Abstract
Ladderanes are hydrocarbon chains with three or five linearly concatenated cyclobutane rings that are uniquely produced as membrane lipid components by anammox (anaerobic ammonia-oxidizing) bacteria. By virtue of their angle and torsional strain, ladderanes are unusually energetic compounds, and if produced biochemically by engineered microbes, could serve as renewable, high-energy-density jet fuel components. The biochemistry and genetics underlying the ladderane biosynthetic pathway are unknown, however, previous studies have identified a pool of 34 candidate genes from the anammox bacterium, Kuenenia stuttgartiensis, some or all of which may be involved with ladderane fatty acid biosynthesis. The goal of the present study was to establish a systematic means of testing the candidate genes from K. stuttgartiensis for involvement in ladderane biosynthesis through heterologous expression in E. coli under anaerobic conditions. This study describes an efficient means of assembly of synthesized, codon-optimized candidate ladderane biosynthesis genes in synthetic operons that allows for changes to regulatory element sequences, as well as modular assembly of multiple operons for simultaneous heterologous expression in E. coli (or potentially other microbial hosts). We also describe in vivo functional tests of putative anammox homologs of the phytoene desaturase CrtI, which plays an important role in the hypothesized ladderane pathway, and a method for soluble purification of one of these enzymes. This study is, to our knowledge, the first experimental effort focusing on the role of specific anammox genes in the production of ladderanes, and lays the foundation for future efforts toward determination of the ladderane biosynthetic pathway. Our substantial, but far from comprehensive, efforts at elucidating the ladderane biosynthetic pathway were not successful. We invite the scientific community to take advantage of the considerable synthetic biology resources and experimental results developed in this study to elucidate the biosynthetic pathway that produces unique and intriguing ladderane lipids.
Introduction
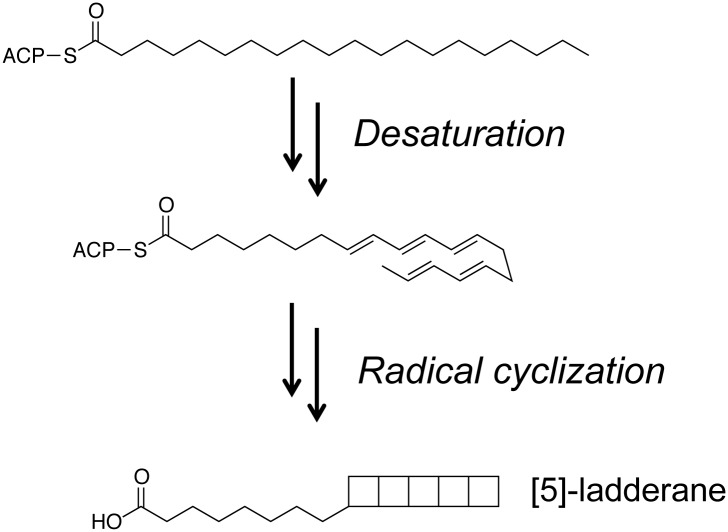
Ladderanes (e.g., Fig 1) are hydrocarbon chains with three or five fused cyclobutane rings that are uniquely produced as membrane lipid components by anammox (anaerobic ammonia-oxidizing) bacteria [1–3]. Ladderanes are unusually energetic compounds by virtue of their angle and torsional strain [4, 5]. While renewable, microbially produced fuels derived from conventional fatty acids, such as fatty acid ethyl esters or medium-chain methyl ketones, have recently been developed and have favorable properties as diesel fuel blending agents [6–10], it is plausible that fuels derived from ladderane fatty acids would have excellent jet fuel properties, in particular, high volumetric energy density. For example, estimations of volumetric energy densities of ladderane structures suggest that a [5]-ladderane could have ~46% greater volumetric energy density than conventional jet fuel used in the U.S. (S1 Table), which would lead to greater energy efficiency.
Fig 1. Structure of C20 [5]-ladderane fatty acid, and the proposed major steps of the ladderane biosynthetic pathway.
desaturation of acyl-ACPs to form polyunsaturated (all-trans) intermediates and cyclization via a radical cascade mechanism (adapted from [11]).
The primary challenge in producing ladderane-derived fuels in a renewable fashion, for example, using engineered microbes to make them from cellulosic sugars, is that the underlying biochemistry and genetics of ladderane biosynthesis are unknown. Ladderane synthesis has been an enigmatic topic of fascination to synthetic organic chemists as well as biochemists [5, 11–13]. Although an elegant chemical synthesis of [5]-ladderane fatty acid (pentacycloanammoxic acid) was accomplished [12], the synthesis was laborious, low yielding (~2%), and required unconventional chemical feedstocks, making it poorly suited to scale-up. Biosynthesis of linearly concatenated cyclobutane rings has no precedent among known enzymatic reactions. However, Rattray and co-workers [11] have identified a pool of 34 gene candidates, some or all of which may be involved with ladderane biosynthesis. These 34 genes (occurring in four gene clusters) were identified in the reconstructed genome of the anammox bacterium Kuenenia stuttgartiensis, which dominated an enrichment culture whose metagenome was sequenced [13]; note that anammox bacteria have not yet been isolated in pure culture. The selection of the 34 candidate genes [11] was based in part on key hypothesized reactions in ladderane fatty acid biosynthesis, namely desaturation of an acyl-ACP to form a polyunsaturated (all-trans) intermediate and radical-mediated polycyclization of that intermediate to form the ladderane structure (Fig 1). Arguments that this group of genes could be involved in ladderane biosynthesis included the following: (i) the four gene clusters include canonical and non-canonical versions of fatty acid biosynthetic genes, suggesting their involvement in synthesis of fatty acid derivatives (such as ladderane fatty acids); (ii) the clusters include homologs of key putative gene products, namely phytoene desaturases and S-adenosylmethionine (SAM) radical enzymes, which could be involved in desaturation of acyl-ACPs and radical cascade cyclization, respectively; (iii) computational chemistry studies have indicated that ladderane cyclization could be mediated by radical chemistry, although polycyclization of a hydrocarbon with the unsaturation pattern shown in Fig 1 (middle) would be endothermic, and cationic polycyclization has also been proposed (by analogy to terpene cyclization) [5, 14]; and (iv) searches for polyketide synthases (PKSs), which could potentially catalyze synthesis of all-trans polyunsaturated compounds, in the K. stuttgartiensis genome yielded no positive results.
The purpose of the present study was to establish a means of testing the 34 candidate genes from K. stuttgartiensis for involvement in ladderane bioysnthesis through synthesis of the codon-optimized genes and heterologous expression in Escherichia coli under anaerobic conditions. E. coli was chosen as a host because it is a facultative anaerobe that can be grown fermentatively and because, to date, no anammox bacterial species have been isolated in pure culture and there is no genetic system established for any anammox bacterium [2, 15]. Furthermore, anammox bacteria are obligate anaerobes that have a generation time of approximately two weeks and require nonstandard cultivation conditions for growth [15], making these organisms intractable for biochemical studies of the proposed scope and magnitude. This study describes the design and assembly of synthetic operons including candidate ladderane pathway genes, as well as a method for soluble purification of one of the crucial enzymes in the hypothesized pathway, a desaturase. This study is, to our knowledge, the first experimental effort focusing on the role of specific anammox genes in the production of ladderanes, and lays the foundation for future efforts toward determination of the ladderane biosynthetic pathway.
Materials and Methods
Bacterial strains, plasmids, and reagents
Bacterial strains and plasmids used in this study are listed in Table 1. Strains and plasmids along with their associated information (annotated GenBank-format sequence files) have been deposited in the public instance of the JBEI Registry [16] (https://public-registry.jbei.org/folders/222; JBEI strain and plasmid numbers along with JPUB part IDs are given in Table 1) and are physically available from the authors and/or Addgene (http://www.addgene.org) upon request. Phusion DNA polymerase, restriction enzymes, and T4 DNA ligase were purchased from Thermo Scientific (Waltman, MA). Plasmid extractions were carried out using Qiagen Miniprep Kits (Valencia, CA). All organic solvents were purchased from Sigma-Aldrich (St. Louis, MO) and were pesticide-residue-analysis grade.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | JPUB Part IDa | Relevant Characteristics | Source or reference |
|---|---|---|---|
| E. coli strains | |||
| BL21(DE3) | F- ompT gal dcm lon hsdSB(rB- mB-) λ(DE3) | [17] | |
| DH10B | F- mcrA endA1 recA1 galE15 galK16 nupG rpsL ΔlacX74 Φ80lacZΔM15 araD139 Δ(ara leu)7697 Δ(mrr-hsdRMS-mcrBC) λ- | [18] | |
| DH5αZ1 | attB::[lacIq PN25-tetR SpR] F- Δ(argF-lac)169 Φ80dlacZ58(M15) ΔphoA8 glnX44(AS) λ- deoR481 gyrA96(NalR) recA1 endA1 thiE1 hsdR17 | [19] | |
| MG1655 | F- λ- rph-1 | [20] | |
| ladd-initial | JPUB_006782 | DH5αZ1with pPJ176 | This study |
| op3 final | JPUB_006764 | DH5αZ1with with pPJ167 | This study |
| op4 final | JPUB_006766 | DH5αZ1with with pPJ168 | This study |
| op5 final | JPUB_006768 | DH5αZ1with with pPJ169 | This study |
| op6 final | JPUB_006770 | DH5αZ1with with pPJ170 | This study |
| op7 final | JPUB_006772 | DH5αZ1with with pPJ171 | This study |
| op8 final | JPUB_006774 | DH5αZ1with with pPJ172 | This study |
| op10 final | JPUB_006776 | DH5αZ1with with pPJ173 | This study |
| Lyc-no-CrtI | JPUB_006743 | MG1655 with pPJ179 | This study |
| Lyc36 | JPUB_006744 | MG1655 with pPJ177 | This study |
| Lyc07 | JPUB_006745 | MG1655 with pPJ178 | This study |
| Lyc | JPUB_006810 | MG1655 with pLyc | [21] |
| MBP-3336 | JPUB_006784 | BL21(DE3) with pPJ180 | This study |
| MBP-3607 | JPUB_006786 | BL21(DE3) with pPJ181 | This study |
| MBP-CrtI | JPUB_006788 | BL21(DE3) with pPJ182 | This study |
| N-His-3336 | JPUB_006790 | BL21(DE3) with pPJ183 | This study |
| N-His-3607 | JPUB_006792 | BL21(DE3) with pPJ184 | This study |
| N-His-CrtI | JPUB_006794 | BL21(DE3) with pPJ185 | This study |
| C-His-3336 | JPUB_006796 | BL21(DE3) with pPJ186 | This study |
| C-His-3607 | JPUB_006798 | BL21(DE3) with pPJ187 | This study |
| C-His-CrtI | JPUB_006800 | BL21(DE3) with pPJ188 | This study |
| N-StrepII-3607 | JPUB_006806 | BL21(DE3) with pPJ219 | This study |
| C-StrepII-3607 | JPUB_006808 | BL21(DE3) with pPJ220 | This study |
| N-8xHis-StrepII-MBP-3607 | JPUB_006802 | BL21(DE3) with pPJ217 | This study |
| Plasmids | |||
| pFAB217 | KmR; p15a vector | [22] | |
| pBbA0k | KmR; p15a vector | [23] | |
| pBbE0k | KmR; ColE1 vector | [23] | |
| pBbE0a_mut | AmpR; ColE1 vector; BsaI site removed by mutagenesis | [23] | |
| pET24 | KmR; T7 promoter | Novagen | |
| pET28a | KmR; T7 promoter | Novagen | |
| pET28a-MBP | KmR; T7 promoter; maltose-binding protein (MBP) tag sequence | Footnote b | |
| pSKB3-EL3 | KmR; T7 promoter; 8xHis, StrepII, and MBP tag sequences | Footnote c | |
| pPJ174 | JPUB_006779 | KmR; operon 1 genes (kuste3603, kuste3605, kuste3606) with unique Ptet promoter, BCD, and terminator in pFAB217 | This study |
| pPJ175 | JPUB_006781 | KmR; operon 2 genes (kuste3604, kuste3607, kuste3608) with unique Ptet promoter, BCD, and terminator in pBbA0k | This study |
| pPJ176 | JPUB_006783 | KmR; EcoRI- and BamHI-digestion fragment of pPJ174 ligated into EcoRI and BglII sites of pPJ175 | This study |
| pPJ158 | JPUB_006747 | KmR; operon 3 genes (kuste2803, kuste2804) in pBbE0k | This study |
| pPJ159 | JPUB_006749 | KmR; operon 4 genes (kuste2802, kuste2805) in pBbE0k | This study |
| pPJ160 | JPUB_006751 | AmpR; operon 5 genes (kuste3340, kuste3350) in pBbE0a_mut | This study |
| pPJ161 | JPUB_006753 | KmR; operon 6 genes (kuste3346, kuste3348, kuste3349) in pBbE0k | This study |
| pPJ162 | JPUB_006755 | AmpR; operon 7 genes (kuste3338, kuste3347) in pBbE0a_mut | This study |
| pPJ163 | JPUB_006757 | AmpR; operon 8 genes (kuste3342, kuste3343) in pBbE0a_mut | This study |
| pPJ164 | JPUB_006759 | AmpR; operon 9 genes (kuste3345, kuste3351, kuste3352) in pBbE0a_mut | This study |
| pPJ165 | JPUB_006761 | KmR; operon 10 genes (kuste3336, kuste3339, kuste3341) in pBbE0k | This study |
| pPJ166 | JPUB_006763 | AmpR; operon 11 genes (kuste3335, kuste3344) in pBbE0a_mut | This study |
| pPJ149 | JPUB_006726 | KmR; new unique Ptet promoter and terminator for operon 3 in pBbA0k | This study |
| pPJ150 | JPUB_006728 | KmR; new unique Ptet promoter and terminator for operon 4 in pBbA0k | This study |
| pPJ151 | JPUB_006730 | KmR; new unique Ptet promoter and terminator for operon 5 in pBbA0k | This study |
| pPJ152 | JPUB_006732 | KmR; new unique Ptet promoter and terminator for operon 6 in pBbA0k | This study |
| pPJ153 | JPUB_006722 | KmR; new unique Ptet promoter and terminator for operon 7 in pBbA0k | This study |
| pPJ154 | JPUB_006734 | KmR; new unique Ptet promoter and terminator for operon 8 in pBbA0k | This study |
| pPJ146 | JPUB_006724 | KmR; old Ptet promoter, BCD, and terminator for operon 9 in pBbA0k | This study |
| pPJ156 | JPUB_006736 | KmR; new unique Ptet promoter and terminator for operon 10 in pBbA0k | This study |
| pPJ167 | JPUB_006765 | KmR; operon 3 genes in pPJ149 | This study |
| pPJ168 | JPUB_006767 | KmR; operon 4 genes in pPJ150 | This study |
| pPJ169 | JPUB_006769 | KmR; operon 5 genes in pPJ151 | This study |
| pPJ170 | JPUB_006771 | KmR; operon 6 genes in pPJ152 | This study |
| pPJ171 | JPUB_006773 | KmR; operon 7 genes in pPJ153 | This study |
| pPJ172 | JPUB_006775 | KmR; operon 8 genes in pPJ154 | This study |
| pPJ173 | JPUB_006777 | KmR; operon 10 genes in pPJ156 | This study |
| pLyc | CmR; BR322 vector with araC repressor; crtE, crtI, and crtB under PBAD promoter | [21] | |
| pPJ179 | JPUB_006738 | CmR; pLyc with crtI gene removed | This study |
| pPJ177 | JPUB_006740 | CmR; pLyc with kuste3336 replacing crtI | This study |
| pPJ178 | JPUB_006742 | CmR; pLyc with kuste3607 replacing crtI | This study |
| pPJ180 | JPUB_006785 | KmR; kuste3336 with N-terminal MBP tag sequence in pET28a-MBP | This study |
| pPJ181 | JPUB_006787 | KmR; kuste3607 with N-terminal MBP tag sequence in pET28a-MBP | This study |
| pPJ182 | JPUB_006789 | KmR; Pantoea agglomerans crtI with N-terminal MBP tag sequence in pET28a-MBP | This study |
| pPJ183 | JPUB_006791 | KmR; kuste3336 with N-terminal 6xHis tag sequence in pET28a | This study |
| pPJ184 | JPUB_006793 | KmR; kuste3607 with N-terminal 6xHis tag sequence in pET28a | This study |
| pPJ185 | JPUB_006795 | KmR; Pantoea agglomerans crtI with N-terminal 6xHis tag sequence in pET28a | This study |
| pPJ186 | JPUB_006797 | KmR; kuste3336 with C-terminal 6xHis tag sequence in pET24 | This study |
| pPJ187 | JPUB_006799 | KmR; kuste3607 with C-terminal 6xHis tag sequence in pET24 | This study |
| pPJ188 | JPUB_006801 | KmR; Pantoea agglomerans crtI with C-terminal 6xHis tag sequence in pET24 | This study |
| pPJ218 | JPUB_006805 | KmR; E. coli acpP with N-terminal StrepII tag sequence in pET24 | This study |
| pPJ219 | JPUB_006807 | KmR; kuste3607 with N-terminal StrepII tag sequence in pET24 | This study |
| pPJ220 | JPUB_006809 | KmR; kuste3607 with C-terminal StrepII tag sequence in pET24 | This study |
| pPJ217 | JPUB_006803 | KmR; kuste3607 with N-terminal 8xHis, StrepII, and MBP tag sequences in pSKB3-EL3 | This study |
a Accessible via public instance of the JBEI Registry (https://public-registry.jbei.org/folders/222)
b Provided by Andrew Hagen (JBEI).
c Provided by Aindrila Mukhopadhyay (JBEI).
Synthesized DNA sequences, including candidate ladderane biosynthesis genes from anammox bacteria
All 34 K. stuttgartiensis genes identified as candidates for the ladderane biosynthesis pathway [11] were codon-optimized for expression in E. coli using an empirically derived codon usage table [24]. Codon optimization, including restriction site removal, and oligo design (150mers) were performed using GeneDesign [25]. Oligos were pooled for synthesis using acoustic deposition (Labcyte Echo 550), and synthesis was performed at the Joint Genome Institute using a 2-step PCA approach in 2 μL final volume as previously described [26]. PCA products were purified by gel excision and cloned into pENTR (Life Technologies) by In-Fusion cloning (Clontech). Plating and picking were performed using a QPix 400 system (Molecular Devices). Eight colonies per construct were sequence-verified using PACBIO RSII system (Pacific Biosciences). Synthesized gene sequences are listed in S2 Table.
Operon regulatory elements consisted of unique tetracycline-inducible promoters (Ptet), bicistronic design (BCD) elements, and terminator sequences, which were obtained from BIOFAB [22, 27]. Additional DNA parts were purchased as gBlocks or oligonucleotides from Integrated DNA Technologies (Coralville, IA). Oligonucleotides used for DNA assemblies, with the exception of those used for restriction digest and ligation, were designed using j5 software [28, 29]. Operon composition, DNA assembly templates and primers, and gBlock sequences are listed in S3, S4 and S5 Tables, respectively.
Operon plasmid assembly
Candidate ladderane biosynthesis genes were divided among 11 synthetic operons based upon putative function (Table 2). The operon 1 plasmid was assembled as follows. The promoter and BCD sequences were PCR-amplified from a gBlock, while gene CDSs, including intergenic RBSs, were PCR-amplified from JGI shuttle plasmids carrying each respective gene. These parts were assembled with a PCR-amplified pFAB217 backbone (p15A ori, kanR) using a modified Golden Gate [30] assembly method, which involved BsaI digestion of parts overnight, followed by ligation with a high-concentration ligase for 30 min at room temperature. The operon 2 plasmid was assembled in a similar fashion as operon 1, with the exception that a pBbA0k backbone (p15A ori, kanR) [23] was used. The original terminator in the operon 2 plasmid was later replaced with a his[min] terminator by inverse PCR using phosphorylated primers containing the necessary sequence to be added. Each assembly was sequence-verified. Finally, operons 1 and 2 were combined in a single plasmid, pPJ176 (Table 1), by isolating operon 1 through EcoRI and BamHI digestion and inserting the operon 1 fragment into the operon 2 plasmid backbone, which had been digested with EcoRI and BglII.
Table 2. Synthetic operons used in this study.
| Operon | Gene | Putative Function | Notes |
|---|---|---|---|
| 1 | kuste3603 | ACP (acyl carrier protein) | Operon 1 + Operon 2 genes constitute the second most highly expressed candidate gene cluster in K. stuttgartiensis and encode some unique products |
| 1 | kuste3605 | FabF (β-ketoacyl-ACP synthase II) | (see above) |
| 1 | kuste3606 | non-canonical FabF | (see above) |
| 2 | kuste3604 | FabZ (β-hydroxyacyl-ACP dehydratase) | (see above) |
| 2 | kuste3607 | phytoene desaturase | (see above) |
| 2 | kuste3608 | SAM radical enzyme | (see above) |
| 3 | kuste2803 | SAM radical enzyme | Operon 3 + Operon 4 genes constitute a complete cluster in K. stuttgartiensis and encode some unique products |
| 3 | kuste2804 | FabF | (see above) |
| 4 | kuste2802 | ACP synthase | (see above) |
| 4 | kuste2805 | non-canonical FabB (β-ketoacyl-ACP synthase I) | (see above) |
| 5 | kuste3340 | ACP | Activate fatty acid synthesis intermediates |
| 5 | kuste3350 | ACP | (see above) |
| 6 | kuste3346 | FabF | β-ketoacyl-ACP synthase activity |
| 6 | kuste3348 | non-canonical FabF | (see above) |
| 6 | kuste3349 | FabF | (see above) |
| 7 | kuste3338 | SAM methylase | SAM radical reactions are hypothesized to play a role in ladderane cyclization |
| 7 | kuste3347 | SAM methylase | (see above) |
| 8 | kuste3342 | SAM radical enyzme | (see above) |
| 8 | kuste3343 | SAM radical enzyme | (see above) |
| 9 | kuste3345 | unknown | Function of gene product is unknown |
| 9 | kuste3351 | conserved hypothetical protein | (see above) |
| 9 | kuste3352 | unknown | (see above) |
| 10 | kuste3336 | phytoene desaturase | Catalysis of putative desaturation or reduction reactions |
| 10 | kuste3339 | FabZ | (see above) |
| 10 | kuste3341 | FabG | (see above) |
| 11 | kuste3335 | glycosyltransferase | Function not typically associated with fatty acid synthesis |
| 11 | kuste3344 | phenylacetyl-CoA ligase | (see above) |
Plasmids carrying completed operons 3–11 were each assembled from two separate shuttle plasmids, one carrying operon genes and the other carrying regulatory elements (Ptet promoter, BCD, terminator) (Fig 2). For the operon 3, 4, 6, and 10 gene plasmids, respective genes were PCR-amplified from JGI shuttle plasmids and cloned into a pBbE0k backbone (colE1 ori, kanR), which had been digested with BamHI and XhoI, through Gibson assembly [31]. The operon 5, 7, 8, 9, and 11 gene plasmids were assembled in a similar fashion, with the exception that the backbone was derived from BamHI and XhoI digestion of pBbE0a_mut, (colE1 ori, ampR, BsaI site removed through mutagenesis). For operons containing more than two genes (6, 9, 10), two-step assemblies were conducted to assemble the complete gene plasmids. Each assembly reaction was sequence-verified.
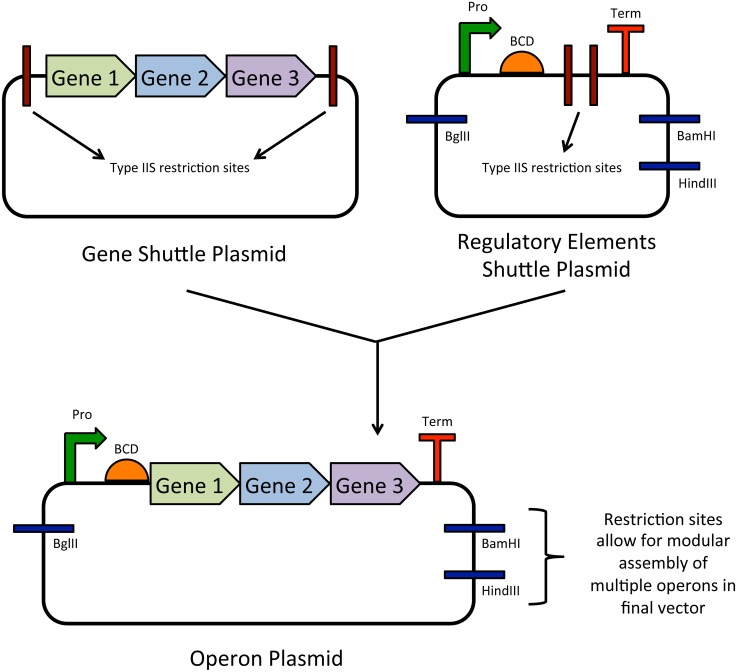
Fig 2. DNA assembly scheme for construction of operons 3–11 (see Table 2 for additional detail).
Each operon has a unique Ptet promoter, bicistronic design (BCD) element, and terminator chosen from the BIOFAB database. Restriction sites in each final operon plasmid allow for efficient, modular assembly of multiple operons in a final vector, such as a bacterial artificial chromosome or fosmid.
The regulatory-element shuttle plasmid for each operon consisted of promoter, BCD, and terminator sequences that were PCR-amplified from gBlocks and assembled in a PCR-amplified pBbA0k backbone, using the CPEC technique [32]. Shuttle plasmids for operons 5, 7, 8, and 9 required an additional assembly step because the complete sequence for promoter or terminator could not be included in the initial CPEC assembly primers, due to possible mispriming. The missing sequences for each of these plasmids were introduced by inverse PCR using phosphorylated primers. Each assembly reaction was sequence-verified. The gene and regulatory elements shuttle plasmids for each operon had complementary Type IIS restriction sites (S4 Table). These sites were used in Golden Gate assemblies to insert each set of operon genes into the respective regulatory elements plasmids, yielding the final operon plasmids.
Operon promoter replacement
The promoters for operons 3–11 were replaced with stronger variants from the BIOFAB registry that were previously characterized [22]. Each of the new promoters was introduced into its respective regulatory elements shuttle plasmid through one-part CPEC assemblies. To simplify analysis of protein expression, BCDs were removed during assembly. The new promoters were combined with the respective gene sets as described above to produce new operons 3–11. Sequences for the new promoters are listed in S3 Table.
DNA assembly for protein expression of putative desaturases
Vectors for expression of each C-terminal 6xHis-tagged desaturase were prepared by PCR-amplifying genes and inserting into an NdeI- and XhoI-digested pET24 vector backbone (Novagen Biosciences, Madison, WI) through Gibson assembly. The kuste3336 and kuste3607 genes were amplified from JGI shuttle plasmids, whereas the Pantoea agglomerans crtI gene was PCR-amplified from the previously described pLyc plasmid [21] (Table 1). Gibson assemblies were also performed to introduce the kuste3607 gene into each of two separate pET24 backbones, which were PCR-amplified from pET24-N-StrepII-EcAcpP (pPJ218; Table 1), to give constructs expressing either N- or C-terminal StrepII-tagged proteins. N-6xHis- and N-6xHis-MBP-tagged expression constructs were prepared by PCR-amplifying kuste3607 from its JGI shuttle plasmid, digesting with NdeI and XhoI, and ligating into either digested pET28 or pET28a-MBP (JBEI ICE Part ID JBx_014631), respectively. The kuste3607 gene was also inserted into pSKB3-EL3 (JBEI-7594) through Gibson assembly to construct a vector expressing N-8xHis-StrepII-MBP-3607, which has a tobacco etch virus (TEV) protease recognition site.
The pLyc-no-CrtI plasmid (pPJ179; Table 1) was constructed through a one-part CPEC assembly [32] that removed the crtI gene from pLyc. pLyc-3336 and pLyc-3607 plasmids (pPJ177 and pPJ178; Table 1) were prepared through Gibson assemblies to replace the pLyc crtI gene with either the kuste3336 or kuste3607 gene, respectively. Protein sequences are listed in S6 Table.
Cell growth and fatty acid production
Anaerobic fatty acid production in E. coli DH5αZ1 transformed with pPJ176 (operons 1 and 2; Table 1) was compared to that of DH5αZ1 carrying the empty pBbA0k vector. For each strain, a 250-mL serum bottle containing 200 mL of EZ Rich medium (Teknova, Hollister, CA) supplemented with 0.2% glucose and 50 μg/mL kanamycin was inoculated from an overnight culture to a starting OD600 of 0.005 and sealed with a butyl rubber stopper. The culture was incubated at 37°C shaking at 200 rpm until the OD600 reached ~0.4, at which point gene expression was induced by addition of 200 nM anhydrotetracycline (ATc) under anaerobic conditions. Anaerobic growth was continued at 37°C overnight. The next day, cells were harvested in 30-mL high-strength glass centrifuge tubes and supernatant was decanted. The cell pellet was flash-frozen in liquid nitrogen and lyophilized overnight in a Labconco lyophilizer (Kansas City, MO). Biomass was stored at room temperature until fatty acid extraction.
The remaining operon strains were individually tested in the same manner, with the exception that cultures were grown in 125-mL serum bottles containing 100 mL media, and 3 mM KNO3 was added under anaerobic conditions at the time of induction, where noted, to obtain higher cell density.
Extraction and GC/MS analysis of fatty acids
All solvents used were pesticide-residue-analysis grade and all glassware was washed with ultrapure acetone. The extraction method was modified from that described by Sinninghe Damsté et al. [33] Briefly, 6 mL MeOH was added to lyophilized biomass in a high-strength glass centrifuge tube, which was then vortexed and sonicated in an ice water bath for 10 min, followed by centrifugation at 5000 x g for 5 min at 20°C and collection of solvent in a 40-mL pre-cleaned glass vial. The extraction process was repeated once with 6 mL of MeOH:CH2Cl2 (1:1) and three times with 6 mL CH2Cl2, resulting in 30 mL extract. The extract was evaporated to dryness using an R-210 rotary evaporator (Buchi, Flawil, Switzerland). The residue was reconstituted in ~2 mL CH2Cl2, transferred to a 10-mL Reacti-Vial, and evaporated to ~100 μL under an ultra high purity nitrogen gas stream. Extracts were then derivatized with ethereal diazomethane prepared in an Aldrich diazomethane-generator (Sigma-Aldrich), followed by reconstitution in 100 μL CH2Cl2.
GC-MS analyses were performed with a model 7890A GC (Agilent, Santa Clara, CA) with a DB-5 fused silica capillary column (30-m length, 0.25-mm inner diameter, 0.25-μm film thickness; J & W Scientific) coupled to an HP 5975C series quadrupole mass spectrometer. One-μL injections were performed by a model 7683B autosampler. The GC oven was programmed from 40°C (held for 2 min) to 130°C at 15°C/min, then to 300°C at 5°C/min and held for 10 min; the injection port temperature was 250°C, and the transfer line temperature was 280°C. The carrier gas, ultra-high-purity helium, flowed at a constant rate of 1 mL/min. Injections were splitless, with the split turned on after 0.5 min. The extraction and GC/MS analysis methods were validated by the detection of ladderane fatty acids (e.g., a C20 [3]-ladderane fatty acid derivatized as a methyl ester) from anammox culture biomass samples graciously provided by Barth F. Smets of the Technical University of Denmark.
Expression and purification of kuste3607
An overnight culture of E. coli BL21(DE3) expressing N-8xHis-StrepII-MBP-3607 (Table 1) was used to inoculate two 2-L baffled flasks, each containing 1 liter of lysogeny broth (LB) supplemented with 50 μg/mL kanamycin. Cultures were grown at 37°C until the OD600 reached ~0.5, at which point protein expression was induced by addition of 50 μM IPTG and growth continued at 18°C overnight. Cells were harvested the next day and stored at -80°C until further processing. The pellet was resuspended in 100 mL of Buffer L consisting of 50 mM sodium phosphate (pH 7.5), 500 mM NaCl, 10% glycerol, 1 mM DTT, 0.2 mg/mL lysozyme, 10 mM MgCl2, 10 μg/mL DNase I, and two Pierce Protease Inhibitor Mini Tablets (Thermo Scientific, Wilmington, DE). Cells were lysed using an EmulsiFlex-C3 high-pressure homogenizer (Avestin, Ottawa, ON, Canada), followed by centrifugation of lysate at 15000 x g for 30 min at 4°C. Soluble lysate was aspirated and subjected to purification on an ÄKTAexplorer FPLC system equipped with a 5-mL StrepTrap HP column (GE Healthcare Life Sciences, Marlborough, MA). Following injection, protein was washed with Buffer A [50 mM sodium phosphate (pH 7.5), 500 mM NaCl, 10% glycerol, 1 mM DTT] and eluted with 6 column volumes (CV) of Buffer B (A + 2.5 mM desthiobiotin). Elution samples were analyzed by SDS-PAGE and fractions containing eluted 3607 were pooled. Approximately 5 mg of protein was obtained in this fashion. 0.1% Igepal CA-630 was added to the protein, which was then concentrated to ~2 mL with a 30-kDa MWCO concentrator. Recombinant TEV protease was added to the protein at a ratio of 1:100 protease:protein and incubated at 4°C overnight to cleave fusion tags from the 3607 enzyme. The cleaved protein was used for assays the following day.
Cell growth for lycopene production
Overnight cultures of E. coli MG1655 expressing pPJ179, pPJ177, pPJ178, or pLyc were used to inoculate 50 mL LB supplemented with 30 μg/mL chloramphenicol. Cultures were grown at 37°C until the OD600 reached ~0.5, at which point 10 μM L-arabinose was added and growth continued at 18°C overnight. The next day, 20 OD x mL of culture was harvested and frozen for proteomic analysis, while the remainder pelleted in high-strength glass centrifuge tubes, which were stored at -80°C until product extraction.
Lycopene extraction and HPLC analysis
To each frozen cell pellet, 1 mL MeOH and 4 mL hexane were added. Samples were vortexed and sonicated in an ice-water bath for 15 min, followed by incubation at room temperature for 10 min and centrifugation at 5000 x g for 15 min at 20°C to separate the organic and aqueous phases. The hexane layer was transferred to a 10-mL Reacti-Vial and concentrated to 50 μL under a gentle nitrogen gas stream.
Extracts were subjected to HPLC analysis using an Agilent 1200 series HPLC system, with a 3 μm, 250-mm x 2.1-mm reverse-phase Inertsil ODS-3 column (GL Sciences, Tokyo, Japan) as previously described [34], with the exception that lycopene was detected at 470 nm.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis
For RT-qPCR analysis of gene expression from operons 1 and 2, 15-mL samples from induced and non-induced cultures were collected and transferred to 50-mL tubes containing 1.9 mL of 5% phenol in ethanol. The cell mixture was incubated on ice for 10 min and then centrifuged at 1600 x g for 10 min at 4°C. Supernatant was decanted and the pellet flash-frozen in liquid nitrogen for storage at -80°C until further processing. mRNA was extracted with a RNeasy Mini Kit (Qiagen, Valencia, CA). Qiagen DNaseI was used for on-column DNA digestion and the Turbo DNA-free Kit (Thermo Fisher Scientific, Waltham, MA) for a second DNase treatment after mRNA purification. RNA concentration was quantified with a NanoDrop 1000 spectrophotometer (Thermo Scientific). cDNA was prepared from 4 μg RNA using the SMARTScribe Reverse Transcriptase Kit (Clontech, Mountain View, CA). qPCR was performed with 4 μL of tenfold-diluted cDNA as template and SsoAdvanced SYBR Green Supermix (Bio-Rad, Hercules, CA) on a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA). qPCR primers were designed with the web-based IDT PrimerQuest tool. The E. coli hcaT gene was used as an endogenous control [35] and standards consisted of pPJ176 at 0.005, 0.05, 0.5, 5, and 50 ng. Analyses were run in triplicate.
Shotgun proteomics
For analysis of protein expression from ladd-initial, 25 mL of cells were harvested at the end of growth and lysed with Thermo Scientific B-PER reagent at 4-mL / gram cell paste. Lysate was buffer-exchanged to 100 mM NH4HCO3 (AMBIC) by three rounds of concentration and dilution with a 3-kDa MWCO concentrator. Samples were then processed as previously described [36].
Analyses of samples from strains individually expressing operons 3–8 and 10 were conducted as for ladd-initial, except 20 OD x mL of cells were used and lysate was sonicated using a Qsonica sonicator (Newtown, CT) equipped with a microtip (5 sec on, 5 sec off, 25 sec processing time, power = 1). Lysate was then clarified by centrifugation at 15000 x g for 5 min at 4°C in a microcentrifuge. Soluble lysate was collected and buffer-exchanged to 100 mM AMBIC, while pelleted material was washed three times with tenfold-diluted B-PER reagent. Both soluble and pellet samples were processed for proteomic analysis as previously described [37]. Briefly, the proteins were extracted by chloroform/methanol precipitation and resuspended in 100 mM AMBIC with 20% acetonitrile. The proteins were reduced with tris(2-carboxyethyl)phosphine (TCEP) for 30 min, followed by incubation with iodoacetamide (IAA; 10 mM final) for 30 min in the dark, and overnight digestion with MS-grade trypsin (1:50 w/w trypsin:protein) at 37°C. Samples were analyzed on an Agilent 1290 UHPLC—6550 QTOF liquid chromatography mass spectrometer (LC-MS/MS; Agilent Technologies) system, with previously described operating parameters [37]. Peptides were separated on a Sigma-Aldrich Ascentis Express Peptide ES-C18 column (2.1-mm x 100-mm, 2.7- μm particle size, operated at 60°C) at a flow rate of 0.4 mL/min. The chromatography gradient conditions were as follows: from the initial starting condition [95% buffer A (100% water, 0.1% formic acid) and 5% buffer B (100% acetonitrile, 0.1% formic acid)] the buffer B composition was increased to 35% over 30 min; then buffer B was increased to 80% over 3 min and held for 7 min, followed by a ramp back down to 5% B over 1 min where it was held for 6 min to re-equilibrate the column to original conditions. Data were analyzed with the Mascot search engine version 2.3.02 (Matrix Science) and filtered and validated using Scaffold v4.3.0 (Proteome Software Inc.), as previously described [37].
Results and Discussion
Gene synthesis and design of synthetic operons
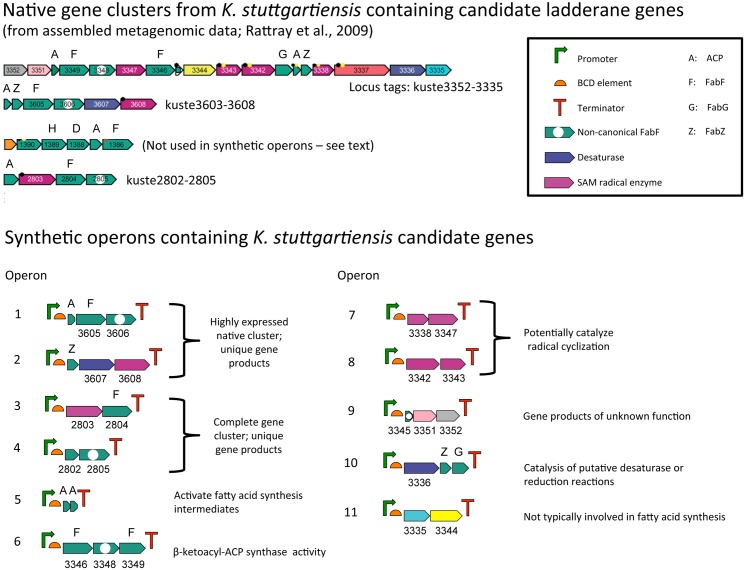
All 34 K. stuttgartiensis genes that were previously identified as potential candidates for the ladderane biosynthetic pathway [11] were codon-optimized for expression in E. coli and synthesized by JGI (S2 Table). To elucidate which of the candidate genes are involved in ladderane production, genes were grouped together in synthetic operons based on putative function (Fig 3, Table 2). This approach supports rational and efficient identification of gene function: changes in fatty acid profiles can be attributed to a putative function (operon), and then to specific gene(s) within the operon. To regulate gene expression at different levels in separate operons and prevent possible homologous recombination within large vectors, unique promoters, translation initiation (bicistronic design, BCD) elements, and terminators were chosen for each operon. Sequences for each of these parts were obtained from the BIOFAB database, which includes characterization data for each part [22, 27]. Previously reported metatranscriptome data [38] were used to select Ptet promoters of various relative strengths for each operon in order to simulate native gene expression levels in K. stuttgartiensis (native expression levels are presented in S7 Table). The Ptet system has the beneficial characteristics of low “leaky” expression and low sensitivity to catabolite repression as compared to other inducible promoter systems [23]. BCD elements were used to prevent premature translation termination caused by RNA secondary structure formation. Operons 1 and 2 were combined to form pPJ176 (Table 1), which contains genes kuste3603-3608. This cluster was the focus of preliminary cell growth and fatty acid analyses, as the encoded genes are from the second most highly expressed of the candidate gene clusters after the canonical type II fatty acid synthesis genes kustd1386-1391 (S7 Table). Moreover, the kuste3603-3608 genes encode unique putative enzymes, such as a non-canonical FabF, a desaturase, and a SAM radical enzyme [11] (Table 2). Each of the final operons 3–11 were assembled from two separately constructed shuttle vectors, one carrying genes and the other carrying regulatory element sequences (Fig 2). This approach aids future replacement of promoters, BCDs, or terminators, as sequence changes can be made in the regulatory element shuttle plasmid without affecting gene sequences. Each of the synthetic operon plasmids also contains restriction sites that allow for efficient, modular assembly of multiple operons in different combinations in a final vector, such as a bacterial artificial chromosome or fosmid.
Fig 3. Candidate ladderane synthesis genes from Kuenenia stuttgartiensis and synthetic operon design.
(Top) Candidate genes identified by Rattray et al. [11] in native gene clusters with locus tags (adapted from [11]). (Bottom) Synthetic operons designed in this study containing candidate genes grouped by putative function or native gene clusters. Locus tags (without the kuste prefix) are shown for most genes in the synthetic operons; more details are given in Table 2.
The kustd1391-1386 cluster consists of putative rpmF, plsX, fabH, fabD, acp, and fabF genes (Fig 3). These genes display homology and synteny to the E. coli rpmF-fabF gene cluster involved in canonical type II fatty acid biosynthesis, with the exception that the K. stuttgartiensis fabG (kuste3341) is located within a separate gene cluster. Because of the homology between the K. stuttgartiensis kustd gene cluster and E. coli fatty acid biosynthesis genes, the kustd gene cluster was not included in the synthetic operon design in order to limit the number of operon combinations to be tested and simplify downstream analyses. The kuste3337 gene, encoding a putative membrane protein of unknown function, was also excluded from operon design. Potential roles for the kustd1386-1391 and kuste3337 genes in ladderane production cannot be entirely ruled out, as the pathway may involve an unconventional mode of fatty acid synthesis. These genes can be included in future synthetic operon designs, following the procedure described above.
Analysis of fatty acids and gene expression in operon strains
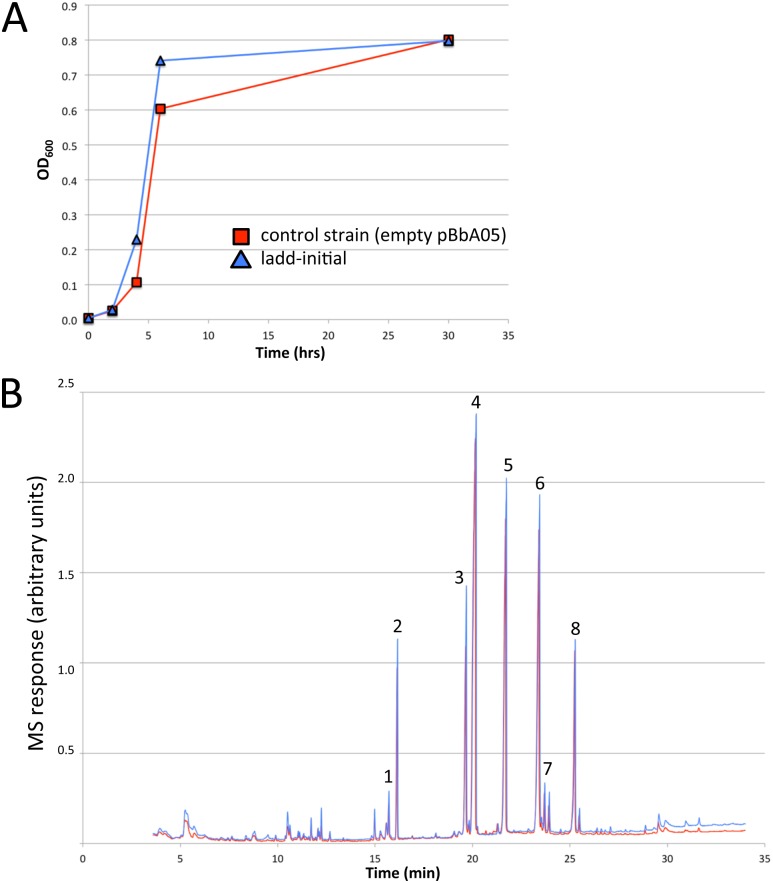
For preliminary analyses, E. coli DH5αZ1 (containing a chromosomal copy of the tetR repressor) [19] carrying pPJ176 (strain ladd-initial; Table 1) was grown under anaerobic conditions to determine whether expression of the kuste3603-3608 genes would lead to changes in fatty acid profile, namely the production of ladderanes or postulated polyunsaturated fatty acid intermediates. The control strain consisted of DH5αZ1 carrying an empty pBbA0k vector. Cell growth rate and final OD600 were similar in both strains (Fig 4A). A detailed inspection of GC/MS spectra from both strains did not show any substantial differences in fatty acid profile or the presence of ladderanes or ladderane intermediates (Fig 4B). To determine whether the absence of novel fatty acids was due to impaired gene expression, RT-qPCR was performed to analyze candidate gene transcription. This analysis indicated that each of the kuste3603-3608 genes was transcribed (S1 Fig). Additionally, shotgun proteomic analysis detected each of the expected proteins in whole lysate samples (S8 Table). These combined analyses suggest that the absence of detected products is not due to poor gene expression, but rather that kuste3603-3608 may not be necessary or sufficient for ladderane production.
Fig 4. Growth and fatty acid profiles for strain expressing operons 1 and 2 and control strain.
(A) Growth curve of ladd-initial and control strains. (B) GC/MS total ion chromatograms (TIC) of fatty acids extracted from ladd-initial and control strains post-cultivation and subjected to methyl ester derivatization. The most prominent fatty acid methyl esters are labeled with numbers: 1, C14:1; 2, C14:0; 3, C16:1; 4, C16:0; 5, C17 cyclopropane fatty acid (CFA); 6, C18:1; 7, C18:0; 8, C19 CFA.
Before testing various operon combinations, each of operons 3–8 and 10 was individually tested to analyze gene expression and any possible changes in fatty acid profile. Operons 9 and 11 were not tested at this point because the respective genes encode products of unknown function or enzymes that are not typically involved in fatty acid biosynthesis (Fig 3, Table 2). None of the individual operon strains led to ladderane or polyunsaturated fatty acid production, or other novel compounds. When samples of each strain were analyzed through shotgun proteomics, none of the expected proteins were detected (data not shown), suggesting the possibility that the promoters were of insufficient strength to yield detectable levels of protein expression. Each of the Ptet promoters for operons 3–11 was thus changed to a stronger variant, based on BIOFAB characterization data [22] (Table 3). Additionally, the BCD elements were removed to preclude these sequences as a confounding factor. New versions of operons 3–8 and 10 (Table 1; op3 final–op8 final; op10 final) were individually tested for gene expression and fatty acid profile changes. No new fatty acid products were observed through GC/MS analysis. The new promoters did lead to improved gene expression: all expected proteins were detected through shotgun proteomic analysis, with the exception of kuste3342, 3343, and 3350 (Table 3). However, the detected proteins were only found in the pellet fractions of lysate samples, except for kuste2802 and kuste3340 proteins, which were detectable at low levels in soluble lysate samples (Table 3). The lack of detection of kuste3350 protein is expected, as the gene encodes a putative, 66-amino acid acyl carrier protein that would be difficult to detect through trypsinization and LC/MS/MS analysis. It is unknown why the kuste3342 and kuste3343 gene products, putative SAM radical, iron-sulfur enzymes, were not detected. One possibility is that inefficient iron-sulfur cluster assembly may have led to degradation or inefficient protein expression [39–41]. The proteomics results raise another possibility, that the lack of detected ladderanes or intermediates is the result of candidate protein insolubility that may arise from inefficient protein folding. Protein insolubility could be addressed by using weaker ribosome binding sites (RBSs) or further optimization of promoter sequences or culture and induction conditions. It is also possible that E. coli is not an optimal host for heterologous expression of anammox genes, in which case another host could be used for future studies. Nevertheless, the operon assembly scheme and gene expression results provide an efficient system to analyze the effects of candidate gene expression on fatty acid profile.
Table 3. Designed operon promoter strength and actual expression based on shotgun proteomic analysisa.
| Operon | Original Promoter Strengthb | New Promoter Strengthb | Gene | % of total MS spectra | Rankc |
|---|---|---|---|---|---|
| 3 | 25 | 93 | kuste2803 | 0.16 | 76 |
| 3 | d | d | kuste2804 | 0.25 | 38 |
| 4 | 26 | 100 | kuste2802 | 0.14 | 91 |
| 4 | d | d | kuste2805 | 0.6 | 6 |
| 5 | 20 | 90 | kuste3340 | 0.04 | 282 |
| 5 | d | d | kuste3350 | NDe | ND |
| 6 | 13 | 86 | kuste3346 | 0.57 | 13 |
| 6 | d | d | kuste3348 | 0.67 | 8 |
| 6 | d | d | kuste3349 | 0.94 | 3 |
| 7 | 4 | 82 | kuste3338 | 0.33 | 26 |
| 7 | d | d | kuste3347 | 0.45 | 14 |
| 8 | 7 | 83 | kuste3342 | ND | ND |
| 8 | d | d | kuste3343 | ND | ND |
| 10 | 19 | 89 | kuste3336 | 0.8 | 3 |
| 10 | d | d | kuste3339 | 0.27 | 38 |
| 10 | d | d | kuste3341 | 0.37 | 24 |
a Samples consist of pellet material after lysate clarification. With the exception of kuste2803 and kuste3340, no target proteins were detected in soluble lysate samples.
b Based on strengths of corresponding constitutive promoter sequences from BIOFAB database, relative to new operon 4 promoter.
c Rank among 740 total detected proteins, based on percentage of total spectra.
d Same as above value for this operon.
e ND, not detected.
Analysis of in vivo lycopene production by putative ladderane desaturases
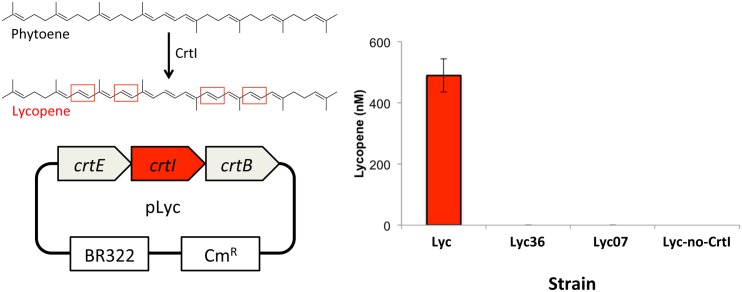
The putative desaturase genes were chosen for further analyses because the encoded enzymes could potentially catalyze unique fatty acid modification activity, and are hypothesized to play a crucial role in ladderane synthesis, namely, formation of polyunsaturated fatty acids. The pool of candidate genes includes two putative desaturases, kuste3336 and kuste3607. Both genes are annotated as encoding phytoene desaturases and display 31 and 33% amino acid sequence identity to the lycopene-forming phytoene desaturase (CrtI) from Methyloglobulus morosus KoM1, respectively. To test whether these gene products catalyze phytoene desaturase activity, the crtI gene in the pLyc vector was separately replaced with kuste3336 and kuste3607 (pPJ177 and pPJ178, respectively, in Table 1). pLyc encodes all the necessary genes to convert farnesyl pyrophosphate to lycopene (including the phytoene desaturase step) [21]. Separate E. coli MG1655 strains expressing pPJ177 and pPJ178 (Lyc36 and Lyc07, respectively; Table 1) were grown for lycopene production analyses in comparison to a pLyc (no change to crtI) strain and a control strain with the crtI gene removed (Lyc-no-CrtI; Table 1). HPLC analyses of extracts from each strain indicated that the Lyc36 and Lyc07 strains did not produce lycopene (Fig 5). These results suggest that, despite moderate sequence similarity, the kuste3336 and kuste3607 gene products do not display CrtI-like activity, and may instead display novel functions or substrate specificities, possibly playing a role as fatty acid desaturases in the hypothesized ladderane biosynthetic pathway [11].
Fig 5. In vivo tests of function in putative phytoene desaturases from K. stuttgartiensis (kuste3336 and kuste3607).
(Left) Phytoene desaturation to lycopene catalyzed by CrtI and schematic of the pLyc vector. (Right) Lycopene production in E. coli MG1655 strains (from left to right): Lyc (positive control), Lyc36 (crtI in pLyc replaced with kuste3336), Lyc07 (crtI in pLyc replaced with kuste3607), and Lyc-no-CrtI (negative control with crtI gene removed) (see Table 1 for details on strains).
Optimization of purification of putative ladderane-related desaturases
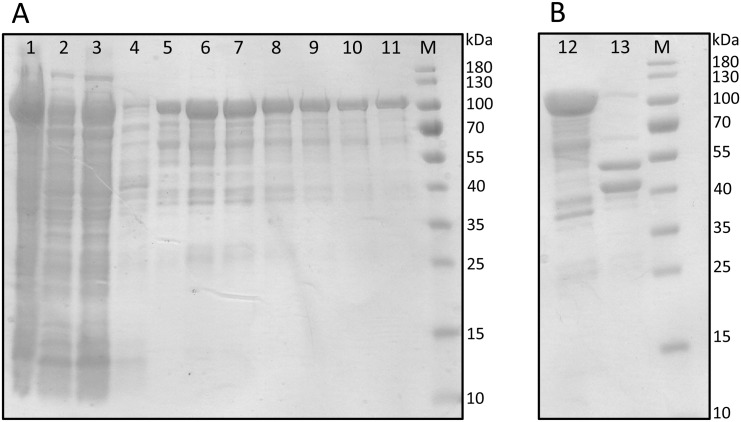
In addition to in vivo analyses of lycopene production, soluble expression of the putative desaturases was desired for in-depth in vitro assays. Based on the previously described structural work on the CrtI phytoene desaturase from Pantoea ananatis [42], plasmids encoding the kuste3336 and kuste3607 genes with C-terminal 6xHis tags were assembled. The kuste3336 protein displayed poor expression and, while kuste3607 expressed, the majority of the expressed protein was insoluble. Optimization of growth, induction, and lysis conditions did not appreciably improve the solubility of the C-6xHis-tagged protein. Plasmids encoding kuste3607 with different tags were then assembled, including N-6xHis-, N-StrepII-, C-StrepII-, and MBP-tagged variants (Table 1). The most soluble version of kuste3607 was obtained as an N-8xHis-StrepII-MBP-tagged protein, which could be purified through StrepTactin-based affinity chromatography and separated from the tags by cleavage with recombinant TEV protease (Fig 6). Since kuste3607 appears not to be a phytoene desaturase based on in vivo studies, but may serve some as-yet unidentified role in ladderane biosynthesis, the ability to express it in soluble form may facilitate future experiments of its actual function.
Fig 6. SDS-PAGE analyses of 8xHis-StrepII-MBP-3607 purification (A) and tobacco etch virus (TEV) protease cleavage (B).
Lanes: M, molecular mass markers; Lane 1, pellet material after lysate clarification; Lane 2, soluble lysate; Lane 3, StrepTrap column flowthrough; Lane 4, protein washed from column; Lanes 5–11, protein elution fractions; Lane 12, pooled elution fractions; Lane 13; protein after digestion with TEV protease. The expected molecular masses of 8xHis-StrepII-MBP-3607, tag-free 3607 protein, and MBP are 100, 56, and 44 kDa, respectively.
Less extensive expression studies were performed with the SAM radical proteins. Some soluble expression was observed for the C-terminally His-tagged versions of kuste2803 and kuste3608 (strains JPUB_006827 and JPUB_006833, respectively; S6 Table).
Conclusions
This study describes an efficient means of assembly of 34 synthesized, codon-optimized candidate ladderane biosynthesis genes in synthetic operons that allows for changes to regulatory element sequences, as well as modular assembly of multiple operons for simultaneous heterologous expression in E. coli (or potentially other microbial hosts). Initial analyses of gene expression indicate that protein insolubility may represent a challenge for fatty acid profile studies, but this can potentially be addressed through further optimization of promoter and RBS sequences or through changes in culture conditions. The lycopene production assay results suggest that the putative desaturases encoded by kuste3336 and kuste3607 do not possess CrtI-like activity, despite sequence similarity among the enzymes and their annotated function. The desaturases may instead display novel functions that are crucial for ladderane synthesis, namely desaturation of fatty acids to form intermediates that are cyclized through downstream radical reactions. The purification scheme for the kuste3607 desaturase described above can be used to prepare soluble protein for in-depth assays aimed at determining the function and substrate specificity of the desaturase. This study is a first step toward elucidating the unique biosynthetic pathway for production of ladderane fatty acids. We invite the scientific community to take advantage of the synthetic biology resources and experimental results developed in this study to elucidate the biosynthetic pathway that produces unique and intriguing ladderane lipids.
Supporting Information
Fold-increase is given as relative quantification (RQ) value, obtained by the ΔΔCT method. Bars represent the range between the minimum and maximum RQ values.
(TIF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We are grateful to Barth F. Smets of the Technical University of Denmark for providing anammox reactor biomass samples. We thank Edward Baidoo for assistance with HPLC operation, and Arthur Fong and Thu Tran for performing PCR and DNA assembly procedures. This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. Work conducted by the Joint Genome Institute was supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. J.D.K. has a financial interest in Amyris and Lygos.
Data Availability
All relevant data are within the paper, its Supporting Information files, and the JBEI public registry (accession numbers in Table 1).
Funding Statement
This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U. S. Department of Energy (http://www.energy.gov/), Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. Work conducted by the Joint Genome Institute was supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jetten MS, Niftrik L, Strous M, Kartal B, Keltjens JT, Op den Camp HJ. Biochemistry and molecular biology of anammox bacteria. Crit Rev Biochem Mol Biol. 2009;44(2–3):65–84. 10.1080/10409230902722783 . [DOI] [PubMed] [Google Scholar]
- 2.Kartal B, van Niftrik L, Keltjens JT, Op den Camp HJ, Jetten MS. Anammox—growth physiology, cell biology, and metabolism. Adv Microb Physiol. 2012;60:211–62. 10.1016/B978-0-12-398264-3.00003-6 . [DOI] [PubMed] [Google Scholar]
- 3.van Niftrik L, Jetten MS. Anaerobic ammonium-oxidizing bacteria: unique microorganisms with exceptional properties. Microbiol Mol Biol Rev. 2012;76(3):585–96. 10.1128/MMBR.05025-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novak I. Ring strain in [n]ladderanes. Journal of Physical Chemistry A. 2008;112(40):10059–63. 10.1021/Jp805377c WOS:000259760300053. [DOI] [PubMed] [Google Scholar]
- 5.Nouri DH, Tantillo DJ. They came from the deep: Syntheses, applications, and biology of ladderanes. Current Organic Chemistry. 2006;10(16):2055–74. 10.2174/138527206778742678 WOS:000241345600003. [DOI] [Google Scholar]
- 6.Beller HR, Lee TS, Katz L. Natural products as biofuels and bio-based chemicals: fatty acids and isoprenoids. Nat Prod Rep. 2015;32(10):1508–26. 10.1039/c5np00068h . [DOI] [PubMed] [Google Scholar]
- 7.Lennen RM, Pfleger BF. Microbial production of fatty acid-derived fuels and chemicals. Curr Opin Biotechnol. 2013;24(6):1044–53. 10.1016/j.copbio.2013.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463(7280):559–62. 10.1038/nature08721 . [DOI] [PubMed] [Google Scholar]
- 9.Goh EB, Baidoo EE, Keasling JD, Beller HR. Engineering of bacterial methyl ketone synthesis for biofuels. Appl Environ Microbiol. 2012;78(1):70–80. 10.1128/AEM.06785-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goh EB, Baidoo EE, Burd H, Lee TS, Keasling JD, Beller HR. Substantial improvements in methyl ketone production in E. coli and insights on the pathway from in vitro studies. Metab Eng. 2014;26:67–76. 10.1016/j.ymben.2014.09.003 . [DOI] [PubMed] [Google Scholar]
- 11.Rattray JE, Strous M, Op den Camp HJ, Schouten S, Jetten MS, Damste JS. A comparative genomics study of genetic products potentially encoding ladderane lipid biosynthesis. Biol Direct. 2009;4:8 10.1186/1745-6150-4-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mascitti V, Corey EJ. Enantioselective synthesis of pentacycloanammoxic acid. J Am Chem Soc. 2006;128(10):3118–9. 10.1021/ja058370g . [DOI] [PubMed] [Google Scholar]
- 13.Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, et al. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature. 2006;440(7085):790–4. 10.1038/nature04647 . [DOI] [PubMed] [Google Scholar]
- 14.Nouri DH, Tantillo DJ. Attack of radicals and protons on ladderane lipids: quantum chemical calculations and biological implications. Org Biomol Chem. 2012;10(29):5514–7. 10.1039/c2ob25717c . [DOI] [PubMed] [Google Scholar]
- 15.Kartal B, de Almeida NM, Maalcke WJ, Op den Camp HJ, Jetten MS, Keltjens JT. How to make a living from anaerobic ammonium oxidation. FEMS Microbiol Rev. 2013;37(3):428–61. 10.1111/1574-6976.12014 . [DOI] [PubMed] [Google Scholar]
- 16.Ham TS, Dmytriv Z, Plahar H, Chen J, Hillson NJ, Keasling JD. Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. Nucleic Acids Res. 2012;40(18):e141 10.1093/nar/gks531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189(1):113–30. . [DOI] [PubMed] [Google Scholar]
- 18.Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138(2):179–207. . [DOI] [PubMed] [Google Scholar]
- 19.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25(6):1203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guyer MS, Reed RR, Steitz JA, Low KB. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45 Pt 1:135–40. . [DOI] [PubMed] [Google Scholar]
- 21.Chou HH, Keasling JD. Programming adaptive control to evolve increased metabolite production. Nat Commun. 2013;4:2595 10.1038/ncomms3595 . [DOI] [PubMed] [Google Scholar]
- 22.Mutalik VK, Guimaraes JC, Cambray G, Lam C, Christoffersen MJ, Mai QA, et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat Methods. 2013;10(4):354–60. 10.1038/nmeth.2404 . [DOI] [PubMed] [Google Scholar]
- 23.Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, et al. BglBrick vectors and datasheets: A synthetic biology platform for gene expression. J Biol Eng. 2011;5:12 10.1186/1754-1611-5-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quan J, Saaem I, Tang N, Ma S, Negre N, Gong H, et al. Parallel on-chip gene synthesis and application to optimization of protein expression. Nat Biotechnol. 2011;29(5):449–52. 10.1038/nbt.1847 . [DOI] [PubMed] [Google Scholar]
- 25.Richardson SM, Nunley PW, Yarrington RM, Boeke JD, Bader JS. GeneDesign 3.0 is an updated synthetic biology toolkit. Nucleic Acids Res. 2010;38(8):2603–6. 10.1093/nar/gkq143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heins RA, Cheng X, Nath S, Deng K, Bowen BP, Chivian DC, et al. Phylogenomically guided identification of industrially relevant GH1 beta-glucosidases through DNA synthesis and nanostructure-initiator mass spectrometry. ACS Chem Biol. 2014;9(9):2082–91. 10.1021/cb500244v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cambray G, Guimaraes JC, Mutalik VK, Lam C, Mai QA, Thimmaiah T, et al. Measurement and modeling of intrinsic transcription terminators. Nucleic Acids Res. 2013;41(9):5139–48. 10.1093/nar/gkt163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hillson NJ, Rosengarten RD, Keasling JD. j5 DNA assembly design automation software. ACS Synth Biol. 2012;1(1):14–21. 10.1021/sb2000116 . [DOI] [PubMed] [Google Scholar]
- 29.Chen J, Densmore D, Ham TS, Keasling JD, Hillson NJ. DeviceEditor visual biological CAD canvas. J Biol Eng. 2012;6(1):1 10.1186/1754-1611-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4(5):e5553 10.1371/journal.pone.0005553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–5. 10.1038/nmeth.1318 . [DOI] [PubMed] [Google Scholar]
- 32.Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009;4(7):e6441 10.1371/journal.pone.0006441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinninghe Damste JS, Rijpstra WI, Geenevasen JA, Strous M, Jetten MS. Structural identification of ladderane and other membrane lipids of planctomycetes capable of anaerobic ammonium oxidation (anammox). FEBS J. 2005;272(16):4270–83. 10.1111/j.1742-4658.2005.04842.x . [DOI] [PubMed] [Google Scholar]
- 34.Kumar S, Hahn FM, Baidoo E, Kahlon TS, Wood DF, McMahan CM, et al. Remodeling the isoprenoid pathway in tobacco by expressing the cytoplasmic mevalonate pathway in chloroplasts. Metab Eng. 2012;14(1):19–28. 10.1016/j.ymben.2011.11.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou K, Zhou L, Lim Q, Zou R, Stephanopoulos G, Too HP. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol Biol. 2011;12:18 10.1186/1471-2199-12-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Batth TS, Keasling JD, Petzold CJ. Targeted proteomics for metabolic pathway optimization. Methods Mol Biol. 2012;944:237–49. 10.1007/978-1-62703-122-6_17 . [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez Fernandez-Nino SM, Smith-Moritz AM, Chan LJ, Adams PD, Heazlewood JL, Petzold CJ. Standard flow liquid chromatography for shotgun proteomics in bioenergy research. Front Bioeng Biotechnol. 2015;3:44 10.3389/fbioe.2015.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kartal B, Maalcke WJ, de Almeida NM, Cirpus I, Gloerich J, Geerts W, et al. Molecular mechanism of anaerobic ammonium oxidation. Nature. 2011;479(7371):127–30. 10.1038/nature10453 . [DOI] [PubMed] [Google Scholar]
- 39.Frazzon J, Fick JR, Dean DR. Biosynthesis of iron-sulphur clusters is a complex and highly conserved process. Biochem Soc Trans. 2002;30(4):680–5. . [DOI] [PubMed] [Google Scholar]
- 40.Grawert T, Kaiser J, Zepeck F, Laupitz R, Hecht S, Amslinger S, et al. IspH protein of Escherichia coli: studies on iron-sulfur cluster implementation and catalysis. J Am Chem Soc. 2004;126(40):12847–55. 10.1021/ja0471727 . [DOI] [PubMed] [Google Scholar]
- 41.Roche B, Aussel L, Ezraty B, Mandin P, Py B, Barras F. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim Biophys Acta. 2013;1827(3):455–69. 10.1016/j.bbabio.2012.12.010 . [DOI] [PubMed] [Google Scholar]
- 42.Schaub P, Yu Q, Gemmecker S, Poussin-Courmontagne P, Mailliot J, McEwen AG, et al. On the structure and function of the phytoene desaturase CRTI from Pantoea ananatis, a membrane-peripheral and FAD-dependent oxidase/isomerase. PLoS One. 2012;7(6):e39550 10.1371/journal.pone.0039550 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fold-increase is given as relative quantification (RQ) value, obtained by the ΔΔCT method. Bars represent the range between the minimum and maximum RQ values.
(TIF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper, its Supporting Information files, and the JBEI public registry (accession numbers in Table 1).