Asexually propagated potato shows greater copy number variation compared with sexually propagated plant species, with a strong connection to environmental response pathways.
Abstract
Clonally reproducing plants have the potential to bear a significantly greater mutational load than sexually reproducing species. To investigate this possibility, we examined the breadth of genome-wide structural variation in a panel of monoploid/doubled monoploid clones generated from native populations of diploid potato (Solanum tuberosum), a highly heterozygous asexually propagated plant. As rare instances of purely homozygous clones, they provided an ideal set for determining the degree of structural variation tolerated by this species and deriving its minimal gene complement. Extensive copy number variation (CNV) was uncovered, impacting 219.8 Mb (30.2%) of the potato genome with nearly 30% of genes subject to at least partial duplication or deletion, revealing the highly heterogeneous nature of the potato genome. Dispensable genes (>7000) were associated with limited transcription and/or a recent evolutionary history, with lower deletion frequency observed in genes conserved across angiosperms. Association of CNV with plant adaptation was highlighted by enrichment in gene clusters encoding functions for environmental stress response, with gene duplication playing a part in species-specific expansions of stress-related gene families. This study revealed unique impacts of CNV in a species with asexual reproductive habits and how CNV may drive adaption through evolution of key stress pathways.
INTRODUCTION
Cultivated potato (Solanum tuberosum) comprises a unique plant species (Gavrilenko et al., 2013; Hirsch et al., 2013; Uitdewilligen et al., 2013), consisting primarily of diverse diploid and tetraploid subspecies that can harbor introgressions from various wild populations (Hawkes, 1990; Spooner et al., 2007). Varieties and landraces are maintained as clones in vitro or by collection and planting of seed tubers, yielding significant potential for accumulating somatic mutations in the genome. The most widely grown variety in North America, Russet Burbank, has been maintained clonally for over 100 years and was itself selected as a somatic mutant of an older variety. The asexual and highly heterozygous nature of potato offers a unique model to examine genome variation compared with homozygous, or seed-propagated, plants, such as Arabidopsis thaliana, soybean (Glycine max), and maize (Zea mays). Without routine meiotic events imposing purifying selection at each generation (Simko et al., 2006), mutations have the potential to be retained at higher levels than in species tolerant of inbreeding and are more likely mitotic in origin. The mutation load in cultivated backgrounds is extremely high (Xu et al., 2011), demonstrated by low fertility in elite clones and severe inbreeding depression observed during selfing (De Jong and Rowe, 1971).
Sequence-level mutations, including single nucleotide polymorphisms (SNPs) and small insertions/deletions, have been widely investigated in several plant species (Morrell et al., 2011). With respect to structural variation, recent genome-wide surveys using array and sequencing technologies have revealed copy number variants and presence/absence variants from hundreds to millions of bases in length are prevalent in plants and animals (Abecasis et al., 2012; Żmieńko et al., 2014), supporting their importance as components of genome diversity in eukaryotes. A growing body of evidence now suggests they play a key role underlying phenotypic diversity. While often associated with likelihood of genetic disorders in mammals (Weischenfeldt et al., 2013), copy number variation (CNV) has been shown to benefit adaptive traits in plants, such as daylength neutrality in wheat (Triticum aestivum; Díaz et al., 2012), and is speculated to be an underlying component of hybrid vigor (Lai et al., 2010). At the functional level, CNV has also been linked to genes involved in stress responses, such as submergence tolerance in rice (Oryza sativa; Xu et al., 2006; Hattori et al., 2009), nematode resistance in soybean (Cook et al., 2012), and aluminum tolerance in maize (Maron et al., 2013). While genome-wide structural variation studies in maize (Chia et al., 2012), soybean (Lam et al., 2010), and Arabidopsis (Cao et al., 2011) have shown that CNV patterns are widespread and exhibit different frequency among sexually reproducing plant species, the impact of structural variation on genome and phenotypic diversity has yet to be explored in any clonally propagated plant.
The richest source of genomic variation for S. tuberosum exists among its native South American progenitors (Ortiz, 2001). SNPs derived from elite North American cultivars show greater variation among South American landraces than modern clones and their wild relatives (Hardigan et al., 2015), demonstrating the diversity in native populations of cultivated potato. Unlike sequence level mutation, the contribution of structural variation to this diversity remains undetermined in this clonally propagated plant species. Limited CNV analyses performed at the cytogenetic level (Iovene et al., 2013) with select BAC-sized regions showed large tracts of the potato genome (>100 kb) are commonly absent from multiple homologous chromosomes of autotetraploids, supporting extensive genome plasticity.
We present an analysis of structural variation in diploid S. tuberosum, an asexually reproducing and obligate outcrossing species, based on next-generation sequencing. This study examined a panel of 12 monoploid/doubled monoploid clones derived from native South American landrace populations, selected for their rare, nonlethal introduction of full homozygosity into this highly heterozygous genome. This panel reflected more structural variation within 12 related S. tuberosum clones than previous plant studies encompassing much larger data sets, suggesting greater tolerance of mutation in populations of asexually reproducing species. The underlying causes could be masking of dysfunctional and deleterious alleles in a heterozygous state and an inability to purge deleterious alleles via meiosis. Thousands of CNVs including duplications, deletions, and presence/absence variation (PAV) were identified in all clones, including those closely related to the reference genotype, with variants larger than 100 kb frequently observed in pericentromeric regions. As these homozygous clones were capable of growth and development ex vitro, we were able to annotate many dispensable genes and estimate the core gene set required for survival. While we observed a low frequency of deletions in genes encoding functions conserved across angiosperms, CNV was shown to be closely associated with loci involved in stress tolerance, supporting the concept of an adaptive role for gene duplication in diversification of plant environmental responses. Finding that nearly half the genes specific to the potato lineage were impacted by duplication or deletion reinforced the connection between CNV and evolution of novel genes at the species level.
RESULTS
Generation of a Monoploid Panel
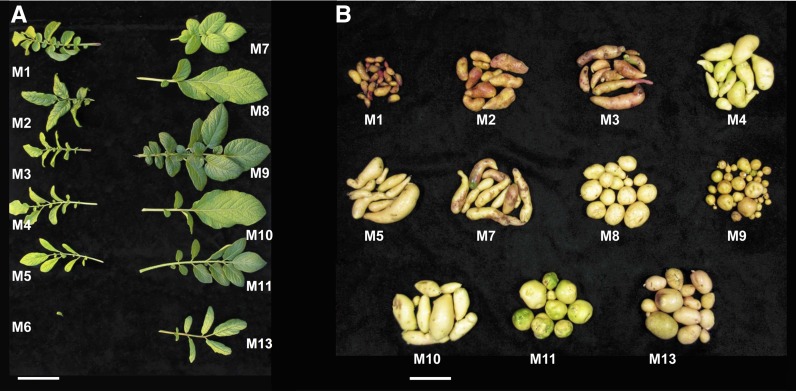
Diploid potato landraces are the progenitors of modern tetraploids, being native to the Andes Mountains of South America and existing as heterozygous populations used in breeding new varieties (Ortiz, 2001; Spooner et al., 2007). A panel of 12 monoploid and doubled monoploid clones (referred to as “monoploids” for simplicity) (Table 1) were generated via anther culture using germplasm primarily composed of S. tuberosum Group Phureja landraces with limited introgression of Group Stenotomum, Group Tuberosum, and Solanum chacoense backgrounds. Clones were derived from three maternal landrace populations randomly pollinated by diploids from a photoperiod-adapted research population (Supplemental Figure 1) (Haynes, 1972). Four clones (M1, M9, M10, and M11) were direct products of landrace family crosses, while others (M2, M3, M6, M7, and M8) were subsequently generated in combination with heterogeneous breeding stocks harboring limited introgression from dihaploids of cultivated tetraploid potato (S. tuberosum Group Tuberosum) or wild S. chacoense. M13 alone was an interspecific hybrid, with introgressions from S. chacoense. Three clones (M2, M3, and M7) were derived from backcross (BC1) progeny of the doubled monoploid Group Phureja clone DM1-3 516 R44 (hereafter referred to as DM) used to generate the potato reference genome (Xu et al., 2011), offering reference points as closely related germplasm. These clones were selected for introduction of full homozygosity into a naturally heterozygous genome, without lethality and with limited floral or tuber developmental defects (Figure 1). Floral phenotype was affected in several clones; M2 and M10 displayed fused stamen and carpel whorls and M13 lacked stamens entirely. M3 and M5 showed premature abortion of flower buds, although occasionally wild-type flowers were produced. M6 alone did not flower, rarely produced a few small tubers (<0.5 cm) with no plant yielding more than 0.5 g, and showed dramatic reduction in whole plant vigor, suggesting deleterious mutation of core genes. Hence, while several clones demonstrated morphological defects as a result of significant mutation load, all but M6 were able to mature and initiate tuber and floral development and therefore represent the minimal gene set required for development and reproduction of cultivated potato.
Table 1. Summary of Genetic Background Composition, Sequencing Data, and Variant Calls Associated with Clones in the Monoploid Panel.
| Genetic Background (%) | Variant Counts |
|||||||
|---|---|---|---|---|---|---|---|---|
| Clone | Phurejaa | Tuberosumb | Wildc | Ploidyd | CNVs (Total) | Duplications | Deletions | SNPs |
| DM | 100 | 0 | 0 | 2x | 0 | 0 | 0 | 0 |
| M1 | 100 | 0 | 0 | 1x | 8,837 | 2,577 | 6,260 | 3,433,063 |
| M2 | 92 | 5 | 3 | 1x | 4,996 | 1,565 | 3,431 | 1,557,476 |
| M3 | 92 | 5 | 3 | 1x | 2,978 | 897 | 2,081 | 800,333 |
| M4e | >50 | – | – | 1x | 8,424 | 2,572 | 5,852 | 3,242,070 |
| M5e | >50 | – | – | 1x | 9,194 | 2,887 | 6,307 | 3,664,157 |
| M6 | 85 | 9 | 6 | 1x | 8,627 | 2,864 | 5,763 | 3,632,667 |
| M7 | 92 | 8 | 0 | 1x | 4,062 | 1,222 | 2,840 | 1,186,135 |
| M8 | 92 | 8 | 0 | 1x | 8,716 | 2,617 | 6,099 | 3,625,031 |
| M9 | 100 | 0 | 0 | 1x | 8,496 | 2,703 | 5,793 | 3,989,158 |
| M10 | 100 | 0 | 0 | 2x | 8,640 | 2,645 | 5,995 | 3,718,500 |
| M11 | 100 | 0 | 0 | 2x | 8,962 | 2,639 | 6,323 | 3,648,940 |
| M13e | ∼40–50 | ∼0–10 | 50 | 1x | 10,532 | 3,468 | 7,064 | 4,764,182 |
Genetic input from diploid South American landrace populations of S. tuberosum Groups Phureja and Stenotomum.
Genetic input from dihaploids of S. tuberosum Group Tuberosum (tetraploid cultivated potato).
Genetic input from S. chacoense, a diploid wild species sexually compatible with cultivated potato species.
Ploidy is reported from initial flow cytometry results; several clones spontaneously doubled in culture (M1, M5, M7, M8, and M9).
Direct or indirect product of somatic fusions from diverse germplasm with primarily diploid landrace background.
Figure 1.
Phenotypic Variation in a Homozygous Potato Panel.
Leaf (A) and tuber (B) variation observed in the monoploid panel. M6 tubers are not available. Bars = 5 cm.
Sequencing and Variant Detection
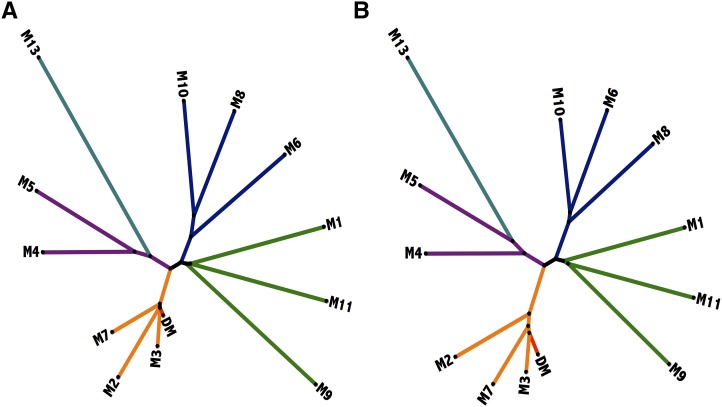
Genome resequencing was conducted to provide coverage of 30-69x for comprehensive SNP and CNV analysis in the monoploid panel (Supplemental Table 1). We aligned reads to an improved version of the DM potato reference genome (v4.04; see Methods) that includes 55.7 Mb of previously unassembled sequence. The DM v4.04 assembly was repeat-masked to limit analysis of structural variation to low-copy sequence. The number of SNPs relative to DM ranged from 800,333 in M3 to 4,764,182 in M13 (Table 1), reflective of the pedigree relationships between the clones and reference genotype (Supplemental Figure 1). To confirm SNP calling accuracy, we compared variant calls from read alignments of 10 clones to variant calls generated using the Infinium 8303 potato array (Felcher et al., 2012), resulting in 98.5% concordance. Of the SNPs, 2.4 to 4.4% were located in coding regions and 70.1 to 75.7% were intergenic, with 0.67 to 0.84 ratios of synonymous to nonsynonymous changes in coding SNPs (Supplemental Table 2). A SNP phylogeny measuring genetic distance between the monoploids closely supported their known pedigrees (Figure 2A).
Figure 2.
Phylogenetic Trees of Monoploid Panel Clones Including the DM Reference Genotype.
Branch colors indicate genetic background of clones; DM reference genotype (red; DM), backcross progeny of DM (orange; M2, M3, and M7), direct progeny of nonreference landrace populations (green; M1, M9, and M11), landraces containing introgressions from non-landrace germplasm (blue; M6, M8, and M10), descended from intercrossed somatic hybrids (purple; M4 and M5), and wild/landrace interspecific hybrid (turquoise; M13).
(A) Tree based on 12 million genome-wide SNP markers.
(B) Tree based on copy number status of potato genes relative to the DM reference annotation.
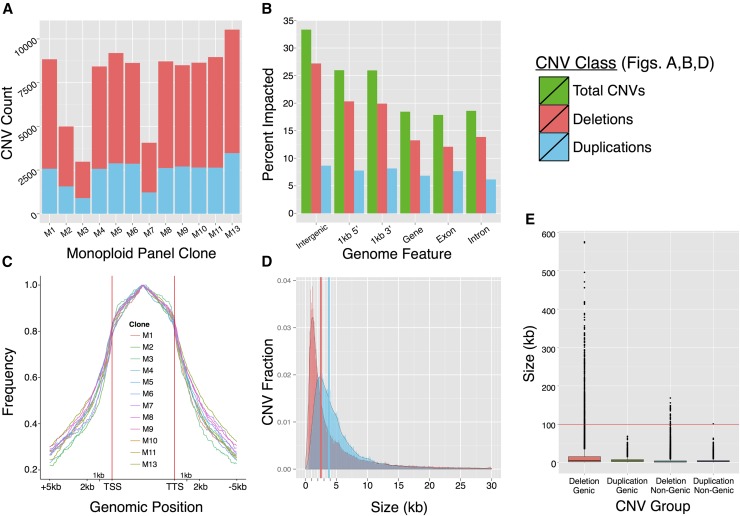
Copy number variant detection was implemented in 100-bp genomic windows using CNVnator (Abyzov et al., 2011). With read depth coverage of 30-69x per clone (Supplemental Table 1), CNV detection, breakpoint precision, and copy number accuracy were well supported. For this analysis, CNVs were defined as duplications when exhibiting more copies relative to the reference genome or deletions if containing fewer copies than the reference. Several thousand CNVs were called in each monoploid ranging from 500 bp (minimum length) to 575 kb, with total CNV calls per individual varying from 2978 to 10,532 (Table 1, Figure 3A; Supplemental Table 3), indicating a wide range of structural variation among the clones and the reference genome. We compared CNVnator calls to those derived using a read depth method similar to other published plant CNV studies (Cao et al., 2011; Xu et al., 2012). For the 12 clones, we observed 95 and 84% support of total CNVnator deletion and duplication calls, respectively, by the read depth method (Supplemental Table 4). CNVnator was significantly more conservative in calling CNVs; few calls were unique to CNVnator (range of 0.6 to 1 Mb for deletions and 1.3 to 2.4 Mb for duplications), whereas the read depth method generated substantially more unique variant calls (range of 79 to 151 Mb for deletions and 37 to 120 Mb for duplications). PCR validation supported 100 and 74% of the predicted copy number variants (46 target deletions and 42 target duplications) for primer pairs in which a single product of the predicted size was observed in both the reference genotype DM and at least one clone predicted to be single copy at that locus (Supplemental Figure 2). The lack of full concordance between the computational predictions and the experimental validation results are due in part to technical limitations including sequence divergence in the primer binding sites between the clones as indicated by an inability to amplify the target locus in all variant and nonvariant clones and insertions/deletions within the target amplification regions observed across the panel (Supplemental Figure 2). Based on the concordance observed both with read depth estimations and experimental results, we feel that CNVnator provides a robust assessment of structural variation within our panel.
Figure 3.
Summary Statistics of Monoploid Panel CNVs.
(A) Frequency of CNV per clone. The total number of filtered duplications (blue) and deletions (red) for each clone.
(B) CNV representation within potato genome features. The percentage of sequence classes impacted by duplication and deletion in the monoploid panel. The number of CNVs is nonadditive due to overlap between duplication and deletion regions.
(C) Distribution of CNV frequency (per clone) relative to position of all duplicated genes (required minimum 50% gene model overlap with a duplicated sequence).
(D) CNV size distribution. Relative frequency of all CNV sizes up to 30 kb. Solid lines indicate median size for duplications and deletions.
(E) Box plot of size of CNV for genic and non-genic duplications and deletions.
Like SNPs, CNV rates reflected the expected divergence of clones from the DM reference genotype. The greatest extent of CNV was observed in M13, a hybrid of landrace diploids and wild S. chacoense, and therefore was most likely to show different patterns of genome evolution. By contrast, backcross progeny of the DM reference genotype (M2, M3, and M7) exhibited lower CNV frequencies, although several thousand CNVs were found in each clone. To assess the ability of the CNV calls to reflect genetic relationships in the monoploid panel, we generated a second phylogeny based on gene level CNV (see below) using copy status (duplicated, deleted, and non-CNV) as allelic states for annotated reference genes. The resulting CNV tree closely reflected relationships estimated using SNPs (Figure 2B). This demonstrated the CNV calls were accurate at the gene level and that, like SNPs, they can effectively predict genetic relationships, supporting previous findings that CNVs are shared across accessions and reflect natural population structure (Cao et al., 2011).
Extent and Distribution of CNV in the Diploid Potato Genome
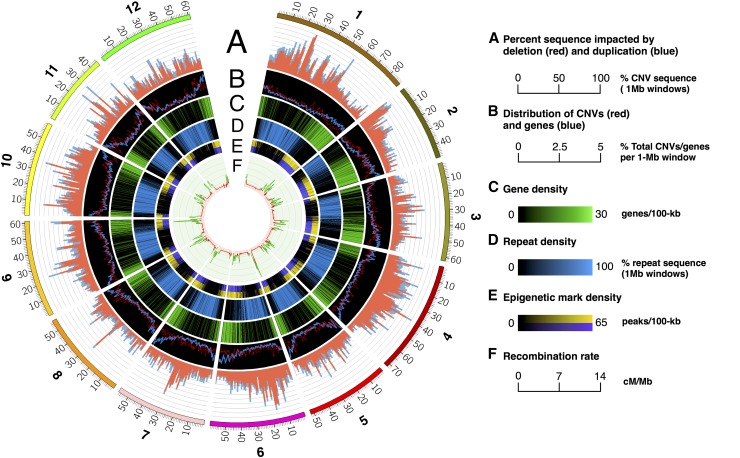
A total of 92,464 CNVs were identified in the panel (Supplemental Data Set 1), collectively impacting 30.2% of non-gap sequence in the DM v4.04 reference genome. Many CNVs were conserved among the clones, sharing close breakpoints or corresponding to identical regions. Ratios of duplication and deletion were highly conserved, with duplications comprising 29.2 to 33.2% of total CNVs per clone. Similar bias in detection of deletions has been observed in previous comparative genomic hybridization and next generation sequencing-based studies (Żmieńko et al., 2014). Structural variation was most common in intergenic sequence and on a genome scale was often more prevalent in pericentromeric regions with lower frequency observed in the gene-dense euchromatic arms, particularly in regions with high rates of recombination (Figure 4). This is consistent with a comprehensive examination of CNV in humans where CNV was enriched within pericentromeric regions (Lu et al., 2015; Zarrei et al., 2015). In maize, as shown using genotyping-by-sequencing, PAVs were enriched in the pericentromere (Lu et al., 2015) and negatively correlated with recombination rate, whereas a transcript-based PAV study (Hirsch et al., 2014) revealed PAVs were distributed throughout the maize genome with a lower frequency in pericentromeric regions. Thus, structural variation may differ for genic versus nongenic segments of a genome and our detection of CNV enrichment in the pericentromere reflects the use of whole genome resequencing data to assess structural variation.
Figure 4.
Chromosomal Distribution of CNVs, Genes, Repetitive Sequence, and Recombination Rates in the Diploid Potato Genome.
(A) Percentage of total non-gap sequence (0 to 100%) impacted by deletion (red) and duplication (blue) in 1-Mb nonoverlapping windows.
(B) Distribution of CNV counts (red) and gene counts (blue) (% total chromosome count in 1-Mb bins, 0.2-Mb step size).
(C) Gene density (genes per 1-Mb window, 0.2-Mb step size).
(D) Repeat density (% repetitive sequence in 1-Mb windows, 0.2-Mb step size).
(E) Heat map of gene activating histone mark density (peaks per 1-Mb window, 0.2-Mb step size; yellow = H3K4me2 and purple = H4K5ac).
(F) Recombination rate (0 to 14 cM/Mb) based on a biparental F1 mapping population (Manrique-Carpintero et al., 2015).
The frequency of bases impacted by duplication was only slightly reduced (∼1.8%) in genes compared with intergenic space (Figure 3B; Supplemental Data Set 2). By comparison, rates of deletion were reduced in gene flanking sequence and 15% lower in coding sequence, suggesting a degree of selection against deleterious impacts on gene function (Figure 3B). While total gene sequence displayed similar rates of duplication and less deletion than whole-genome sequence, genes that were impacted by CNV (minimum 50% gene model overlap) showed signs of nonrandom targeting by CNV mechanisms. These genes displayed peak CNV frequencies within their gene bodies and a marked decrease of CNV frequency in the sequences bordering their 5′ and 3′ ends (Figure 3C; Supplemental Figure 3). The reduced impact of CNV on overall coding sequence may result from selection against deleterious effects on expression of core gene functions, supported by a more substantial disparity in deletion compared with duplication rates with duplications being less likely to impair gene function.
Large Structural Variants Are Common in Potato
Copy number variants were typically several kilobases or smaller, with a 3.0-kb median size in the panel (Figure 3D). Duplications (median 3.8 kb) tended to be larger than deletions (median 2.5 kb), although the fraction of CNVs represented by duplication diminished at larger size ranges (Supplemental Figure 4). Size distribution was highly conserved among clones in the panel, suggesting similar patterns of formation and retention in the population (Supplemental Figure 5).
Large-scale structural variation was also found to impact the diploid potato genome. A subset of variants was greater than 100 kb in length, the largest reaching 575 kb and present in clones M2 and M8, which lacked a known relationship. These CNVs (619 corresponding to 233 distinct regions) comprised 0.67% of total calls and were almost exclusively deletions (99.8%), which accounted for the majority of outlier CNV sizes (Figure 3E). Large CNVs may arise from different mechanisms than smaller, more common variants. Most CNVs are several kilobases or less, potentially resulting from nonallelic homologous recombination in regions containing segmental homology (Lu et al., 2012) or in regions without low-copy repeats as a result of microhomology and replication errors (Stankiewicz and Lupski, 2010; Arlt et al., 2012). Other CNVs may arise from retrotransposon activity, a common driver of structural variation in grass genomes (Morgante et al., 2007). However, a study of BAC-level (100 kb+) CNV in potato showed CNVs of this size are not segmental variants (Iovene et al., 2013), instead showing presence/absence across clones or between homologous chromosomes within a clone. BAC-sized regions were commonly found to be missing on one to three homologous chromosomes of autotetraploids (Iovene et al., 2013). These variants likely correspond to the large CNVs identified in this study based on read depth, supporting the near exclusive detection of large CNVs as deletions in the monoploid panel. Large regions of the reference genome absent in the panel appear as deletions, while clone-specific regions not present in the DM v4.04 assembly are undetectable by read depth, requiring independent assembly as PAVs.
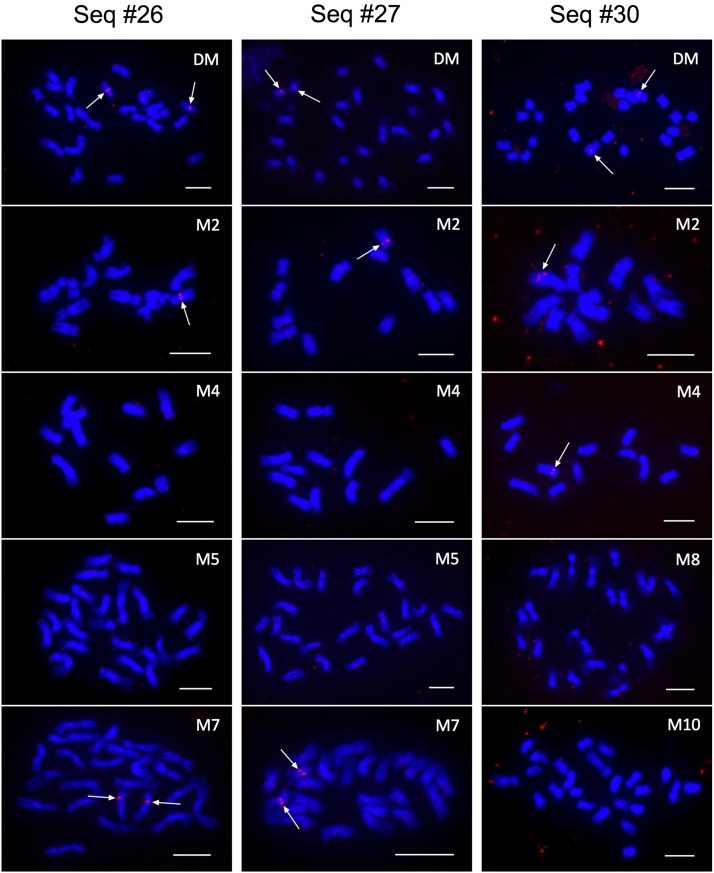
To confirm the computational identification of these large CNVs, we performed fluorescence in situ hybridization (FISH) of three selected large CNVs (Seq26, Seq27, and Seq30), which span 105, 137.6, and 102.9 kb, respectively. Seq26 and Seq27 are at 28,282,100 to 28,387,100 bp and 30,733,700 to 30,871,300 bp on chromosome 7, respectively, and Seq30 is located on 22,656,700 to 22,759,600 bp on chromosome 9. Primers were designed to amplify four to five single copy DNA fragments for each CNV locus (Supplemental Data Set 3), and DNA fragments amplified from the same CNV locus were pooled and labeled as a FISH probe. All three probes generated consistent FISH signals on a pair of DM chromosomes (Figure 5). The signals from the Seq26 and Seq27 probes were located close to the centromere of the target chromosome. In fact, most of the FISH signals overlapped with the primary constriction of the chromosome. Seq30 mapped to the middle of the long arm of its target chromosome. We then performed FISH using each probe on four monoploid/doubled monoploid clones selected based on computational prediction of presence/absence. The presence/absence of the FISH signals were concordant with the computational analysis (Figure 5) supporting our computational CNV calling method.
Figure 5.
FISH of the Reference Genotype DM and Monoploid/Doubled Monoploid Clones Using Probes Targeting CNV.
Probes designed to multiple segments within three 100-kb+ computationally predicted CNV regions (Sequence 26 [∼28.2 Mb Chromosome 7], Sequence 27 [∼30.7 Mb Chromosome 7], and Sequence 30 [∼22.7 Mb Chromosome 9]) were labeled with digoxigenin-11-dUTP (red; arrows) and hybridized to chromosomes from the reference genotype (DM) and a subset of the monoploid/doubled monoploids (M2, M4, M5, M7, M8, and M10). Chromosomes were prepared from root tip cells and were counterstained with 4′,6-diamidino-2-phenylindole (blue). Perfect concordance between the computational prediction of CNV and the FISH signals was observed. Bars = 5 μm.
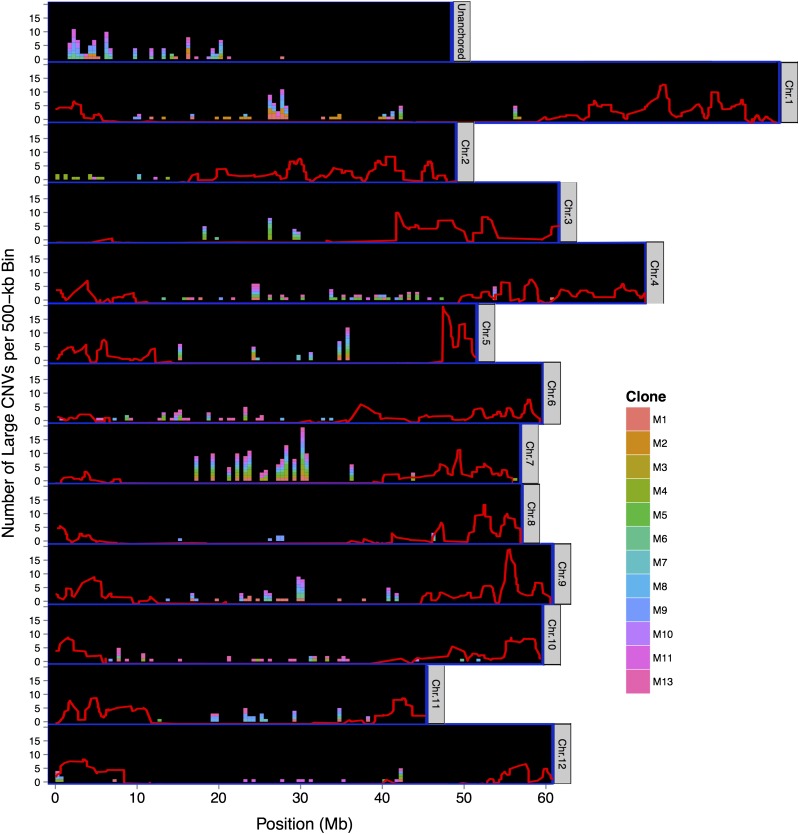
Large CNVs tended to be heterochromatic or located in the pericentromeres (Figure 6), underscoring the deleterious effects they can introduce to critical genes enriched in the euchromatic arms. Many corresponded to similar regions in different clones, with highly conserved breakpoints (Supplemental Data Set 4). Chromosomes 5 and 7 contained numerous large CNVs shared by clones lacking a recent common ancestor, with a CNV on chromosome 5 reflecting deletion of a 100-kb sequence in all clones except M3 (BC1 progeny of DM) and breakpoints conserved to within 100 bp in most clones. Such conservation in germplasm from distinct progenitors suggests these variants descend from shared ancestral CNV events. Patterns of large-scale CNV also differed among chromosomes. Chromosomes 2 and 8 contained few large deletions, most being clone specific. More than half the large CNVs on chromosome 10 were specific to the hybrid M13, reflecting greater structural variation between cultivated potato and its wild relative S. chacoense on this chromosome. Notably, the only duplication larger than 100 kb was a 6x increase of repeats in the subtelomeric region on the short arm of chromosome 12 in the hybrid M13, indicating large-scale differences in genome structure between sexually compatible wild and landrace potato species.
Figure 6.
Positions of Large (>100-kb) Copy Number Variants in the Potato Reference Genome Assembly by Counts per Clone in Nonoverlapping 500-kb Bins.
Variants are color coded for each clone. Red lines show chromosome-wide estimates of recombination frequency (cM/Mb) indicating the euchromatic arms (scale = 0 to 14 cM/Mb) (Manrique-Carpintero et al., 2015). “Unanchored” track represents all scaffolds that could not be anchored to the 12 main chromosomes.
Although large CNVs were uncommon in the euchromatic arms (Figure 6), the majority of these variants encompassed genes; 1110 genes were deleted by large CNVs, while 875 (∼81%) encoded proteins of unknown function or were associated with transposable elements (TEs). Few overlapped regulatory genes with the exception of F-box proteins, for which CNV is common in plants (Xu et al., 2009). Despite low rates of CNV impacting core gene functions, many potato genes were in fact subject to structural variation in the monoploid panel.
Role for CNV in Potato Adaptation
In total, 11,656 potato genes (29.7%) overlapped CNV calls, with 9001 genes (22.9%) affected in at least half their annotated gene model (Supplemental Data Set 5). To limit functional analysis to genes confidently affected by CNV, we used this second group to define the CNV gene set. Within the CNV gene set, ∼11% consisted of TEs, ribosomal DNA, or nuclear organellar insertions, while 48% encoded proteins of unknown function, supporting association of CNV with genes that may be dispensable. Many CNV-impacted genes were also linked to pathogen resistance and abiotic stress tolerance. Gene Ontology (Ashburner et al., 2000) associations revealed several functions significantly enriched in the CNV gene set (Supplemental Data Sets 6 and 7), and many related directly (defense response, hypersensitive response, and response to UV-B) or indirectly (flavonol and trehalose biosynthesis and calcium transport) to stress tolerance, consistent with reports of CNV impacting stress-related pathways in other plant species. CNVs have been shown to influence phenotypes including modified reproductive habits and acquired tolerance to a range of harmful environmental factors, with gene duplication conferring herbicide resistance (Gaines et al., 2010), nematode resistance (Cook et al., 2012), as well as tolerance of frost (Knox et al., 2010), submergence (Xu et al., 2006), and aluminum and boron toxicity (Sutton et al., 2007; Maron et al., 2013).
To investigate if this relationship was supported in regions of the potato genome enriched in CNV activity, we counted copy number variable genes in 200-kb windows to identify regions containing high rates of gene level CNV (Supplemental Data Set 8). Gene annotations in the 10 most highly enriched regions were examined in detail to determine functional relationship. Each contained tandem clusters of genes with conserved functions related to stress response, supporting the role of CNV in potato adaptation.
SAURs
The region most enriched for CNV genes was located on chromosome 11 at 0.83 to 1.23 Mb, containing 19 auxin-induced SAURs (small auxin-up RNA) located in tandem arrays, with 17 of 19 duplicated in at least one clone. Additional CNV-enriched clusters were found on chromosomes 1, 4, and 12. SAURs comprise a large family of auxin-induced genes that exhibit species-specific expansion in both monocots and dicots (Jain et al., 2006). A study of this gene family in Solanum identified 99 SAURs in tomato (Solanum lycopersicum) and 134 in potato, showing greater expansion in Solanum species relative to Arabidopsis, rice, and sorghum (Sorghum bicolor; Wu et al., 2012). Phylogenetic analysis revealed expansion of multiple Solanaceae-specific subgroups, with upstream regulatory sequences containing cis-elements related to auxin signaling, light signaling, drought stress, salt stress, heat shock, and calcium response, while most tomato SAURs were induced by auxin and regulated by abiotic stress (Wu et al., 2012). Diploid potato contains more SAURs than several well-annotated monocot and dicot species, including its close relative tomato. To determine if recent duplications within diploid populations contributed to the Solanum-specific expansion of SAURs seen in potato, we generated a phylogenetic tree using protein sequences of SAURs identified by Wu et al. (2012) in rice, Arabidopsis, tomato, and potato (Supplemental Figure 6). Potato SAURs displaying CNV were enriched in two large clades reflecting the most significant Solanum-specific expansions of this gene family, offering evidence for the impact of duplication on gene family diversification in these species. Our results suggest that SAURs continue to undergo duplication within closely related populations of diploid cultivated potato, highlighting the role of CNV in the rapid evolution of a gene family involved in abiotic stress response. The large number of potato genes compared with tomato in these clades, along with high rates of CNV within related Group Phureja clones, support ongoing SAUR gene expansion in potato.
Disease Resistance
The second highest density of CNV genes was found on chromosome 11 at 42.59 to 43.05 Mb, containing a cluster of 16 genes encoding nucleotide binding site leucine-rich repeat (NBS-LRR) disease resistance proteins, of which, 14 showed variation in copy number. This is consistent with previous studies conducting genetic mapping of potato resistance quantitative trait loci, showing they are often clustered in the genome (Gebhardt and Valkonen, 2001). Resistance genes are typically found in clusters or hot spots in the genomes of many plant species and are known to be fast evolving as a result of local gene duplications (Bergelson et al., 2001). Three genes conferring race-specific resistance to Phytophthora infestans (R3, R6, and R7) and a root cyst nematode resistance gene (Gro1.3) were previously mapped to this locus (Gebhardt and Valkonen, 2001). Notably, three other regions among the 10 most highly enriched for CNV genes were also disease resistance clusters, highlighting the rapid evolution of gene families required for response to changing disease pressure. These were located on chromosomes 4, 7, and 9, with the cluster on chromosome 4 corresponding to the R2 locus for late blight resistance (Gebhardt and Valkonen, 2001).
Secondary Metabolites
A third locus at ∼85 Mb on chromosome 1 contained 21 Methylketone Synthase 1 (MKS1) genes, 18 showing CNV in the panel. Methylketones are secondary metabolites produced in the glandular trichomes of solanaceous species such as tomato and potato and, in particular, their wild relatives (Bonierbale et al., 1994; Antonious, 2001). In response to insects, these compounds are secreted onto the leaf surface, conferring resistance to a variety of pests. MKS1 expression has been directly correlated with methylketone levels and leaf gland density (Fridman et al., 2005), confirming their role in defense against herbivory. Studies of its function suggest MKS1 emerged recently in its gene family and may be Solanum specific (Yu et al., 2010). Similar to patterns observed in microbial resistance genes, plant genes offering defense against insect attack may be fast evolving in order to generate new sources of genetic resistance. Their tandem clustering reflects grouping of other insect defense pathway genes in the Solanaceae, including steroidal glycoalkaloid biosynthesis (Itkin et al., 2013). Phylogenetic clustering of genes with sequence homology to the five tomato MKS1 genes showed they fall within a Solanum-specific clade containing only potato and tomato orthologs (Supplemental Figure 7). Other plants, including the asterid Mimulus guttatus, lacked close orthologs, confirming the likelihood that MKS1 function emerged recently in the genus Solanum. The Solanum-specific clade containing MKS1 also showed greater diversification in the diploid potato genome than tomato, with over twice as many potato homologs. Almost all potato MKS1 genes showed CNV in the monoploid panel, supporting a role of duplication in species-specific expansion of gene families involved in plant stress pathways.
Chromosome 9 contained 10 copies of the gene encoding desacetoxyvindoline 4′-hydroxylase (D4H), the indole alkaloid biosynthetic pathway enzyme used in synthesis of vindoline. Indole alkaloids have been associated with response to fungal elicitors, insect herbivory, and UV light exposure (St-Pierre et al., 2013), and vindoline acts as a primary substrate to form the cytotoxic chemotherapeutic vinblastine in Catharanthus roseus (Vazquez-Flota and De Luca, 1998). While this enzymatic function is not likely conserved in potato, its diversification may result in production of other defensive compounds. Another CNV-enriched locus on chromosome 5 contained a cluster of eight flavonol 4′-sulfotransferases. Flavonols, one of the most abundant classes of flavonoids in plants, have antioxidant properties and play a major role in plant response to abiotic stress, particularly UV light damage (Gill and Tuteja, 2010), and sulfate conjugation of secondary metabolites can affect their function within plant systems (Varin et al., 1997; Klein and Papenbrock, 2004). The remaining clusters contained duplicated genes encoding mannan endo-1,4-β-mannosidase and GH3 indole-3-acetic acid-amido synthetase, respectively, each with roles in cell wall modification already implicated in pathogen response (Ding et al., 2008; Westfall et al., 2010).
Association of CNV with disease resistance genes is well established in plants (Ellis et al., 2000). The extensive CNV observed in SAURs, MKS1, and other gene families in closely related germplasm suggests these are also rapidly evolving, supported by their lineage-specific expansions (Supplemental Figures 6 and 7). Whole-genome duplication is proposed to be a mechanism supporting adaptive evolution and speciation (De Bodt et al., 2005). It appears local gene duplication introduces similar potential for diversification and subfunctionalization in potato. Our finding that the most highly enriched CNV clusters harbor genes implicated in biotic and abiotic stress response furthers the hypothesis that evolution through local gene duplication can be adaptive, allowing plants to develop genetic resistance to changing environmental pressure from pests, disease, and abiotic stress such as drought.
Gene Expression as a Predictor of CNV
Gene-level CNV revealed an association with stress-related functions, as well as TEs and proteins of unknown function, some of which may not be essential for development. We investigated whether gene expression patterns support this connection, using an atlas of RNA-seq libraries representing a tissue series, as well as abiotic and biotic stress treatments for the DM reference genotype (Xu et al., 2011), to categorize the potato gene set into expression classes (Supplemental Table 5). The frequency of genes in each expression class was compared in the duplicated and deleted versus non-CNV gene sets on a per clone basis to determine how gene expression relates to likelihood of CNV. Classes included confidently expressed genes (fragments per kilobase per million mapped reads (FPKM) ≥ 10 for multiple tissue types), lowly expressed genes (FPKM < 1 in all tissues), and genes showing response to hormone or stress treatments (5-fold FPKM induction). Abiotic stress treatments included salt, mannitol, drought, abscisic acid (ABA), and heat, while biotic stress treatments included P. infestans, benzothiadiazole (salicylic acid analog), and β-aminobutyric acid (jasmonic acid analog). Hormone treatments included auxin, cytokinin, ABA, and gibberellic acid.
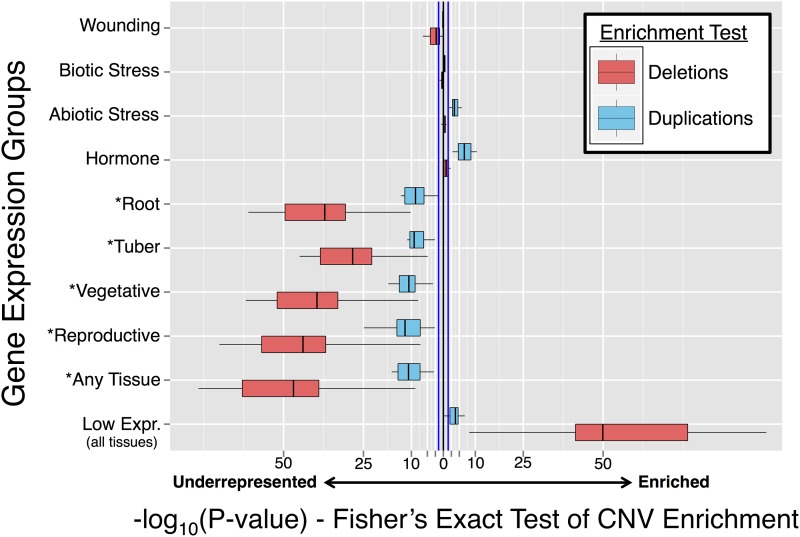
Genes with expression induced by at least one form of abiotic stress or hormone treatment were significantly enriched among duplications (P ≤ 0.05; Figure 7), supporting the relationship of duplication with genes involved in environmental response and adaptation. Individual abiotic stress treatments were unequally represented; salt-induced genes were most prevalent in the duplicated gene set, followed by drought-induced genes (Supplemental Figure 8). Mannitol, heat, and ABA-responsive genes were more common among duplicated genes, but less significantly (P ≤ 0.05). For hormone-responsive genes, those induced by cytokinin were more significantly duplicated than any other stress or hormone induced class. Biotic stress response classes (induced by P. infestans, benzothiadiazole, and β-aminobutyric acid) were not significantly enriched or underrepresented in either CNV group (Figure 7). While plant defense genes are known to be fast-evolving (Ellis et al., 2000), classic NBS-LRR disease resistance genes are lowly expressed and not typically induced by pathogen or elicitor treatment. Genes induced by wounding that mimic herbivory were significantly underrepresented among deletions in most clones (Figure 7), suggesting selection against loss of genes required for response to physical stress.
Figure 7.
Representation of Genes from Various Expression Groups in the Duplicated and Deleted Gene Sets Relative to Genes Not Impacted by CNV.
Scores are based on negative log-10 transformation of P values from a Fisher’s exact test of count data, with enrichment indicating increased representation in the copy number variant gene sets and underrepresentation indicating lower prevalence in the CNV gene sets. Blue lines indicates significance threshold (P = 0.05). An asterisk denotes confidently expressed genes as defined as having a FPKM value > 10.
Expression analysis further supported the association of CNV with dispensable genes and selection against impacting core functions. Genes with low expression in all tissues were highly enriched in the deleted gene set and to a lesser extent in duplicated genes (Figure 7), suggesting low selection against mutation. The mean representation of lowly expressed genes in the deleted set was 56.4% per clone, higher in non-CNV genes (29.1%), or the frequency of weakly expressed genes in the DM reference genome (30.4%). Genes with high expression levels in any major tissue category (aboveground vegetative, reproductive, root, and tuber) were strongly underrepresented among duplications and deletions, reflecting the greater likelihood of highly expressed genes serving core functions (Figure 7). These genes were less likely to experience deletion than duplication, reinforcing its greater potential for deleterious effect. For each major tissue type (leaves, flowers, roots, tubers, and whole in vitro plant) CNV rates became lower at increasing FPKM levels, with strong correlation across tissues (Supplemental Figure 9). Consistent with expression data, we observed that two histone marks associated with permissive transcription (H3K4me2 and H4K5,8,12,16ac) in DM leaves and tubers were preferentially associated with genes not impacted by CNV (Holoch and Moazed, 2015), while CNV frequency was increased in genes lacking one or both activating marks (Table 2).
Table 2. Extent of CNV for DM Reference Genes Associated with Transcription-Activating Histone Marks.
| Percentage of Genes Impacted by CNVa |
|||||
|---|---|---|---|---|---|
| DM Histone Markb | Total Genes | Non-CNV | Total CNV | Duplicated | Deleted |
| H3K4me2-leaf | 24,637 | 86.1 | 13.9 | 6.4 | 8.7 |
| H3K4me2-tuber | 6,206 | 75.4 | 24.6 | 10.8 | 16.3 |
| H4K5ac-leaf | 11,974 | 90.9 | 9.1 | 4.4 | 5.6 |
| H4K5ac-tuber | 22,344 | 87.6 | 12.4 | 5.8 | 7.8 |
| No leaf Mark | 14,316 | 61.4 | 38.6 | 11.8 | 30.3 |
| No tuber Mark | 14,531 | 62.1 | 37.9 | 11.6 | 29.6 |
| No activating Mark | 11,975 | 59.0 | 41.0 | 11.8 | 32.7 |
Values indicate the percent of genes in the DM reference affected by CNV as observed in the monoploid panel.
Genes were required to share 50% gene model overlap with a histone mark for association.
Core and Dispensable Gene Set
Genome resequencing studies have revealed plant and animal species contain core sets of genes required for growth and development, as well as dispensable genes that are missing in individuals (Li et al., 2010; Hirsch et al., 2014), leading to the concept of the pan-genome. Dispensable genes have been speculated to be involved in heterosis in outcrossing species (Lai et al., 2010; Ding et al., 2012) and stress adaptation (DeBolt, 2010; Żmieńko et al., 2014) and are thought to contribute to species diversification and development of novel gene functions (Wang et al., 2006). Thousands of deleted genes were identified in the monoploid panel. Despite an abundance of missing genes, each homozygous clone (except M6) was able to flower and tuberize (Figure 1), suggesting they possessed the core gene set required for development and reproduction. Dispensable genes were defined as those affected by deletion in at least one flowering and tuberizing clone, with the CNV spanning at least half an exon within the gene. Of 8888 (22.6%) genes overlapping deletions among these clones, 7183 were classified as dispensable. An additional 1429 nondeleted genes were predicted to contain SNPs encoding premature stop codons, indicating at least 8612 (21.9%) genes in DM may be dispensable. We defined the core potato gene set of 30,401 genes (77.4%), as all annotated DM genes not impacted by deletion or premature stop in the study panel. As each monoploid/doubled monoploid clone had to survive the monoploid sieve (Wenzel et al., 1979) to be included in this study, we have most likely underestimated the number of haplotypes containing deleterious/dysfunctional alleles and deletions present in the progenitor diploid clones. Improvements in the cost and ease of whole-genome sequencing and assembly of heterozygous diploid and tetraploid genomes will permit refinement of the composition of the core genome of potato in the future.
M6 displayed heavily restricted vegetative growth and rare tuberization and was unable to flower, indicating clone-specific mutation(s) in the core potato gene set. We examined CNV and SNP alleles unique in M6 to identify putative genes essential for development and flowering in potato (Supplemental Data Set 9). One candidate gene was a partial deletion of the putative homolog (78% amino acid sequence identity) of Arabidopsis RADICAL-INDUCED CELL DEATH1 (PGSC0003DMG400014419), which encodes a protein that interacts with over 20 transcription factors and is required for development (Jaspers et al., 2009). In Arabidopsis, rcd1 mutants had extremely stunted phenotypes with deformed leaves, developmental defects, and inhibited flowering (Jaspers et al., 2009), similar to the M6 phenotype. M6 harbored additional clone-specific deletion of genes encoding an inhibitor of growth protein (PGSC0003DMG400011588) and a kinetochore protein involved in cell division (PGSC0003DMG400010002).
PAV represents a form of CNV in which genes lack copies in the reference but are present in nonreference individuals. To estimate the contribution of transcript-level PAV to the dispensable gene set, unmapped RNA sequences from the monoploids were pooled and assembled into putative PAV transcripts, yielding 1169 sequences with 1263 isoforms. DM genomic sequence reads were aligned to the genome and PAV transcripts to identify potential unassembled reference sequences missing from the DM v4.04 assembly. In total, 1256 putative PAVs lacking high-quality read coverage from DM were classified as true PAVs (Supplemental Data Set 10). Only 224 PAVs could be assigned a protein function. As with genes affected by CNV, many were related to TEs, resistance proteins, and proteins of unknown function (Supplemental Data Set 11). This is likely a significant underrepresentation of gene level PAV in potato, as it was based on transcripts derived from only two tissues and will fail to capture PAV transcripts expressed in other tissues or transcripts that are weakly expressed.
Evolution of Dispensable Genes
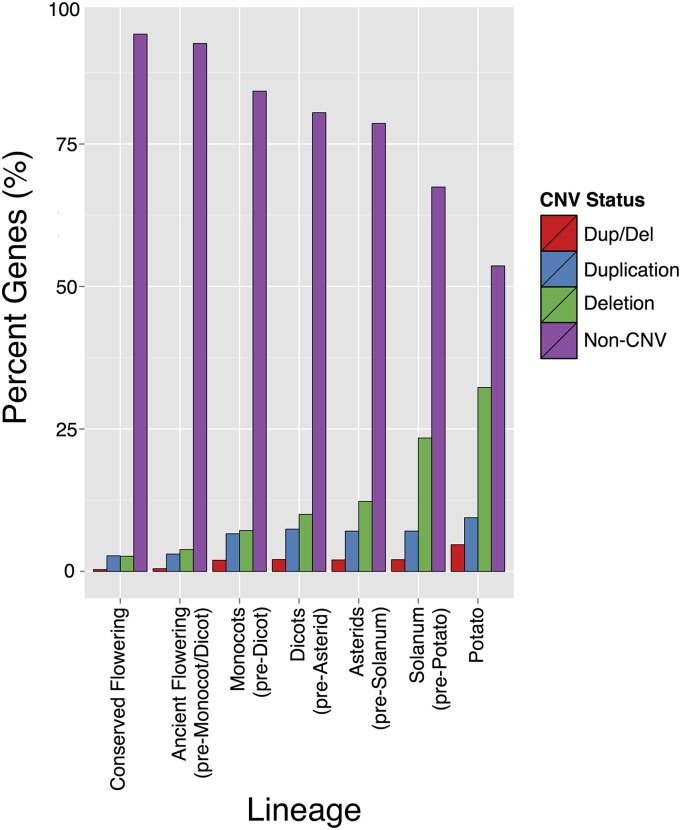
We evaluated CNV in genes arising at different levels of the potato lineage to study the origin of its dispensable genome. Orthologous gene clusters were generated for nine angiosperm species, including closely related tomato (S. lycopersicum), non-Solanaceae asterid M. guttatus, core eudicot Aquilegia coerulea, monocot rice, and the basal angiosperm Amborella trichopoda. Based on ortholog clustering, genes were classified as lineage specific in potato (3584), Solanum (11,604), asterids (12,205), and eudicots (14,892) or conserved in flowering plants (10,392) (Supplemental Figure 10). Relatively few genes (601) in potato seem to have appeared in asterids prior to separation of the genus Solanum from its other species, after which many (11,604) appeared in the Solanum lineage. Most of these genes (8020) arose before speciation of potato, whereas 3584 are potato specific. This suggests major gene diversification occurred after Solanum separated from other asterids, with further expansion at the species level in potato, possibly due to an increase in rapidly evolving genes with high rates of sequence divergence and/or a high birth/death rate in Solanum-lineage specific genes. This may explain their lack of similarity with genes of known function. CNV frequency, particularly deletion, was progressively higher in more recent lineages (Figure 8), supporting the association of dispensable genomes with recently evolved genes observed in species such as maize (Morgante et al., 2007). Genes arising in the Solanum lineage were more likely to be dispensable and 32% of potato species-specific genes were missing in at least one monoploid, whereas genes with conserved orthologs in angiosperms had extremely low rates of CNV. It is important to note the genomes used in our evolutionary analyses were annotated separately, such that genes associated with CNV may not be equally represented within the annotated proteome of each genome. However, this bias is unlikely to be large enough to explain the observed differences in variation, particularly in light of the relatively few clones needed to observe such genome variation in potato. Overall, these results support a relationship of CNV with gene diversification at the species level and highlight the potentially disruptive force of deletion, and to a lesser extent duplication, on genes serving core functions in flowering plants.
Figure 8.
CNV Frequency among Potato Genes Arising at Different Levels of the Green Plant Lineage.
“Potato” contains S. tuberosum Group Phureja species-specific genes. “Solanum” contains Solanum-specific potato genes predating potato speciation. “Asterid” contains Asterid-specific potato genes predating Solanum. “Dicots” contains eudicot-specific potato genes predating asterids. “Monocots” contains potato genes found in monocots and eudicots predating the differentiation of eudicots. “Ancient Flowering” includes all potato genes that arose before monocots. “Core Flowering” includes potato genes with orthologs in all flowering plant species.
DISCUSSION
The extent of CNV in the monoploid panel supports diploid potato possessing a greater degree of structural variation than reported in several sexually reproducing species. Overall, CNV impacted 30% of the genome and 11,656 genes, underscoring the heterogeneous nature of haplotypes within diploid potato compared with most sexually reproducing diploids. In contrast, a study on the core and dispensable gene set of soybean (G. max) explored the genomes of seven wild Glycine soja ecotypes (Li et al., 2014) with read-depth analysis, identifying only 1978 of 54,175 soybean genes (3.7%) impacted by CNV, significantly fewer than in our study. Other primarily inbreeding species, including Arabidopsis, cucumber (Cucumis sativus), and rice, also show limited structural variation relative to potato. Cao et al. (2011) resequenced 80 Arabidopsis lines from eight geographically distinct populations across Europe and Central Asia. Using a read-depth approach, 1059 CNVs (minimum length 1 kb) were identified across all lines, impacting ∼500 protein coding genes (<2%) and 2.2 Mb (∼1.6%) of the assembled genome. In a recent study including a panel of 115 cucumber accessions, fewer structural variants were discovered than in Arabidopsis (Zhang et al., 2015). A similar analysis of 50 rice accessions, including 10 wild species, detected 1327 gene loss events (2.4%) and 865 gene-associated duplications (Xu et al., 2012).
This study shows CNV is a major component of the significant genomic diversity of clonally propagated potato. Like potato, maize is another outcrossing heterozygote containing significant diversity at a structural level (Żmieńko et al., 2014), with breeders relying on heterosis as an essential component of plant vigor. Extensive CNV and PAV between maize inbreds have been speculated as components of heterosis, in which the CNV and PAVs permit complementation of missing genes and greater phenotypic diversity (Lai et al., 2010; Hansey et al., 2012). Maize contains a large pan-genome contributing to its diversity, and it is estimated that the B73 maize reference contains 74% of the low-copy gene fraction present in all inbreds (Lu et al., 2015). Chia et al. (2012) resequenced 103 maize lines, including a mixture of wild, predomesticated, and elite germplasm and concluded 32% of genes in the B73 reference were affected by CNV. In this study of 12 related clones derived from only a few native populations, ∼30% of potato genes overlapped CNVs, with ∼23% affected in over half their gene model, suggesting clonally propagated potato tolerates greater rates of mutation than many sexually reproducing species. Passage through the monoploid sieve (via anther culture) freed the panel of lethal alleles and structural variants present in their heterozygous diploid progenitors, with the clones representing rare combinations of nonlethal alleles. In comparison to maize inbreds selected for vigor and fertility, we applied much less pressure as our only selective criteria were surviving the monoploid sieve and capacity for growth in vitro. As a consequence, the spectrum of dispensable genes identified in this study may not be directly comparable with dispensable genes identified in species such as maize. However, the abundance of variants able to be retained and identified in this study implies that CNVs and other somatic mutations may be less likely to be removed from the genomes of cultivated clones.
It was observed that CNV is more likely to impact species-specific gene groups and dispensable genes, suggesting recent genome expansions in species will influence their degree of structural variation. Plants with whole-genome duplications, or genomes enlarged by TE activity such as maize (Fu and Dooner, 2002; Brunner et al., 2005), have greater potential for genes to be impacted by CNV, whether by reduced selection on duplicated coding sequences (Tang et al., 2008; Mun et al., 2009; Schnable et al., 2009) or targeting by mobile elements (Kidwell and Lisch, 1997; Slotkin and Martienssen, 2007). Low rates of sexual reproduction may also contribute to distinct patterns of structural variation, with fewer nonallelic homologous recombination events occurring during meiosis and a higher rate of nonrecurrent mitotic CNVs formed during DNA replication. This may explain the negative relationship between structural variation and recombination frequency observed on the arms of several potato chromosomes, a feature separating it from the distribution of CNV in maize (Springer et al., 2009). Gene density is also greater in the arms of potato chromosomes, such that selection against deleterious mutation in these regions could result in lower retention. Comparing structural variation within wild potato populations with higher rates of sexual reproduction and asexually propagated clones may help to elucidate the long-term impacts of asexual reproduction on plant genome variation. This study supports earlier observations of large-scale CNV in potato (Iovene et al., 2013). We can now speculate that the structural variation observed in tetraploid potato is not due to polyploidy alone because substantial genome heterogeneity is also present in diploid potato. Overall, this study adds a new dimension to our understanding of intraspecies genome variation. In contrast to sexually reproducing species such Arabidopsis and maize, where meiotic events routinely purge recessive deleterious alleles in successive generations and in which inbreeding and outcrossing may affect CNV frequency, diploid and tetraploid potato retain a heavy genetic load that remains masked due to asexual reproduction and heterozygosity.
METHODS
Germplasm
The potato clones in this study were anther-culture generated monoploids and doubled monoploids derived primarily from three accessions of a long photoperiod adapted population of diploid Solanum tuberosum Group Phureja landraces (Haynes, 1972) with limited introgression from wild Solanum chacoense and dihaploids of cultivated S. tuberosum Group Tuberosum (Supplemental Figure 1). All but M6 were able to grow under normal greenhouse conditions and produced both flowers and tubers. Ploidy is reported based on original flow cytometry analysis. Several monoploids (M1, M5, M7, M8, and M9) underwent spontaneous chromosome doubling in tissue culture since initial ploidy confirmation and are now doubled monoploids.
Improved Assembly of the Potato Reference Genome Sequence (DM v4.04)
Genomic DNA was isolated from DM stem and leaf tissue using the cetyltrimethyl ammonium bromide method, sheared to 300 bp using a Covaris ultrasonicator, end repaired, A-tailed, ligated to Illumina compatible adaptors, and PCR amplified for eight cycles. Cleaned DM genomic reads that did not map to the DM v4.03 assembly (31.5 million pairs and 1.4 million singletons; Sharma et al., 2013) were assembled into contigs using Velvet (v1.2.10) (Zerbino and Birney, 2008) using a k-mer size of 61 and minimum contig length of 200 bp. Contigs were searched against the v4.03 assembly using WUBLAST and excluded if they aligned with ≥97% identity and ≥30% coverage. Remaining contigs represented novel DM sequences absent in the v4.03 assembly (Sharma et al., 2013). These were searched using BLAST against the NCBI nr database to remove contaminants. The final, filtered contigs represent 55.7 Mb of novel DM sequence and were concatenated by order of length into a pseudomolecule “chrUn” with 500-bp gaps. The new DM v4.04 assembly is the addition of the chrUn pseudomolecule to the existing v4.03 genome assembly (Sharma et al., 2013). Contigs were annotated using the MAKER pipeline (r112) (Cantarel et al., 2008).
Monoploid and Doubled Monoploid Genomic, Transcriptomic, and Chromatin Immunoprecipitation Data Sets
DNA was isolated from monoploid and doubled monoploid leaf tissue using the Qiagen DNeasy Plant Mini kit, sheared to ∼200 bp and 600 to 700 bp using a Covaris ultrasonicator, and Illumina TruSeq libraries were constructed. For M6, Illumina compatible libraries were constructed as described above for DM. Libraries were sequenced in paired-end mode generating 100-nucleotide reads on the Illumina HiSeq platform, yielding a combined coverage of ∼30 to 69X for each clone (Supplemental Table 1). Total RNA was extracted from monoploid and doubled monoploid leaf and tuber tissues using the Qiagen RNeasy Plant Mini kit, and RNA-seq libraries were prepared using the TruSeq mRNA kit. RNA-seq libraries were sequenced in the single-end mode on the Illumina HiSeq platform generating 50-nucleotide reads, yielding 26 to 57 M reads per clone. ChIP-seq data were generated from the DM reference genotype using antibodies for two histone marks associated with transcribed genes, H3K4me2 and H4K5,8,12,16ac as previously described (Yan et al., 2008). Immunoprecipitated DNA samples from mature leaf and tuber tissue were used for library construction with the same steps as other DNA libraries (with the exception of 13 PCR cycles) and sequenced on an Illumina HiSeq in paired-end mode with 100-nucleotide reads.
Variant Calling
Whole-genome sequence and RNA-seq reads were cleaned using Cutadapt (v1.2.1) (Martin, 2011), using a minimum base quality of 10 and a minimum read length of 30 bp after trimming. The first 10 bases were trimmed from the 5′ ends of genomic DNA reads and the first base from the 5′ ends of RNA-seq to remove sequence bias. Genomic reads were mapped to the DM v4.04 potato genome assembly in paired-end mode using BWA-MEM (v0.7.8) (Li, 2013) with default parameters. Duplicates were marked using PicardTools (v1.106; http://broadinstitute.github.io/picard). GATK IndelRealigner (v2.8.1) (McKenna et al., 2010) was used to refine alignments, and SAMTools (v0.1.19) (Li et al., 2009) was used to merge the 200- and 600-bp library BAM files for downstream SNP and CNV calling. RNA-seq reads were mapped to the DM v4.04 assembly using TopHat (v1.4.1) (Trapnell et al., 2009) with minimum and maximum intron lengths of 10 and 15,000 bp, respectively, allowing for up to three mismatches in the seed alignment.
SNP calls were generated with SAMTools mpileup and converted to VCF format with bcftools (v0.1.19; http://samtools.github.io/bcftools/); calls were filtered in VCFtools (v0.1.11) (Danecek et al., 2011) using criteria D=100/Q=20/q=10/d=5/r and refiltered on a per-sample basis with maximum SNP read coverage set to each sample’s theoretical coverage. A custom script was used to select homozygous calls with a minimum SNP quality of 100 and minimum genotype quality of 80. SNP function was predicted using Annovar (Wang et al., 2010). SNP calls were compared with allele calls on the same clones using the Infinium 8303 potato array (Felcher et al., 2012).
CNVs were called from genomic BAM files based on read depth using CNVnator (Abyzov et al., 2011) with a window size of 100 bp. Raw CNV calls were filtered using quality scores generated by the software with a cutoff P value of 0.05, removing many small deletions (<500 bp) with low support. As quality scores were much lower for small intergenic CNVs, those below 500 bp were removed. CNV regions containing an N-content above 10% in the reference sequence were also removed. To account for mapping bias and errors in the reference assembly, we generated CNV calls by mapping reads from the DM reference genotype to its own assembly. In total, 139 genes were found missing based on DM self-CNV analysis and excluded as annotation artifacts. Copy number estimates generated from the DM reference genotype that were above or below a single copy were considered as mapping bias or errors in the reference assembly, and custom scripts were used to adjust copy number estimates in the monoploid panel based on these values. To limit analysis of variants to a set of high confidence calls, we considered regions with a copy number estimate between 0.8 and 1.4 indistinguishable from single copy regions and excluded from further analysis. BEDTools (Quinlan and Hall, 2010) and custom scripts were used to determine CNV-gene overlaps and assign gene copy number. For confident association, a CNV had to span at least half the gene model. Genes for which a CNV covered at least half an exon but less than half the gene model were considered partially duplicated or deleted.
To assess the sensitivity and specificity of CNVnator to identify structural variants, we performed a custom read depth analysis. Median read depths were calculated in 100-bp windows and divided by whole-genome median coverage to obtain relative window coverage. Window estimates were then normalized based on DM mapping bias. Adjacent windows with high or low coverage were concatenated to form CNV blocks, merging nearby blocks within 200 bp. Genotypes were calculated as the mean of all individual window estimates within a block. CNV blocks were removed if they contained 10% N-content, were shorter than 500 bp, and if they occurred in regions where >80% of samples were called as CNVs (regions with significant mapping bias). For validation, CNVnator calls were required to have at least 50% coverage by CNVs of the same class from the read depth method. To experimentally validate structural variant calls, deletions were randomly assessed using PCR with multiple computationally predicted single-copy and variant (duplicate or deletion) clones (Supplemental Data Set 12). Reaction conditions were 10 ng template DNA, 0.2 μM each primer, 0.2 μM deoxynucleotide triphosphate, and 0.625 units Taq DNA polymerase (New England Biolabs) in 1× reaction buffer [20 mM Tris-HCl, pH 8.8, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, and 0.1% Triton X-100]. Duplications were cycled at 95°C for 4 min, 25 cycles of 95°C 30 s, 53°C 45 s, 68°C 1 min, with a final extension of 68°C for 5 min. For deletions, the reactions were at 95°C for 4 min, 30 cycles of 95°C for 30 s, 55°C for 45 s, 68°C for 1 min, with a final extension of 68°C for 5 min. Reactions were run on a 1.2% agarose gel.
Unmapped RNA-seq reads from each clone were pooled to generate de novo transcript assemblies using Trinity (Grabherr et al., 2011). Contigs were aligned to the DM v4.04 assembly with GMAP (Wu and Watanabe, 2005) and excluded if they had greater than 85% coverage and sequence identity to the reference genome. Sequences below 500 bp were also excluded. Transcripts were then aligned with BLASTX to the Uniref100 database to remove contaminants and the remaining set aligned to NCBI nr protein database for functional annotation. To validate putative PAV transcripts, we mapped genomic DNA sequences from DM to both the reference and PAV transcripts, filtered for high-quality alignments (MapQ ≥ 20), and removed PAVs with median read depth above half their theoretical coverage (30×).
FISH Analysis
Root tips for FISH analysis were obtained from greenhouse-grown plants. Chromosome preparation and FISH were performed following published protocols (Cheng et al., 2002). PCR-amplified DNA fragments (Supplemental Data Set 3) were pooled and labeled with digoxigenin-11-dUTP (Roche Diagnostics) using a standard nick translation reaction. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole in VectaShield antifade solution (Vector Laboratories). FISH images were processed using Meta Imaging Series 7.5 software, and the final contrast of the images was processed using Adobe Photoshop CC 2014 software.
Chromatin Immunoprecipitation Sequencing Analysis
Chromatin immunoprecipitation sequencing reads were cleaned using Cutadapt (Martin, 2011) with minimum base quality 10 and minimum read length of 10 nucleotides. Reads were mapped to the DM v4.04 assembly in paired-end mode using Bowtie (v1.0.0) (Langmead, 2010). Peaks were called with HOMER (v4.3) (Heinz et al., 2010) using default parameters with minimum peak size of 150 bp and minimum peak distance of 300 bp.
Phylogenetic Analysis
Genetic distances were estimated from SNP and gene level CNV data using PHYLIP (http://evolution.genetics.washington.edu/phylip.html). For each type, 1000 bootstrap data sets were used to generate a consensus tree. Distances from the original data sets were used to add branch lengths to consensus trees. Tree diagrams were generated using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). CNV-based relationships were determined using copy status (duplicated, deleted, and non-CNV) as allele states for potato genes. SAUR and MKS1 trees were created using PHYLIP with multiple-protein alignments generated using ClustalW (Thompson et al., 2002). Alignments are available as Supplemental Data Sets 13 and 14.
Gene Lineage and Functional Analysis
Gene lineage was determined based on ortholog clustering of the predicted proteomes of nine species (http://phytozome.jgi.doe.gov; Aquilegia coerulea v1.1, Arabidopsis thaliana TAIR10, Mimulus guttatus v2.0, Oryza sativa v7.0, Populus trichocarpa v3.0, Solanum lycopersicum iTAG2.3, Solanum tuberosum v3.4, Vitis vinifera 12x; Amborella trichopoda v1.0; http://amborella.huck.psu.edu/data) using OrthoMCL (v1) (Li et al., 2003). TE-related genes were identified based on the existing DM functional annotations, PFAM domains (Bateman et al., 2004) associated with repetitive DNA, and alignment against the RepBase gene database (Jurka et al., 2005) (cutoff 1E-10), finding 2886 TE genes in the DM gene set. Gene Ontology assignments were obtained from SpudDB (ftp://ftp.plantbiology.msu.edu/pub/data/SGR/GO_annotations/) and a Fisher’s exact test was used to test enrichment in CNV duplicates and deletions.
Copy Number Variable Enriched Gene Clusters
To determine regions of the genome with high frequency of copy number variable genes, we split the reference assembly into overlapping 200-kb bins with a step size of 10 kb and counted the number of genes showing CNV in each bin. Bins containing significant numbers of CNV genes were determined using a minimum threshold based on the mean of all genomic windows plus three standard deviations. Consecutive bins showing enrichment were combined into single regions and ranked by average number of CNV genes per bin.
Recombination Frequency
Recombination rates were estimated using SNPs from an F1 potato mapping population that used the DM reference genotype as a parent (Manrique-Carpintero et al., 2015). Marey maps were generated by plotting genetic positions of markers against their physical position (Chakravarti, 1991) and then a 0.1 cubic spline interpolation fitted curve was calculated. The slope of the line connecting adjacent markers was used as a local estimate of recombination rate (cM/Mb).
Accession Numbers
Sequence data from this article can be found in the National Center for Biotechnology Information Sequence Read Archive under the BioProject accession number PRJNA287005. The updated assembly of the reference genome can be downloaded from SpudDB (http://potato.plantbiology.msu.edu/pgsc_download.shtml) or from the DRYAD repository (http://dx.doi.org/10.5061/dryad.vm142). The high-confidence SNP variant calls and the transcript-derived PAVs are available for download from the DRYAD repository under accession number http://dx.doi.org/10.5061/dryad.vm142.
Supplemental Data
Supplemental Figure 1. Pedigree information for the monoploid panel clones.
Supplemental Figure 2. Experimental PCR validation of 15 randomly selected duplication and deletion loci.
Supplemental Figure 3. Distribution of copy number variation frequency (per clone) relative to the position of all genes impacted by deletion.
Supplemental Figure 4. Fraction of copy number variants represented by duplication and deletion binned by size.
Supplemental Figure 5. Copy number variation size distribution by clone.
Supplemental Figure 6. Phylogenetic tree based on protein alignment of annotated small auxin upregulated RNA (SAUR) genes from rice, Arabidopsis, tomato, and potato proteomes.
Supplemental Figure 7. Phylogenetic tree based on protein alignment of genes with sequence homology to five tomato methylketone synthase 1 (MKS1) genes from Amborella, rice, Arabidopsis, Mimulus guttatus, tomato, and potato.
Supplemental Figure 8. Box plot of copy number variation enrichment for individual stress and hormone response expression classes.
Supplemental Figure 9. Summary of copy number variation rates in genes with different expression levels based on fragments per kilobase per million mapped reads values from leaf, flower, root, tuber, and whole in vitro plant tissues.
Supplemental Figure 10. Overview of potato gene lineage categories generated based on orthologous gene clustering.
Supplemental Table 1. Whole-genome resequencing data generated for the monoploid panel.
Supplemental Table 2. Information on single nucleotide polymorphisms identified in the monoploid panel.
Supplemental Table 3. Number of copy number variants identified in the monoploid panel.
Supplemental Table 4. Comparison of structural variation identified by CNVnator and through read depth analyses.
Supplemental Table 5. Gene expression categories assessed for enrichment in the CNV gene set.
Supplemental Data Set 1. Information on all copy number variant regions identified in the monoploid panel.
Supplemental Data Set 2. Extent of potato genome features impacted by copy number variation in the monoploid panel.
Supplemental Data Set 3. Primers used to amplify fluorescent in situ hybridization probes to validate large copy number variants.
Supplemental Data Set 4. Information on 100-kb+ copy number variant regions identified in the monoploid panel.
Supplemental Data Set 5. Copy number estimates for genes confidently associated with copy number variation in the monoploid panel.
Supplemental Data Set 6. Significance values for differential representation of Gene Ontology terms in the duplicated and nonduplicated gene sets (based on Fisher’s exact test).
Supplemental Data Set 7. Significance values for differential representation of Gene Ontology terms in the deletion and nondeletion gene sets (based on Fisher’s exact test).
Supplemental Data Set 8. Genomic regions significantly enriched for genes impacted by copy number variation.
Supplemental Data Set 9. M6-specific alleles including gene level copy number variants and potentially deleterious single nucleotide polymorphisms.
Supplemental Data Set 10. Putative presence/absence transcript assembly coverage validation.
Supplemental Data Set 11. Putative presence/absence transcript PFAM domains.
Supplemental Data Set 12. Primers used in experimental validation of CNVnator structural variants.
Supplemental Data Set 13. Text file of protein alignments used for phylogenetic analysis of SAURs genes.
Supplemental Data Set 14. Text file of protein alignments used for phylogenetic analysis of MKS1 genes.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Science Foundation (ISO-1237969) to C.R.B., D.S.D., J.J., and R.E.V. and Hatch Project VA-135853 to R.E.V.
AUTHOR CONTRIBUTIONS
M.A.H., J.P.H., G.M.P., N.C.M.-C., C.P.L., and Z.Z. analyzed data. P.L. and R.E.V. constructed the monoploids and doubled monoploids. E.C., L.N, X.Y., and Z.Z. isolated DNA and RNA, constructed libraries, and performed FISH. B.V. supervised sequencing and data submission. E.C. mapped sequences to provide sample data. D.S.D., J.J., R.E.V., and C.R.B. designed and provided oversight of the experiments. M.A.H., E.C., J.K., J.P.H., P.L., C.P.L., G.M.P., Z.Z., J.J., R.E.V., and C.R.B. wrote the manuscript. All authors approved the manuscript.
Glossary
- SNP
single nucleotide polymorphism
- CNV
copy number variation
- PAV
presence/absence variation
- FISH
fluorescence in situ hybridization
- TE
transposable element
- FPKM
fragments per kilobase per million mapped reads
- ABA
abscisic acid
Footnotes
Articles can be viewed online without a subscription.
References
- Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A.; 1000 Genomes Project Consortium (2012). An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abyzov A., Urban A.E., Snyder M., Gerstein M. (2011). CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 21: 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonious G.F. (2001). Production and quantification of methyl ketones in wild tomato accessions. J. Environ. Sci. Health B 36: 835–848. [DOI] [PubMed] [Google Scholar]
- Arlt M.F., Wilson T.E., Glover T.W. (2012). Replication stress and mechanisms of CNV formation. Curr. Opin. Genet. Dev. 22: 204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., et al. ; The Gene Ontology Consortium (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32: D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J., Kreitman M., Stahl E.A., Tian D. (2001). Evolutionary dynamics of plant R-genes. Science 292: 2281–2285. [DOI] [PubMed] [Google Scholar]
- Bonierbale M.W., Plaisted R.L., Pineda O., Tanksley S.D. (1994). QTL analysis of trichome-mediated insect resistance in potato. Theor. Appl. Genet. 87: 973–987. [DOI] [PubMed] [Google Scholar]
- Brunner S., Fengler K., Morgante M., Tingey S., Rafalski A. (2005). Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17: 343–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B.L., Korf I., Robb S.M., Parra G., Ross E., Moore B., Holt C., Sánchez Alvarado A., Yandell M. (2008). MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18: 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., et al. (2011). Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963. [DOI] [PubMed] [Google Scholar]
- Chakravarti A. (1991). A graphical representation of genetic and physical maps: the Marey map. Genomics 11: 219–222. [DOI] [PubMed] [Google Scholar]
- Cheng Z., Buell C.R., Wing R.A., Jiang J. (2002). Resolution of fluorescence in-situ hybridization mapping on rice mitotic prometaphase chromosomes, meiotic pachytene chromosomes and extended DNA fibers. Chromosome Res. 10: 379–387. [DOI] [PubMed] [Google Scholar]
- Chia J.-M., et al. (2012). Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44: 803–807. [DOI] [PubMed] [Google Scholar]
- Cook D.E., et al. (2012). Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338: 1206–1209. [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., McVean G., Durbin R.; 1000 Genomes Project Analysis Group (2011). The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S., Maere S., Van de Peer Y. (2005). Genome duplication and the origin of angiosperms. Trends Ecol. Evol. (Amst.) 20: 591–597. [DOI] [PubMed] [Google Scholar]
- DeBolt S. (2010). Copy number variation shapes genome diversity in Arabidopsis over immediate family generational scales. Genome Biol. Evol. 2: 441–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong H., Rowe P.R. (1971). Inbreeding in cultivated diploid potatoes. Potato Res. 14: 74–83. [Google Scholar]
- Díaz A., Zikhali M., Turner A.S., Isaac P., Laurie D.A. (2012). Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS One 7: e33234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D., Wang Y., Han M., Fu Z., Li W., Liu Z., Hu Y., Tang J. (2012). MicroRNA transcriptomic analysis of heterosis during maize seed germination. PLoS One 7: e39578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Cao Y., Huang L., Zhao J., Xu C., Li X., Wang S. (2008). Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20: 228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J., Dodds P., Pryor T. (2000). Structure, function and evolution of plant disease resistance genes. Curr. Opin. Plant Biol. 3: 278–284. [DOI] [PubMed] [Google Scholar]
- Felcher K.J., Coombs J.J., Massa A.N., Hansey C.N., Hamilton J.P., Veilleux R.E., Buell C.R., Douches D.S. (2012). Integration of two diploid potato linkage maps with the potato genome sequence. PLoS One 7: e36347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman E., Wang J., Iijima Y., Froehlich J.E., Gang D.R., Ohlrogge J., Pichersky E. (2005). Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell 17: 1252–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Dooner H.K. (2002). Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99: 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines T.A., et al. (2010). Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc. Natl. Acad. Sci. USA 107: 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilenko T., Antonova O., Shuvalova A., Krylova E., Alpatyeva N., Spooner D.M., Novikova L. (2013). Genetic diversity and origin of cultivated potatoes based on plastid microsatellite polymorphism. Genet. Resour. Crop Evol. 60: 1997–2015. [Google Scholar]
- Gebhardt C., Valkonen J.P. (2001). Organization of genes controlling disease resistance in the potato genome. Annu. Rev. Phytopathol. 39: 79–102. [DOI] [PubMed] [Google Scholar]
- Gill S.S., Tuteja N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48: 909–930. [DOI] [PubMed] [Google Scholar]
- Grabherr M.G., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansey C.N., Vaillancourt B., Sekhon R.S., de Leon N., Kaeppler S.M., Buell C.R. (2012). Maize (Zea mays L.) genome diversity as revealed by RNA-sequencing. PLoS One 7: e33071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardigan M.A., Bamberg J., Buell C.R., Douches D.S. (2015). Taxonomy and genetic differentiation among wild and cultivated germplasm of Solanum sect. Petota. Plant Genome 8: 10.3835/plantgenome2014.06.0025. [DOI] [PubMed] [Google Scholar]
- Hattori Y., et al. (2009). The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030. [DOI] [PubMed] [Google Scholar]
- Hawkes J.G. (1990). The Potato: Evolution, Biodiversity and Genetic Resources. (London: Belhaven Press; ). [Google Scholar]
- Haynes F.L. (1972). The use of cultivated diploid Solanum species in potato breeding. In Prospects for the Potato in the Developing World: An International Symposium on Key Problems and Potentials for Greater Use of the Potato in the Developing World, Lima, Peru, E.R. French, ed (Lima, Peru: International Potato Center), pp. 100–110. [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C.N., Hirsch C.D., Felcher K., Coombs J., Zarka D., Van Deynze A., De Jong W., Veilleux R.E., Jansky S., Bethke P. (2013). Retrospective view of North American potato (Solanum tuberosum L.) breeding in the 20th and 21st centuries. G3 (Bethesda) 3: 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C.N., et al. (2014). Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26: 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D., Moazed D. (2015). RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovene M., Zhang T., Lou Q., Buell C.R., Jiang J. (2013). Copy number variation in potato - an asexually propagated autotetraploid species. Plant J. 75: 80–89. [DOI] [PubMed] [Google Scholar]
- Itkin M., et al. (2013). Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341: 175–179. [DOI] [PubMed] [Google Scholar]
- Jain M., Tyagi A.K., Khurana J.P. (2006). Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88: 360–371. [DOI] [PubMed] [Google Scholar]
- Jaspers P., Blomster T., Brosché M., Salojärvi J., Ahlfors R., Vainonen J.P., Reddy R.A., Immink R., Angenent G., Turck F., Overmyer K., Kangasjärvi J. (2009). Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 60: 268–279. [DOI] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. (2005). Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110: 462–467. [DOI] [PubMed] [Google Scholar]
- Kidwell M.G., Lisch D. (1997). Transposable elements as sources of variation in animals and plants. Proc. Natl. Acad. Sci. USA 94: 7704–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M., Papenbrock J. (2004). The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. J. Exp. Bot. 55: 1809–1820. [DOI] [PubMed] [Google Scholar]
- Knox A.K., Dhillon T., Cheng H., Tondelli A., Pecchioni N., Stockinger E.J. (2010). CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor. Appl. Genet. 121: 21–35. [DOI] [PubMed] [Google Scholar]
- Lai J., et al. (2010). Genome-wide patterns of genetic variation among elite maize inbred lines. Nat. Genet. 42: 1027–1030. [DOI] [PubMed] [Google Scholar]
- Lam H.-M., et al. (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42: 1053–1059. [DOI] [PubMed] [Google Scholar]
- Langmead B. (2010). Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics 32: 11.17.11–11.17.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. http://arxiv.org/abs/1303.3997.
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C.J. Jr., Roos D.S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., et al. (2010). Building the sequence map of the human pan-genome. Nat. Biotechnol. 28: 57–63. [DOI] [PubMed] [Google Scholar]
- Li Y.H., et al. (2014). De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 32: 1045–1052. [DOI] [PubMed] [Google Scholar]
- Lu F., et al. (2015). High-resolution genetic mapping of maize pan-genome sequence anchors. Nat. Commun. 6: 6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Han X., Qi J., Yang J., Wijeratne A.J., Li T., Ma H. (2012). Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res. 22: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrique-Carpintero N.C., Coombs J.J., Cui Y., Veilleux R.E., Buell C.R., Douches D. (2015). Genetic map and quantitative trait locus analysis of agronomic traits in a diploid potato population using single nucleotide polymorphism markers. Crop Sci. 55: 2566–2579. [Google Scholar]
- Maron L.G., et al. (2013). Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc. Natl. Acad. Sci. USA 110: 5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17: 10–12. [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M., De Paoli E., Radovic S. (2007). Transposable elements and the plant pan-genomes. Curr. Opin. Plant Biol. 10: 149–155. [DOI] [PubMed] [Google Scholar]
- Morrell P.L., Buckler E.S., Ross-Ibarra J. (2011). Crop genomics: advances and applications. Nat. Rev. Genet. 13: 85–96. [DOI] [PubMed] [Google Scholar]
- Mun J.-H., et al. (2009). Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 10: R111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz R. (2001). The State of the Use of Potato Genetic Diversity. Broadening the Genetic Base of Crop Production. (Wallingford, UK: CABI Publishing; ), pp. 181–200. [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Sharma S.K., Bolser D., de Boer J., Sønderkær M., Amoros W., Carboni M.F., D’Ambrosio J.M., de la Cruz G., Di Genova A., Douches D.S. (2013). Construction of reference chromosome-scale pseudomolecules for potato: Integrating the potato genome with genetic and physical maps. G3 (Bethesda) 3: 2031–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simko I., Haynes K.G., Jones R.W. (2006). Assessment of linkage disequilibrium in potato genome with single nucleotide polymorphism markers. Genetics 173: 2237–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Martienssen R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8: 272–285. [DOI] [PubMed] [Google Scholar]
- Spooner D.M., Núñez J., Trujillo G., Herrera Mdel.R., Guzmán F., Ghislain M. (2007). Extensive simple sequence repeat genotyping of potato landraces supports a major reevaluation of their gene pool structure and classification. Proc. Natl. Acad. Sci. USA 104: 19398–19403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N.M., et al. (2009). Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5: e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P., Lupski J.R. (2010). Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61: 437–455. [DOI] [PubMed] [Google Scholar]
- St-Pierre B., Besseau S., Clastre M., Courdavault V., Courtois M., Creche J., Ducos E., de Bernonville T.D., Dutilleul C., Glevarec G. (2013). Deciphering the evolution, cell biology and regulation of monoterpene indole alkaloids. Adv. Bot. Res. 68: 73–109. [Google Scholar]
- Sutton T., Baumann U., Hayes J., Collins N.C., Shi B.-J., Schnurbusch T., Hay A., Mayo G., Pallotta M., Tester M., Langridge P. (2007). Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 318: 1446–1449. [DOI] [PubMed] [Google Scholar]
- Tang H., Bowers J.E., Wang X., Ming R., Alam M., Paterson A.H. (2008). Synteny and collinearity in plant genomes. Science 320: 486–488. [DOI] [PubMed] [Google Scholar]
- Xu X., et al. ; Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T., Higgins D.G. (2002). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics, http://dx.doi.org/10.1002/0471250953.bi0203s00. [DOI] [PubMed]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitdewilligen J.G., Wolters A.-M.A., D’hoop B.B., Borm T.J., Visser R.G., van Eck H.J. (2013). A next-generation sequencing method for genotyping-by-sequencing of highly heterozygous autotetraploid potato. PLoS One 8: e62355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varin L., Marsolais F., Richard M., Rouleau M. (1997). Sulfation and sulfotransferases 6: Biochemistry and molecular biology of plant sulfotransferases. FASEB J. 11: 517–525. [DOI] [PubMed] [Google Scholar]
- Vazquez-Flota F.A., De Luca V. (1998). Developmental and light regulation of desacetoxyvindoline 4-hydroxylase in Catharanthus roseus (L.) G. Don. Evidence Of a multilevel regulatory mechanism. Plant Physiol. 117: 1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., et al. (2006). High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell 18: 1791–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J., Symmons O., Spitz F., Korbel J.O. (2013). Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet. 14: 125–138. [DOI] [PubMed] [Google Scholar]
- Wenzel G., Schieder O., Przewozny T., Sopory S.K., Melchers G. (1979). Comparison of single cell culture derived Solanum tuberosum L. plants and a model for their application in breeding programs. Theor. Appl. Genet. 55: 49–55. [DOI] [PubMed] [Google Scholar]
- Westfall C.S., Herrmann J., Chen Q., Wang S., Jez J.M. (2010). Modulating plant hormones by enzyme action: the GH3 family of acyl acid amido synthetases. Plant Signal. Behav. 5: 1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Liu S., He Y., Guan X., Zhu X., Cheng L., Wang J., Lu G. (2012). Genome-wide analysis of SAUR gene family in Solanaceae species. Gene 509: 38–50. [DOI] [PubMed] [Google Scholar]
- Wu T.D., Watanabe C.K. (2005). GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875. [DOI] [PubMed] [Google Scholar]
- Xu G., Ma H., Nei M., Kong H. (2009). Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc. Natl. Acad. Sci. USA 106: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Xu X., Fukao T., Canlas P., Maghirang-Rodriguez R., Heuer S., Ismail A.M., Bailey-Serres J., Ronald P.C., Mackill D.J. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708. [DOI] [PubMed] [Google Scholar]
- Xu X., et al. (2012). Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30: 105–111. [DOI] [PubMed] [Google Scholar]
- Yan H., Talbert P.B., Lee H.-R., Jett J., Henikoff S., Chen F., Jiang J. (2008). Intergenic locations of rice centromeric chromatin. PLoS Biol. 6: e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Nguyen T.T., Guo Y., Schauvinhold I., Auldridge M.E., Bhuiyan N., Ben-Israel I., Iijima Y., Fridman E., Noel J.P., Pichersky E. (2010). Enzymatic functions of wild tomato methylketone synthases 1 and 2. Plant Physiol. 154: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrei M., MacDonald J.R., Merico D., Scherer S.W. (2015). A copy number variation map of the human genome. Nat. Rev. Genet. 16: 172–183. [DOI] [PubMed] [Google Scholar]
- Zerbino D.R., Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Mao L., Chen H., Bu F., Li G., Sun J., Li S., Sun H., Jiao C., Blakely R. (2015). Genome-wide mapping of structural variations reveals a copy number variant that determines reproductive morphology in cucumber. Plant Cell 27: 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żmieńko A., Samelak A., Kozłowski P., Figlerowicz M. (2014). Copy number polymorphism in plant genomes. Theor. Appl. Genet. 127: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.