rapid degradation of class I proteins. Upper left, nonconformed mK3. Upper right, mK3 forms a complex with TAP/tapasin in a class I-independent manner. The association of mK3 with TAP/tapasin is primarily mediated through the C-terminal domain of mK3, and this interaction may induce a conformational change within mK3 that not only stabilizes mK3 but also restricts the mobility of its RING-CH domain in the N terminus to achieve the proximity, conformation, and/or orientation required for its E3 ligase function. In this model, tapasin, TAP1, and TAP2 are all essential components of this functional interaction with mK3, although our results suggest that TAP1 and -2 are the direct binding partners of mK3. Bottom, an ER-related E2-conjugating enzyme and/or another necessary factor(s) are recruited by the interactions with the N-terminal region of mK3 (both within and outside the RING-CH domain). These interactions result in the conjugation of Ub to the class I molecules. The Ub conjugation is specific for class I proteins due to a required proximity and/or conformation or orientation imposed by mK3 binding to tapasin/TAP1-tapasin/TAP2 complexes. Thus, substrate specificity is determined by interaction with TAP/tapasin, rather than a specific targeting sequence in the tail of class I.
The finding that kK5RING-swap retains partial Ub ligase activity yet the entire N-terminal domain swap (kK5/mK3/mK3) completely abolishes the target ubiquitination suggests that the region flanking the RING-CH domain is involved in the recruitment of ubiquitination components such as E2-conjugating enzymes. This finding is in agreement with general structure-function properties shared by all single-subunit RING E3 ligases examined thus far, where E2 binding depends on interactions with flanking residues as well as with the RING domain (28). Indeed, the crystal structure of the c-Cb1-UbcH7 (a RING finger E3 and its cognate E2) complex revealed that a linker between the RING domain and the substrate binding domain forms part of the interface with the E2 (49). It is interesting that the entire kK5 N-terminal domain is nonfunctional in the context of mK3. This is distinct from the KSHV system, where the entire N-terminal domain of kK5 can be replaced with the corresponding domain from kK3 without a loss in function (33). The kK5 RING-CH domain can function to downregulate human B7.2 in the context of the wt kK5 molecule in mouse cells (Fig. 1A). This demonstrates that the kK5 RING-CH domain is capable of efficiently exploiting at least some of the mouse ubiquitination components. Thus, the inefficiency of the kK5 RING-CH in the context of mK3 could reflect a distinct requirement related to the different sites where mK3- and kK5-mediated ubiquitination takes place (ER versus post-ER). More specifically, this inefficiency could be attributed to a defect in the interaction of the kK5 RING-CH domain with ER-related E2 enzymes (e.g., MmUbc6 and MmUbc7) (39).
How a substrate, once ubiquitinated, gets sentenced to destruction in different subcellular locations is still a puzzle for Ub-induced protein degradation. It is evident that the fates of ubiquitinated proteins depend in some cases on the types of Ub conjugates formed. For instance, poly-Ub chains linked through the lysine 48 residue of Ub usually target proteins to proteasome (14, 27, 44), whereas a mono-Ub tag usually links targets to diverse proteasome-independent cellular functions, including endocytosis and lysosome degradation (16). In addition, intracellular location helps to determine the fate of ubiquitinated proteins: ubiquitination in the nucleus might not have the same consequence as that in the cytosol, and ubiquitination of a TM protein at the ER membrane might have a different result from that at the plasma membrane (14, 44). Here, we show that the majority of ubiquitinated H chains in the presence of the kK5 RING-CH chimera are endo H sensitive and the pattern of ubiquitinated H chains is indistinguishable from that seen in wt mK3-expressing cells. This similarity would indicate that the RING-CH domain is probably not the factor determining the ultimate fate (site of degradation) of the target molecules. We therefore speculate that the distinct strategy for substrate selection used by mK3, versus that for kK3 and kK5, is important in deciding the fate of its ubiquitinated target. Understanding the mechanisms involved in this decision making will provide crucial insight into not only viral protection but also diverse cell biological processes regulated by ubiquitination.
In summary, our data support a model (Fig. 8) wherein sequences in the C-terminal domain of mK3 are critical for its interaction with TAP/tapasin and mK3 interaction with TAP/tapasin determines its substrate specificity for class I and not B7.2 proteins. Our findings suggest that mK3 binds directly to TAP1 and -2 and not tapasin, since TAP1- and TAP2-deficient cells fail to support significant expression levels of mK3, even though they can express high levels of tapasin. In this case, tapasin is probably required to (i) stabilize the TAP complex, leading to increased mK3 levels, and (ii) recruit class I into the peptide-loading complex. The fact that association with TAP/tapasin stabilizes mK3 (20) represents a very clever mechanism whereby mK3 levels can be coordinated with the levels of the class I assembly machinery present in cells under different conditions. While mK3 is anchored to TAP primarily through its C-terminal domain, the RING-CH domain in the N-terminal region of mK3 remains free to catalyze or facilitate the ubiquitination of the cytoplasmic tail of the class I protein. Indeed, our data further support the notion that the proximity of the targeted class I tail to the RING-CH domain of mK3 is critical for Ub conjugation and that this required proximity is determined by mK3 binding to TAP. Consistent with a proximity model, we observed that mK3 must functionally interact with both TAP1 and TAP2 subunits. Interaction of mK3 with both TAP subunits may be required for attaining the necessary proximity to class I, since TAP1 and TAP2 are each associated with two class I molecules (23).
The importance of TAP/tapasin for mK3-mediated degradation by the proteasome raises the intriguing question of whether other K3 family members like kK3 and kK5 may also rely on host proteins for their substrate selection and degradation in the lysosome. Based on our findings with mK3, definition of the host proteins with which various K3 family members interact will provide key mechanistic insights into their mechanisms of substrate recognition and sites of substrate degradation. In the specific case of mK3, further characterization of the downstream pathways initiated by this molecule should provide insights into normal ER-associated degradation pathways.
Acknowledgments
We thank Herbert W. Virgin IV for his valuable support throughout this study and for his critical review of the manuscript. We also thank Yik Yeung Lawrence Yu for his technical advice and help with initiating this project.
This work was funded by NIH grants AI19687, AI46553, AI42793, and T32AI07063.
REFERENCES
- 1.Androlewicz, M. J., B. Ortmann, P. M. van Endert, T. Spies, and P. Cresswell. 1994. Characteristics of peptide and major histocompatibility complex class I/beta 2-microglobulin binding to the transporters associated with antigen processing (TAP1 and TAP2). Proc. Natl. Acad. Sci. USA 91:12716-12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boname, J. M., B. D. de Lima, P. J. Lehner, and P. G. Stevenson. 2004. Viral degradation of the MHC class I peptide loading complex. Immunity 20:305-317. [DOI] [PubMed] [Google Scholar]
- 3.Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15:627-636. [DOI] [PubMed] [Google Scholar]
- 4.Cannon, M. J., C. K. Osborn, R. A. Nazaruk, V. Grigoriev, and M. D. Crew. 1999. Diminished recognition of HLA-A2 proteins lacking a cytoplasmic domain (CY) by A2-restricted, EBV-specific CTLs: possible role of the CY in TAP association. Immunogenetics 49:346-350. [DOI] [PubMed] [Google Scholar]
- 5.Carbone, F. R., and M. J. Bevan. 1989. Induction of ovalbumin-specific cytotoxic T cells by in vivo peptide immunization. J. Exp. Med. 169:603-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coscoy, L., and D. Ganem. 2003. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. 13:7-12. [DOI] [PubMed] [Google Scholar]
- 7.Coscoy, L., D. J. Sanchez, and D. Ganem. 2001. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 155:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fruh, K., E. Bartee, K. Gouveia, and M. Mansouri. 2002. Immune evasion by a novel family of viral PHD/LAP-finger proteins of gamma-2 herpesviruses and poxviruses. Virus Res. 88:55-69. [DOI] [PubMed] [Google Scholar]
- 10.Fruh, K., A. Gruhler, R. M. Krishna, and G. J. Schoenhals. 1999. A comparison of viral immune escape strategies targeting the MHC class I assembly pathway. Immunol. Rev. 168:157-166. [DOI] [PubMed] [Google Scholar]
- 11.Gewurz, B. E., R. Gaudet, D. Tortorella, E. W. Wang, and H. L. Ploegh. 2001. Virus subversion of immunity: a structural perspective. Curr. Opin. Immunol. 13:442-450. [DOI] [PubMed] [Google Scholar]
- 12.Grandea, A. G., III, T. N. Golovina, S. E. Hamilton, V. Sriram, T. Spies, R. R. Brutkiewicz, J. T. Harty, L. C. Eisenlohr, and L. Van Kaer. 2000. Impaired assembly yet normal trafficking of MHC class I molecules in tapasin mutant mice. Immunity 13:213-222. [DOI] [PubMed] [Google Scholar]
- 13.Harris, M. R., L. Lybarger, Y. Y. Yu, N. B. Myers, and T. H. Hansen. 2001. Association of ERp57 with mouse MHC class I molecules is tapasin dependent and mimics that of calreticulin and not calnexin. J. Immunol. 166:6686-6692. [DOI] [PubMed] [Google Scholar]
- 14.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 15.Hewitt, E. W., L. Duncan, D. Mufti, J. Baker, P. G. Stevenson, and P. J. Lehner. 2002. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 21:2418-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 17.Laney, J. D., and M. Hochstrasser. 1999. Substrate targeting in the ubiquitin system. Cell 97:427-430. [DOI] [PubMed] [Google Scholar]
- 18.Li, S., H. O. Sjogren, U. Hellman, R. F. Pettersson, and P. Wang. 1997. Cloning and functional characterization of a subunit of the transporter associated with antigen processing. Proc. Natl. Acad. Sci. USA 94:8708-8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lie, W. R., N. B. Myers, J. M. Connolly, J. Gorka, D. R. Lee, and T. H. Hansen. 1991. The specific binding of peptide ligand to Ld class I major histocompatibility complex molecules determines their antigenic structure. J. Exp. Med. 173:449-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lybarger, L., X. Wang, M. R. Harris, H. W. Virgin, and T. H. Hansen. 2003. Virus subversion of the MHC class I peptide-loading complex. Immunity 18:121-130. [DOI] [PubMed] [Google Scholar]
- 21.Mansouri, M., E. Bartee, K. Gouveia, B. T. Hovey Nerenberg, J. Barrett, L. Thomas, G. Thomas, G. McFadden, and K. Fruh. 2003. The PHD/LAP-domain protein M153R of myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J. Virol. 77:1427-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Means, R. E., S. Ishido, X. Alvarez, and J. U. Jung. 2002. Multiple endocytic trafficking pathways of MHC class I molecules induced by a herpesvirus protein. EMBO J. 21:1638-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ortmann, B., J. Copeman, P. J. Lehner, B. Sadasivan, J. A. Herberg, A. G. Grandea, S. R. Riddell, R. Tampe, T. Spies, J. Trowsdale, and P. Cresswell. 1997. A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science 277:1306-1309. [DOI] [PubMed] [Google Scholar]
- 24.Ozato, K., T. H. Hansen, and D. H. Sachs. 1980. Monoclonal antibodies to mouse MHC antigens. II. Antibodies to the H-2Ld antigen, the products of a third polymorphic locus of the mouse major histocompatibility complex. J. Immunol. 125:2473-2477. [PubMed] [Google Scholar]
- 25.Ozato, K., N. Mayer, and D. H. Sachs. 1980. Hybridoma cell lines secreting monoclonal antibodies to mouse H-2 and Ia antigens. J. Immunol. 124:533-540. [PubMed] [Google Scholar]
- 26.Petersen, J. L., C. R. Morris, and J. C. Solheim. 2003. Virus evasion of MHC class I molecule presentation. J. Immunol. 171:4473-4478. [DOI] [PubMed] [Google Scholar]
- 27.Pickart, C. M. 2000. Ubiquitin in chains. Trends Biochem. Sci. 25:544-548. [DOI] [PubMed] [Google Scholar]
- 28.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 29.Powis, S. J. 1997. Major histocompatibility complex class I molecules interact with both subunits of the transporter associated with antigen processing, TAP1 and TAP2. Eur. J. Immunol. 27:2744-2747. [DOI] [PubMed] [Google Scholar]
- 30.Pretell, J., R. S. Greenfield, and S. S. Tevethia. 1979. Biology of simian virus 40 (SV40) transplantation antigen (TrAg). V. In vitro demonstration of SV40 TrAg in SV40 infected nonpermissive mouse cells by the lymphocyte mediated cytotoxicity assay. Virology 97:32-41. [DOI] [PubMed] [Google Scholar]
- 31.Raghuraman, G., P. E. Lapinski, and M. Raghavan. 2002. Tapasin interacts with the membrane-spanning domains of both TAP subunits and enhances the structural stability of TAP1 · TAP2 complexes. J. Biol. Chem. 277:41786-41794. [DOI] [PubMed] [Google Scholar]
- 32.Sadasivan, B., P. J. Lehner, B. Ortmann, T. Spies, and P. Cresswell. 1996. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity 5:103-114. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez, D. J., L. Coscoy, and D. Ganem. 2002. Functional organization of MIR2, a novel viral regulator of selective endocytosis. J. Biol. Chem. 277:6124-6130. [DOI] [PubMed] [Google Scholar]
- 34.Smith, J. D., N. B. Myers, J. Gorka, and T. H. Hansen. 1993. Model for the in vivo assembly of nascent Ld class I molecules and for the expression of unfolded Ld molecules at the cell surface. J. Exp. Med. 178:2035-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smotrys, J. E., and M. E. Linder. 2004. Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73:559-587. [DOI] [PubMed] [Google Scholar]
- 36.Stevenson, P. G., S. Efstathiou, P. C. Doherty, and P. J. Lehner. 2000. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. USA 97:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevenson, P. G., J. S. May, X. G. Smith, S. Marques, H. Adler, U. H. Koszinowski, J. P. Simas, and S. Efstathiou. 2002. K3-mediated evasion of CD8(+) T cells aids amplification of a latent gamma-herpesvirus. Nat. Immunol. 3:733-740. [DOI] [PubMed] [Google Scholar]
- 38.Swanson, R., M. Locher, and M. Hochstrasser. 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Mat alpha 2 repressor degradation. Genes Dev. 15:2660-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiwari, S., and A. M. Weissman. 2001. Endoplasmic reticulum (ER)-associated degradation of T cell receptor subunits. Involvement of ER-associated ubiquitin-conjugating enzymes (E2s). J. Biol. Chem. 276:16193-16200. [DOI] [PubMed] [Google Scholar]
- 40.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 41.Townsend, A., C. Ohlen, J. Bastin, H. G. Ljunggren, L. Foster, and K. Karre. 1989. Association of class I major histocompatibility heavy and light chains induced by viral peptides. Nature 340:443-448. [DOI] [PubMed] [Google Scholar]
- 42.Van Parijs, L., Y. Refaeli, J. D. Lord, B. H. Nelson, A. K. Abbas, and D. Baltimore. 1999. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity 11:281-288. [DOI] [PubMed] [Google Scholar]
- 43.Virgin, H. W., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2:169-178. [DOI] [PubMed] [Google Scholar]
- 45.Williams, A., C. A. Peh, and T. Elliott. 2002. The cell biology of MHC class I antigen presentation. Tissue Antigens 59:3-17. [DOI] [PubMed] [Google Scholar]
- 46.Yewdell, J. W., and A. B. Hill. 2002. Viral interference with antigen presentation. Nat. Immunol. 3:1019-1025. [DOI] [PubMed] [Google Scholar]
- 47.Yu, Y. Y., M. R. Harris, L. Lybarger, L. A. Kimpler, N. B. Myers, H. W. Virgin, and T. H. Hansen. 2002. Physical association of the K3 protein of gamma-2 herpesvirus 68 with major histocompatibility complex class I molecules with impaired peptide and β2-microglobulin assembly. J. Virol. 76:2796-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, Y. Y., H. R. Turnquist, N. B. Myers, G. K. Balendiran, T. H. Hansen, and J. C. Solheim. 1999. An extensive region of an MHC class I alpha 2 domain loop influences interaction with the assembly complex. J. Immunol. 163:4427-4433. [PubMed] [Google Scholar]
- 49.Zheng, N., P. Wang, P. D. Jeffrey, and N. P. Pavletich. 2000. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102:533-539. [DOI] [PubMed] [Google Scholar]