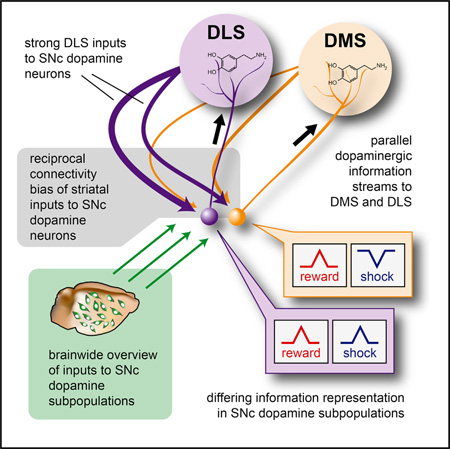
SUMMARY
Recent controversy regarding the diversity of midbrain dopamine neurons has highlighted the importance—and the challenges—of defining mammalian neuronal cell types. Although neurons may be best categorized using inclusive criteria spanning biophysical properties, wiring of inputs, wiring of outputs, and activity during behavior, linking all of these measurements to cell types within the intact brains of living mammals has been difficult. Here, using an array of intact-brain circuit interrogation tools, including CLARITY, optogenetics, viral tracing, and fiber photometry, we explore the diversity of dopamine neurons within the substantia nigra pars compacta (SNc). We identify two parallel nigrostriatal dopamine neuron subpopulations differing in biophysical properties, input wiring, output wiring to dorsomedial striatum (DMS) versus dorsolateral striatum (DLS), and natural activity patterns during free behavior. Our results reveal independently operating nigrostriatal information streams, with implications for understanding the logic of dopaminergic feedback circuits and the diversity of mammalian neuronal cell types.
Graphical abstract
INTRODUCTION
Dopamine (DA) is a neurotransmitter that is crucial for many biological processes relevant to health and disease and is thought to regulate (among other behaviors) voluntary movement, reinforcement learning, and motivation (Bromberg-Martin et al., 2010). Seminal early studies into the information encoded by midbrain DA neurons suggested that a key function of DA is to transmit reward prediction error signals (Mirenowicz and Schultz, 1996; Schultz et al., 1997; Waelti et al., 2001), a hypothesis concordant with temporal difference learning models (Montague et al., 1996, 2004; Steinberg et al., 2013; Sutton, 1988). However, not all midbrain DA neurons appear to encode similar information in their activity patterns (Bromberg-Martin et al., 2010; Lammel et al., 2014; Roeper, 2013). For example, DA neurons have been observed that differ in their responses to aversive stimuli, leading to the hypothesis that the DA neurons, which increase their firing in response to these stimuli, may signal salience rather than value (Brischoux et al., 2009; Bromberg-Martin et al., 2010; Matsumoto and Hikosaka, 2009), though controversy on this point remains (Cohen et al., 2012; Fiorillo et al., 2013, 2013; Ungless et al., 2004).
The concept that there are diverse subsets of midbrain DA neurons transmitting distinct signals naturally leads to the question of whether these subsets are functionally incorporated into different circuits with different roles in the brain. Explorations of DA neurons in the ventral tegmental area (VTA) have revealed that these neurons can be divided into distinct categories based on their projection targets, which include the prefrontal cortex, nucleus accumbens (NAc) core, NAc medial shell, NAc lateral shell, and amygdala. When divided by projection target, different VTA DA neuron classes express varying levels of the DA transporter (DAT), DA D2 autoreceptors, GIRK channels, and HCN channels mediating the Ih current (Lammel et al., 2008, 2011; Margolis et al., 2008), all of which could affect the dynamics of signals represented and transmitted. Furthermore, projection target-defined subpopulations are located in different subregions of the VTA, their excitatory synapses are differentially modulated by rewarding and aversive stimuli, and they receive distinct inputs, which can elicit opposite behaviors when selectively recruited (Lammel et al., 2008, 2011, 2012).
Distinctions among DA neurons have also increasingly been drawn between DA neurons within the VTA and those within the substantia nigra pars compacta (SNc). For example, a recent viral circuit tracing study showed that VTA and SNc DA neurons receive different proportions of inputs from key brain regions (Watabe-Uchida et al., 2012). In particular, this study noted that SNc neurons receive a large proportion of their inputs from the dorsal striatum. A major point of controversy, however, arises from discrepancies with studies attempting to functionally characterize direct striatal projections to midbrain DA neurons using optogenetics and slice recordings that have failed to identify such connections (Chuhma et al., 2011; Xia et al., 2011) or found them to be very weak (Bocklisch et al., 2013). The functional connectivity from striatum to SNc thus remains an open question.
In addition to differences from VTA neurons, there are hints in the literature that SNc DA neurons alone could be further divisible into functionally distinct subsets. One pioneering study demonstrated that DA neurons located more medially within the SNc express higher levels of K-ATP channels, which mediate burst firing (Schiemann et al., 2012). Another study used recordings in awake monkeys to demonstrate a correlation between the depth at which a DA neuron was recorded along the electrode (indicating a more ventromedial location in the midbrain) and the likelihood that it would decrease rather than increase its firing in response to an aversive cue (Matsumoto and Hikosaka, 2009).
How might such differences among SNc DA neurons relate to circuit wiring and function? One intriguing hypothesis is that distinct locations within the SNc give rise to projections targeting distinct locations within the dorsal striatum, such as dorsomedial striatum (DMS) and dorsolateral striatum (DLS); distinct SNc subfields could furthermore be set up to receive, process, and transmit functionally distinct streams of information from elsewhere in the brain. Such circuit organization would have powerful implications. For example, given the high medial SNc expression of K-ATP channels that enable bursting, the prediction could be made that the striatal target field of the medial SNc would receive strong novelty-driven bursts of DA during exploratory behavior that the striatal target field of the lateral SNc could not receive.
However, other lines of research suggest that DAergic projections to the DMS and DLS carry similar information streams because restriction of DA signaling to either subregion has similar effects (Darvas and Palmiter, 2009, 2010). In fact, SNc DA neurons could in principle even be similar to VTA DA neurons in terms of the valence of information represented because mice will self-stimulate for SNc DA neuron activity just as for VTA DA neuron activity (Ilango et al., 2014; Rossi et al., 2013).
In the end, though this is clearly a question of great potential significance, it remains unknown whether different SNc DA neuron populations are fundamentally distinct in their projection targets in striatum, in the types of information represented in their electrical activity, or in the sources of their incoming information across the brain. Here, using an array of intact-brain circuit interrogation tools, including CLARITY, optogenetics, viral tracing, and fiber photometry during behavior, we explore this issue and find that all three conjectures regarding the complexity of SNc functional wiring are borne out, with substantial implications for understanding the logic of DAergic signaling to striatum.
RESULTS
Unbiased Brain-wide Mapping of Inputs to Striatum Subfield-Projecting SNc Neurons
To directly explore the hypothesis that distinct SNc neuron populations receive and deliver separable information streams, we began with an unbiased approach for globally mapping the input/output relationships of SNc neurons. We mapped inputs onto two subpopulations of SNc DA neurons defined by their outputs to the DMS and DLS. These regions of the dorsal striatum have been functionally distinguished by previous studies (Castañé et al., 2010; Faure et al., 2005; Featherstone and McDonald, 2004; Yin and Knowlton, 2004; Yin et al., 2004, 2005a, 2009, 2005b), leading us to hypothesize that SNc projections to these areas might also participate in separable circuits.
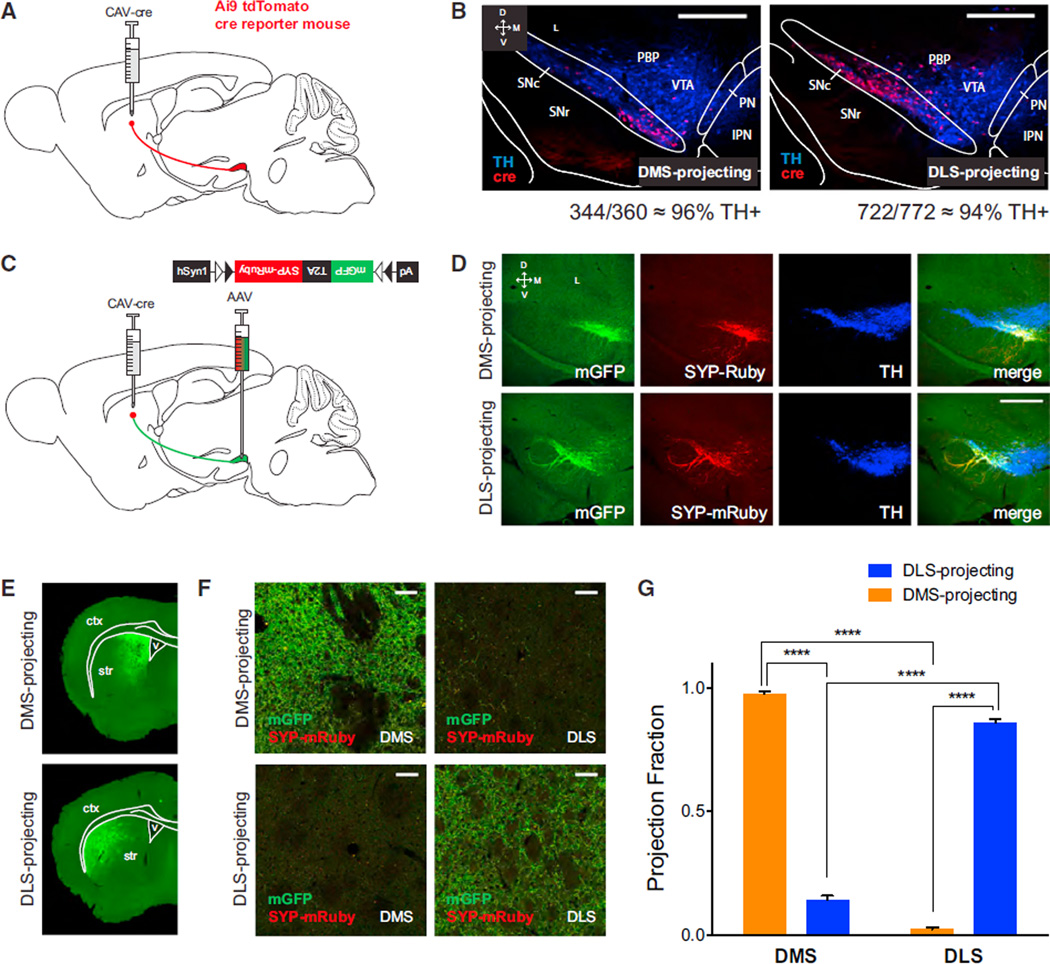
We achieved unbiased global visualization of SNc inputs and outputs using a combination of CLARITY and CLARITY-optimized light-sheet microscopy (COLM) (Chung et al., 2013; Tomer et al., 2014), along with a variation of rabies-based circuit mapping (Schwarz et al., 2015); the latter method operates similarly to previously published rabies-based circuit mapping technologies, utilizing a GFP-expressing rabies virus (RVdG-GFP) that both lacks the glycoprotein needed to spread trans-synaptically and is pseudotyped with EnvA (so that it can infect only neurons expressing TVA, an avian receptor protein normally absent in mammalian cells [Wickersham et al., 2007]).Glycoprotein and TVA can then be expressed using cre-dependent vectors to direct the rabies virus to infect and spread to connected inputs from a cre-defined subset of neurons. In TRIO, cre is delivered from the retrograde CAV-cre virus (Hnasko et al., 2006; Soudais et al., 2001), which transduces axon terminals and thereby defines “starter cells” (from which rabies tracing will occur) by virtue of their projection target. We injected CAV-cre into either the DMS or DLS and then injected AAVs expressing cre-dependent rabies glycoprotein (G) and TC, a high efficiency version of the TVA receptor fused to mCherry (Miyamichi et al., 2013), into the SNc. Two weeks later, we injected RVdG-GFP into the SNc (Figure 1A). Control experiments revealed that resulting putative starter cells in the SNc, as defined by neurons that expressed both RVdG-GFP and TC, were predominantly DAergic (TH+; 96% ± 3% after CAV-cre injection into the DMS, n = 4 mice; 98% ± 2% after CAV-cre injection into the DLS, n = 4 mice; Figures 1B–1D). Additional controls demonstrated that the RVdG-GFP was properly EnvA-pseudotyped and that long-range tracing of inputs was cre dependent (Figure S1A and S1B).
Figure 1. Whole-Brain Mapping of Inputs to DMS-Projecting and DLS-Projecting SNc DA Neurons.
(A) Viral injection strategy for whole-brain input mapping (TRIO). CAV-cre is injected into the striatum (DMS or DLS), and AAVs expressing cre-dependent TC and G are injected into the SNc. Two weeks later, RVdG-GFP is injected into the SNc, where it infects TC-expressing cells and spreads one synapse upstream from G-expressing cells.
(B) Low-magnification (5×) images of DMS- and DLS-projecting SNc “starter cells,” from which tracing likely occurred. Green shows RVdG-GFP expression, red shows TC expression, and blue shows (TH) immunostaining.
(C) Quantification of the percentage of starter cells that were TH+. Error bars are SEM.
(D) High-magnification (40×) images from the sections shown in (B).
(E) Examples of GFP labeling of ipsilateral inputs to DMS- and DLS-projecting SNc DA neurons from various brain regions. S1, somatosensory cortex; NAc, nucleus accumbens; DS, dorsal striatum; BNST, bed nucleus of the stria terminalis; GPe, globus pallidus external segment; HY, hypothalamus; CeA, central amygdala; LHb, lateral habenula; DR, dorsal raphe; MRN, medial raphe nucleus; PAG, periaqueductal gray.
(F) Quantification of ipsilaterally labeled inputs to DMS- and DLS-projecting SNc DA neurons, shown as a percentage of all ipsilateral inputs. Error bars are SEM. ***p < 0.001 and ****p < 0.0001. Other abbreviations: motor cortex (M1/2), somatosensory cortex (S1), fundus of the striatum/interstitial nucleus of the posterior limb of the anterior commissure (FS/IPAC), substantia innominata/ventral pallidum (SI/VP), superior colliculus (SC), inferior colliculus (IC), substantia nigra pars compacta (SNc), substantia nigra pars reticulata (SNr), ventral tegmental area (VTA), pedunculopontine nucleus (PPN), and parabrachial nucleus (PB).
We began our analysis of GFP-labeled inputs to DMS- and DLS-projecting SNc DA neurons by optically clarifying the labeled brains (Chung et al., 2013; Tomer et al., 2014) and visualizing using COLM (Tomer et al., 2014). We reasoned that, although thin sectioning approaches used for analysis of anatomical tracing have yielded important insights, they are not ideal for visualization of brain-wide patterns. Thin sectioning provokes particular caveats for the interpretation of negative results: slices may tear, fragment, or otherwise be damaged or lost, and strategies to minimize overcounting of somata split across adjacent section (such as counting only separated sections—e.g., every third or sixth) may underestimate cell counts, particularly in small brain regions. Whole-brain CLARITY/ COLM circumvents these difficulties, allowing an intact global overview of brain-wide structural patterns.
Using CLARITY/COLM, we noted an interesting GFP expression pattern in the striatum (Movies S1, S2, S3, and S4). When inputs to DMS-projecting SNc DA neurons were labeled, we observed relatively strong labeling in the NAc and DMS (Movies S1 and S2), whereas when inputs to DLS-projecting SNc DA neurons were labeled, we observed relatively strong labeling in the DLS, particularly in the caudal tail of the striatum (Movies S3 and S4). Moreover, fine details of local neuronal architecture in GFP-labeled neurons could be observed when regions of interest were viewed at full resolution and high magnification (Movie S5).
To quantify these divergent input patterns to SNc subfields depending on their projection target, we followed up with detailed targeted slicing approaches and quantified the inputs from each brain region to DMS- and DLS-projecting SNc DA neurons as a proportion of the total inputs observed (Figures 1E and 1F and Table S1). Existing atlases used to define major brain regions were further refined to define the boundaries of the DMS and DLS (Figures S1C and S1D). Because the inputs to SNc DA neurons are almost entirely ipsilateral (DMS-proj 96.4% ± 0.9%, n = 4; DLS-proj 96.5% ± 0.8%, n = 4, p = 0.91), we focused on the injected hemisphere (contralateral inputs were examined separately; Figures S1E and S1F and Table S1), noting first that brain-wide inputs to SNc DA neurons, regardless of projection target, broadly matched those observed by Watabe-Uchida et al. (2012) using RVdG-GFP tracing from the SNc of DAT-cre mice. However, despite gross similarities between groups, a two-way ANOVA revealed a significant interaction between starter cell projection target and input area (p < 0.001). Multiple comparisons revealed significant differences in inputs from the DMS (p < 0.001) and the DLS (p < 0.0001; Figure 1F), indicating a marked preferential reciprocal connectivity of dorsal striatal subregions to their specific DAergic SNc input subregions. Injections in D1-tomato bacterial artificial chromosome (BAC) transgenic mice further revealed that striatal inputs to SNc DA neurons arise from DA D1 receptor-expressing neurons (Figure S1G; 92/97 DMS-projecting neurons and 210/214 DLS-projecting neurons), consistent with the idea that D1 (but not D2) striatal neurons project directly to the midbrain and with hypotheses regarding patch/matrix organization of striatum (Crittenden and Graybiel, 2011). These anatomical studies suggested a fundamental distinction in circuit incorporation for projection-defined SNc subregions and provided a roadmap for a more detailed investigation of the circuit.
Organization of SNc DA Neurons Projecting to Distinct Regions of Dorsal Striatum
To quantify the spatial patterns of cre expression following injections of CAV-cre into the striatum, we injected CAV-cre into the DMS or DLS of Ai9 tdTomato cre reporter mice (Figure 2A). Viral spread in the striatum was contained within the targeted subregion (Figure S2A). In the SNc, two key observations were made. First, cre expression was largely restricted to DA neurons, identified by tyrosine hydroxylase (TH) immunostaining, following injection in both the DMS (344/360 neurons) and the DLS (722/772 neurons). Although a few scattered TH+ cells were labeled in the VTA in both groups, adjacent non-DAergic regions did not contain cre-expressing cells (Figure 2B). Second, the patterns of cre expression following DMS and DLS injection differed. DMS-projecting neurons were observed in the medial SNc, whereas DLS-projecting neurons were observed in the lateral SNc. This differing anatomical localization of DMS- and DLS-projecting SNc DA neurons supports the idea of parallel nigrostriatal subcircuits within the SNc.
Figure 2. Medial-Lateral Distribution of SNc DA Neurons Projecting to Medial and Lateral Subregions of Dorsal Striatum.
(A) CAV-cre was injected into the striatum of Ai9 td-Tomato cre reporter mice, labeling SNc neurons projecting to the injected region in red.
(B) DMS- and DLS-projecting SNc neurons are labeled with td-Tomato (red). Blue shows TH+ neurons. Both DMS- and DLS-projecting SNc neurons were found to be predominantly DAergic. Scale bars are 0.5 mm.
(C) To map the outputs of DMS- and DLS-projecting SNc DA neurons, CAV-cre was injected into the striatum (DMS or DLS) of wild-type mice. Cre was then used to drive expression of mGFP-T2A–SYP-mRuby, labeling processes in green and presynaptic sites in red.
(D) DMS- and DLS-projecting SNc DA neurons expressing mGFP and SYP-mRuby. Staining for TH (blue) shows overlap with DA neuron cell bodies. Scale bars are 0.5 mm.
(E) Example images (53) of patterns of mGFP expression in the striatal projections of DMS- or DLS-projecting DA neurons. Ctx, cortex; Str, striatum; V, ventricle.
(F) Example images (403) of regions of interest (DMS, DLS) in mice expressing mGFP and SYP-mRuby in DMS- and DLS-projecting SNc DA neurons. Scale bars are 50 µm.
(G) Projection fraction of DMS- and DLS-projecting SNc DA neurons in DMS and DLS. Error bars are SEM. ****p < 0.0001. See also Figure S2.
We next asked whether DMS- and DLS-projecting SNc DA neurons projected only to the DMS or DLS, or instead collateralized substantially. To visualize the boundaries of SNc DA neuron axonal arborizations in the striatum, we injected CAV-cre into the striatum and an adeno-associated virus (AAV) for cre-dependent expression of membrane-bound GFP (mGFP) and synaptophysin-mRuby (SYP-mRuby) into the SNc (Beier et al., 2015 [in this issue of Cell]; Figure 2C). With this approach, axons were labeled in green, and putative presynaptic sites were labeled in red. In the SNc, both green and red labeling were visible as anticipated (Figure 2D). The areas of highest fluorescence colocalized with TH in a pattern concordant with the locations of DMS- and DLS-projecting SNcDAneurons observed in Ai9 mice (Figure 2B). In the striatum, axons of DMS-projecting SNc DA neurons remained within the DMS, and axons of DLS-projecting SNc DA neurons remained within the DLS (Figures 2E–2G and S2B).
To quantify this differential axon distribution, we acquired systematic image series from each mouse corresponding to the full anterior-posterior span of the striatum (Figure S2B). Projection fraction was calculated as the axonal coverage area in one region (DMS or DLS) divided by the total area covered across the entire striatum (Figure 2H). The entire striatum was defined to include the NAc, but we observed negligible contributions of projections to any of the NAc subregions across all conditions (Figure S2C). No GFP-labeled axons were observed in the pre-frontal cortex or amygdala (Figure S2D). In dorsal striatum, an overwhelming fraction of DMS-projecting axons was localized within the bounds of the DMS (DMS 0.98 ± 0.01, n = 2, versus DLS 0.02 ± 0.01, n = 2; two-way ANOVA, brain area 3 projection target interaction, p < 0.0001; post hoc Tukey’s multiple comparisons, p < 0.0001); similarly, DLS-projecting axons were localized largely within the bounds of the DLS (DMS 0.14 ± 0.02, n = 2, versus DLS 0.86 ± 0.02, n = 2; post hoc Tukey’s multiple comparisons, p < 0.0001). These data confirmed that the SNc projections to the DMS and DLS are largely parallel, defining separable nigrostriatal subcircuits with the potential to convey distinct and independent signals to the DMS and DLS.
Distinct Properties of SNc DA Neurons Depending upon Projection Target
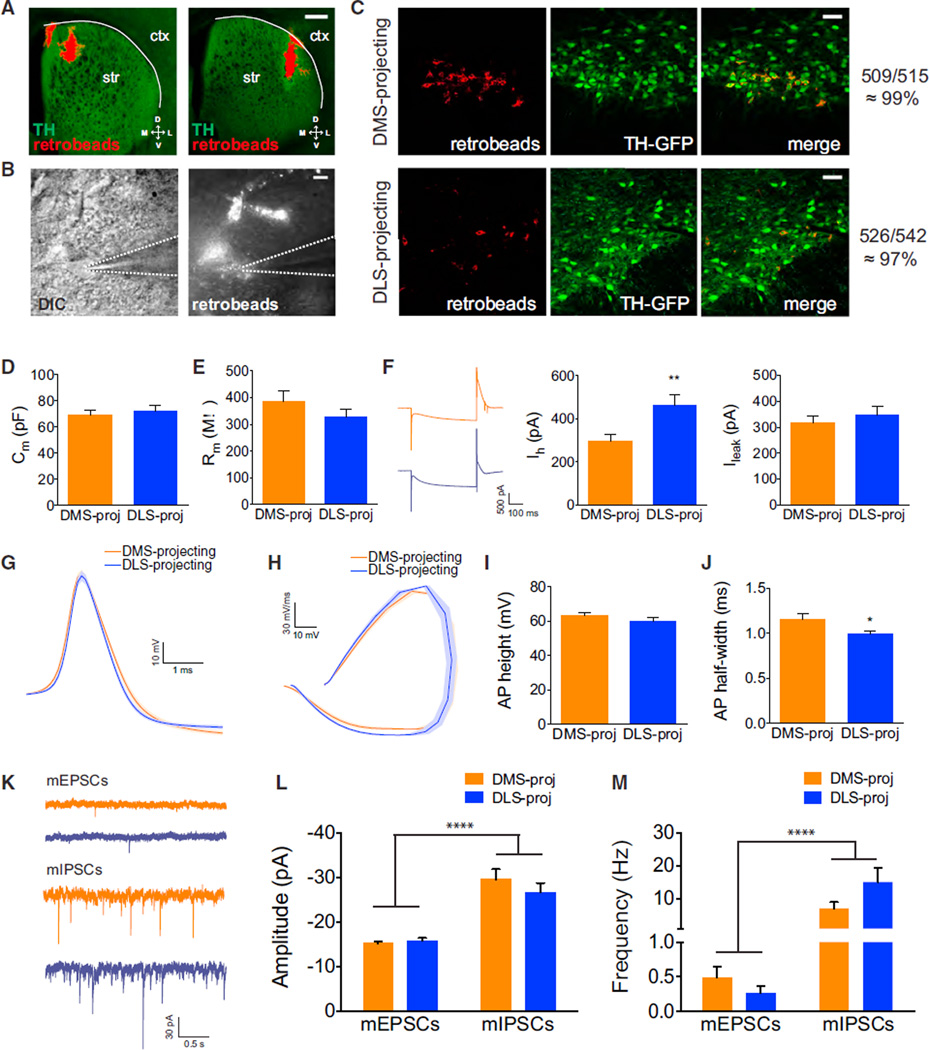
We next explored whether projection-defined SNc DA neurons could be distinguished by intrinsic electrophysiological properties. To mark neurons for recording, we injected red-fluorescent retrobeads into the DMS or DLS of TH::GFP mice. Retrobead-labeled SNc neurons were also primarily GFP+ (Figure 3C; 509/515 DMS-projecting neurons and 526/542 DLS-projecting neurons). Although the specificity of the TH::GFP line has been questioned (Lammel et al., 2015), we found that GFP expression is specific for TH+ neurons in the SNc (Figure S3). Retrobead injections remained well localized to the injection site (Figure 3A) and were readily identifiable for patching (Figure 3B).
Figure 3. Intrinsic Properties of DMS-Projecting and DLS-Projecting SNc DA Neurons.
(A) Retrobeads (red) injected into the striatum of TH::GFP mice. Ctx, cortex; Str, striatum. Scale bars are 0.5 mm.
(B) DIC and red fluorescent images of a patched retrobead-containing neuron. Dotted lines highlight the position of the recording electrode. Scale bars are 10 µm.
(C) Colocalization of retrobead-containing neurons (red) with TH-GFP-labeled neurons (green) in the SNc. Colocalization rates were ~99% and ~97% in DMS-and DLS-projecting neurons, respectively.
(D) Membrane capacitance (Cm) of DMS- and DLS-projecting SNc DA neurons. Error bars are SEM.
(E) Membrane resistance (Rm) of DMS- and DLS-projecting SNc DA neurons. Error bars are SEM.
(F) Left, example responses of DMS-projecting SNc neurons (orange) and DLS-projecting SNc neurons (blue) to a hyperpolarizing current injection. Right, Ih and Ileak current measurements in response to hyperpolarizing current injection from DMS- and DLS-projecting SNc DA neurons. Error bars are SEM. **p < 0.01.
(G) Average action potential waveforms from DMS- and DLS-projecting SNc DA neurons. Area of light shading is SEM.
(H) Phase plots of average action potential waveforms from DMS- and DLS-projecting SNc DA neurons. Area of light shading is SEM.
(I) Average action potential height from DMS- and DLS-projecting SNc DA neurons. Error bars are SEM.
(J) Average action potential half widths from DMS- and DLS-projecting SNc DA neurons. Error bars are SEM. *p < 0.05.
(K) Example traces of excitatory (top) and inhibitory (bottom) miniature postsynaptic currents from DMS-projecting (orange) and DLS-projecting (blue) SNc DA neurons.
(L) Amplitudes of mEPSCs and mIPSCs recorded from DMS-projecting and DLS-projecting SNc DA neurons. Error bars are SEM. ****p < 0.0001.
(M) Frequencies of mEPSCs and mIPSCs from recorded DMS-projecting and DLS-projecting SNc DA neurons. Error bars are SEM. ****p < 0.0001.
See also Figure S3.
Whole-cell patch-clamp analysis of retrobead+ and GFP+ SNc neurons revealed similar membrane capacitance, resistance, and leak currents of DMS-projecting and DLS-projecting DA neurons (Figures 3D and 3E; Cm, DMS-proj 69.24 ± 3.589 pF, n = 21 versus DLS-proj 71.94 ± 4.563 pF, n = 16, p = 0.64; Rm, DMS-proj 384.7 ± 41.09 MU, n = 21 versus DLS-proj 327.4 ± 30.03 MU, n = 16, p = 0.30; Ileak, DMS-proj 317.1 ± 25.51 pA, n = 21 versus DLS-proj 345.6 ± 35.81 pA, n = 16, p = 0.51). However, we identified distinctly larger Ih currents in DLS-projecting DA neurons (Figure 3F; Ih, DMS-proj 296.2 ± 29.25 pA, n = 21 versus DLS-proj 460.8 ± 49.69 pA, n = 16, p < 0.01). All recorded neurons displayed significant Ih currents and broad action potential waveforms consistent with reliable identification of DA neurons across groups. A small but significant difference was observed in action potential half-width of DMS- versus DLS-projecting neurons (Figures 3G–3J; AP heights, DMS-proj 63.34 ± 1.901 mV, n = 21 versus DLS-proj 59.95 ± 2.395 mV, n = 16, p = 0.27;AP half widths, DMS-proj 1.152 ± 0.06 ms, n = 21 versus DLS-proj 0.9938 ± 0.03 ms, n = 16, p < 0.05).
SNc DA Neurons Are Embedded within a Largely Inhibitory Network
Projection-defined SNc DA neurons exhibit different intrinsic properties, project to non-overlapping striatal subregions, and receive differential inputs, together suggesting fundamentally different roles in the circuit. However, since the nature of the signals carried by the differing afferents remained unclear, we next functionally investigated these afferents using slice electrophysiology. As a first assessment of global functional afferent input, we recorded miniature excitatory and inhibitory postsynaptic currents (mEPSCs and mIPSCs) (Figure 3K). Although no significant differences were observed between DMS- and DLS-projecting SNc DA neurons, both the amplitudes and frequencies of mIPSCs were higher than for mEPSCs across both groups (Figures 3L and 3M; mEPSC amplitude, DMS-proj −15.27 ± 0.50 pA, n = 15 versus DLS-proj −15.79 ± 0.68 pA, n = 15; mIPSC amplitude, DMS-proj −29.69 ± 2.23 pA, n = 15 versus DLS-proj −26.71 ± 2.13 pA, n = 15; two-way ANOVA, effect of mEPSCs versus mIPSCs, p < 0.0001; mEPSC frequency DMS-proj 0.49 ±0.16 Hz, n = 15 versus DLS-proj 0.27 ±0.10 Hz, n = 15; mIPSC frequency, DMS-proj 7.11 ± 1.86 Hz, n = 15 versus DLS-proj 15.07 ± 4.29 pA, n = 15; two-way ANOVA, effect of mEPSCs versus mIPSCs, p < 0.0001). This finding indicates that SNc DA neurons in general are embedded within a largely inhibitory network, in agreement with our findings and the findings of Watabe-Uchida et al. (2012) that SNc DA neurons receive afferents from many areas known to have GABAergic projection neurons, including the striatum.
Functional Optogenetic Characterization of Dorsal Striatal Inputs to DMS- versus DLS-Projecting SNc DA Neurons
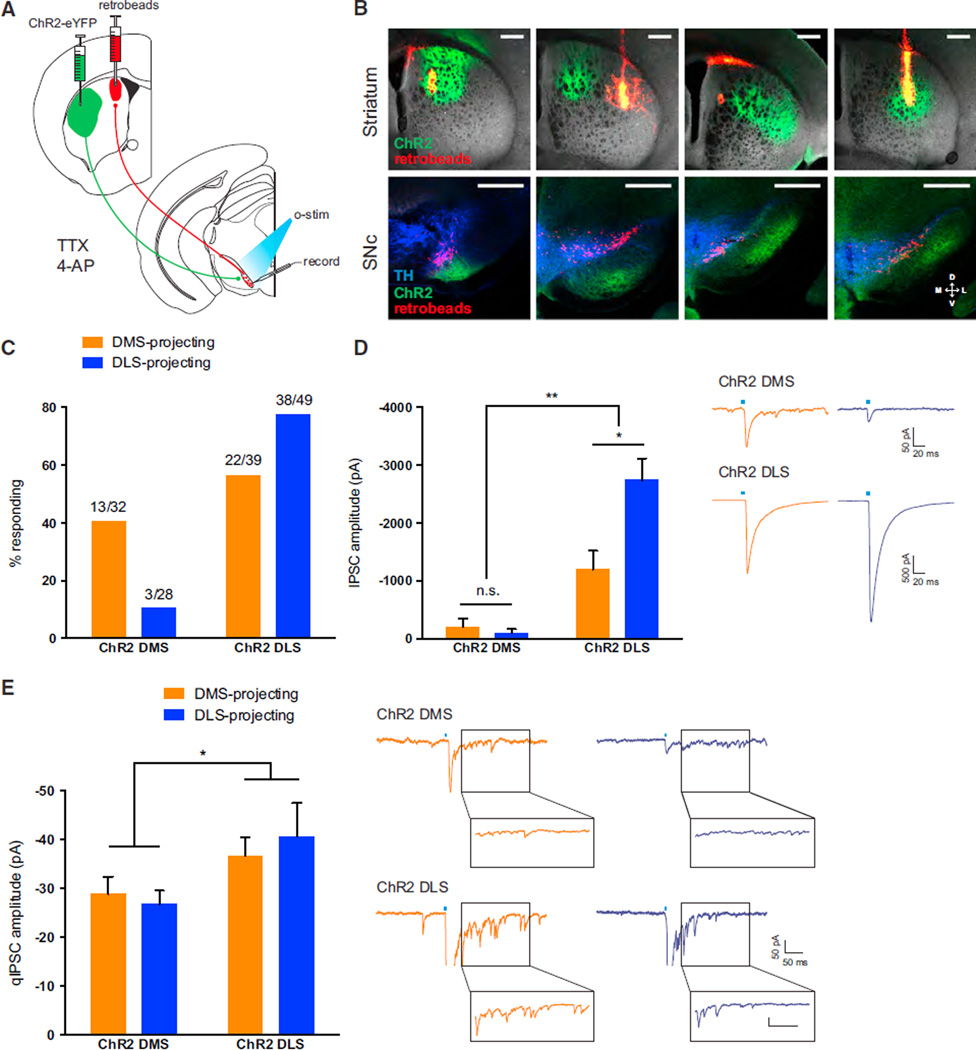
To extend these findings, we employed targeted optogenetics to test the functionality of synapses from dorsal striatum to SNc, where we had specifically observed differences in inputs onto projection-defined SNc neurons using TRIO (Figure 1F). First, we expressed ChR2-eYFP in either DMS or DLS. Then, we injected retrobeads into one of these regions to enable patching of projection-defined SNc neurons. Blue light pulses stimulated inputs specifically from the striatal subregion expressing ChR2 (Figure 4A). Examples of injection sites for ChR2 and retrobeads are shown in Figure 4B, along with the resulting distributions of red and green fluorescence in SNc (green fibers were observed both in the SNc and in the underlying SNr, where many direct pathway striatal neurons project).
Figure 4. Optogenetic Mapping of Dorsal Striatal Inputs to DMS- and DLS-Projecting SNc DA Neurons: Reciprocal Connectivity Bias and Divergent Strength.
(A) ChR2-eYFP was expressed in either the DMS or the DLS. Red retrobeads labeled inputs to either the DMS or the DLS. Retrobead-containing cells in the SNc were patched, and blue light was flashed to activate ChR2-expressing terminals nearby. Responses were recorded in the presence of TTX (1 µM) and 4-AP (100 µM) to isolate monosynaptic inputs.
(B) Example images of striatal injection sites in the four experimental groups (top row) and of retrobead and ChR2expression in the SNc (bottom row). Blue shows TH immunostaining. Images are of slices used for recordings. Scale bars are 0.5 mm.
(C) Percentage of patched neurons responding to ChR2 stimulation of striatal terminals in the four experimental groups.
(D) IPSC amplitudes of responding neurons from (C). Example traces from each group are shown on the right. The blue bar indicates the time of blue light stimulation. Error bars are SEM. **p < 0.01 and ****p < 0.0001.
(E) Quantification of the quantal IPSC (qIPSC) amplitude observed in DMS- and DLS-projecting SNc DA neurons in response to stimulation of DMS or DLS ChR2-expressing terminals. Example traces of evoked asynchronous IPSCs are shown on the right, with the period of qIPSC event collection highlighted in a pop out box. The blue bar indicates the time of stimulation. Error bars are SEM. *p < 0.05.
Because striatal projection neurons are GABAergic, the glutamate receptor antagonists NBQX (5 µM) and APV (50 µM) were included in the extracellular solution to isolate inhibitory postsynaptic currents (IPSCs), and a high-chloride internal solution was used to facilitate event detection. We also included the voltage-gated sodium channel antagonist TTX (1 µM) and potassium channel antagonist 4-AP (100 µM) to isolate monosynaptic inputs by preventing disynaptic disinhibitory responses through the GABAergic cells of the SNr (with TTX) while enabling ChR2 to drive neurotransmitter release in the absence of action potentials (with 4-AP) (Petreanu et al., 2009). In all mice, recorded neurons were identified that directly responded to blue light stimulation, with response rates varying by condition. Connections from DMS to DMS-projecting SNc DA neurons and connections from DLS to DLS-projecting SNc DA neurons were clearly favored (Figure 4C; X2 p < 0.0001), mirroring our TRIO results (Figure 1) and functionally confirming a striking reciprocal connectivity between dorsal striatal subregions and their DAergic inputs.
DLS Inputs to SNc DA Neurons Are Stronger than DMS Inputs
Broad anatomical methods, while useful, cannot resolve key aspects of function, such as distinguishing very strong from very weak connections. In contrast, targeted optogenetic experiments can provide information not only about connection probabilities, but also about input strength. We next found, via IPSC amplitude quantification for SNc neurons that responded to stimulation of striatal afferents, that responses detected in both DMS- and DLS-projecting SNc neurons were much larger when DLS inputs were stimulated (Figure 4D; ChR2 DMS/ DMS-proj −205.27 ± 146.09 pA, n = 13; ChR2 DMS/DLS-proj −96.23 ± 69.28 pA, n = 3; ChR2 DLS/DMS-proj −1193.10 ± 324.30 pA, n = 22; ChR2 DLS/DLS-proj −2737.96 ± 369.82 pA, n = 38; two-way ANOVA, effect of ChR2 injection site, p < 0.01). Additionally, DLS-projecting SNc neurons had larger responses to DLS stimulation than did DMS-projecting SNc neurons (Tukey’s multiple comparisons, p < 0.05).
The observed differences in IPSC amplitudes between SNc neurons receiving inputs from the DMS and DLS were not fully explained by differences in the numbers of inputs arising from the two striatal subregions (Figures 1 and 4C), suggesting additional possible differences in quantal IPSC amplitude, the number of synapses per input cell, and/or release probability. To test for differences in quantal amplitude, we replaced calcium in the extracellular recording solution with strontium to induce asynchronous release and found that the average quantal IPSC amplitude was larger for inputs arising from the DLS versus the DMS (Figure 4E; ChR2 DMS/DMS-proj −28.86 ± 3.48 pA, n = 10; ChR2 DMS/DLS-proj −26.77 ± 2.75 pA, n = 8; ChR2 DLS/DMS-proj −36.64 ± 3.77 pA, n = 11; ChR2 DLS/DLS-proj −40.60 ± 4.94 pA, n = 10, two-way ANOVA, effect of ChR2 injection site, p < 0.05), providing an additional partial explanation for the observed differences in IPSC amplitude.
Selective Opposite-Valence Encoding of Aversive Stimuli by DMS- versus DLS-Projecting SNc DA Neurons
Parallel nigrostriatal SNc DA subcircuits clearly have the potential to deliver different kinds of information to the DMS and DLS. To formally test this intriguing possibility, we monitored activity from projection-defined SNc DA neurons during behavior using fiber photometry, a method for collecting population intracellular [Ca2+] fluorescent signals from a genetically encoded Ca2+ indicator such as GCaMP through a single chronic fiber optic implant (Gunaydin et al., 2014). We expressed GCaMP6f (Chen et al., 2013) in DMS- or DLS-projecting SNc DA neurons by injecting CAV-cre into the DMS or the DLS, followed by injection of a cre-dependent GCaMP6f construct into the SNc. Through a fiber optic implanted into SNc at the GCaMP6f injection site (Figure 5A), we delivered excitation light at 490 nm to stimulate GCaMP6f fluorescence in a Ca2+-dependent manner and at 405 nm, a wavelength that we determined was the excitation isosbestic point of GCaMP6f and that allowed us to perform ratiometric measurements of GCaMP6f activity, thereby correcting for bleaching and artifactual signal fluctuations (Figure S4A–S4D).
Figure 5. DMS- and DLS-Projecting SNc DA Neurons Respond Similarly to Appetitive but Differentially to Aversive Stimulation.
(A) CAV-cre was injected into either the DMS or DLS. AAV-expressing cre-dependent GCaMP6f was injected into the SNc. A 400 µm fiber optic implant was placed in the SNc to allow for measurement of GCaMP6f signals.
(B) Fiber photometry rig schematic. See Experimental Procedures for a more detailed description.
(C) Example of responses to reward observed in a mouse expressing GCaMP6f in DMS-projecting SNc DA neurons. Time 0 is aligned to either rewarded or non-rewarded port entries in an operant chamber. Arrow is placed at time 0. Area of light shading is SEM.
(D) Example of responses to reward observed in a mouse expressing GCaMP6f in DLS-projecting SNc DA neurons.
(E) Quantification of the peak ΔF/F observed in response to reward in mice expressing GCaMP6f in DMS-projecting SNc DA neurons or in mice expressing GCaMP6f in DLS-projecting SNc DA neurons.
(F) Quantification of the ΔF/F observed at time 0 in response to a non-rewarded port entry in mice expressing GCaMP6f in DMS-projecting SNc DA neurons or in mice expressing GCaMP6f in DLS-projecting SNc DA neurons.
(G) Example of responses to shock observed in a mouse expressing GCaMP6f in DMS-projecting SNc DA neurons. Yellow bar indicates the time of the shock (0–0.5 s). Area of light shading is SEM. (H) Example of responses to shock observed in a mouse expressing GCaMP6f in DLS-projecting SNc DA neurons.
(I) Quantification of the peak (extreme, whether negative or positive) ΔF/F observed during the shock in mice expressing GCaMP6f in DMS-projecting SNc DA neurons or in mice expressing GCaMP6f in DLS-projecting SNc DA neurons. ****p < 0.0001.
(J) Quantification of the mean ΔF/F observed 1–5 s post-shock in mice expressing GCaMP6f in DMS-projecting SNc DA neurons or in mice expressing GCaMP6f in DLS-projecting SNc DA neurons. ***p < 0.001. The same mice are shown in (C)–(J). See also Figure S4.
We recorded the activity of DMS- or DLS-projecting SNc DA neurons following either appetitive or aversive stimuli. To record appetitive signals, we trained mice to lever press for a sucrose reward retrieved from a reward port. Each press had a 10% chance of delivering reward, a contingency that allowed us to record during both rewarded and unrewarded port entries within the same behavioral session for comparison. DMS- and DLS-projecting SNc DA neurons reacted similarly to rewarding outcomes (Figures 5C–5E); a similar peak ΔF/F was observed on rewarded port entries for both sets of mice (DMS-proj 2.164 ± 0.549%, n = 7 versus DLS-proj 1.753 ± 0.247%, n = 9, p = 0.47). Peaks were not observed for non-rewarded port entries (Figures 5C, 5D, and 5F; DMS-proj 0.043 ± 0.195%, n = 7 versus DLS-proj −0.274 ± 0.083%, n = 9, p = 0.13).
To record responses to aversive stimuli, we subjected the same set of GCaMP6f-expressing mice to mild, unpredicted electrical shocks. Strikingly, DMS- and DLS-projecting SNc DA neurons responded oppositely to this aversive stimulus. DMS-projecting neurons showed a marked dip in activity at the time of shock, whereas DLS-projecting neurons showed an increase (Figures 5G–5I; peak ΔF/F i.e., positive or negative extreme during shock, DMS-proj −2.628 ± 0.513%, n = 8 versus DLS-proj 1.527 ± 0.358%, n = 9, p < 0.0001). Additionally, DLS-projecting neurons were returned immediately to baseline, whereas more persistent changes were observed in DMS-projecting neurons (Figure 5J; mean ΔF/F between seconds 1 and 5, DMS-proj 2.403 ± 0.525%, n = 8 versus DLS-proj −0.3146 ± 0.176%, n = 9, p < 0.001). Post hoc histology revealed that GCaMP6f-expressing axons were located in the DMS or DLS as expected, that fiber optic implants were placed appropriately in the SNc relative to GCaMP6f-expressing cell bodies, and that GCaMP6f-expressing cell bodies were TH+ (Figure S4E). We also verified that differences in movement between groups during the reward and shock sessions could not account for the observed differences in the photometry signal (Figures S4F–S4K). Together, these results suggest that the projections from subsets of SNc DA neurons to subregions of the dorsal striatum are set up so that the DMS and DLS receive fundamentally different signals; the DMS may receive a value signal (DAergic population firing signals the valence of the outcome), whereas the DLS may receive a salience signal (DAergic firing alerts the system to adaptation-relevant events, whether appetitive or aversive).
DISCUSSION
Here, we have developed and applied anatomical and functional methodologies to provide a deeper understanding of striatoni-grostriatal circuitry. We began by demonstrating that novel circuit-tracing and -recording methodologies can be effectively combined with whole-brain CLARITY/COLM imaging (Chung et al., 2013; Tomer et al., 2014) to provide a uniquely informative overview of richly defined cell types and their global connectivity motifs within the intact experimental subject brain. Further development of high-throughput processing of these whole-brain datasets will be needed (Chung and Deisseroth, 2013; Kim et al., 2013, 2015) to fully capitalize on this opportunity for the rapid advancement of understanding structure-function relationships in the brain. For example, looking to the future, it will be vital to increase experimental-subject group sizes despite the immensely large datasets acquired for each experimental subject. Although our current methods successfully captured the large though complex connectivity differences observed here, larger group sizes would broaden the potential to identify biologically and potentially clinically important differences with smaller effect sizes, including, perhaps, effects of age, gender, and life experience.
Additionally, we have shown that, despite the utility of anatomical tracing techniques such as TRIO, following up findings from their use with other experimental modalities examining the functionality of identified connections is essential. By stimulating inputs from the striatum to SNc DA neurons optogenetically, we discovered that DLS inputs are an order of magnitude stronger than DMS inputs, a finding that could not have been predicted anatomically but is nevertheless likely to be crucial in thinking about SNc circuit function. Our observations of unequivocal inputs from the dorsal striatum to the SNc contrast with certain earlier reports, which failed to find connections using similar techniques (Chuhma et al., 2011; Xia et al., 2011). Although it is often difficult to explain negative findings, these previous studies used younger animals than this study did, which is consistent with a possible role of age- or experience-dependent plasticity; moreover, we have shown that striatal projections to DA neurons may be very difficult to find if the subregions of the striatum and the SNc are not well matched (highlighting the value of the CLARITY/COLM/TRIO approach). For example, if ChR2 expression were predominantly in DMS and laterally located (DLS-projecting) DA neurons were being patched, we would expect connections to be extremely sparse, small and difficult to detect.
Our result regarding the in vivo activity of DMS- and DLS-projecting SNc DA neurons is concordant with the hypothesis put forward by Matsumoto and Hikosaka (2009) that DA neurons located more ventrolaterally within the midbrain increase firing in response to aversive stimuli. By targeting SNc DA neurons for observation based on their striatal projection target, we directly tested and substantially extended this hypothesis, demonstrating that the differences in the representation of aversive stimuli among subsets of SNc DA neurons relate to projection target. Additionally, we observed a sustained activity increase in DMS-projecting SNc DA neurons following shock. We speculate that this population of DA neurons may also encode for a heightened sensitivity or motivational state following an unfamiliar outcome (unlike the reward stimulus, the mice had no previous experience of shock prior to test day) in order to promote future action-outcome learning.
Regarding anatomy of efferents alone, our finding that the projections from SNc DA neurons to the DMS and DLS arise from distinct locations within the SNc is not inconsistent with several previous findings. For example, in rats that had the DA neuron toxin 6-OHDA injected into the DLS, degeneration of DA neurons was observed primarily in the lateral SNc (Faure et al., 2005). Similarly, for mice in which DA neuron TH expression was restricted to neurons projecting to either the DLS or the ventromedial striatum (VMS), DLS-projecting neurons were located more laterally than VMS-projecting neurons (Darvas and Palmiter, 2009; 2010; see also Schiemann et al., 2012).
Information transmitted along these separable activity streams will depend on intrinsic properties of the DA neurons and on their afferents. Indeed, combined with other recent lines of evidence (Lammel et al., 2008, 2011; Margolis et al., 2008), we have found that projection-defined subpopulations of DA neurons display different intrinsic properties. For example, increasing-amplitude Ih currents are observed progressing medially to laterally within the midbrain DA system; DA neurons in the medial paranigral nucleus of the VTA, which project to the prefrontal cortex, amygdala, NAc core, and medial shell, express little to no Ih current (Lammel et al., 2011), whereas DA neurons in the lateral SNc, which project to the DLS, express the largest Ih currents. Differences in Ih currents are strongly correlated with action potential rebound delays and can influence pacemaking activity (Neuhoff et al., 2002), particularly in the case of calbindin-negative SNc DA neurons (which constitute the majority of the SNc DA neuron population).
Afferents to projection-defined SNc DA neurons also differ; in particular, we here identify new principles of afferent-projection complexity in SNc subcircuits (see Beier et al., 2015) for additional illuminating information regarding VTA subcircuits). A prominent finding is that the DMS and DLS are preferentially reciprocally connected with the very same DA neurons that project back to these areas. This information notably enriches the ascending spiral model proposed by Haber et al. (2000), which could not make definitive statements about striatal inputs onto DAergic versus non-DAergic midbrain cells. We also observed strong DLS projections to DMS-projecting DA neurons; this finding implies a route of lateral to medial information flow that is not explored in the Haber model.
In summary, our identification of two distinct nigrostriatal DA circuits—separable by the combined evaluation of inputs, outputs, biophysical properties, and environmental information representations—both reveals independently controlled information representations streaming through SNc and provides a generalizable framework for brain-wide mapping of diverse populations of neurons defined by multiple independent types of features. Particularly in the case of DA neurons, this type of approach may improve our understanding of the circuit mechanisms underlying normal brain function, as well as diseases such as depression, addiction, and schizophrenia.
EXPERIMENTAL PROCEDURES
Animals
All experiments were approved by the Stanford University IACUC committee, protocol 10747. All mice (Ai9, TH-GFP, D1-tdTomato, and wild-type) were on a C57BL6/6J background and were 2–4 months old.
Stereotaxic Injections
Viruses and retrobeads were injected and optical implants placed at the following coordinates, relative to bregma: DMS +0.75 AP, 1.5 ML, −2.8 DV; DLS +0.25 AP, 2.5 ML, −3.4 DV; medial SNc −3.1, 0.8 ML, −4.7 DV; lateral SNc −3.1, 1.3 ML, −4.2 DV.
Histology
TH staining was done with 1:500 primary antibody overnight at 4°C and 1:500 secondary antibody coupled to Alexa Fluor 647 for 2–3 hr at room temperature. GFP staining was done with 1:500 primary antibody coupled directly to either Alexa Fluor 488 or Alexa Fluor 647 overnight at 4°C. Counterstaining was done with Neurotrace 435/455 Blue Fluorescent Nissl Stain (1:500) and/or DAPI (300 nM).
CLARITY
Brains were perfused and incubated with CLARITY monomer solution containing 1% acrylamide, 0.0125% bis-acrylamide, and 4% PFA and then polymerized at 37°C for 6–7 hr. Brains were passively cleared in SDS Borate Buffer (pH 8.5) at 37°C for 4–5 weeks, equilibrated in Focusclear for imaging and imaged using COLM methods (Tomer et al., 2014).
Slice Electrophysiology
300 mm coronal sections were prepared in an NMDG-based solution at room temperature. Striatal slices were fixed in 4% PFA and saved for verification of injection sites. Whole-cell recordings were performed in standard aCSF at 30–32°C. Where indicated, TTX (1 µm), 4-AP (100 µM), NBQX (5 µM), APV (50 µM), and picrotoxin (50 µM) were added. In some experiments, extracellular Ca2+ was replaced with Sr2+ to induce asynchronous release. K-gluconate internal was used for recording action potentials and Ih currents. EPSCs were recorded using CeMeSO3 internal and IPSCs were recorded using high chloride CsCl internal. 5 ms blue light pulses (475 nm, ~10 mW/mm2) were used to stimulate ChR2.
Reward Behavior
Mice were trained to lever press for 20% sucrose reward on an RR10 schedule, earning a maximum of 40 25 µl reward in a 1 hr session.
Shock Behavior
Mice received 15 mild foot shocks (0.4 mA, 0.5 s) on an RI60 schedule.
Fiber Photometry
A 490 nm LED was sinusoidally modulated at 211 Hz and passed through a GFP excitation filter. A 405 nm LED was modulated at 531 Hz and passed through a 405 nm bandpass filter. Both light streams were coupled to a high NA (0.48), large core (400 mm) optical fiber patch cord, which was mated to a matching brain implant in each mouse. GCaMP6f fluorescence was collected by the same fiber, passed through a GFP emission filter, and focused onto a photoreceiver. Custom software running on a real-time signal processor controlled the LEDs and independently demodulated the fluorescence brightness due to 405 nm and 490 nm excitation. The timing of behavioral variables was recorded by the same system. To calculate ΔF/F, a least-squares linear fit was applied to the 405 nm signal to align it to the 490 nm signal, producing a fitted 405 nm signal that was used to normalize the 490 nm as follows: ΔF/F = (490 nm signal − fitted 405 nm signal)/fitted 405 nm signal.
Statistics
Unpaired t tests were used for comparisons between two groups (DMS- and DLS-projecting SNc DA neurons). Two-way ANOVAs were used to assess how the properties or responses of DMS- and DLS-projecting SNc DA neurons were affected by other factors (e.g., input area). When a statistically significant effect was observed using a two-way ANOVA, post hoc testing with correction for multiple comparisons was performed using Tukey’s or Sidak’s multiple comparisons test.
Supplementary Material
Highlights.
Intact, inclusive approaches to classifying neuronal cell types
Differential brain-wide circuit incorporation of SNc dopamine neuron subpopulations
Opposite valence encoding of shock by projection target-defined SNc neurons
Independently controlled information streams from the SNc to the DMS and DLS
Acknowledgments
We thank L. Ye and K. Engberg for advice on CLARITY; C. Ramakrishnan, S. Pak, A. Shi On Hong, A. Wang, A. Lei, and H. Swanson for assistance with animal husbandry and reagent production and acquisition; P. Rothwell and L. Steinberg for advice and sharing of transgenic mouse lines; E. Callaway for advice on rabies tracing; and the entire K.D. and L.L. labs for invaluable discussion. We also thank E. Kremer (IGMM) for supplying CAV-cre virus, M. Kay (Stanford) for providing the pRC-DJ plasmid used to produce AAVDJ, the Stanford Viral and Vector Core for packaging AAVDJ virus, and D. Kim for supplying pGP-CMV-GCaMP6f (Addgene plasmid #40755), which was re-cloned in our lab by C. Ramakrishnan. We thank the UNC vector core for supplying the AAV5 and AAV8 viruses used in these studies and the Salk Vector Core for supplying rabies (amplified in-house by the L.L. lab). T.N.L. was supported by a Stanford Dean’s Postdoctoral Fellowship and by an NRSA Postdoctoral Fellowship (1F32MH105053-01). K.A.Z. was supported by an NRSA Predoctoral Fellowship (1F31MH105151-01). This work was also supported by the Hughes Collaborative Innovation Award (HCIA) and the NIMH Silvio Conte Center at Stanford (P50 MH086403 to R.C.M.). K.D. is supported by the DARPA Neuro-FAST program, NIMH, NIDA, NSF, the Simons Foundation, the Gatsby Foundation, the Wiegers http://www.Family Fund, and the Grosfeld, Reeves, Snyder, Woo, and Albert Yu and Mary Bechman Foundations. All optogenetics (http://www.optogenetics.org), CLARITY (http://clarityresourcecenter.org), and COLM (http://clarityresourcecenter.com/COLM.html) reagents and protocols are distributed and supported freely.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, four figures, one table, and five movies and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2015.07.014.
AUTHOR CONTRIBUTIONS
T.N.L. and K.D. designed experiments with input from L.L. and K.T.B on viral tracing and with input from R.T. on CLARITY and COLM implementation. L.L. and K.T.B. provided unpublished TRIO procedures. T.N.L. performed and analyzed all experiments with assistance from K.T.B. on viral tracing, assistance from R.T. and A.K.C. on whole-brain COLM imaging experiments and related image analysis, assistance from K.E.E. on image quantification, and assistance from C.S., T.J.D., and K.A.Z. on fiber photometry methods development, implementation, and combination with freely moving behavior. T.N.L. and K.D. wrote the paper with editorial input from all authors. K.D. supervised all aspects of the work.
REFERENCES
- Beier KT, Steinberg EE, DeLoach KE, Xie S, Miyamichi K, Schwarz L, Gao XJ, Kremer EJ, Malenka RC, Luo L. Circuit architecture of VTA dopamine neurons revealed by systematic input-output mapping. Cell. 2015;162 doi: 10.1016/j.cell.2015.07.015. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocklisch C, Pascoli V, Wong JCY, House DRC, Yvon C, de Roo M, Tan KR, Lüscher C. Cocaine disinhibits dopamine neurons by potentiation of GABA transmission in the ventral tegmental area. Science. 2013;341:1521–1525. doi: 10.1126/science.1237059. [DOI] [PubMed] [Google Scholar]
- Brischoux F, Chakraborty S, Brierley DI, Ungless MA. Phasic excitation of dopamine neurons in ventral VTA by noxious stimuli. Proc. Natl. Acad. Sci. USA. 2009;106:4894–4899. doi: 10.1073/pnas.0811507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg-Martin ES, Matsumoto M, Hikosaka O. Dopamine in motivational control: rewarding, aversive, and alerting. Neuron. 2010;68:815–834. doi: 10.1016/j.neuron.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañé A, Theobald DEH, Robbins TW. Selective lesions of the dorsomedial striatum impair serial spatial reversal learning in rats. Behav. Brain Res. 2010;210:74–83. doi: 10.1016/j.bbr.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuhma N, Tanaka KF, Hen R, Rayport S. Functional connectome of the striatal medium spiny neuron. J. Neurosci. 2011;31:1183–1192. doi: 10.1523/JNEUROSCI.3833-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nat. Methods. 2013;10:508–513. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim S-Y, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JY, Haesler S, Vong L, Lowell BB, Uchida N. Neuron-type-specific signals for reward and punishment in the ventral tegmental area. Nature. 2012;482:85–88. doi: 10.1038/nature10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden JR, Graybiel AM. Basal Ganglia disorders associated with imbalances in the striatal striosome and matrix compartments. Front. Neuroanat. 2011;5:59. doi: 10.3389/fnana.2011.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvas M, Palmiter RD. Restriction of dopamine signaling to the dorsolateral striatum is sufficient for many cognitive behaviors. Proc. Natl. Acad. Sci. USA. 2009;106:14664–14669. doi: 10.1073/pnas.0907299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvas M, Palmiter RD. Restricting dopaminergic signaling to either dorsolateral or medial striatum facilitates cognition. J. Neurosci. 2010;30:1158–1165. doi: 10.1523/JNEUROSCI.4576-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure A, Haberland U, Condé F, El Massioui N. Lesion to the nigrostriatal dopamine system disrupts stimulus-response habit formation. J. Neurosci. 2005;25:2771–2780. doi: 10.1523/JNEUROSCI.3894-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone RE, McDonald RJ. Dorsal striatum and stimulus-response learning: lesions of the dorsolateral, but not dorsomedial, striatum impair acquisition of a stimulus-response-based instrumental discrimination task, while sparing conditioned place preference learning. Neuroscience. 2004;124:23–31. doi: 10.1016/j.neuroscience.2003.10.038. [DOI] [PubMed] [Google Scholar]
- Fiorillo CD. Two dimensions of value: dopamine neurons represent reward but not aversiveness. Science. 2013;341:546–549. doi: 10.1126/science.1238699. [DOI] [PubMed] [Google Scholar]
- Fiorillo CD, Yun SR, Song MR. Diversity and homogeneity in responses of midbrain dopamine neurons. J. Neurosci. 2013;33:4693–4709. doi: 10.1523/JNEUROSCI.3886-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Ad-hikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, et al. Natural neural projection dynamics underlying social behavior. Cell. 2014;157:1535–1551. doi: 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber SN, Fudge JL, McFarland NR. Striatonigrostriatal pathways in primates form an ascending spiral from the shell to the dorsolateral striatum. J. Neurosci. 2000;20:2369–2382. doi: 10.1523/JNEUROSCI.20-06-02369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnasko TS, Perez FA, Scouras AD, Stoll EA, Gale SD, Luquet S, Phillips PEM, Kremer EJ, Palmiter RD. Cre recombinase-mediated restoration of nigrostriatal dopamine in dopamine-deficient mice reverses hypophagia and bradykinesia. Proc. Natl. Acad. Sci. USA. 2006;103:8858–8863. doi: 10.1073/pnas.0603081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilango A, Kesner AJ, Keller KL, Stuber GD, Bonci A, Ikemoto S. Similar roles of substantia nigra and ventral tegmental dopamine neurons in reward and aversion. J. Neurosci. 2014;34:817–822. doi: 10.1523/JNEUROSCI.1703-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-Y, Chung K, Deisseroth K. Light microscopy mapping of connections in the intact brain. Trends Cogn. Sci. 2013;17:596–599. doi: 10.1016/j.tics.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Kim Y, Venkataraju KU, Pradhan K, Mende C, Taranda J, Turaga SC, Arganda-Carreras I, Ng L, Hawrylycz MJ, Rockland KS, et al. Mapping social behavior-induced brain activation at cellular resolution in the mouse. Cell Rep. 2015;10:292–305. doi: 10.1016/j.celrep.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Hetzel A, Häckel O, Jones I, Liss B, Roeper J. Unique properties of mesoprefrontal neurons within a dual mesocorticolimbic dopamine system. Neuron. 2008;57:760–773. doi: 10.1016/j.neuron.2008.01.022. [DOI] [PubMed] [Google Scholar]
- Lammel S, Ion DI, Roeper J, Malenka RC. Projection-specific modulation of dopamine neuron synapses by aversive and rewarding stimuli. Neuron. 2011;70:855–862. doi: 10.1016/j.neuron.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Lim BK, Ran C, Huang KW, Betley MJ, Tye KM, Deisser-oth K, Malenka RC. Input-specific control of reward and aversion in the ventral tegmental area. Nature. 2012;491:212–217. doi: 10.1038/nature11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Lim BK, Malenka RC. Reward and aversion in a heterogeneous midbrain dopamine system. Neuropharmacology. 2014;76(Pt B):351–359. doi: 10.1016/j.neuropharm.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Steinberg EE, Földy C, Wall NR, Beier K, Luo L, Malenka RC. Diversity of transgenic mouse models for selective targeting of midbrain dopamine neurons. Neuron. 2015;85:429–438. doi: 10.1016/j.neuron.2014.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis EB, Mitchell JM, Ishikawa J, Hjelmstad GO, Fields HL. Midbrain dopamine neurons: projection target determines action potential duration and dopamine D(2) receptor inhibition. J. Neurosci. 2008;28:8908–8913. doi: 10.1523/JNEUROSCI.1526-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Hikosaka O. Two types of dopamine neuron distinctly convey positive and negative motivational signals. Nature. 2009;459:837–841. doi: 10.1038/nature08028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirenowicz J, Schultz W. Preferential activation of midbrain dopamine neurons by appetitive rather than aversive stimuli. Nature. 1996;379:449–451. doi: 10.1038/379449a0. [DOI] [PubMed] [Google Scholar]
- Miyamichi K, Shlomai-Fuchs Y, Shu M, Weissbourd BC, Luo L, Mizrahi A. Dissecting local circuits: parvalbumin interneurons underlie broad feedback control of olfactory bulb output. Neuron. 2013;80:1232–1245. doi: 10.1016/j.neuron.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague PR, Dayan P, Sejnowski TJ. A framework for mesencephalic dopamine systems based on predictive Hebbian learning. J. Neurosci. 1996;16:1936–1947. doi: 10.1523/JNEUROSCI.16-05-01936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague PR, Hyman SE, Cohen JD. Computational roles for dopamine in behavioural control. Nature. 2004;431:760–767. doi: 10.1038/nature03015. [DOI] [PubMed] [Google Scholar]
- Neuhoff H, Neu A, Liss B, Roeper J. I(h) channels contribute to the different functional properties of identified dopaminergic subpopulations in the midbrain. J. Neurosci. 2002;22:1290–1302. doi: 10.1523/JNEUROSCI.22-04-01290.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L, Mao T, Sternson SM, Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeper J. Dissecting the diversity of midbrain dopamine neurons. Trends Neurosci. 2013;36:336–342. doi: 10.1016/j.tins.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Rossi MA, Sukharnikova T, Hayrapetyan VY, Yang L, Yin HH. Operant self-stimulation of dopamine neurons in the substantia nigra. PLoS ONE. 2013;8:e65799. doi: 10.1371/journal.pone.0065799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiemann J, Schlaudraff F, Klose V, Bingmer M, Seino S, Magill PJ, Zaghloul KA, Schneider G, Liss B, Roeper J. K-ATP channels in dopamine substantia nigra neurons control bursting and novelty-induced exploration. Nat. Neurosci. 2012;15:1272–1280. doi: 10.1038/nn.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz W, Dayan P, Montague PR. A neural substrate of prediction and reward. Science. 1997;275:1593–1599. doi: 10.1126/science.275.5306.1593. [DOI] [PubMed] [Google Scholar]
- Schwarz LA, Miyamichi K, Gao XJ, Beier KT, Weissbourd B, DeLoach KE, Ren J, Ibanes S, Malenka RC, Kremer EJ, Luo L. Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature. 2015 doi: 10.1038/nature14600. http://dx.doi.org/10.1038/nature14600. [DOI] [PMC free article] [PubMed]
- Soudais C, Laplace-Builhe C, Kissa K, Kremer EJ. Preferential transduction of neurons by canine adenovirus vectors and their efficient retrograde transport in vivo. FASEB J. 2001;15:2283–2285. doi: 10.1096/fj.01-0321fje. [DOI] [PubMed] [Google Scholar]
- Steinberg EE, Keiflin R, Boivin JR, Witten IB, Deisseroth K, Janak PH. A causal link between prediction errors, dopamine neurons and learning. Nat. Neurosci. 2013;16:966–973. doi: 10.1038/nn.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RS. Learning to predict by the methods of temporal differences. Mach. Learn. 1988;3:9–44. [Google Scholar]
- Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungless MA, Magill PJ, Bolam JP. Uniform inhibition of dopamine neurons in the ventral tegmental area by aversive stimuli. Science. 2004;303:2040–2042. doi: 10.1126/science.1093360. [DOI] [PubMed] [Google Scholar]
- Waelti P, Dickinson A, Schultz W. Dopamine responses comply with basic assumptions of formal learning theory. Nature. 2001;412:43–48. doi: 10.1038/35083500. [DOI] [PubMed] [Google Scholar]
- Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzel-mann K-K, Young JAT, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Driscoll JR, Wilbrecht L, Margolis EB, Fields HL, Hjelmstad GO. Nucleus accumbens medium spiny neurons target non-dopaminergic neurons in the ventral tegmental area. J. Neurosci. 2011;31:7811–7816. doi: 10.1523/JNEUROSCI.1504-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HH, Knowlton BJ. Contributions of striatal subregions to place and response learning. Learn. Mem. 2004;11:459–463. doi: 10.1101/lm.81004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HH, Knowlton BJ, Balleine BW. Lesions of dorsolateral striatum preserve outcome expectancy but disrupt habit formation in instrumental learning. Eur. J. Neurosci. 2004;19:181–189. doi: 10.1111/j.1460-9568.2004.03095.x. [DOI] [PubMed] [Google Scholar]
- Yin HH, Knowlton BJ, Balleine BW. Blockade of NMDA receptors in the dorsomedial striatum prevents action-outcome learning in instrumental conditioning. Eur. J. Neurosci. 2005a;22:505–512. doi: 10.1111/j.1460-9568.2005.04219.x. [DOI] [PubMed] [Google Scholar]
- Yin HH, Ostlund SB, Knowlton BJ, Balleine BW. The role of the dorsomedial striatum in instrumental conditioning. Eur. J. Neurosci. 2005b;22:513–523. doi: 10.1111/j.1460-9568.2005.04218.x. [DOI] [PubMed] [Google Scholar]
- Yin HH, Mulcare SP, Hilário MRF, Clouse E, Holloway T, Davis MI, Hansson AC, Lovinger DM, Costa RM. Dynamic reorganization of striatal circuits during the acquisition and consolidation of a skill. Nat. Neurosci. 2009;12:333–341. doi: 10.1038/nn.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.