A collection of green algal mutants with known insertion sites is presented and used to identify a Chlamydomonas gene important in green algal triacylglycerol biosynthesis.
Abstract
The green alga Chlamydomonas reinhardtii is a leading unicellular model for dissecting biological processes in photosynthetic eukaryotes. However, its usefulness has been limited by difficulties in obtaining mutants in specific genes of interest. To allow generation of large numbers of mapped mutants, we developed high-throughput methods that (1) enable easy maintenance of tens of thousands of Chlamydomonas strains by propagation on agar media and by cryogenic storage, (2) identify mutagenic insertion sites and physical coordinates in these collections, and (3) validate the insertion sites in pools of mutants by obtaining >500 bp of flanking genomic sequences. We used these approaches to construct a stably maintained library of 1935 mapped mutants, representing disruptions in 1562 genes. We further characterized randomly selected mutants and found that 33 out of 44 insertion sites (75%) could be confirmed by PCR, and 17 out of 23 mutants (74%) contained a single insertion. To demonstrate the power of this library for elucidating biological processes, we analyzed the lipid content of mutants disrupted in genes encoding proteins of the algal lipid droplet proteome. This study revealed a central role of the long-chain acyl-CoA synthetase LCS2 in the production of triacylglycerol from de novo-synthesized fatty acids.
INTRODUCTION
Plants are the origin of most of our food and many of our basic materials and have enormous potential as a source of renewable energy and industrial chemicals. Advances in our understanding of plant biology will enable future increases in crop yields and the development of next-generation biofuels and will allow for more informed policy decisions as human activities increasingly impact our environment.
A major obstacle in effectively advancing the use of plants to support human activities is that the functions of thousands of plant genes remain unknown. Arabidopsis thaliana is the best-characterized organism in plant biology research, yet over one-third of its 27,000 genes have not been assigned a molecular function (Kourmpetis et al., 2011) (The Arabidopsis Information Resource website, genome snapshot, retrieved 11/20/15 from http://www.arabidopsis.org/portals/genAnnotation/genome_snapshot.jsp). A much greater fraction of gene products eludes mechanistic and structural understanding. Therefore, new tools are needed to accelerate our understanding of plant gene function. One significant opportunity lies in developing advanced resources for analyzing unicellular, photosynthetic model systems.
The unicellular green alga Chlamydomonas reinhardtii has been an invaluable model organism for elucidating gene functions in photosynthetic organisms (Harris, 2001; Gutman and Niyogi, 2004; McDonald, 2009). Its success is in large part due to its powerful genetic and biochemical properties. Chlamydomonas is haploid during vegetative growth, making mutant phenotypes immediately apparent. All three of its genomes (nuclear, chloroplast, and mitochondrial) have been sequenced and can be transformed (Boynton et al., 1988; Kindle et al., 1989; Randolph-Anderson et al., 1993; Maul et al., 2002; Grossman et al., 2003; Merchant et al., 2007). This alga is particularly suitable for studies of photosynthesis because it can grow heterotrophically with acetate as a carbon and energy source, allowing the isolation of mutants deficient in photosynthesis (Sager and Zalokar, 1958; Levine, 1960; Kates and Jones, 1964; Grossman et al., 2010; Karpowicz et al., 2011; Calderon et al., 2013; Heinnickel et al., 2013; Goodenough, 2015).
In recent years, Chlamydomonas has been increasingly used to study additional biological processes, including lipid biosynthesis (Hu et al., 2008; Wang et al., 2009; Moellering and Benning, 2010; Merchant et al., 2012; Liu and Benning, 2013), pigment biosynthesis and regulation (Lohr et al., 2005; Beale, 2009; Lohr, 2009; Voss et al., 2011), carbon-concentrating mechanisms (Badger et al., 1980; Wang et al., 2011; Brueggeman et al., 2012; Fang et al., 2012), growth during nutrient deprivation (González-Ballester et al., 2010; Miller et al., 2010; Castruita et al., 2011; Boyle et al., 2012; Urzica et al., 2012, 2013; Blaby et al., 2013; Hemschemeier et al., 2013; Toepel et al., 2013; Aksoy et al., 2014; Schmollinger et al., 2014), responses to heat stress (Hemme et al., 2014), photoreception (Beel et al., 2012), fermentation biology and hydrogen gas production (Ghirardi et al., 2007; Mus et al., 2007; Hemschemeier et al., 2008; Dubini et al., 2009; Grossman et al., 2011; Catalanotti et al., 2012, 2013; Magneschi et al., 2012; Murthy et al., 2012; Yang et al., 2014), mating (Umen, 2011; Geng et al., 2014; Liu et al., 2015), the cell cycle (Tulin and Cross, 2014; Cross and Umen, 2015), and cellular quiescence (Tsai et al., 2014).
In addition to its usefulness in studying plant biology, the biochemical similarity of Chlamydomonas flagella to mammalian cilia has made Chlamydomonas a leading model for elucidating cilia structure and function (Jarvik and Rosenbaum, 1980; Tam and Lefebvre, 1993; Li et al., 2004; Mitchell, 2004; Yang et al., 2006; Wirschell et al., 2008), with discoveries including the characterization of intraflagellar transport (Kozminski et al., 1993; Baldari and Rosenbaum, 2010). In addition, Chlamydomonas motility mutants have helped reveal the molecular basis for many human diseases that result from cilia dysfunction, such as polycystic kidney disease (Lewin, 1953; Mintz and Lewin, 1954; Pazour et al., 2000; Qin et al., 2001; Fliegauf et al., 2007). Despite its powerful attributes, the utility of Chlamydomonas as a model organism has been greatly limited by the difficulty of obtaining mutants for specific genes. Gene disruption by homologous recombination is not a viable tool in Chlamydomonas as a consequence of the low frequency of homologous recombination relative to random insertion of exogenous DNA (Slaninová et al., 2008). Furthermore, specific gene modification through zinc-finger nucleases has not proven practical, as it requires time-consuming screens for zinc finger arrays with high specificity for sequences of interest (Sizova et al., 2013). Cas9 coupled with specific guide sequences has been used in multiple systems for high-efficiency genome editing (Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013), but to date has not been successfully used to target gene disruptions in Chlamydomonas (Jiang et al., 2014). While RNA silencing through the use of artificial microRNAs has been developed for Chlamydomonas, suppression of transcript levels is usually incomplete (Molnar et al., 2009; Zhao et al., 2009). Insertional mutagenesis is a well-established procedure for generating mutants of Chlamydomonas but has been mostly used for forward genetic screens (Galván et al., 2007). Methods for isolating mutants with insertions in specific target genes have been developed, but these methods have limited throughput and require a large number of PCRs to identify lesions in genes of interest (Pootakham et al., 2010; Gonzalez-Ballester et al., 2011).
Large-scale mapped mutant libraries have transformed research in bacteria (Baba et al., 2006; Goodman et al., 2009), yeast (Winzeler et al., 1999; Giaever et al., 2002), animals (Venken et al., 2011; Varshney et al., 2013), and land plants (Alonso et al., 2003; May et al., 2003; McCarty et al., 2005; Zhang et al., 2006; Hsing et al., 2007; Bragg et al., 2012; Belcher et al., 2015). An analogous stably maintained library of Chlamydomonas insertional mutants with known insertion sites would have a similarly transformative impact on the ease of elucidating gene functions in this organism.
Multiple major challenges are associated with generating an indexed mutant library for Chlamydomonas. This alga is traditionally propagated by manual transfer, limiting the number of mutants that can be maintained. Cryopreservation has been demonstrated (Hwang and Hudock, 1971; Piasecki et al., 2009), but only for individual strains, making cryopreservation of large collections prohibitively laborious. Until recently, methods available for obtaining sequences flanking the insertion cassette (flanking sequence tags) in Chlamydomonas had insufficient throughput to map the thousands of insertion sites needed for establishing a mapped, indexed library. Conventional methods, including plasmid rescue (Tam and Lefebvre, 1993), thermal asymmetric interlaced PCR (Dent et al., 2005; González-Ballester et al., 2005b), restriction enzyme site-directed amplification PCR (González-Ballester et al., 2005a), inverse PCR and GenomeWalker PCR (Philipps et al., 2011), 3′ rapid amplification of cDNA ends (Meslet-Cladière and Vallon, 2012), and SiteFinding PCR (Li et al., 2012b; Dent et al., 2015), can identify insertion sites for ∼10 mutants in a week. Recently, conventional methods were used to map 459 insertion sites in a collection of photosynthesis-deficient mutants (Dent et al., 2015). Despite this success, the throughput of conventional methods remains low in comparison to the throughput of methods developed for mapping genome-wide libraries in land plants (Alonso et al., 2003; Strizhov et al., 2003; Williams-Carrier et al., 2010; Urbański et al., 2012; McCarty et al., 2013; Hunter et al., 2014), making conventional methods impractical for mapping large-scale Chlamydomonas mutant collections.
We recently significantly increased the throughput of mutant mapping by developing Chlamydomonas MmeI-based insertion site sequencing (ChlaMmeSeq), a protocol that can reveal insertion sites in a pool of tens of thousands of mutants by massively parallel sequencing of 20- to 21-bp sequences immediately flanking the insertion cassettes (Zhang et al., 2014). However, the accuracy of insertion site mapping is challenged by the insertion of random fragments of genomic DNA from distant loci between the cassette and the “true” genomic flanking sequence (Zhang et al., 2014). In those mutants, the flanking sequence originates from the inserted fragment and often maps to the genome, but identifies an incorrect insertion site. At present, there is no high-throughput means for identifying such incorrectly mapped mutants.
Strategies for determining the physical coordinates of individual insertions within a large collection of mutants using combinatorial pooling approaches coupled with next-generation sequencing have been recently developed (Urbański et al., 2012) (TDNA-Seq; https://www.araport.org). The largest reported library mapped using combinatorial pooling approaches contained ∼7000 mutants (Goodman et al., 2009). However, a saturating random insertion library of Chlamydomonas mutants will require mapping the insertion sites for >100,000 mutants, and this would demand an impractical number of pipetting operations using established combinatorial pooling methods.
In this study, we describe the development of broadly applicable technologies for insertion site mapping, combinatorial pooling, and deconvolution of insertion positions within an arrayed library of mutants. Moreover, we present protocols for efficient propagation of tens of thousands of Chlamydomonas mutants on agar and their cryopreservation in microtiter plates. Using these technologies, we generated an indexed library of Chlamydomonas insertional mutants for which the sites of cassette insertion have been determined. We demonstrated the utility of the library by phenotyping candidate mutants for triacylglycerol (TAG) accumulation under nitrogen deprivation conditions. This phenotyping allowed us to identify the long-chain acyl-CoA synthetase LCS2 as a key component of the poorly characterized pathway that incorporates newly synthesized fatty acids into TAG. This library is available to the public through the Chlamydomonas Resource Center; the library can be searched for specific strains and the strains directly ordered from the website https://www.chlamylibrary.org/.
RESULTS
Generation, Maintenance, and Cryopreservation of a Chlamydomonas Mutant Library
To generate a library of Chlamydomonas mutants, we employed random insertional mutagenesis with an insertion cassette conferring paromomycin resistance (Sizova et al., 2001; Zhang et al., 2014). We chose CC-4533 (mt−; http://chlamycollection.org/strain/cc-4533-cw15-mt-jonikas-cmj030/) as the background strain because of its multiple desirable properties, such as normal swimming and the ability to grow heterotrophically (Zhang et al., 2014). Because CC-4533 is derived from but not identical to commonly used lab strains, we compared its genome sequence to those of the commonly used strains (Gallaher et al., 2015). Whole-genome sequencing of CC-4533 revealed only three single nucleotide polymorphisms that may disrupt gene function and are not found in other common laboratory strains, as well as 466 additional single nucleotide polymorphisms and 26 structural variants that are not expected to alter gene function significantly (Supplemental Tables 1 and 2 and Supplemental Data Sets 1 and 2). To facilitate further genetic studies using mutants from this library, we crossed CC-4533 (mt−) with CC-125 (mt+) and then backcrossed mt+ progeny to CC-4533 five times. This procedure yielded a mt+ strain designated CC-5155, which should be nearly isogenic to CC-4533.
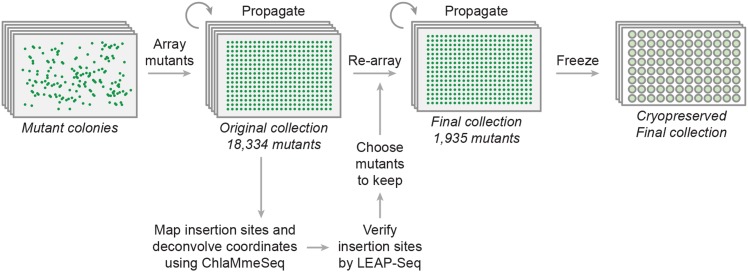
We developed high-throughput robotic procedures for colony picking, mutant propagation, cryopreservation, and pooling mutants to map insertion sites (Figure 1). A robot picked mutant colonies and arrayed them in a 384-colony format on agar-solidified medium. The original collection contained a total of 18,430 colonies on 53 plates (Supplemental Figure 1A). To propagate and maintain the collection, we developed a robust protocol in which the colonies were robotically replicated onto fresh solid medium every four weeks.
Figure 1.
High-Throughput Technologies to Generate, Propagate, and Map Insertions in the Indexed Mutant Library.
Individual transformants were robotically picked into a collection of arrays of 384 colonies on agar and were propagated while insertion sites were being mapped and confirmed. Once the coordinates of high-confidence mutants were identified, these mutants were then arrayed into a final collection that was cryopreserved and is available for distribution.
To evaluate the efficiency of propagation, we quantified frequencies of individual colony gain and loss. Over a period of 18 months, 4% of the colonies were lost due to whole-plate contamination or inefficient transfer (Supplemental Figure 1B). Furthermore, the library initially had 1922 blank spots, and of these, 67 (3.5%) acquired a colony after 18 months, possibly caused by splashing of cells from neighboring colonies during replication. This finding underscores the need to streak mutants from the collection to obtain single colonies for making pure cultures and to confirm the site of insertion prior to physiological mutant characterization. Overall, these data indicate that the mutant library can be robustly propagated on agar for over a year, but also demonstrates a need for a more stable form of storage over longer periods. Therefore, we developed a method for high-throughput cryogenic freezing and recovery of strains in a 96-well plate format. To test the protocol, we thawed eight 96-well plates of library strains and recovered 98 to 100% of the strains in each plate (Supplemental Table 3). Three copies of the library have been cryogenically stored using this method.
We Developed an Advanced Scheme for Mapping Insertion Sites
To map the insertion sites present in pools of mutants, we used ChlaMmeSeq, a recently developed protocol (Zhang et al., 2014). Illumina sequencing of ChlaMmeSeq products yields a 20- to 21-bp flanking sequence tag indicating the genomic sequence flanking each 5′ or 3′ cassette end in a pool of mutants.
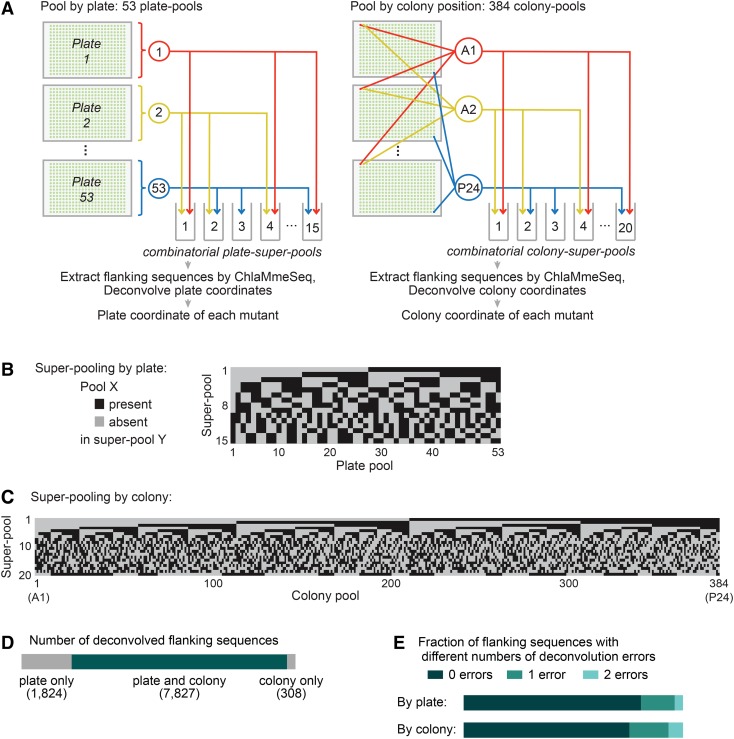
To determine the plate and colony (row and column) coordinates of each mapped insertion, we designed a combinatorial pooling scheme based on one that was previously developed for a bacterial mutant library (Goodman et al., 2009). The latter scheme mapped ∼7000 mutants with ∼80,000 pipetting operations but cannot be practically applied to libraries of tens or hundreds of thousands of mutants because the number of pipetting operations grows more rapidly than the number of mutants to be mapped. To accommodate the larger number of mutants in our library, we modified the scheme to determine the plate coordinate of each insertion separately from the colony coordinate (Figure 2A). This method of dual combinatorial mapping markedly reduces the number of liquid handling operations during the combinatorial pooling stage, enabling mapping of 200,000 mutants with only ∼10,000 pipetting operations.
Figure 2.
Plate and Colony Coordinates of Each Flanking Sequence Were Separately Determined.
(A) To determine plate coordinates, the mutants were pooled by plate into 53 plate-pools. We combinatorially distributed the plate-pools into 15 plate-super-pools and obtained the flanking sequences in each plate-super-pool by ChlaMmeSeq. We then determined the flanking sequences in each plate by deconvolving the plate-super-pool data. We employed a similar process to identify the flanking sequences present at each colony position. This process revealed the plate and colony coordinates for each flanking sequence.
(B) and (C) The patterns of presence and absence of pools in super-pools are shown for super-pooling by plate (B) and by colony (C).
(D) Most flanking sequences were mapped to both a plate and a colony position.
(E) Most flanking sequence coordinates were identified with no errors. An error indicates that a flanking sequence is present in an unexpected super-pool, or absent from an expected super-pool, but the flanking sequence can still be uniquely mapped to a pool due to the error tolerance of our method.
To determine the plate coordinate of each mutant, we first combined colonies into 53 pools by plate (hereafter referred to as “plate-pool,” each plate-pool contained all strains from a given plate). We then dispensed each of the 53 plate-pools into a subset of the 15 plate-super-pools, according to a pattern of presence and absence that was unique to each plate-pool (Figure 2B; Supplemental Data Set 3). We identified flanking sequences present in each plate-super-pool by applying ChlaMmeSeq to each super-pool. The pattern of presence and absence of each flanking sequence across the 15 plate-super-pools allowed us to determine which of the 53 plates contains the mutant harboring the corresponding insertion.
Similarly, to determine the colony coordinates of each mutant, we first combined colonies into 384 pools by colony position on the plate (hereafter referred to as “colony-pool”). Each colony-pool contained all strains at the corresponding position on the 384-colony array (e.g., colony pool A1 contained cells from position A1 of all 53 plates). We then determined the flanking sequences present at each colony position using a super-pooling scheme (Figure 2C; Supplemental Data Set 4) similar to that described above for determining the plate position. Overall, >80% of flanking sequences that were mapped to a plate were also mapped to a colony, indicating that our approach is robust.
We Assigned Flanking Sequences to 6258 Colonies
We deconvolved plate and colony locations for each flanking sequence based on presence and absence of the sequence across the super-pools (Supplemental Figure 2 and Supplemental Data Set 5). In total, we successfully mapped 7827 nuclear genomic flanking sequences to 6258 colonies in the library (Figures 2D and 2E; Supplemental Figure 3), consistent with previously observed success rates in mapping Chlamydomonas insertion sites (Dent et al., 2005, 2015; Zhang et al., 2014; see Discussion). The accuracy of coordinate assignments was initially supported by the strong agreement between positions that lacked colonies and positions that were not assigned flanking sequences (Supplemental Figure 4). To assess the deconvolution accuracy in a quantitative manner, we performed ChlaMmeSeq on two sets of 96 randomly selected mutants and found that 95 and 87% of the flanking sequences assigned to these mutants could be recovered (Supplemental Data Set 6).
The Majority of Mutants Contain Complex Insertions
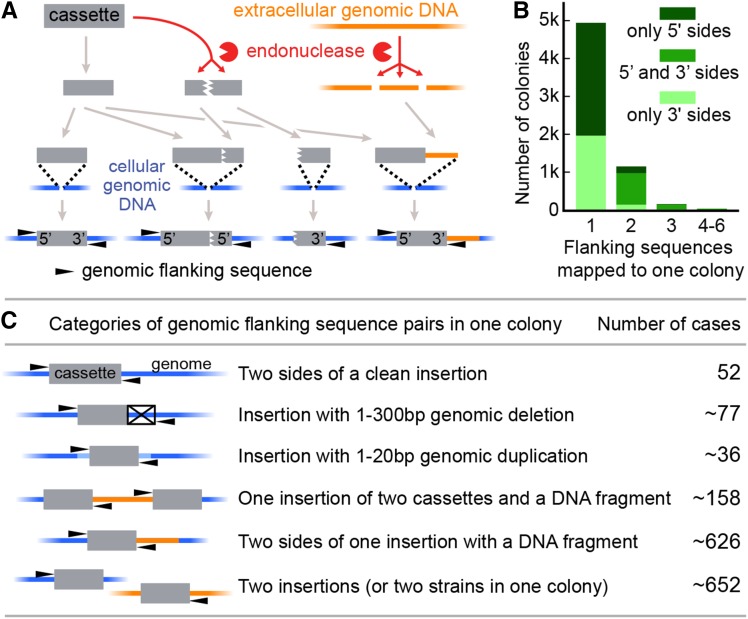
We sought to use the unprecedented number of mutants with multiple flanking sequences in our data set to gain insights into the characteristics of genomic insertions in Chlamydomonas. In previous studies it was observed that insertions can contain complex rearrangements (Figure 3A), including deletions of genomic DNA (Tam and Lefebvre, 1993; Matsuo et al., 2008), cassette truncation (Dent et al., 2005, 2015), and concatenation of cassettes with fragments of genomic DNA potentially generated as a consequence of cell lysis in the culture (Zhang et al., 2014). Consistent with frequent cassette truncation, the majority of mutants (4923/6258) yielded a single flanking sequence (Figure 3B).
Figure 3.
Most Insertions in the Collection Are Complex.
(A) Our previous work suggested a model where transforming DNA is fragmented, with multiple cassette fragments and/or exogenous genomic DNA sequences sometimes inserted into the same site in the Chlamydomonas genome (adapted from Zhang et al. [2014]).
(B) Only one flanking sequence was assigned to most colonies, and two or more flanking sequences were assigned to some colonies. The plot also shows how many of the colonies have only 5′-side flanking sequences, only 3′, or both 5′ and 3′. Two 5′ or two 3′ ends can be derived from a single insertion of two cassettes in opposite orientations, as well as from two independent insertions.
(C) Data from pairs of flanking sequences mapped to the same colony were used to estimate how often different insertion categories were detected in the collection (see Supplemental Figure 5 and Supplemental Methods).
We analyzed the relationship between pairs of flanking sequences assigned to the same colony to evaluate the occurrence of various classes of insertions in the collection (Figure 3C; Supplemental Figure 5). We observed 52 cases of clean insertions, where the flanking sequences from two sides of the insertion aligned to adjacent nucleotides in the genome. Of the 1605 flanking sequence pairs, we estimate (see Methods; Supplemental Methods) that ∼77 were derived from an insertion with a genomic deletion smaller than 300 bp, and ∼36 were derived from an insertion with a 1- to 20-bp genomic duplication. We estimate that ∼158 flanking sequence pairs flank a 36- to 983-bp genomic DNA fragment between two cassettes (Figure 3C; Supplemental Figure 6). In ∼80% of the 1605 sequence pairs, the two flanking sequences aligned to distant genomic regions. This could be explained by the presence of multiple insertions in a single mutant, multiple mutants growing within the same colony, or the introduction of DNA from a distant locus on one or both sides of the cassette. We conclude that clean insertions are rare, small genomic deletions and duplications are often observed, and the majority of mutants contain compound insertions including genomic DNA fragments from distant loci and/or multiple cassettes.
LEAP-Seq Improves Confidence in the Genomic Insertion Site
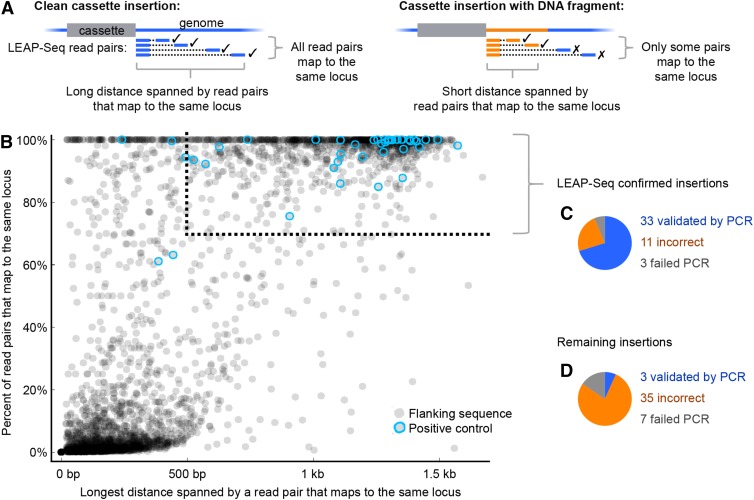
We sought to develop a method to increase our confidence in the assignment of insertion sites. As mentioned above, we frequently observed insertions of short genomic DNA fragments (typically <500 bp) between the cassette and the DNA of the true genomic location. In those cases, the 20 to 21 bp of flanking sequence identified by ChlaMmeSeq would not identify the disrupted locus because the sequence would originate from the DNA inserted between the cassette and the genome. We reasoned that we could identify such cases by evaluating whether the flanking DNA sequence ∼1 kb away from the end of the cassette still corresponds to the same locus as that identified by ChlaMmeSeq. To enable such evaluations, we developed a method that we call linear and exponential amplification of insertion site sequence coupled with paired-end sequencing (LEAP-Seq) (Supplemental Figure 7 and Supplemental Table 4). LEAP-Seq is based on a method developed for mapping insertion sites in mammalian cells (Carette et al., 2011). While the original method only reads 36 bp of genomic sequence immediately adjacent to the cassette, LEAP-Seq generates paired-end sequence reads for which the distal read can be up to 1.5 kb from the cassette, a distance sufficient to read beyond most DNA that is inserted between the cassette and genome.
We performed LEAP-Seq on a pool containing all mutants in the library, sequencing both 5′ and 3′ sides of the cassette. We assigned LEAP-Seq reads to each insertion by matching each LEAP-Seq proximal read to a ChlaMmeSeq read. To distinguish between correctly and incorrectly mapped insertions, we compared genomic mapping sites of LEAP-Seq proximal and distal read pairs (Figure 4A; see Methods; Supplemental Methods). In cases where there was no insertion of a DNA fragment, all distal reads should align to the same locus as the proximal read, and some read pairs would span a large distance (500 to 1500 bp of genomic DNA). If a short DNA fragment is present between the cassette and the true genomic position, a significant fraction of the distal reads will map to another locus because of LEAP-Seq products extending beyond the fragment. Additionally, the proximal and distal read pairs mapping to the same locus will span a much shorter distance, limited by the size of the inserted DNA fragment. To visualize this data, we plotted for each flanking sequence the percent of LEAP-Seq read pairs that mapped to the same locus versus the longest distance spanned by a read pair that mapped to that locus (Figure 4B; Supplemental Figure 8). Consistent with our expectations, we observed a bimodal distribution in both dimensions. Moreover, control mutants with high-confidence insertions showed a high value on both axes.
Figure 4.
LEAP-Seq Improves Confidence in Insertion Sites.
(A) LEAP-Seq reads are used to identify cases where there is a short insertion of genomic DNA from another locus (designated “junk” DNA) between the cassette and the flanking genomic DNA. LEAP-Seq reads are paired-end, with each pair consisting of a proximal and distal read (relative to the cassette). Short insertions of junk DNA will typically produce a low percentage of distal reads mapped to the same locus as the proximal read, and the read pairs will span a short distance at this locus.
(B) For each 5′ flanking sequence with at least 10 reads, the percentage of distal reads mapped to the same locus as the proximal read is plotted against the maximum distance spanned by read pairs at that locus. Positive control flanking sequences, indicated by blue circles, are 5′ flanking sequences that have perfectly matching 3′ flanking sequences mapped to the same colony. Some control flanking sequences showed a small fraction of distal reads mapping to distant loci as a result of apparent artifacts (see Supplemental Methods). The dotted line shows the cutoffs for insertions that we consider confirmed by LEAP-Seq.
(C) PCR validation of LEAP-Seq-confirmed insertions. Results show that 33/44 LEAP-Seq-confirmed insertions were correctly mapped; a further three confirmed insertions could not be checked due to PCR failure (Supplemental Figure 9).
(D) PCR validation for non-LEAP-Seq-confirmed insertions. Results show that 4/42 were correctly mapped; seven could not be checked due to PCR failure.
We classified insertions as confirmed if they met two criteria: (1) >70% of the mapped LEAP-Seq read pairs from that insertion were mapped to the same locus and (2) proximal and distal reads spanned at least 500 bp at this locus. Additionally, we considered insertions to be confirmed if two flanking sequences from the same mutant indicated a clean insertion or a short genomic deletion or duplication, even if the insertion did not satisfy the above criteria (see details in Supplemental Methods). Insertion site and LEAP-Seq data for each mutant are provided in Supplemental Data Sets 7 and 8. LEAP-Seq confirmation greatly enriched the collection for correctly mapped mutants, as shown below.
75% of the LEAP-Seq-Confirmed Insertion Junctions Are Correctly Mapped
To examine the overall quality of our mapping data, we performed PCR to characterize 47 LEAP-Seq-confirmed and 45 non-LEAP-Seq-confirmed insertions (Figures 4C and 4D; Supplemental Data Set 9). Insertions for this analysis were chosen at random from all insertions that were mapped to genes. For the LEAP-Seq-confirmed set, we found that our insertion site mapping was correct in 33 of the insertions, based on two lines of evidence. First, we were able to obtain a PCR product spanning the insertion site with the wild type but not mutant template genomic DNA (Supplemental Figure 9). Second, we could PCR amplify across the expected cassette-genome junctions from the mutant (Li et al., 2007). For 11 other insertions, we observed a wild-type product when using mutant DNA as template, indicating that there was no insertion of the cassette in the expected site. For the remaining three insertions, we were unable to obtain PCR products from either of the PCR assays. We conclude that ∼75% of LEAP-Seq-confirmed insertions are correctly mapped. In contrast, using the same criteria, we found that only 7.9% of the non-LEAP-Seq-confirmed insertions were correctly mapped, underlining the strong enrichment for correctly mapped insertions provided by LEAP-Seq confirmation (Supplemental Figures 8 and 10).
To determine the number of insertions in each mutant, we performed DNA gel blot hybridizations on 23 randomly selected mutants containing a LEAP-Seq-confirmed insertion (Supplemental Figure 11). For 17 of the mutants (74%), we observed a single insertion; the remaining mutants showed two insertions.
The LEAP-Seq-Confirmed Library Includes Insertions in 1562 Genes
We compared the set of LEAP-Seq-confirmed mutants to several gene sets of interest to the scientific community. Of 17,737 Chlamydomonas genes predicted in the v5.3 genome assembly, 1562 genes (∼9%) are represented by insertions in the LEAP-Seq-confirmed library. The library includes 836 Chlamydomonas genes with Arabidopsis homologs, 46 encoding GreenCut2 proteins (Karpowicz et al., 2011), 56 genes encoding proteins in the chloroplast proteome (Terashima et al., 2011), 65 genes encoding proteins in flagella (Pazour et al., 2005), and 430 genes with no functional annotation (Merchant et al., 2007) (Table 1).
Table 1. Many Genes of Interest Are Represented in the Collection of Confirmed Mutants.
| Data Set | No. of Genes with LEAP-Seq-Confirmed Mutants | Total Genes in Data Set |
|---|---|---|
| All Chlamydomonas genes | 1,562 | 17,737 |
| Chlamydomonas genes with Arabidopsis homologs | 836 | 8,309 |
| GreenCut2 | 46 | 537 |
| Chloroplast proteome | 56 | 704 |
| Flagellar proteome | 65 | 772 |
| CiliaCut | 24 | 173 |
| Lipid droplet proteome | 28 | 433 |
| No functional annotation | 430 | 6,767 |
The number of genes with confirmed mutants available is shown for several sets of genes of interest.
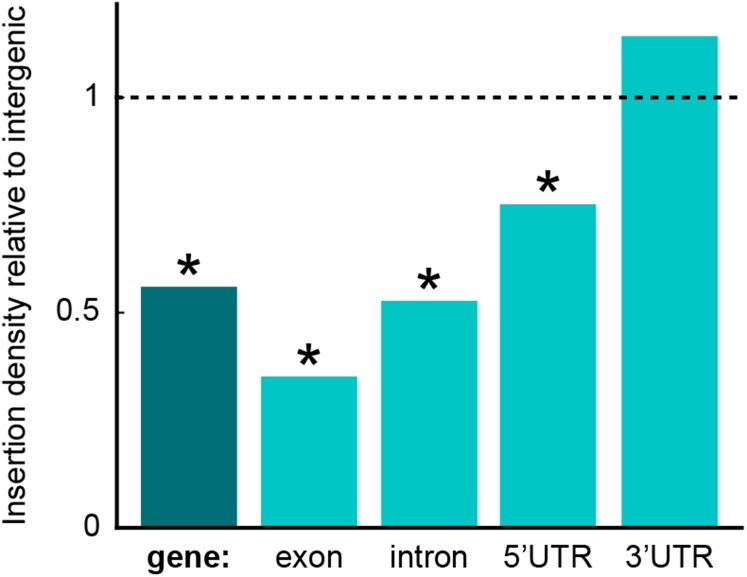
We examined the distribution of LEAP-Seq-confirmed insertions across the genome. Consistent with our previous work (Zhang et al., 2014), the genomic distribution of insertions is largely indistinguishable from random, with <8% of insertions contributing to hot spots and <1% of the genome in cold spots (Supplemental Figure 12 and Supplemental Data Set 10). At a finer scale, insertions in genes are less frequent than insertions into intergenic regions (Figure 5). Within genes, insertion frequency is highest for 3′ untranslated regions (UTRs) and lowest for exons. These observations could be explained by a reduced likelihood of viability of mutants with insertions in introns and exons or by these features having lower likelihood of cassette insertion or AphVIII expression.
Figure 5.
The Density of Confirmed Insertions Differs Between Gene Features and Intergenic Regions.
The data represent 265 intergenic insertions and 1855 insertions into genes. Categories with insertion densities statistically significantly different from the intergenic density are marked with asterisks (P values < 0.001). The densities are adjusted to compensate for differences in the density of uniquely mappable insertion positions between features (see Methods and Supplemental Methods).
LEAP-Seq-Confirmed Mutants Can Be Ordered through a Searchable Web Database
We consolidated the LEAP-Seq-confirmed mutants into seven new 384-colony arrays and cryopreserved the strains in 96-well microtiter plates. The rearrayed mutants are being propagated at the Chlamydomonas Resource Center (CRC) in Minnesota, where they are available for distribution to the public. Mutants can be searched and ordered through the searchable Chlamydomonas Library Project (CLiP) website (https://www.chlamylibrary.org/), which also provides information on the site of the gene disruption and a summary of LEAP-Seq results. Mutants will be sent on slants from the CRC for a distribution fee (initially $100/strain for academia and $1000/strain for industry), which will serve to make the propagation of the mutants at the Center perpetually self-sustaining.
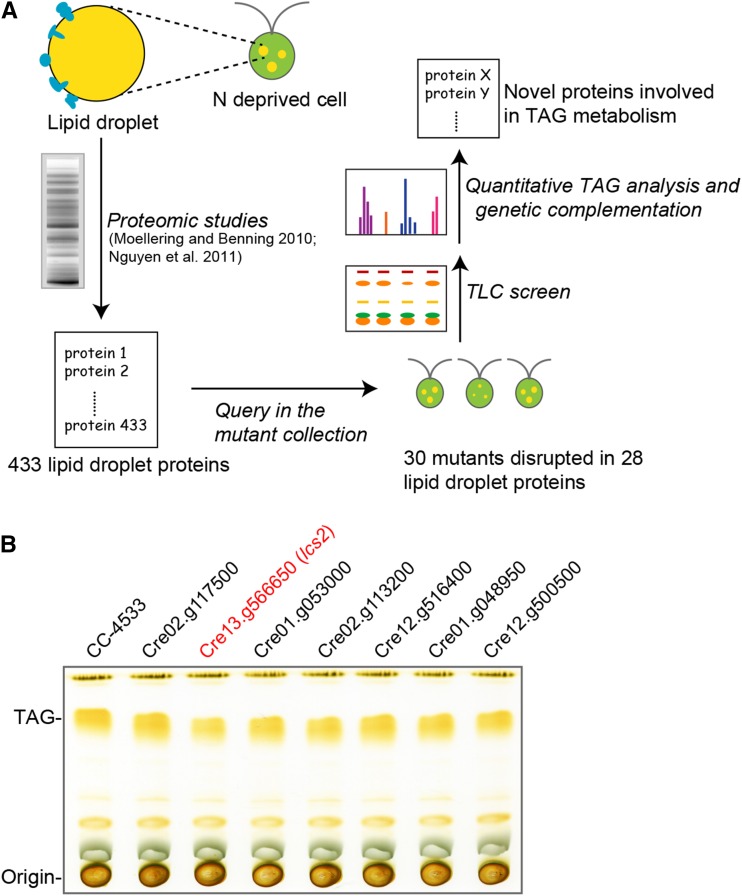
We Used the LEAP-Seq-Confirmed Library to Characterize Mutants Disrupted in Genes Encoding Lipid Droplet Proteins
To demonstrate the utility of the library, we used a candidate-based approach to identify genes involved in accumulation of the storage lipid TAG, which accumulates in organelles called lipid droplets (Goold et al., 2015). In both animals and plants, several proteins that impact TAG synthesis or degradation were found to localize to lipid droplets (Athenstaedt and Daum, 2003, 2005; Eastmond, 2006; Kurat et al., 2006; Farese and Walther, 2009; Wilfling et al., 2013). We therefore chose to screen mutants deficient in genes encoding proteins in the Chlamydomonas lipid droplet proteome (Moellering and Benning, 2010; Nguyen et al., 2011) for perturbed TAG content (Figure 6A). Our mutant library contains LEAP-Seq-confirmed insertions in 28 genes encoding proteins identified in these proteomes (Table 1). We isolated the corresponding mutants, verified their insertion sites by PCR (Supplemental Data Set 11), and analyzed TAG levels following nitrogen deprivation by thin-layer chromatography (TLC) (Figure 6B; Supplemental Figure 13). Of the 27 mutants tested, 23 (85%) were determined by PCR to be correctly mapped, consistent with the 75% confirmation rate observed above for randomly selected mutants. One of these mutants, LMJ.SG0182.015843, hereafter referred to as lcs2 as explained below, showed a defect in TAG accumulation following nitrogen deprivation. We confirmed this defect with biological replicates (Supplemental Figure 14) and chose lcs2 for in-depth characterization.
Figure 6.
Screening of Mutants in Genes Encoding the Lipid Droplet Proteome for Defects in TAG Accumulation.
(A) Lipid droplet proteins. In lipid droplets, which store the cell’s triacylglycerol, 433 proteins have been identified by proteomics. Thirty mutants disrupted in 28 of those proteins were found in the library.
(B) The TAG content in LEAP-Seq confirmed mutants disrupted in genes encoding lipid droplet proteins was analyzed. This revealed a defect in TAG accumulation in the lcs2 mutant (shown in red). One plate with seven mutants is presented here; other mutants are presented in Supplemental Figure 13. Lipids extracted from cells (normalized by the same amount of chlorophyll) were loaded into each lane. Each mutant is labeled with the v5.3 ID of the gene the insertion was mapped to. Detailed information is shown in Supplemental Data Set 11.
LCS2 Is a Novel Component of the TAG Biosynthesis Pathway
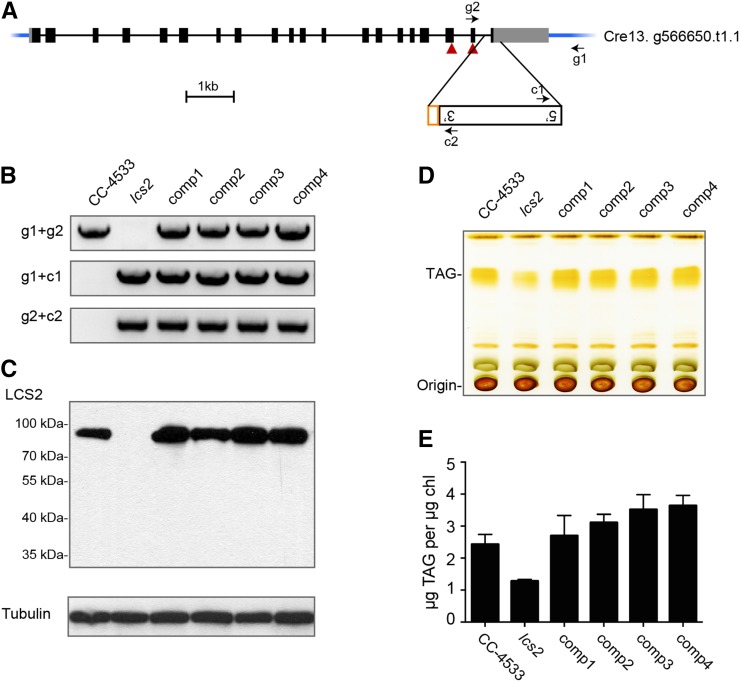
The lcs2 mutant has an insertion that disrupts the 3′ UTR of a gene (Cre13.g566650 in the Chlamydomonas v5.3 genome) encoding a putative long-chain acyl-CoA synthetase (LACS; Figure 7A). This gene was previously designated LCS2 in bioinformatic analyses of the Chlamydomonas genome (Riekhof et al., 2005; Li-Beisson et al., 2015). DNA gel blot analysis using the AphVIII gene as a probe (Supplemental Figure 11) identified a single hybridizing DNA fragment larger than 10 kb, a result consistent with an insertion in lcs2 (a 14.2-kb band is expected from the insertion site). Two splice forms were predicted for LCS2 in the Chlamydomonas v5.3 genome: Cre13.g566650.t1.1 has one fewer exon than Cre13.g566650.t2.1. RT-PCR suggests that Cre13.g566650.t1.1 is the major transcript under our experimental conditions (Supplemental Figure 15 and Supplemental Table 5).
Figure 7.
LCS2 Is Required for TAG Accumulation.
(A) The cassette insertion site in the lcs2 mutant is shown on the LCS2 gene model from the Phytozome v5.3 Chlamydomonas genome. Black solid boxes indicate exons, thin lines indicate introns, and UTRs are shown as gray boxes. A fragment of chloroplast DNA is indicated in orange. Primer locations for PCR are shown as arrows. The two red triangles indicate the location of the peptide against which the antibody was raised, which is translated from two neighboring exons.
(B) Examination of the genotypes of lcs2 and complemented lines (comp1-4) by PCR from genomic DNA. PCR primers are indicated to the left of the panel.
(C) Use of immunoblotting to measure LCS2 protein levels of lcs2 and complemented lines. α-Tubulin was used as a loading control.
(D) Use of TLC to measure TAG content in lcs2 and complemented lines.
(E) To confirm the results, LC-MS was used to quantify TAG content for CC-4533, lcs2, and complemented lines. Error bars indicate standard deviations from three biological replicates.
We confirmed the insertion site in lcs2 by PCR and DNA sequencing. PCR across the predicted insertion site with primer pair g1 + g2 (Supplemental Table 5) yielded a product if CC-4533 genomic DNA was used as a template but not when the template was genomic DNA from lcs2, consistent with a disruption in the mutant (Figure 7B). We confirmed the cassette-genome junction reported by ChlaMmeSeq and LEAP-Seq by PCR with primer pair g1 + c1. We then characterized the junction between the 3′ side of the cassette and the genome. Sequencing of the PCR product obtained from lcs2 with primer pair g2 + c2 indicated an intact cassette 3′ end and an insertion of a 204-bp fragment of chloroplast DNA between the cassette and the last LCS2 intron. Insertions of Chlamydomonas nuclear, chloroplast, and mitochondrial DNA between the cassette and genome sequence have been observed previously (Zhang et al., 2014). In this case, the insertion of chloroplast DNA explains why only the 5′ insertion junction was identified and used for deconvolution. The insertion also resulted in a 215-bp deletion in LCS2. The deleted sequence includes part of the last intron, the entire last exon, and part of the 3′ UTR (Figure 7A).
We readily detected the LCS2 protein by immunoblot analysis in CC-4533 but not in lcs2 (Figure 7C). The size of the LCS2 protein based on migration in the gel is consistent with its predicted molecular mass (74 kD based on Cre13.g566650.t1.1).
To complement the lcs2 mutant, we took advantage of a BAC library of the Chlamydomonas genome (Nguyen et al., 2005). We digested a bacterial artificial chromosome (BAC) containing LCS2 to generate a fragment containing the complete LCS2 genomic sequence and no other open reading frame. We purified and cotransformed this fragment into lcs2 with a selectable marker conferring resistance to hygromycin B (Berthold et al., 2002) and screened the transformants for the presence of an intact LCS2 3′ UTR using the primer pair g1 + g2 and for LCS2 protein accumulation. Four of the transformants (comp1-4) contained the introduced LCS2 gene (Figure 7B) and also accumulated LCS2 protein (Figure 7C).
We analyzed the TAG content of the lcs2 mutant and complemented lines by TLC and liquid chromatography-mass spectrometry (LC-MS). TLC analysis indicated that the four complemented strains contained a similar level of TAG as the CC-4533 parental strain (Figure 7D). Quantitative LC-MS measurements showed that the lcs2 strain contained ∼50% of the amount of TAG in CC-4533 (Figure 7E). The complemented lines showed full recovery of wild-type TAG levels, or slightly higher TAG than the wild type. These data demonstrate that the decreased TAG content in lcs2 is caused by disruption of the LCS2 gene.
LCS2 Is Involved in Incorporating de Novo-Synthesized Fatty Acids into TAG
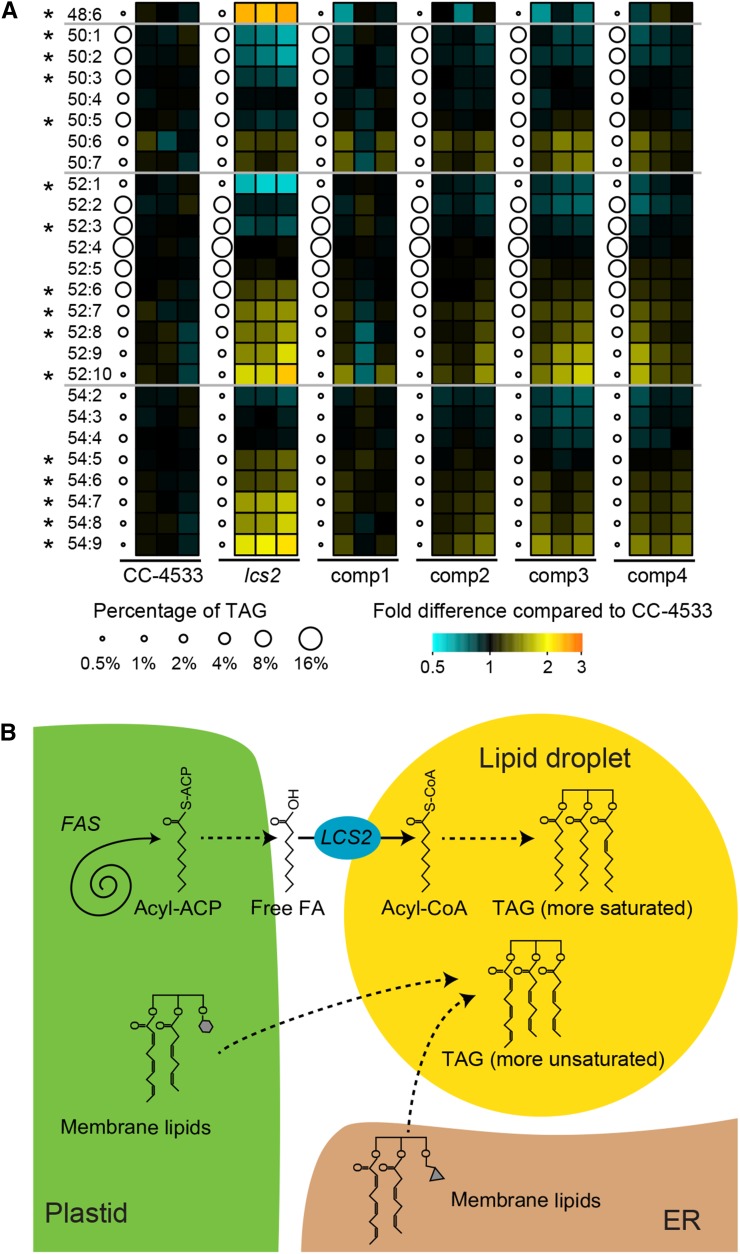
We sought to more precisely establish LCS2 function with respect to TAG metabolism. Homology of LCS2 to long-chain acyl-CoA synthetases suggests that it activates free fatty acids by converting them into acyl-CoAs, substrates for TAG synthesis. If LCS2 has this activity, its free fatty acid substrates could originate from one of two sources: (1) degradation of membrane lipids, which yields large amounts of polyunsaturated fatty acids; and (2) de novo synthesis, which produces more saturated species (Fan et al., 2011; Li et al., 2012b). We reasoned that we could distinguish between these two possibilities by examining the saturation profile of TAG in the lcs2 mutant.
We used LC-MS to analyze TAG composition of lcs2 and the complemented lines in comparison to CC-4533 (Figure 8A). TAG in lcs2 exhibited a smaller fraction of more saturated species (50:1, 50:2, 50:3, 52:1, and 52:3) and a larger fraction of more unsaturated species (48:6, 52:6, 52:7, 52:8, 52:10, 54:5, 54:6, 54:7, 54:8, and 54:9) than CC-4533 and the complemented lines. These results suggest that LCS2 is primarily involved in incorporating de novo-synthesized acyl groups (more saturated fatty acids) into TAG rather than the acyl groups recycled from membrane lipids (Figure 8B).
Figure 8.
LCS2 Is a Key Factor in Fatty Acid Export from the Chloroplast for TAG Synthesis.
(A) LC-MS was used to quantify different molecular species of TAG in the lcs2 mutant and complemented lines in three biological replicates. Each TAG species is denoted as N:M, where N is the number of carbon atoms in acyl chains and M is the number of C=C double bonds. In each strain, the average percentage of total TAG represented by each species is indicated by the size of the corresponding circle. Next to each circle, the percentage abundance of that species in each biological replicate is shown relative to the average percentage abundance of that species in CC-4533, as a heat map. A Mann-Whitney U-test was used to test whether the values in the heat map of lcs2 deviate significantly from the values in strains expressing LCS2 (CC-4533 plus the complemented lines) (asterisk indicates false discovery rate corrected P < 0.02).
(B) A model is proposed for the role of LCS2 in TAG synthesis under nitrogen deprivation. Enzymes and processes are italicized. Dashed arrows indicate processes that may involve multiple steps. Two major sources of acyl groups are represented: (1) LCS2 activates de novo synthesized fatty acids exported from the plastid and provides acyl-CoA for TAG synthesis; and (2) degradation of existing membrane lipids at the plastid or endoplasmic reticulum (ER) can provide acyl groups to TAG through LCS2-independent pathways. ACP, acyl carrier protein; FA, fatty acid; FAS, fatty acid synthesis.
DISCUSSION
A Strategy to Overcome Technical Challenges Associated with Constructing an Indexed Library
In this work, we overcame the challenges of propagation, storage, and rapid mapping of thousands of Chlamydomonas mutants. We developed protocols for propagation of libraries of tens of thousands of Chlamydomonas mutants as arrays of 384 colonies on agar using robotic systems commonly used in yeast genomics. These protocols included redundancy measures that protect the library against frequently encountered fungal contamination (Supplemental Figure 1). For longer term storage, we developed a highly efficient 96-well format freezing protocol (98 to 100% recovery; Supplemental Table 3). These types of technical advances enable efficient propagation and storage of tens of thousands of mutants by genomics researchers and centers like the Chlamydomonas Resource Center. Additionally, we developed an advanced pooling-based deconvolution scheme that allowed us to determine the physical coordinates of colonies harboring specific insertions in a collection of 18,430 colonies (Figures 2A to 2C). Importantly, all of these protocols are by design compatible with larger libraries of hundreds of thousands of mutants, which will be necessary for saturating coverage of the Chlamydomonas genome. These methods should be broadly applicable to studies in other organisms.
The greatest challenge that we faced in mapping the mutants was the complex nature of insertion sites (Figure 3A). The truncation of cassettes during transformation decreases the number of mappable flanking sequences (Zhang et al., 2014) and likely explains why many colonies did not yield a flanking sequence and few gave more than one (Figure 3B). When two flanking sequences were observed from one colony, they frequently appeared to originate from the two ends of one cassette but mapped to different genomic sites (see Methods; Supplemental Methods), suggesting that at least one side of the cassette was flanked by a short insertion of genomic DNA from another locus (Figure 3C). Therefore, many of the flanking sequences obtained from ChlaMmeSeq in our original collection do not report the true genomic insertion site, but instead report the sequence of a short DNA insertion between the cassette and flanking DNA. To overcome this challenge and establish a set of high-confidence mutants, we developed LEAP-Seq, which allowed us to identify mutants with short DNA insertions (Figures 4A and 4B). The mapping sites of the LEAP-Seq-confirmed insertions are accurate in ∼75% of cases (Figure 4C). As a result of the complex nature of insertions, only ∼10% of our initial colonies yielded confirmed flanking sequences. We are developing methods to significantly increase this fraction for future libraries (see below).
The Mutant Library Opens the Door to Novel Biology
Elucidation of biological processes in plants has benefited enormously from the availability of large-scale mutant collections in several model systems (Alonso et al., 2003; May et al., 2003; Zhang et al., 2006; Urbański et al., 2012; Belcher et al., 2015). The resource we generated here combines the power of an indexed, mapped library of insertion mutants with the advantages of Chlamydomonas, a unicellular, rapidly growing, photosynthetic organism.
This collection facilitates the study of a wide range of conserved biological processes that occur at the single-cell level, including the conversion of solar energy to chemical bond energy and the function of cilia in human disease. Our collection represents a library of high confidence mutants in which 1562 genes have defined disruptions. This resource will accelerate the characterization of candidate genes that were identified by phylogenetic analyses (Li et al., 2004; Merchant et al., 2007; Grossman et al., 2010; Karpowicz et al., 2011), proteomic characterization of organelles (Keller et al., 2005; Pazour et al., 2005; Moellering and Benning, 2010; Nguyen et al., 2011; Terashima et al., 2011), and transcriptome analyses under environmental conditions of interest (González-Ballester et al., 2010; Miller et al., 2010; Boyle et al., 2012; Brueggeman et al., 2012; Fang et al., 2012; Urzica et al., 2012; Albee et al., 2013; Blaby et al., 2013; Hemschemeier et al., 2013; Schmollinger et al., 2014).
In addition to enabling the study of candidate genes, the entire library can be screened for phenotypes of interest more easily and rapidly than existing resources in multicellular organisms, providing a tool complementary to those under development for land plants (O’Malley and Ecker, 2010). The library’s current format of arrayed colonies on solid medium is compatible with high-throughput robotic screens using colony size as a powerful and versatile phenotype, as illustrated in yeast (Costanzo et al., 2010). The library is also compatible with a range of other quantitative phenotyping methods, including measurement of pigment levels, chlorophyll fluorescence analysis of arrays on agar (Johnson et al., 2009), and motility screens in 96-well plates (Engel et al., 2011).
All of the strains in the library are compatible with the ChlaMmeSeq protocol (Zhang et al., 2014), which enables tracking of mutant abundances in pools. ChlaMmeSeq generates a unique 20-bp sequence from each mutant. Thus, the library can be pooled, cells exhibiting a phenotype of interest can be selected (e.g., by exposure to a stress or though fluorescence-activated cell sorting), and the identities of individual strains isolated from the pool can be determined. The quantitative nature of ChlaMmeSeq should enable yeast-style pooled screens, in which the growth rates of individual mutants are tracked over several generations (Giaever et al., 2002).
We Are Working Toward a Genome-Saturating Collection
The Chlamydomonas mutant library will become increasingly powerful for screens as more mutants are generated and more alleles are isolated for each gene (O’Malley and Ecker, 2010). Currently, it is not possible to conclude that an observed phenotype in a mutant is due to the annotated insertion because either the flanking sequence could be incorrect or the phenotype could be caused by a second site mutation (Dent et al., 2005). However, the ability to compare the phenotypes of multiple independent alleles for each gene will provide increased confidence in the causal relationship between genotype and phenotype. Toward this end, we are using the approaches established here to map insertion sites in 200,000 mutants, from which we aim to select ∼50,000 mutants to generate a stable collection that covers nearly all nonessential genes. We will only pick mutants with high-confidence flanking sequences and will favor insertions mapped to coding regions within genes. Our protocols remain unchanged, except for three refinements: (1) A collection of 200,000 is being propagated and mapped all at once, instead of the collection of 20,000 presented here. Deconvolution of colony positions in a collection of 200,000 mutants requires 21 plate-super-pools and 20 colony-super-pools. (2) We optimized the transformation protocol to increase the fraction of mutants yielding confirmed flanking sequences. Specifically, we removed the cell-settling step and directly collect cells by centrifugation. We also added a washing step after cell collection and shortened the time during which cells were incubated with cassette DNA before electroporation, in an effort to decrease the possibility of insertions of genomic DNA from lysed cells. (3) Each transformation cassette has been enhanced to include unique built-in DNA barcodes, which can be amplified using common primers annealing to the cassette (Shoemaker et al., 1996) and facilitate extraction of unique identity tags for each mutant. While the cassette is still compatible with ChlaMmeSeq, PCR amplification of these barcodes is being used instead of ChlaMmeSeq for determining the precise location of a mutant within a library. Mutant propagation, cryogenic freezing, combinatorial super-pooling of mutants, and the use of LEAP-Seq to validate insertion sites remain unchanged. We estimate that we will complete mapping and genetic characterizations of this new collection in the next two years if no major hurdles are encountered. Mutants from the complete collection will be made available as soon as possible through the CLiP website and the CRC.
LCS2 Is a Key Component of the Poorly Characterized Pathway That Incorporates de Novo-Synthesized Fatty Acids into TAG
The usefulness of the library presented here was illustrated by the ease with which we were able to rapidly screen candidate genes for roles in TAG metabolism and to identify LCS2 as an important gene in this process. Genes involved in TAG biosynthesis are central to our understanding of lipid metabolism and represent ideal targets for genetic engineering efforts to enhance biofuel production (Hu et al., 2008). Chlamydomonas is arguably the best-characterized alga, but only a handful of lesions causing defects in TAG biosynthesis have been identified, including pgd1 (Li et al., 2012b), pdat1 (Boyle et al., 2012; Yoon et al., 2012), nrr1 (Boyle et al., 2012), and tar1 (Kajikawa et al., 2015). By screening mutants disrupted in lipid droplet protein-encoding genes, we found that an lcs2 mutant has reduced levels of TAG compared with the parental strain (Figures 6B, 7D, and 7E; Supplemental Figure 14). Rescue of the TAG phenotype of lcs2 with wild-type LCS2 genomic DNA (Figures 7D and 7E) demonstrates that LCS2 is required for biosynthesis of ∼50% of the TAG in nitrogen-deprived cells.
Characterization of the lcs2 mutant fills in a key missing link in TAG metabolism of photosynthetic eukaryotes. In photosynthetic eukaryotes, fatty acids are synthesized in the plastid, and the complete pathway leading to their export from the plastid and incorporation into cytosolic lipid droplets remains unknown (Ohlrogge et al., 1979; Riekhof et al., 2005). Previous work has shown that two of the pathways of export of de novo-synthesized fatty acids provide free fatty acids at the outer envelope of the chloroplast (Andrews and Keegstra, 1983; Koo et al., 2004; Li et al., 2012b). It is unclear how these free fatty acids are then activated and incorporated into TAG. It has been hypothesized that LACS enzymes activate free fatty acid products by conversion into acyl-CoA on the path to TAG. However, LACS mutants in vascular plants show only modest effects on TAG accumulation (Fulda et al., 2002, 2004; Shockey et al., 2002; de Azevedo Souza et al., 2009; Lü et al., 2009; Weng et al., 2010; Zhao et al., 2010; Jessen et al., 2011, 2015; Shockey and Browse, 2011). Our work indicates that LCS2, a lipid droplet-localized LACS enzyme (Moellering and Benning, 2010; Nguyen et al., 2011), is required for incorporation of de novo-synthesized fatty acids into TAG. The results suggest a simple model in which free fatty acids produced at the outer envelope of the chloroplast are activated by LCS2 on adjacent lipid droplets for incorporation into TAG (Figure 8B).
Our data are consistent with a model in which TAG is produced by two parallel mechanisms: (1) the LCS2-dependent production of TAG from de novo-synthesized fatty acids and (2) the largely LCS2-independent conversion of membrane lipids into TAG. This second mechanism would provide fatty acids for polyunsaturated TAG synthesis by one of two pathways: (1) the head groups of membrane lipids could be cleaved off to provide diacylglycerol for acyl-CoA:diacylglycerol acyltransferase (Boyle et al., 2012; Deng et al., 2012; La Russa et al., 2012; Hung et al., 2013; Sanjaya et al., 2013) or phospholipid:diacylglycerol acyltransferase (PDAT) (Boyle et al., 2012; Yoon et al., 2012) reactions; or (2) the membrane lipids could directly serve as the polar lipid substrate for PDAT reactions.
More broadly, our library contains mutants in a large number of genes of interest, including dozens of genes implicated in photosynthesis and cilia function, as well as 430 genes with no functional annotation (Table 1). It is our hope that this library, and subsequent ones generated based on the tools presented here, will serve as a resource for the research community and will impact studies ranging from the production of renewable fuels to curing ciliary diseases.
METHODS
More detailed experimental procedures are available in Supplemental Methods.
Mutant Generation and Propagation
The mutants were generated from the Chlamydomonas reinhardtii CC-4533 background strain using electroporation as previously described (Zhang et al., 2014) with one minor difference (Supplemental Methods). After 2 weeks of growth, colonies were picked robotically and arrayed into microplates in a 384-colony format. Two copies of the library were maintained by monthly replications to fresh agar-solidified Tris-acetate-phosphate (TAP) medium with modified trace elements (Kropat et al., 2011) using a RoToR robot (Singer Instruments). Plates were stored at room temperature in low light (∼5 μmol photons m−2 s−1). Colonies lost or gained were revealed by computationally comparing images of library plates before and after 18 months of replications, followed by visual confirmation.
Cryopreservation
For cryopreservation, cells from 384-colony arrays on agar medium were inoculated into 96-well microtiter plates containing 120 μL liquid TAP medium per well and grown for ∼2 weeks. To each well, 100 μL of TAP with 5% (v/v) methanol was added for cryoprotection. Samples were incubated in foam boxes at 80°C for 90 min before being transferred to a vapor-phase liquid nitrogen freezer.
To recover cryopreserved strains, the 96-well plates were incubated at 30°C for 8 min immediately following removal from the liquid nitrogen freezer. Cells were centrifuged, washed with TAP medium, and inoculated into TAP medium. After 2 weeks of growth, cells were pelleted by centrifugation and robotically pinned back to agar plates. Missing colonies were visually identified.
Combinatorial Super-Pooling Scheme
The plate and colony-super-pooling schemes were designed based on binary error-correcting codes. The main features of the schemes are as follows: (1) Each pool is present in a unique combination of super-pools. (2) There are at least six differences between those combinations for any pair of pools, ensuring robustness against either sequencing and liquid-handling errors or low sequencing coverage. (3) The colony-super-pooling combinations were chosen to enable detection of cases where a transformant divides before plating, yielding two “sister colonies” with the same insertion. This leads to the same flanking sequence being part of two pools and therefore being detected in super-pools that contain either of the two pools. Without this precaution, such “sister colony” cases could result in misleading mapping of the flanking sequence. The super-pooling schemes are illustrated in Figures 2B and 2C and provided as Supplemental Data Sets 3 and 4.
Pooling and Flanking Sequence Extraction
Super-pools by plate and by colony were each generated from an independent set of 1-week-old library plates. To generate pools by plate, cells were scraped from agar plates and resuspended in liquid TAP medium. To generate pools by colony, cells from each 384-colony agar plate of the library were inoculated into a single set of four 96-well microtiter plates containing liquid TAP medium. A BioMek liquid-handling robot (Beckman Coulter) was used to generate super-pools using the scheme described above. DNA extraction, flanking sequence extraction by ChlaMmeSeq, and sequencing were performed as described previously (Zhang et al., 2014).
Flanking Sequence Deconvolution
The ChlaMmeSeq data from each super-pool was analyzed as described previously (Zhang et al., 2014), without merging flanking sequences that mapped to adjacent positions in the genome. Only flanking sequences that aligned to the nuclear genome were included in the analysis. The read counts for each flanking sequence were normalized to reads per million. Optimized adaptive cutoffs were used to convert the read counts to 0/1 values denoting the absence/presence of each flanking sequence in each super-pool. The plate and colony locations of each flanking sequence were determined by comparing the observed super-pool combination of each flanking sequence to the expected super-pool combinations corresponding to each plate and colony, and assigning the flanking sequence to the closest plate and colony match with at most 2 errors. See Supplemental Figure 2 for more details.
This method was applied separately to colony-super-pool and plate-super-pool data, from the 5′ and 3′ sides of the cassette. Each data set was processed in three iterations, first focusing on obtaining the highest quality results and then relaxing the parameters to increase the number of deconvolved flanking sequences. The results are shown in Supplemental Figure 3.
A total of 8009 flanking sequences were deconvolved to both a plate and a colony. But because, at the time of analysis (6/20/2014), 182 of these flanking sequences mapped to an empty colony (possibly due to colony loss), only 7827 flanking sequences are reported (Figure 2D).
LEAP-Seq
LEAP-Seq was adapted from Carette et al. (2011), with modifications that both achieve better amplification of Chlamydomonas DNA and allow paired-end sequencing (Supplemental Figure 7). The procedure consists of primer extension from the cassette (linear amplification), bead binding, adapter ligation, exponential PCR, and paired-end sequencing. Briefly, DNA was extracted from the entire library as a pooled sample. For primer extension, a biotinylated primer (P1) that anneals near the 5′ or 3′ end of the cassette was added together with deoxynucleotide triphosphate mixture and Phusion Hotstart II polymerase (F549L; Thermo Fisher Scientific) in order to allow extension of the primer into the flanking sequence. Eighty thermal cycles of denaturation, annealing, and extension were performed. Extended single-stranded DNAs were then captured by streptavidin-coated magnetic beads (60101; Life Technologies). A single-stranded adapter was added for ligation to the extended DNA molecules. The ligation products were exponentially amplified using a primer that anneals to the cassette (P2, closer to the end than P1) and a primer that anneals to the ligation adapter (P3). PCR products were subjected to paired-end sequencing.
LEAP-Seq Data Analysis
The raw paired-end LEAP-Seq data are composed of a proximal read (containing some cassette sequence and the genomic flanking sequence) and a distal read (containing distant genomic flanking sequence). The cassette sequence was removed from the proximal reads; the proximal reads were trimmed to 21 bp (the length provided by ChlaMmeSeq data), and the distal reads were trimmed to 30 bp. Both sides were independently aligned to the Chlamydomonas v5.3 genome and to the cassette sequence allowing one mismatch; nonunique alignments were discarded. Proximal reads aligning to the same position were combined, as they are expected to represent single insertion flanking sequences. Flanking sequences that were not present in the ChlaMmeSeq-based deconvolution results were discarded at this stage. All distal reads corresponding to each proximal flanking sequence were collected and used to calculate the fraction of distal reads aligning to the same locus as the proximal read, and the highest distance spanned by a read pair aligning to the same locus.
Analysis of Flanking Sequence Pairs Mapped to the Same Colony
Flanking sequence pairs that mapped to the same colony were analyzed in order to determine which pairs were derived from the same insertion versus different independent insertions. The pairs derived from the same insertion were used to examine the characteristics of the insertion sites, such as the presence and size of genomic deletions.
Briefly, the pairs were categorized based on their relative orientation and distance (Supplemental Figure 5). Within each distance range, the numbers of pairs in the different relative orientations were compared with the ratio expected for pairs derived from independent insertions (50% same-facing, 25% toward-facing, and 25% away-facing). Cases of significant departures from that ratio were categorized as different insertion models: clean insertions, deletions, genomic duplications, or multifragment insertions. Note that cases categorized as 1- to 2-bp deletions or duplications could also be errors during PCR, sequencing, or data processing. Additionally, the percentage of LEAP-Seq high-confidence flanking sequences and the percentage of 5′+3′ cassette-end pairs (versus 5′+5′ or 3′+3′) in each category were used to corroborate the conclusions.
Because they mapped to locations either greater than 1 kb apart or on different chromosomes, 1278 pairs of flanking sequences were not classified into the above categories. These pairs could represent (1) flanking sequences derived from two independent insertions or (2) flanking sequences derived from two sides of a single insertion, where at least one side additionally contained a short insertion of genomic DNA from a different locus. We estimated that 49% of the 1278 flanking sequence pairs were derived from the same insertion, based on the observed fraction of pairs that were 5′+3′ versus 5′+5′ or 3′+3′.
Because we observed random insertion of genomic DNA fragments contiguous with the cassette, we used the sequence information from ChlaMmeSeq and LEAP-Seq, from both sides of the cassette, to estimate the length distribution of these insertions of random DNA. The results are shown in Supplemental Figure 6: 92% of the random DNA fragments are below 500 bp, and 98% are below 700 bp.
Insertion Distribution and Genome Coverage Analysis
The data sets described in Table 1 are from the following sources: Pazour et al. (2005), Merchant et al. (2007), Moellering and Benning (2010), Karpowicz et al. (2011), Nguyen et al. (2011), and Terashima et al. (2011). The protein IDs for some of the data sets had to be converted from older versions to the current genome version (using http://pathways.mcdb.ucla.edu/algal/id_conversion.html). Data could not be obtained for some genes, and these have been omitted from the table. For the set of all Chlamydomonas genes, genes with Arabidopsis thaliana homologs, and genes with no functional annotation, the Chlamydomonas genome v5.3 from Phytozome was used directly. Genes with Arabidopsis homologs are genes that have a best Arabidopsis hit name, symbol, or defline listed in the Phytozome downloadable annotation file. Genes with no functional annotation are genes that lack any annotation in the Phytozome downloadable annotation or defline files.
The genome/gene/feature mappable lengths were calculated as in (Zhang et al., 2014). To normalize the data, the number of insertions in each feature was divided by the overall mappable length of that feature. Insertions in genes with multiple splice variants were not assigned to any feature. P-values comparing the insertion densities in each pair of features were calculated using the chi-square test of independence (McDonald, 2009).
The simulated data sets and hot/cold spot analysis were done as described (Zhang et al., 2014). The full list of hot/cold spots is provided as Supplemental Data Set 10. The hot/cold spot analysis was also performed on simulated data sets to estimate how many of the hot/cold spots detected by our analysis are likely to be due to the fact that our method slightly under-corrects P values for multiple testing. Out of 20 simulated data sets, 13 had no hot/cold spots with P values <0.05, two had two hot spots, four had one hot spot, and one had one cold spot.
DNA Gel Blotting
Purified genomic DNA was digested with StuI (New England Biolabs), and the resulting fragments were resolved on 0.7% agarose gels. DNA was transferred onto a Zeta-probe membrane (Bio-Rad) and UV cross-linked. The membrane was hybridized to a probe consisting of the AphVIII sequence, which is the only part of the cassette sequence that is not present in the genome.
Nitrogen Deprivation and Lipid Analysis
Exponentially growing cells in TAP medium at 30 to 40 μmol photons m−2 s−1 continuous light were pelleted by centrifugation at 1500g for 2 min, resuspended in TAP-N, pelleted again, and resuspended in TAP-N. After 24 h of incubation in medium devoid of N, aliquots of cells containing the same amount of chlorophyll were collected for lipid extraction and TLC as previously described (Li et al., 2012a). To identify and quantify the TAG species, LC-MS and subsequent tandem mass spectrometry were performed on total lipid extracts as previously described (Terashima et al., 2015).
Complementation of the lcs2 Strain
For complementation of lcs2, a BAC clone was restriction digested to yield a fragment containing the LCS2 gene (including 5′ and 3′ UTRs). This fragment was cotransformed into lcs2 cells with linearized pHyg3 plasmid, which confers resistance hygromycin B. Transformants selected on hygromycin B-containing agar plates were screened initially for presence of wild-type LCS2 genomic DNA by PCR and then for presence of LCS2 protein by immunoblotting. A primary antibody generated against a peptide epitope of LCS2 (CRRPQLQAKYQAKLD-amide) by Yenzym was employed to detect LCS2. As a loading control, the membrane was stripped and reprobed with an α-tubulin antibody raised in mouse (T6074; Sigma-Aldrich).
Custom Software
Software developed in support of this project is available at http://github.com/Jonikas-Lab/Li-Zhang-Patena-2015.
Accession Numbers
Sequence data for the LCS2 gene presented in this article can be found in the Phytozome database under accession number Cre13.g566650. The other genes for which the mutants were analyzed in TAG content are Cre02.g117500, Cre01.g053000, Cre02.g113200, Cre12.g516450, Cre01.g048950, Cre12.g500500, Cre10.g442800, Cre01.g015300, g5059, Cre10.g427700, Cre01.g015300, Cre12.g492900, Cre10.g427700, Cre10.g436800, Cre01.g044650, Cre17.g721000, Cre16.g647534, Cre17.g723850, Cre12.g517000, Cre12.g559250, Cre07.g321400, Cre07.g336950, Cre08.g372100, Cre03.g164750, g2932, and Cre05.g247350. Sequence data for the insertion cassette are found in GenBank under accession number KJ572788. Chloroplast and mitochondrial genomes were added as separate chromosomes from the National Center for Biotechnology Information website: NC_005353 and NC_001638, respectively. Raw data for Supplemental Data Set 8 are available from the Sequence Read Archive under accession number SRP068433.
Supplemental Data
Supplemental Figure 1. Mutants can be robustly maintained as arrays of colonies on agar.
Supplemental Figure 2. Flanking sequences for each super-pool are computationally deconvolved into plate and colony coordinates.
Supplemental Figure 3. Details of flanking sequence deconvolution results separated into three iterations.
Supplemental Figure 4. Validation of deconvolution results.
Supplemental Figure 5. Pairs of flanking sequences mapped to the same colony can be divided into insertion scenarios based on their distance and relative orientation, cassette sides, and LEAP-Seq confidence data.
Supplemental Figure 6. The distribution of lengths of putative genomic DNA fragments surrounded by two cassettes was estimated from data shown in Supplemental Figure 5.
Supplemental Figure 7. LEAP-Seq is employed to obtain longer flanking sequences to confirm cassette insertion sites.
Supplemental Figure 8. Full LEAP-Seq confidence plots for 5′ and 3′ sides of the cassette, with the same confirmed mutant set cutoffs as in Figure 4B.
Supplemental Figure 9. The insertion sites of randomly chosen individual mutants were checked by PCR.
Supplemental Figure 10. The read count in LEAP-Seq and ChlaMmeSeq has little impact on the validation of insertion sites by PCR.
Supplemental Figure 11. Seventeen out of 23 (74%) of mutants in the LEAP-Seq-confirmed set harbor a single insertion.
Supplemental Figure 12. The distribution of insertions in the genome is largely random.
Supplemental Figure 13. Mutants deficient in lipid droplet proteins were screened for TAG deficiency using thin-layer chromatography.
Supplemental Figure 14. We confirmed the TAG deficiency in lcs2 by TLC.
Supplemental Figure 15. RT-PCR supports the Cre13.g566650.t1.1 model of LCS2.
Supplemental Table 1. Summary of variants of our CC-4533 strain relative to the reference Chlamydomonas v5.3 genome and several other strains sequenced by Gallaher et al. (2015).
Supplemental Table 2. Details of high-impact variants unique to CC-4533.
Supplemental Table 3. Cryopreserved mutants can be recovered at a success rate greater than 98%.
Supplemental Table 4. Sequences of oligonucleotide primers used for LEAP-Seq.
Supplemental Table 5. Sequences of oligonucleotide primers used for genotyping and RT-PCR of CC-4533 and lcs2.
Supplemental Data Set 1. Structural variants relative to reference and other strains
Supplemental Data Set 2. High-confidence genomic differences unique to CC-4533.
Supplemental Data Set 3. Binary codes for plate super-pooling.
Supplemental Data Set 4. Binary codes for colony super-pooling.
Supplemental Data Set 5. Read counts of each flanking sequence in each super-pool.
Supplemental Data Set 6. ChlaMmeSeq data for validation of deconvolution.
Supplemental Data Set 7. Full library data: colony positions, insertion sites, deconvolution and LEAP-Seq summary, and gene annotation.
Supplemental Data Set 8. LEAP-Seq read details for each flanking sequence.
Supplemental Data Set 9. Summary of PCR results to check the insertion sites of randomly picked mutants from the mutant library.
Supplemental Data Set 10. Statistically significant insertion hot spots and cold spots.
Supplemental Data Set 11. Insertions in genes encoding lipid droplet proteins were characterized by PCR.
Supplementary Material
Acknowledgments
We thank Zhi-Yong Wang for suggesting the idea to generate this indexed mutant library; Olivier Vallon, Ronan O’Malley, and Joe Ecker for helpful discussions; Matt Laudon for distribution of mutants from the Chlamydomonas Resource Center; Eva Huala, Bob Muller, Shimantika Sharma, and Garret Huntress for developing and improving the CLiP website; Jan Carette and Caleb Marceau for help in developing LEAP-Seq; Sabeeha Merchant, Matteo Pellegrini, and Sean Gallaher for help with genome analysis of the background strain; Shriya Ghosh for recycling plates; Tyler Wittkopp, Munevver Aksoy, Yang Bai, and Robert Jinkerson for help with experiments or data plotting; Xuhuai Ji at the Stanford Functional Genomics Facility and Ziming Weng at the Stanford Center for Genomics and Personalized Medicine for Illumina sequencing services; Theresa McLaughlin at the Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University Mass Spectrometry (http://mass-spec.stanford.edu) for analyzing lipid samples; Glenn Ruiz at YenZym for peptide antibody service; Jay Yang and Ian Morgan at Singer Instruments for technical assistance; Kathryn Barton and Winslow Briggs for providing lab space; John Nguyen for help with the check PCR; and Friedrich Fauser, Wolf Frommer, Robert Jinkerson, and Luke Mackinder for constructive suggestions on the manuscript. This project was supported by a grant from the National Science Foundation (MCB-1146621) awarded to M.C.J. and A.R.G. Genome analysis of the CC-4533 was supported by a grant awarded to Sabeeha Merchant by the National Institutes of Health (R24 GM092473) and a grant awarded to Matteo Pellegrini by the Office of Science of the U.S. Department of Energy (DE-FC02-02ER63421).
AUTHOR CONTRIBUTIONS
M.C.J. and A.R.G. designed the project. S.T.F.-G. performed the CC-4533 genome comparison to other standard laboratory strains. S.S.G. and S.R.B. performed transformation and colony picking to generate the mutant library. N.I., S.R.B., and S.S.G. propagated the mutant library. W.P. designed the scheme for super-pooling. S.R.B. and N.I. performed pooling and super-pooling; R.Z. performed ChlaMmeSeq on the entire library sample. S.R.B., R.Z., and X.L. performed ChlaMmeSeq on super-pools. X.L. developed LEAP-Seq and implemented it for the library. W.P. performed large-scale analyses of insertion positions, mutant deconvolution, and analyses of LEAP-Seq data. W.P., X.L., and J.M.R. validated the accuracy of flanking sequence deconvolution. R.Z. and N.I. performed PCR to check mapping correctness. W.P. generated the program for robotic rearray of mutants. N.I. performed the rearray of the mutant library. W.P., X.L., and R.Z. designed the mutant distribution website. P.A.L. and N.I. developed protocols for mutant distribution. R.Y. performed the DNA gel blot analysis. S.S.G. and R.Y. developed the protocol for cryopreservation and performed cryopreservation. J.M.R., X.L., R.Z., and N.I. performed the molecular characterizations of lipid droplet protein mutants. X.L. performed biochemical characterizations of lipid droplet protein mutants. X.L., N.I., and R.Y. performed complementation of the lcs2 mutant. X.L., W.P., R.Z., N.I., R.Y., A.R.G., and M.C.J. wrote the article.
Glossary
- TAG
triacylglycerol
- UTR
untranslated region
- TLC
thin-layer chromatography
- LACS
long-chain acyl-CoA synthetase
- LC-MS
liquid chromatography-mass spectrometry
- TAP
Tris-acetate-phosphate
Footnotes
Articles can be viewed online without a subscription.
References
- Aksoy M., Pootakham W., Grossman A.R. (2014). Critical function of a Chlamydomonas reinhardtii putative polyphosphate polymerase subunit during nutrient deprivation. Plant Cell 26: 4214–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albee A.J., Kwan A.L., Lin H., Granas D., Stormo G.D., Dutcher S.K. (2013). Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtti. G3 (Bethesda) 3: 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Andrews J., Keegstra K. (1983). Acyl-CoA synthetase is located in the outer membrane and acyl-CoA thioesterase in the inner membrane of pea chloroplast envelopes. Plant Physiol. 72: 735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K., Daum G. (2003). YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J. Biol. Chem. 278: 23317–23323. [DOI] [PubMed] [Google Scholar]
- Athenstaedt K., Daum G. (2005). Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J. Biol. Chem. 280: 37301–37309. [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger M.R., Kaplan A., Berry J.A. (1980). Internal inorganic carbon pool of Chlamydomonas reinhardtii: Evidence for a carbon dioxide-concentrating mechanism. Plant Physiol. 66: 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldari C.T., Rosenbaum J. (2010). Intraflagellar transport: it’s not just for cilia anymore. Curr. Opin. Cell Biol. 22: 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale S.I. (2009). Biosynthesis of chlorophylls and hemes. In The Chlamydomonas Sourcebook, E.H. Harris, D.B. Stern, and G.B. Witman, eds (San Diego, CA: Academic Press), pp. 731–798. [Google Scholar]
- Beel B., Prager K., Spexard M., Sasso S., Weiss D., Müller N., Heinnickel M., Dewez D., Ikoma D., Grossman A.R., Kottke T., Mittag M. (2012). A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell 24: 2992–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcher S., Williams-Carrier R., Stiffler N., Barkan A. (2015). Large-scale genetic analysis of chloroplast biogenesis in maize. Biochim. Biophys. Acta 1847: 1004–1016. [DOI] [PubMed] [Google Scholar]
- Berthold P., Schmitt R., Mages W. (2002). An engineered Streptomyces hygroscopicus aph 7" gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153: 401–412. [DOI] [PubMed] [Google Scholar]
- Blaby I.K., et al. (2013). Systems-level analysis of nitrogen starvation-induced modifications of carbon metabolism in a Chlamydomonas reinhardtii starchless mutant. Plant Cell 25: 4305–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle N.R., et al. (2012). Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287: 15811–15825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J.E., et al. (1988). Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240: 1534–1538. [DOI] [PubMed] [Google Scholar]
- Bragg J.N., Wu J., Gordon S.P., Guttman M.E., Thilmony R., Lazo G.R., Gu Y.Q., Vogel J.P. (2012). Generation and characterization of the Western Regional Research Center Brachypodium T-DNA insertional mutant collection. PLoS One 7: e41916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggeman A.J., Gangadharaiah D.S., Cserhati M.F., Casero D., Weeks D.P., Ladunga I. (2012). Activation of the carbon concentrating mechanism by CO2 deprivation coincides with massive transcriptional restructuring in Chlamydomonas reinhardtii. Plant Cell 24: 1860–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon R.H., García-Cerdán J.G., Malnoë A., Cook R., Russell J.J., Gaw C., Dent R.M., de Vitry C., Niyogi K.K. (2013). A conserved rubredoxin is necessary for photosystem II accumulation in diverse oxygenic photoautotrophs. J. Biol. Chem. 288: 26688–26696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J.E., Guimaraes C.P., Wuethrich I., Blomen V.A., Varadarajan M., Sun C., Bell G., Yuan B., Muellner M.K., Nijman S.M., Ploegh H.L., Brummelkamp T.R. (2011). Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat. Biotechnol. 29: 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castruita M., Casero D., Karpowicz S.J., Kropat J., Vieler A., Hsieh S.I., Yan W., Cokus S., Loo J.A., Benning C., Pellegrini M., Merchant S.S. (2011). Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23: 1273–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotti C., Dubini A., Subramanian V., Yang W., Magneschi L., Mus F., Seibert M., Posewitz M.C., Grossman A.R. (2012). Altered fermentative metabolism in Chlamydomonas reinhardtii mutants lacking pyruvate formate lyase and both pyruvate formate lyase and alcohol dehydrogenase. Plant Cell 24: 692–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotti C., Yang W., Posewitz M.C., Grossman A.R. (2013). Fermentation metabolism and its evolution in algae. Front. Plant Sci. 4: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., et al. (2010). The genetic landscape of a cell. Science 327: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F.R., Umen J.G. (2015). The Chlamydomonas cell cycle. Plant J. 82: 370–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo Souza C., Kim S.S., Koch S., Kienow L., Schneider K., McKim S.M., Haughn G.W., Kombrink E., Douglas C.J. (2009). A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X.D., Gu B., Li Y.J., Hu X.W., Guo J.C., Fei X.W. (2012). The roles of acyl-CoA: diacylglycerol acyltransferase 2 genes in the biosynthesis of triacylglycerols by the green algae Chlamydomonas reinhardtii. Mol. Plant 5: 945–947. [DOI] [PubMed] [Google Scholar]
- Dent R.M., Haglund C.M., Chin B.L., Kobayashi M.C., Niyogi K.K. (2005). Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 137: 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R.M., Sharifi M.N., Malnoë A., Haglund C., Calderon R.H., Wakao S., Niyogi K.K. (2015). Large-scale insertional mutagenesis of Chlamydomonas supports phylogenomic functional prediction of photosynthetic genes and analysis of classical acetate-requiring mutants. Plant J. 82: 337–351. [DOI] [PubMed] [Google Scholar]
- Dubini A., Mus F., Seibert M., Grossman A.R., Posewitz M.C. (2009). Flexibility in anaerobic metabolism as revealed in a mutant of Chlamydomonas reinhardtii lacking hydrogenase activity. J. Biol. Chem. 284: 7201–7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond P.J. (2006). SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18: 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B.D., Ishikawa H., Feldman J.L., Wilson C.W., Chuang P.T., Snedecor J., Williams J., Sun Z., Marshall W.F. (2011). A cell-based screen for inhibitors of flagella-driven motility in Chlamydomonas reveals a novel modulator of ciliary length and retrograde actin flow. Cytoskeleton (Hoboken) 68: 188–203. [DOI] [PubMed] [Google Scholar]
- Fan J., Andre C., Xu C. (2011). A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. 585: 1985–1991. [DOI] [PubMed] [Google Scholar]
- Fang W., Si Y., Douglass S., Casero D., Merchant S.S., Pellegrini M., Ladunga I., Liu P., Spalding M.H. (2012). Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. Plant Cell 24: 1876–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese R.V. Jr., Walther T.C. (2009). Lipid droplets finally get a little R-E-S-P-E-C-T. Cell 139: 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., Omran H. (2007). When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8: 880–893. [DOI] [PubMed] [Google Scholar]
- Fulda M., Schnurr J., Abbadi A., Heinz E., Browse J. (2004). Peroxisomal Acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 16: 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda M., Shockey J., Werber M., Wolter F.P., Heinz E. (2002). Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid beta-oxidation. Plant J. 32: 93–103. [DOI] [PubMed] [Google Scholar]
- Gallaher S.D., Fitz-Gibbon S.T., Glaesener A.G., Pellegrini M., Merchant S.S. (2015). Chlamydomonas genome resource for laboratory strains reveals a mosaic of sequence variation, identifies true strain histories, and enables strain-specific studies. Plant Cell 27: 2335–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galván A., González-Ballester D., Fernández E. (2007). Insertional mutagenesis as a tool to study genes/functions in Chlamydomonas. Adv. Exp. Med. Biol. 616: 77–89. [DOI] [PubMed] [Google Scholar]
- Geng S., De Hoff P., Umen J.G. (2014). Evolution of sexes from an ancestral mating-type specification pathway. PLoS Biol. 12: e1001904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi M.L., Posewitz M.C., Maness P.C., Dubini A., Yu J., Seibert M. (2007). Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms. Annu. Rev. Plant Biol. 58: 71–91. [DOI] [PubMed] [Google Scholar]
- Giaever G., et al. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- González-Ballester D., Casero D., Cokus S., Pellegrini M., Merchant S.S., Grossman A.R. (2010). RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22: 2058–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ballester D., de Montaigu A., Galván A., Fernández E. (2005a). Restriction enzyme site-directed amplification PCR: a tool to identify regions flanking a marker DNA. Anal. Biochem. 340: 330–335. [DOI] [PubMed] [Google Scholar]
- González-Ballester D., de Montaigu A., Higuera J.J., Galván A., Fernández E. (2005b). Functional genomics of the regulation of the nitrate assimilation pathway in Chlamydomonas. Plant Physiol. 137: 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ballester D., Pootakham W., Mus F., Yang W., Catalanotti C., Magneschi L., de Montaigu A., Higuera J.J., Prior M., Galván A., Fernandez E., Grossman A.R. (2011). Reverse genetics in Chlamydomonas: a platform for isolating insertional mutants. Plant Methods 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U. (2015). Historical perspective on Chlamydomonas as a model for basic research: 1950-1970. Plant J. 82: 365–369. [DOI] [PubMed] [Google Scholar]
- Goodman A.L., McNulty N.P., Zhao Y., Leip D., Mitra R.D., Lozupone C.A., Knight R., Gordon J.I. (2009). Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold H., Beisson F., Peltier G., Li-Beisson Y. (2015). Microalgal lipid droplets: composition, diversity, biogenesis and functions. Plant Cell Rep. 34: 545–555. [DOI] [PubMed] [Google Scholar]
- Grossman A.R., Catalanotti C., Yang W., Dubini A., Magneschi L., Subramanian V., Posewitz M.C., Seibert M. (2011). Multiple facets of anoxic metabolism and hydrogen production in the unicellular green alga Chlamydomonas reinhardtii. New Phytol. 190: 279–288. [DOI] [PubMed] [Google Scholar]
- Grossman A.R., Harris E.E., Hauser C., Lefebvre P.A., Martinez D., Rokhsar D., Shrager J., Silflow C.D., Stern D., Vallon O., Zhang Z. (2003). Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot. Cell 2: 1137–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A.R., Karpowicz S.J., Heinnickel M., Dewez D., Hamel B., Dent R., Niyogi K.K., Johnson X., Alric J., Wollman F.A., Li H., Merchant S.S. (2010). Phylogenomic analysis of the Chlamydomonas genome unmasks proteins potentially involved in photosynthetic function and regulation. Photosynth. Res. 106: 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman B.L., Niyogi K.K. (2004). Chlamydomonas and Arabidopsis. A dynamic duo. Plant Physiol. 135: 607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.H. (2001). Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 363–406. [DOI] [PubMed] [Google Scholar]
- Heinnickel M.L., Alric J., Wittkopp T., Yang W., Catalanotti C., Dent R., Niyogi K.K., Wollman F.A., Grossman A.R. (2013). Novel thylakoid membrane GreenCut protein CPLD38 impacts accumulation of the cytochrome b6f complex and associated regulatory processes. J. Biol. Chem. 288: 7024–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemme D., et al. (2014). Systems-wide analysis of acclimation responses to long-term heat stress and recovery in the photosynthetic model organism Chlamydomonas reinhardtii. Plant Cell 26: 4270–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemschemeier A., Casero D., Liu B., Benning C., Pellegrini M., Happe T., Merchant S.S. (2013). Copper response regulator1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia. Plant Cell 25: 3186–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemschemeier A., Jacobs J., Happe T. (2008). Biochemical and physiological characterization of the pyruvate formate-lyase Pfl1 of Chlamydomonas reinhardtii, a typically bacterial enzyme in a eukaryotic alga. Eukaryot. Cell 7: 518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing Y.I., et al. (2007). A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol. Biol. 63: 351–364. [DOI] [PubMed] [Google Scholar]
- Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., Darzins A. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54: 621–639. [DOI] [PubMed] [Google Scholar]
- Hung C.H., Ho M.Y., Kanehara K., Nakamura Y. (2013). Functional study of diacylglycerol acyltransferase type 2 family in Chlamydomonas reinhardtii. FEBS Lett. 587: 2364–2370. [DOI] [PubMed] [Google Scholar]
- Hunter C.T., Suzuki M., Saunders J., Wu S., Tasi A., McCarty D.R., Koch K.E. (2014). Phenotype to genotype using forward-genetic Mu-seq for identification and functional classification of maize mutants. Front. Plant Sci. 4: 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S.W., Hudock G.A. (1971). Stability of Chlamydomonas reinhardtii in liquid nitrogen storage. J. Phycol. 7: 300–303. [Google Scholar]
- Jarvik J.W., Rosenbaum J.L. (1980). Oversized flagellar membrane protein in paralyzed mutants of Chlamydomonas reinhardrii. J. Cell Biol. 85: 258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen D., Olbrich A., Knüfer J., Krüger A., Hoppert M., Polle A., Fulda M. (2011). Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis. Plant J. 68: 715–726. [DOI] [PubMed] [Google Scholar]
- Jessen D., Roth C., Wiermer M., Fulda M. (2015). Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis. Plant Physiol. 167: 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Brueggeman A.J., Horken K.M., Plucinak T.M., Weeks D.P. (2014). Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot. Cell 13: 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. (2013). RNA-programmed genome editing in human cells. eLife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X., Vandystadt G., Bujaldon S., Wollman F.A., Dubois R., Roussel P., Alric J., Béal D. (2009). A new setup for in vivo fluorescence imaging of photosynthetic activity. Photosynth. Res. 102: 85–93. [DOI] [PubMed] [Google Scholar]
- Kajikawa M., Sawaragi Y., Shinkawa H., Yamano T., Ando A., Kato M., Hirono M., Sato N., Fukuzawa H. (2015). Algal dual-specificity tyrosine phosphorylation-regulated kinase, triacylglycerol accumulation regulator1, regulates accumulation of triacylglycerol in nitrogen or sulfur deficiency. Plant Physiol. 168: 752–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz S.J., Prochnik S.E., Grossman A.R., Merchant S.S. (2011). The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J. Biol. Chem. 286: 21427–21439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kates J.R., Jones R.F. (1964). Fluoroacetate inhibition of amino acids during photosynthesis of Chlamydomonas reinhardti. Science 143: 145–146. [DOI] [PubMed] [Google Scholar]
- Keller L.C., Romijn E.P., Zamora I., Yates J.R. III, Marshall W.F. (2005). Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15: 1090–1098. [DOI] [PubMed] [Google Scholar]
- Kindle K.L., Schnell R.A., Fernández E., Lefebvre P.A. (1989). Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol. 109: 2589–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo A.J., Ohlrogge J.B., Pollard M. (2004). On the export of fatty acids from the chloroplast. J. Biol. Chem. 279: 16101–16110. [DOI] [PubMed] [Google Scholar]
- Kourmpetis Y.A., van Dijk A.D., van Ham R.C., ter Braak C.J. (2011). Genome-wide computational function prediction of Arabidopsis proteins by integration of multiple data sources. Plant Physiol. 155: 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K.G., Johnson K.A., Forscher P., Rosenbaum J.L. (1993). A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA 90: 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S.S., Malasarn D. (2011). A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66: 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat C.F., Natter K., Petschnigg J., Wolinski H., Scheuringer K., Scholz H., Zimmermann R., Leber R., Zechner R., Kohlwein S.D. (2006). Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J. Biol. Chem. 281: 491–500. [DOI] [PubMed] [Google Scholar]
- La Russa M., Bogen C., Uhmeyer A., Doebbe A., Filippone E., Kruse O., Mussgnug J.H. (2012). Functional analysis of three type-2 DGAT homologue genes for triacylglycerol production in the green microalga Chlamydomonas reinhardtii. J. Biotechnol. 162: 13–20. [DOI] [PubMed] [Google Scholar]
- Levine R.P. (1960). Genetic control of photosynthesis in Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 46: 972–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin R.A. (1953). Studies on the flagella of algae. Il. Formation of flagella by Chlamydomonas in light and darkness. Ann. N. Y. Acad. Sci. 56: 1091–1093. [DOI] [PubMed] [Google Scholar]
- Li J.B., et al. (2004). Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117: 541–552. [DOI] [PubMed] [Google Scholar]
- Li X., Benning C., Kuo M.H. (2012a). Rapid triacylglycerol turnover in Chlamydomonas reinhardtii requires a lipase with broad substrate specificity. Eukaryot. Cell 11: 1451–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Moellering E.R., Liu B., Johnny C., Fedewa M., Sears B.B., Kuo M.H., Benning C. (2012b). A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24: 4670–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Rosso M.G., Viehoever P., Weisshaar B. (2007). GABI-Kat SimpleSearch: an Arabidopsis thaliana T-DNA mutant database with detailed information for confirmed insertions. Nucleic Acids Res. 35: D874–D878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y., Beisson F., Riekhof W. (2015). Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J. 82: 504–522. [DOI] [PubMed] [Google Scholar]
- Liu B., Benning C. (2013). Lipid metabolism in microalgae distinguishes itself. Curr. Opin. Biotechnol. 24: 300–309. [DOI] [PubMed] [Google Scholar]
- Liu Y., Pei J., Grishin N., Snell W.J. (2015). The cytoplasmic domain of the gamete membrane fusion protein HAP2 targets the protein to the fusion site in Chlamydomonas and regulates the fusion reaction. Development 142: 962–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr M. (2009). Carotenoids. In The Chlamydomonas Sourcebook, H. E.H., S. E.B., and W. G.B., eds (Academic Press), pp. 799–817. [Google Scholar]
- Lohr M., Im C.S., Grossman A.R. (2005). Genome-based examination of chlorophyll and carotenoid biosynthesis in Chlamydomonas reinhardtii. Plant Physiol. 138: 490–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü S., Song T., Kosma D.K., Parsons E.P., Rowland O., Jenks M.A. (2009). Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 59: 553–564. [DOI] [PubMed] [Google Scholar]
- Magneschi L., Catalanotti C., Subramanian V., Dubini A., Yang W., Mus F., Posewitz M.C., Seibert M., Perata P., Grossman A.R. (2012). A mutant in the ADH1 gene of Chlamydomonas reinhardtii elicits metabolic restructuring during anaerobiosis. Plant Physiol. 158: 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. (2013). RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T., Okamoto K., Onai K., Niwa Y., Shimogawara K., Ishiura M. (2008). A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev. 22: 918–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul J.E., Lilly J.W., Cui L., dePamphilis C.W., Miller W., Harris E.H., Stern D.B. (2002). The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14: 2659–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May B.P., Liu H., Vollbrecht E., Senior L., Rabinowicz P.D., Roh D., Pan X., Stein L., Freeling M., Alexander D., Martienssen R. (2003). Maize-targeted mutagenesis: A knockout resource for maize. Proc. Natl. Acad. Sci. USA 100: 11541–11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D.R., Latshaw S., Wu S., Suzuki M., Hunter C.T., Avigne W.T., Koch K.E. (2013). Mu-seq: sequence-based mapping and identification of transposon induced mutations. PLoS One 8: e77172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D.R., et al. (2005). Steady-state transposon mutagenesis in inbred maize. Plant J. 44: 52–61. [DOI] [PubMed] [Google Scholar]
- McDonald J.H. (2009). Handbook of Biological Statistics, 2nd ed. (Baltimore, MD: Sparky House Publishing; ). [Google Scholar]
- Merchant S.S., Kropat J., Liu B., Shaw J., Warakanont J. (2012). TAG, you’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr. Opin. Biotechnol. 23: 352–363. [DOI] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meslet-Cladière L., Vallon O. (2012). A new method to identify flanking sequence tags in chlamydomonas using 3′-RACE. Plant Methods 8: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R., et al. (2010). Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154: 1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz R.H., Lewin R.A. (1954). Studies on the flagella of algae. V. Serology of paralyzed mutants of Chlamydomonas. Can. J. Microbiol. 1: 65–67. [DOI] [PubMed] [Google Scholar]
- Mitchell D.R. (2004). Speculations on the evolution of 9+2 organelles and the role of central pair microtubules. Biol. Cell 96: 691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering E.R., Benning C. (2010). RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 9: 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A., Bassett A., Thuenemann E., Schwach F., Karkare S., Ossowski S., Weigel D., Baulcombe D. (2009). Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 58: 165–174. [DOI] [PubMed] [Google Scholar]
- Murthy U.M., Wecker M.S., Posewitz M.C., Gilles-Gonzalez M.A., Ghirardi M.L. (2012). Novel FixL homologues in Chlamydomonas reinhardtii bind heme and O(2). FEBS Lett. 586: 4282–4288. [DOI] [PubMed] [Google Scholar]
- Mus F., Dubini A., Seibert M., Posewitz M.C., Grossman A.R. (2007). Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 282: 25475–25486. [DOI] [PubMed] [Google Scholar]
- Nguyen H.M., Baudet M., Cuiné S., Adriano J.M., Barthe D., Billon E., Bruley C., Beisson F., Peltier G., Ferro M., Li-Beisson Y. (2011). Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics 11: 4266–4273. [DOI] [PubMed] [Google Scholar]
- Nguyen R.L., Tam L.W., Lefebvre P.A. (2005). The LF1 gene of Chlamydomonas reinhardtii encodes a novel protein required for flagellar length control. Genetics 169: 1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J.B., Kuhn D.N., Stumpf P.K. (1979). Subcellular localization of acyl carrier protein in leaf protoplasts of Spinacia oleracea. Proc. Natl. Acad. Sci. USA 76: 1194–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley R.C., Ecker J.R. (2010). Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J. 61: 928–940. [DOI] [PubMed] [Google Scholar]
- Pazour G.J., Agrin N., Leszyk J., Witman G.B. (2005). Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170: 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Dickert B.L., Vucica Y., Seeley E.S., Rosenbaum J.L., Witman G.B., Cole D.G. (2000). Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151: 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps G., Krawietz D., Hemschemeier A., Happe T. (2011). A pyruvate formate lyase-deficient Chlamydomonas reinhardtii strain provides evidence for a link between fermentation and hydrogen production in green algae. Plant J. 66: 330–340. [DOI] [PubMed] [Google Scholar]
- Piasecki B.P., Diller K.R., Brand J.J. (2009). Cryopreservation of Chlamydomonas reinhardtii: a cause of low viability at high cell density. Cryobiology 58: 103–109. [DOI] [PubMed] [Google Scholar]
- Pootakham W., Gonzalez-Ballester D., Grossman A.R. (2010). Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol. 153: 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Rosenbaum J.L., Barr M.M. (2001). An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr. Biol. 11: 457–461. [DOI] [PubMed] [Google Scholar]
- Randolph-Anderson B.L., Boynton J.E., Gillham N.W., Harris E.H., Johnson A.M., Dorthu M.P., Matagne R.F. (1993). Further characterization of the respiratory deficient dum-1 mutation of Chlamydomonas reinhardtii and its use as a recipient for mitochondrial transformation. Mol. Gen. Genet. 236: 235–244. [DOI] [PubMed] [Google Scholar]
- Riekhof W.R., Sears B.B., Benning C. (2005). Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: discovery of the betaine lipid synthase BTA1Cr. Eukaryot. Cell 4: 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Zalokar M. (1958). Pigments and photosynthesis in a carotenoid-deficient mutant of Chlamydomonas. Nature 182: 98–100. [DOI] [PubMed] [Google Scholar]
- Sanjaya M., Miller R., Durrett T.P., Kosma D.K., Lydic T.A., Muthan B., Koo A.J., Bukhman Y.V., Reid G.E., Howe G.A., Ohlrogge J., Benning C. (2013). Altered lipid composition and enhanced nutritional value of Arabidopsis leaves following introduction of an algal diacylglycerol acyltransferase 2. Plant Cell 25: 677–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmollinger S., et al. (2014). Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 26: 1410–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey J., Browse J. (2011). Genome-level and biochemical diversity of the acyl-activating enzyme superfamily in plants. Plant J. 66: 143–160. [DOI] [PubMed] [Google Scholar]
- Shockey J.M., Fulda M.S., Browse J.A. (2002). Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 129: 1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker D.D., Lashkari D.A., Morris D., Mittmann M., Davis R.W. (1996). Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat. Genet. 14: 450–456. [DOI] [PubMed] [Google Scholar]
- Sizova I., Fuhrmann M., Hegemann P. (2001). A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277: 221–229. [DOI] [PubMed] [Google Scholar]
- Sizova I., Greiner A., Awasthi M., Kateriya S., Hegemann P. (2013). Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. Plant J. 73: 873–882. [DOI] [PubMed] [Google Scholar]
- Slaninová M., Hroššová D., Vlček D., Wolfgang W. (2008). Is it possible to improve homologous recombination in Chlamydomonas reinhardtii? Biologia 63: 941–946. [Google Scholar]
- Strizhov N., Li Y., Rosso M.G., Viehoever P., Dekker K.A., Weisshaar B. (2003). High-throughput generation of sequence indexes from T-DNA mutagenized Arabidopsis thaliana lines. Biotechniques 35: 1164–1168. [DOI] [PubMed] [Google Scholar]
- Tam L.W., Lefebvre P.A. (1993). Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics 135: 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima M., Freeman E.S., Jinkerson R.E., Jonikas M.C. (2015). A fluorescence-activated cell sorting-based strategy for rapid isolation of high-lipid Chlamydomonas mutants. Plant J. 81: 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima M., Specht M., Hippler M. (2011). The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr. Genet. 57: 151–168. [DOI] [PubMed] [Google Scholar]
- Toepel J., Illmer-Kephalides M., Jaenicke S., Straube J., May P., Goesmann A., Kruse O. (2013). New insights into Chlamydomonas reinhardtii hydrogen production processes by combined microarray/RNA-seq transcriptomics. Plant Biotechnol. J. 11: 717–733. [DOI] [PubMed] [Google Scholar]
- Tsai C.H., Warakanont J., Takeuchi T., Sears B.B., Moellering E.R., Benning C. (2014). The protein Compromised Hydrolysis of Triacylglycerols 7 (CHT7) acts as a repressor of cellular quiescence in Chlamydomonas. Proc. Natl. Acad. Sci. USA 111: 15833–15838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin F., Cross F.R. (2014). A microbial avenue to cell cycle control in the plant superkingdom. Plant Cell 26: 4019–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen J.G. (2011). Evolution of sex and mating loci: an expanded view from Volvocine algae. Curr. Opin. Microbiol. 14: 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbański D.F., Małolepszy A., Stougaard J., Andersen S.U. (2012). Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. Plant J. 69: 731–741. [DOI] [PubMed] [Google Scholar]
- Urzica E.I., Casero D., Yamasaki H., Hsieh S.I., Adler L.N., Karpowicz S.J., Blaby-Haas C.E., Clarke S.G., Loo J.A., Pellegrini M., Merchant S.S. (2012). Systems and trans-system level analysis identifies conserved iron deficiency responses in the plant lineage. Plant Cell 24: 3921–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urzica E.I., Vieler A., Hong-Hermesdorf A., Page M.D., Casero D., Gallaher S.D., Kropat J., Pellegrini M., Benning C., Merchant S.S. (2013). Remodeling of membrane lipids in iron-starved Chlamydomonas. J. Biol. Chem. 288: 30246–30258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney G.K., et al. (2013). A large-scale zebrafish gene knockout resource for the genome-wide study of gene function. Genome Res. 23: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K.J., Schulze K.L., Haelterman N.A., Pan H., He Y., Evans-Holm M., Carlson J.W., Levis R.W., Spradling A.C., Hoskins R.A., Bellen H.J. (2011). MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8: 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss B., Meinecke L., Kurz T., Al-Babili S., Beck C.F., Hess W.R. (2011). Hemin and magnesium-protoporphyrin IX induce global changes in gene expression in Chlamydomonas reinhardtii. Plant Physiol. 155: 892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Duanmu D., Spalding M.H. (2011). Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii: inorganic carbon transport and CO2 recapture. Photosynth. Res. 109: 115–122. [DOI] [PubMed] [Google Scholar]
- Wang Z.T., Ullrich N., Joo S., Waffenschmidt S., Goodenough U. (2009). Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 8: 1856–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng H., Molina I., Shockey J., Browse J. (2010). Organ fusion and defective cuticle function in a lacs1 lacs2 double mutant of Arabidopsis. Planta 231: 1089–1100. [DOI] [PubMed] [Google Scholar]
- Wilfling F., et al. (2013). Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell 24: 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R., Stiffler N., Belcher S., Kroeger T., Stern D.B., Monde R.A., Coalter R., Barkan A. (2010). Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant J. 63: 167–177. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Wirschell M., Zhao F., Yang C., Yang P., Diener D., Gaillard A., Rosenbaum J.L., Sale W.S. (2008). Building a radial spoke: flagellar radial spoke protein 3 (RSP3) is a dimer. Cell Motil. Cytoskeleton 65: 238–248. [DOI] [PubMed] [Google Scholar]
- Yang P., Diener D.R., Yang C., Kohno T., Pazour G.J., Dienes J.M., Agrin N.S., King S.M., Sale W.S., Kamiya R., Rosenbaum J.L., Witman G.B. (2006). Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 119: 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., et al. (2014). Alternative acetate production pathways in Chlamydomonas reinhardtii during dark anoxia and the dominant role of chloroplasts in fermentative acetate production. Plant Cell 26: 4499–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K., Han D., Li Y., Sommerfeld M., Hu Q. (2012). Phospholipid:diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell 24: 3708–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li C., Wu C., Xiong L., Chen G., Zhang Q., Wang S. (2006). RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Res. 34: D745–D748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Patena W., Armbruster U., Gang S.S., Blum S.R., Jonikas M.C. (2014). High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell 26: 1398–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Katavic V., Li F., Haughn G.W., Kunst L. (2010). Insertional mutant analysis reveals that long-chain acyl-CoA synthetase 1 (LACS1), but not LACS8, functionally overlaps with LACS9 in Arabidopsis seed oil biosynthesis. Plant J. 64: 1048–1058. [DOI] [PubMed] [Google Scholar]
- Zhao T., Wang W., Bai X., Qi Y. (2009). Gene silencing by artificial microRNAs in Chlamydomonas. Plant J. 58: 157–164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.