Abstract
The ORF2 gene of Gill-associated virus (GAV) of Penaeus monodon prawns resides 93 nucleotides downstream of the ORF1a-ORF1b gene and encodes a 144-amino-acid hydrophilic polypeptide (15,998 Da; pI, 9.75) containing 20 basic (14%) and 13 acidic (9%) residues and 19 prolines (13%). Antiserum to a synthetic ORF2 peptide or an Escherichia coli-expressed glutathione S-transferase-ORF2 fusion protein detected a 20-kDa protein in infected lymphoid organ and gill tissues in Western blots. The GAV ORF2 fusion protein antiserum also cross-reacted with the p20 nucleoprotein in virions of the closely related Yellow head virus. By immuno-gold electron microscopy, it was observed that the ORF2 peptide antibody localized to tubular GAV nucleocapsids, often at the ends or at lateral cross sections. As GAV appears to contain only two structural protein genes (ORF2 and ORF3), these data indicate that GAV differs from vertebrate nidoviruses in that the gene encoding the nucleocapsid protein is located upstream of the gene encoding the virion glycoproteins.
Gill-associated virus (GAV) of Penaeus monodon prawns is a type species of the genus Okavirus in the Roniviridae of the order Nidovirales (5, 7, 22). Chronic GAV infection, in which replication is restricted to the foci of hypertrophied cells in the lymphoid organ (LO), is ubiquitous in wild and farmed P. monodon prawns on the east coast of Australia (8, 33, 39). Acute-phase infection, in which GAV spreads to a wide range of tissues, has been linked to farm disease outbreaks since at least 1996 (33, 34, 35). The tubular helical nucleocapsids and rod-shaped, enveloped virions of GAV are morphologically identical to those of Yellow head virus (YHV), which has caused mass deaths in P. monodon prawns cultured in Asia, and both viruses cause similar cytopathologies (2, 3, 20, 34, 35, 36). Sequence similarity levels in the ORF1b gene and the ORF3 glycoprotein gene indicate that GAV and YHV are closely related geographic topotypes (6, 16, 28).
The complete sequencing of the 26,235-nucleotide (nt) RNA genome of GAV has identified five genes ordered 5′-ORF1a/ORF1b-ORF2-ORF3-ORF4-(A)n-3′ (5, 7). YHV also has a long (>22-kb) (plus-strand) single-stranded RNA genome (24, 36, 43), but sequences have been reported only for the region from the ORF1b gene to the 3′ poly(A) tail (16, 28, 29). In GAV, ORF1a contains a 3C-like proteinase with VxHE↓(L,V) cleavage site specificity (44); ORF1b contains SDD-type polymerase, zinc finger, and helicase motifs; and the polyprotein pp1ab is C terminally extended from pp1a by −1 ribosomal frameshifting at an AAAUUUU slippage site preceding an RNA pseudoknot (7). GAV transcribes two 3′-coterminal subgenomic mRNAs (sgmRNAs) with 5′ AC termini that map to sites within conserved intergenic sequences upstream of ORF2 and ORF3 (9). Unlike coronaviruses (27) and arteriviruses (37), but as identified in the shorter three of the four sgmRNAs of the Berne equine torovirus (31, 38), the two GAV sgmRNAs do not contain 5′ genomic leader sequences.
Purified YHV virions contain three structural proteins, gp116 (110 to 135 kDa), gp64 (63 to 67 kDa), and p20 (20 to 22 kDa) (16, 24, 40). The gp116 and gp64 glycoproteins are likely to form the envelope projections (32) and are generated by posttranslational cleavage of the ORF3 polyprotein (5, 16). Here we report evidence that the highly charged 144-amino-acid (aa) protein encoded by the GAV ORF2 gene is equivalent to YHV p20 and is the viral nucleocapsid (N) protein.
A 4.3-kb region downstream of the GAV ORF1b gene was amplified from LO total RNA by reverse transcription-PCR (7, 8) using the Expand Long Template PCR system (Roche), a primer targeting a terminal ORF1b region (GAV18, 5′-GGTGAGTGCCCATTCATTCCACA-3′), and an antisense primer (GAV20, 5′-TTGCCTGACAAGATGGTGAAGCC-3′) targeted elsewhere that primed at a sequence in ORF3. The 4.3-kb region of DNA was purified using a QIAquick column (QIAGEN) and cloned into pGEM-T (Promega) (26), and three clones (pP18/20-1, pP18/20-9, and pP18/20-16) were sequenced in both directions using the Big-Dye dye terminator system (ABI).
The ORF2 coding sequence was amplified from clone pP18/20-16 by PCR using the primers GAV117 (5′-GCCTAGGATCCTGCAATGAACCGCCGCGCA-3′) and GAV116 (5′-AATTGGATCCTTAGGGTTGAGTGTCACCTTC-3′) and cloned into pQE10 (QIAGEN) or was amplified using the primers GAV173 (5′-GACGGATCCATGAACCGCCGCGCACGCC-3′) and GAV116 and cloned into pGEX-KG (13). Two portions of the ORF2 coding sequence N-terminally truncated to internal in-frame Met (i.e., M11 or M61) codons (Fig. 1) were amplified using the PCR primer GAV193 (5′-CTAAGGATCCCATGCCCGTCCGGCGCCCT-3′) or GAV194 (5′-TCACGGATCCTATGCAAGCATCACCATTCATT-3) in combination with GAV116 and cloned into pQE10. Primer BamHI cloning sites are underlined, and Met or termination codons are in bold. Plasmids were transformed into Escherichia coli DH5-α cells (26), clone insert orientations were determined by directional PCR, and four clones of each construct were sequenced to confirm the insert integrity.
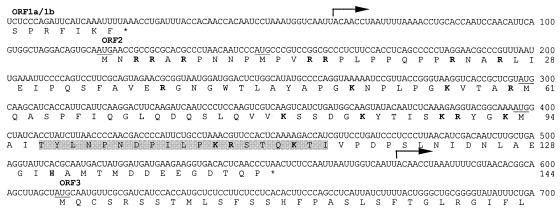
FIG. 1.
Nucleotide and predicted amino acid sequences of the GAV ORF2 gene and genome regions extending into the upstream ORF1a-ORF1b and downstream ORF3 genes. The sequence is the consensus of three clones, pP18/20-1, pP18/20-9, and pP18/20-16. Indicated are the 5′-terminal positions of the 3′-coterminal sgmRNAs (arrows), the ORFa/1b and ORF2 gene termination codons (*), the in-frame AUG codons (underlined) and basic amino acids (bold) in ORFZ, and the sequence of the synthetic peptide PN1 (shaded).
The plasmids pGEX-ORF2, pQE-ORF2, pQE-ORF2-M11, and pQE-ORF2-M61 were transformed into E. coli M15(pREP4) cells (QIAGEN) to express glutathione S-transferase (GST)-ORF2, His6-ORF2, His6-ORF2-M11, and His6-ORF2-M61 fusion proteins, respectively. Briefly, the overnight cultures were diluted to an A600 of 0.2 in SuperBroth medium containing 100 μg of ampicillin/ml and 25 μg of kanamycin/ml and grown at 37°C to an A600 of 0.6 to 0.8, and protein expression was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The cultures were incubated for 2 to 4.5 h and analyzed directly by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or used to purify GST-ORF2 inclusion bodies. Inclusion bodies were purified by repeated Dounce homogenization and sonication in lysis buffers containing 1 mg of lysozyme/ml and 0.1 to 0.5% Triton X-100 and used directly as an antigen in immunizations (25).
The C-terminal ORF2 peptide PN1 (CT97YLNPNDPILPKRSTQKTI115) (Fig. 1) was synthesized by Chiron Technologies, Melbourne, Australia. An N-terminal Cys residue facilitated its covalent linkage to keyhole limpet hemocyanin (KLH) that had been activated with m-maleimidobenzoy-N-hydroxysuccinate ester (Pierce) (14).
Antiserum was produced in crossbred rabbits injected intramuscularly (two sites; 0.5 ml/site; ∼0.5 mg of KLH-PN1/site or ∼0.2 mg of GST-ORF2/site) with GST-ORF2 inclusion bodies or KLH-PN1 peptide emulsified in an equal volume of Montanide ISA70 adjuvant (Seppic, Paris, France). The rabbits were given a booster injection twice at intervals of 2 to 3 weeks, and the specificity and titer of the peptide antiserum were assessed by an enzyme-linked immunosorbent assay (14).
Proteins separated by SDS-PAGE (26) were either stained with 0.2% Coomassie blue R250 or transferred semidry onto Hybond C (Amersham) membranes (14). Western blotting was performed using a 1:200 dilution of rabbit antibody in 5% skim milk powder, and detection was carried out using a 1:1,000 dilution of biotinylated donkey anti-rabbit immunoglobulin (Amersham), a 1:1,000 dilution of streptavidin-biotin-horseradish peroxidase conjugate (Amersham), and 4-chloro-1-naphthol (14).
Small pieces (<1 mm2 in diameter) of LO tissue were fixed in 4% paraformaldehyde-0.1% glutaraldehyde in 66 mM cacodylate buffer (pH 7.4) for 48 h at 4°C, cryopreserved in polyvinylpyrrolidone-sucrose, snap-frozen in liquid nitrogen, and cryosubstituted with methanol containing 0.5% uranyl acetate for 48 h at −85°C. The tissue was warmed to −45°C, embedded in Lowicryl K11 M, and UV polymerized. Ultrathin sections mounted on carbon- and Formvar-coated nickel grids were incubated in a 1:20 dilution of KLH-PN1 or GST-ORF2 antiserum in phosphate-buffered saline (PBS) block solution (PBS, 0.2% bovine serum albumin, 0.2% fish skin gelatin, 20 mM glycine, 0.05% Tween 20) for 2 h. Control sections were incubated in prebleed serum. After being washed in PBS block solution, the sections were incubated for 1 h in goat anti-rabbit immunoglobulin G conjugated with 10-nm-diameter colloidal gold particles (BioCell) diluted 1:40 in PBS block solution and washed twice for 5 min in water. The tissues were stained with 4% uranyl acetate and then with Reynold's lead citrate and examined at 80 kV with a Jeol 1010 transmission electron microscope.
The 435-nt ORF2 gene of GAV encodes a 144-aa polypeptide and is bounded by intergenic sequences of 93 nt to the upstream ORF1a-ORF1b gene and 57 nt to the downstream ORF3 gene (Fig. 1). The ORF2 protein has a deduced molecular mass of 15,998 Da and a pI of 9.75 and is highly hydrophilic, containing 20 basic (14%) and 13 acidic (9%) residues. There is a cluster of 7 arginines in the N-terminal 26 aa, and 5 of the 8 C-terminal residues are acidic. ORF2 also contains no cysteines and 19 prolines (13%), 8 of which are clustered between residues 7 and 22.
BLAST 2.0 (1) searches of the GenPept or translated sequences of the GenBank database identified no polypeptides significantly related to ORF2. This is consistent with alignments of pp1ab motifs showing that GAV is evolutionarily very distant from the vertebrate nidoviruses (7). In size, the 144-aa ORF2 is between the N proteins of arteriviruses (110 to 128 aa) (4, 10, 12, 23) and toroviruses (160 to 167 aa) (11, 18, 30) and significantly smaller than the 377- to 454-aa N proteins of coronaviruses (19). ORF2 displays general structural similarities to torovirus N proteins in the absence of cysteines and a high prevalence of prolines and of basic residues that are likely to facilitate nucleic acid binding (15). It is also noteworthy that the architecture of the helical nucleocapsids and rod-shaped virions of GAV (33, 34) and YHV (2, 3, 24, 40, 43) somewhat resemble torovirus nucleocapsid and virion structures (41, 42). Moreover, curled virions similar in form to the crescent-shaped torovirus particles (42) have been observed in purified YHV (24), suggesting that okavirus nucleocapsids are also quite flexible. As N proteins play a significant role in defining the structures of nucleocapsids and virions, it is not surprising that the GAV N protein might share more general similarities with the cognate proteins of toroviruses than with those of coronaviruses or arteriviruses, which have distinctly different virion architectures.
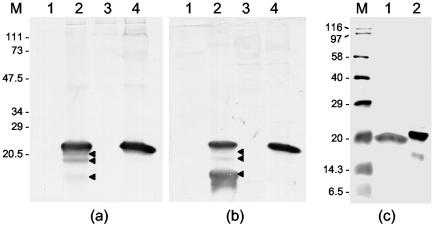
Western blotting with antiserum to either the KLH-PN1 peptide or the GST-ORF2 fusion protein detected the ORF2 gene product (molecular mass, ∼22 kDa) in the LO and gill tissue of a single moribund P. monodon prawn sampled at 6 days after injection (34) with GAV (Fig. 2). In LO, three smaller polypeptides (molecular masses, ∼21, 20, and 17 kDa) were also detected, although the 17-kDa protein was detected weakly by the GST-ORF2 antiserum (Fig. 2a). Since these initial ORF2 size estimates were based on poorly resolved, prestained protein standards, they were reassessed using biotinylated protein standards. In Western blots with the PN1 peptide antiserum (Fig. 2c and 3a), the native ORF2 protein migrated alongside the 20-kDa biotinylated standard, and the relative migration of the His6-ORF2 protein (molecular mass, ∼21.5 kDa) was consistent with the calculated additional mass (1,639 Da) of its N-terminal His6 tag. The estimated size of ORF2 was thus revised to 20 kDa, and the sizes of the three smaller ORF2 derivatives were revised to 19, 17, and 14.5 kDa, respectively.
FIG. 2.
Western blot of uninfected and GAV-infected P. monodon tissues carried out with antiserum to E. coli-expressed GST-ORF2 fusion protein (a) and KLH-linked peptide PN1 (b and c). (a and b) LO (lanes 1 and 2) and gill tissues (lanes 3 and 4) from an uninfected prawn (lanes 1 and 3) and a GAV-infected prawn (lanes 2 and 4). Three proteins smaller than the ORF2 protein detected in LO tissue are indicated (◂). (c) GAV-infected gill tissue (lane 1) and the E. coli-expressed His6-ORF2 fusion protein (lane 2). Proteins were separated by SDS-PAGE in 12% (a and b) or 15% (c) polyacrylamide gels. The molecular masses (in kilodaltons) of prestained protein standards (Bio-Rad) (a and b) and biotinylated protein standards (Amersham) (c) are indicated (lanes M).
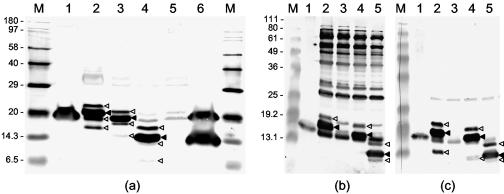
FIG. 3.
Western blot of native ORF2 in GAV-infected LO and gill tissues compared to His6-ORF2 and N-terminally truncated His6-ORF2 fusion proteins expressed in E. coli. (a) GAV-infected gill tissue (lane 1) and LO tissue (lane 6) and IPTG-induced E. coli cultures transformed with pQE-ORF2 (lane 2), pQE-ORF2-M11 (lane 3), pQE-ORF2-M61 (lane 4), and pQE10 (lane 5) were resolved in a 15% polyacrylamide gel and immunodetected with KLH-PN1 peptide antiserum. Lane M contains biotinylated protein standards. (b and c) GAV-infected gill (lane 1) and IPTG-induced E. coli cultures transformed with pQE-ORF2 (lane 2), pQE10 (lane 3), pQE-ORF2-M11 (lane 4), and pQE-ORF2-M61 (lane 5) resolved in a 12% polyacrylamide gel and immunodetected using antiserum to GST-ORF2 (b) or KLH-PN1 peptide (c). Lanes M contain BenchMark prestained protein ladders (Invitrogen). The predominant His6-ORF2 fusion proteins (◂) and minor larger and smaller His6-ORF2 fusion protein forms that were also immunodetected (◃) are indicated.
ScanProcite was used to identify potential posttranslational modification motifs that might explain the difference between the calculated (16 kDa) and estimated (20 kDa) masses of the GAV ORF2 protein. Several phosphorylation sites were identified, as is common in RNA-binding proteins, including the Berne torovirus N protein (15). However, phosphorylation alone would not account for the size disparity, which we suspect is due to the electrophoretic mobility of ORF2 being retarded by its intrinsic structure (14% basic and 13% Pro residues).
Two smaller forms of the Berne torovirus nucleocapsid (N) protein have been detected in infected cells, although there are conflicting reports about whether these result from proteolysis (15) or internal initiation of translation (30). To establish whether the 14.5-kDa ORF2 derivative detected in LO tissue might be due to internal initiation of translation, His6-ORF2 proteins N-terminally truncated upstream of Met11 and Met61 were expressed in E. coli and analyzed in Western blots. The PN1 peptide antiserum detected the His6-ORF2 (∼21.5-kDa), His6-ORF2-M11 (∼19-kDa), and His6-ORF2-M61 (∼15-kDa) proteins in addition to three minor polypeptides that were proportionally larger and smaller than each primary recombinant protein (Fig. 3a and c). Although bacterial proteins were also obvious, all but the smallest minor His6-ORF2-related polypeptides were also detected by the GST-ORF2 antiserum (Fig. 3b). Although the 15-kDa His6-ORF2-M61 protein was similar in size to the 14.5-kDa ORF2 derivative detected in LO tissue (Fig. 2), importantly, unlike the latter, it reacted well with the GST-ORF2 antiserum (compare Fig. 2a and 3b).
We hypothesize, therefore, that the 14.5-kDa ORF2 derivative is more likely the result of proteolysis at a C-terminal position in or beyond the PN1 peptide sequence. However, to be consistent with our data, antibodies to GST-ORF2 would primarily have to target epitopes C terminal to the proteolysis site. In support of this supposition, a protein proportionally smaller than each His6-ORF2 fusion protein, suggesting that it was trimmed at a common C-terminal position, was detected by antiserum to the PN1 peptide but not to GST-ORF2 (Fig. 3b and c). Although the overall homology was poor, a Clustal W multiple sequence alignment (data not shown) identified a trypsin cleavage motif (KR109) in the GAV ORF2 sequence encompassed by the PN1 peptide that was also present at a comparable position in torovirus N proteins (11, 18, 30). Mutagenesis studies will be required to determine the relevance of this or other potential proteolysis sites in the C-terminal regions of these proteins.
It is of note that the LO tissue used in this analysis was from a moribund prawn acutely infected with GAV and nearing death. Although this fact ensured the presence of high virus protein levels, significant LO structural and cellular degeneration occurs late in infection (34). This degeneration likely involves apoptotic cell death, as has been reported with YHV (17), and explains why ORF2 may have been subjected to proteolysis. Late in infection, nucleocapsids and virions appear in high numbers in gill cells, usually in the absence of major cytopathology, and large arrays of GAV particles accumulate at the external boundaries of epidermal cells and the gill cuticles (34), presumably to be eliminated following molting (21). This accumulation of mature virions likely explains why only intact ORF2 was detected in the gill tissues of the same prawn.
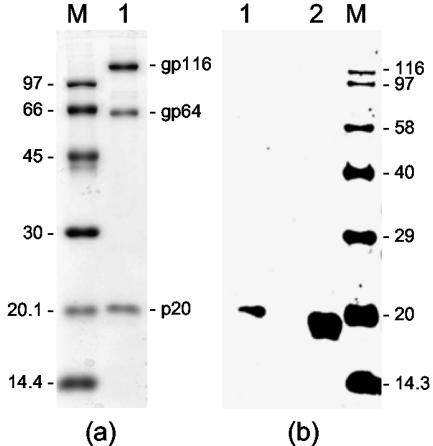
YHV purified from the hemolymph of infected P. monodon prawns by Urografin (Schering) density gradient centrifugation (43) was analyzed by SDS-PAGE and Western blotting to verify that antiserum to the GAV ORF2 gene product would cross-detect the nonglycosylated YHV p20 protein. The three YHV structural proteins (gp116, gp64, and p20) were detected by Coomassie blue staining (Fig. 4a), and the p20 protein, which appeared to be slightly larger than the GAV ORF2 protein synthesized in gill tissue, cross-reacted with the GST-ORF2 antiserum (Fig. 4b).
FIG. 4.
(a) Structural proteins (gp116, gp64, and p20) of purified YHV (lane 1) detected by staining with Coomassie blue. (b) Western blot of purified YHV (lane 1) and GAV-infected gill tissues (lane 2) with GAV GST-ORF2 antiserum. Proteins were resolved in a 12% polyacrylamide gel. Lanes M contain protein standards (a) and biotinylated protein standards (Amersham) (b), with molecular masses (in kilodaltons) indicated.
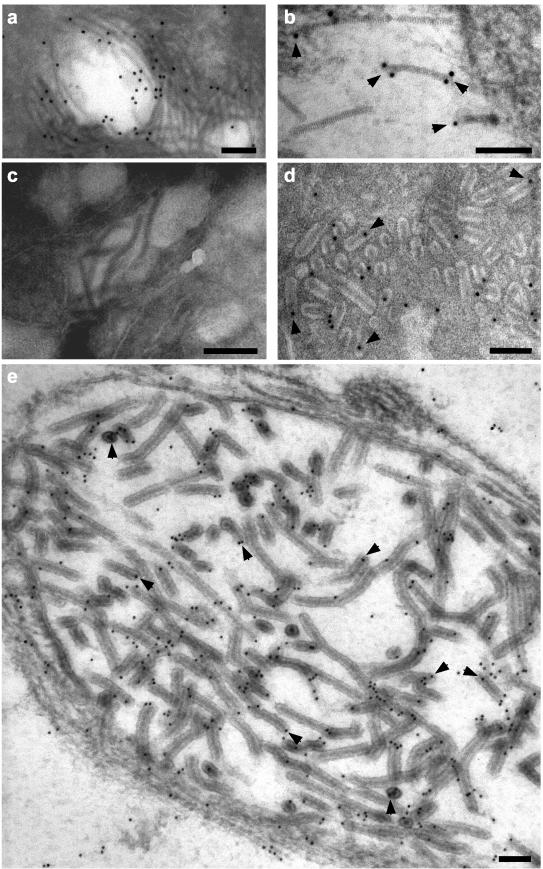
Ultrathin sections of LO cells from GAV-infected P. monodon prawns were immuno-gold particle labeled using the ORF2 PN1 peptide antiserum and examined by electron microscopy (Fig. 5). Gold particles associated with free, striated (∼16-nm-diameter) tubular GAV nucleocapsids within the cell cytoplasm and were often observed at the nucleocapsid ends rather than at positions on their longitudinal surfaces (Fig. 5b). Gold particles also associated with nucleocapsids within mature rod-shaped, enveloped GAV virions (Fig. 5d) and within newly formed virions that had not completely disaggregated into discrete rod-shaped structures and that occurred where nucleocapsids appeared to be actively budding or had recently budded into membranous endoplasmic vesicles (Fig. 5e). Within virions, gold particles were also commonly associated with nucleocapsid ends or with positions where nucleocapsids appeared to have been cross-sectioned transversely. Gold particles were not seen to associate with any specific cellular structures in cytoplasmic regions where GAV nucleocapsids were not evident. Similar results were obtained with the GST-ORF2 antiserum (data not shown), and no association of gold particles with GAV nucleocapsids was observed with preimmune serum (Fig. 5c).
FIG. 5.
Immunoelectron microscopy of ultrathin sections of GAV-infected LO cells using prebleed (c) or KLH-PN1 peptide (a, b, d, and e) antiserum. No gold particles (10 nm in diameter) associated with GAV nucleocapsids with prebleed sera. With the PN1 peptide antiserum, gold particles associated with free striated nucleocapsids (a and b) and nucleocapsids within mature rod-shaped virions (d) and newly formed virions (e) that were observed budding into a membranous endoplasmic vesicle. Good examples of gold particles bound to nucleocapsid ends or to areas where the nucleocapsids had been cross-sectioned laterally are highlighted (arrowheads). Bars represent 100 nm.
The 6.2-kb region of the GAV genome downstream of the ORF1a-ORF1b gene (GenBank accession no. AY039647) contains no discrete genes, other than ORF2, of sufficient length or structure to encode a viral N protein (5). The 5-kb ORF3 genes of GAV and YHV encode a putative precursor glycoprotein containing six transmembrane domains. In YHV, ORF3 is cleaved after transmembrane domains 3 and 5 to generate the structural glycoproteins gp116 and gp64, respectively (16), the larger of which has so far been shown to protrude from the virion envelope (32). No protein of the size (25.4 kDa) predicted for the ORF3 N-terminal, trimembrane-spanning cleavage product of YHV (16) and GAV (5) has been detected in virions. Although it has some structural similarities to the triple-membrane-spanning viral membrane (M) proteins of vertebrate nidoviruses, its function remains unknown. The small (9.6-kDa) ORF4 gene coding sequence is unlikely to be translated at sufficient levels to form a major structural component of virions, since no 3′-coterminal sgmRNA for ORF4 is transcribed in abundance (9). Moreover, the ORF4 region in YHV is interrupted by multiple stop codons, indicating that it is not essential for virus replication (29).
Immuno-gold particle labeling of GAV nucleocapsids by antibodies to the ORF2 synthetic peptide and recombinant GST-ORF2 fusion protein offers strong evidence that the 20-kDa ORF2 gene product is the viral N protein. YHV contains a homolog of the GAV ORF2 gene (28, 29), and it is clear from data presented here that the ORF2 protein is equivalent to the nonglycosylated p20 structural protein of YHV (16). Moreover, a monoclonal antibody reactive for the YHV p20 protein has also recently been shown to bind to nucleocapsids (32). The finding that the ORF2 gene encodes the GAV N protein conspicuously distinguishes this crustacean okavirus from the vertebrate nidoviruses in which the N protein gene resides in the 3′-terminal genome region downstream of genes coding for the virion glycoproteins and membrane protein.
Acknowledgments
We thank Roger Pearson for help with peptide conjugation and antibody production, Ross Tellam and Tony Vuocolo for useful discussions on bacterial expression, Rick Webb and Malcom Jones, University of Queensland, and Russell McCulloch for assistance with immunoelectron microscopy, and Sasimanas Unajak, Mahidol University, Bangkok, Thailand, for supplying purified YHV.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boonyaratpalin, S., K. Supamattaya, J. Kasornchandra, S. Direkbusaracom, U. Aekpanithanpong, and C. Chantanachookin. 1993. Non-occluded baculo-like virus, the causative agent of yellow-head disease in the black tiger shrimp (Penaeus monodon). Fish Pathol. 28:103-109. [Google Scholar]
- 3.Chantanachookin, C., S. Boonyaratpalin, J. Kasornchandra, D. Sataporn, U. Ekpanithanpong, K. Supamataya, S. Riurairatana, and T. W. Flegel. 1993. Histology and ultrastructure reveal a new granulosis-like virus in Penaeus monodon affected by yellow-head disease. Dis. Aquat. Org. 17:145-157. [Google Scholar]
- 4.Chen, Z., L. Kuo, R. R. Rowland, C. Even, K. S. Faaberg, and P. G. Plagemann. 1993. Sequences of 3′ end of genome and of 5′ end of open reading frame 1a of lactate dehydrogenase-elevating virus and common junction motifs between 5′ leader and bodies of seven subgenomic mRNAs. J. Gen. Virol. 74:643-659. [DOI] [PubMed] [Google Scholar]
- 5.Cowley, J. A., and P. J. Walker. 2002. The complete genome sequence of gill-associated virus of Penaeus monodon prawns indicates a gene organisation unique among nidoviruses. Arch. Virol. 147:1977-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowley, J. A., C. M. Dimmock, C. Wongteerasupaya, V. Boonsaeng, S. Panyim, and P. J. Walker. 1999. Yellow head virus from Thailand and gill-associated virus from Australia are closely related but distinct prawn viruses. Dis. Aquat. Org. 36:153-157. [DOI] [PubMed] [Google Scholar]
- 7.Cowley, J. A., C. M. Dimmock, K. M. Spann, and P. J. Walker. 2000. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 81:1473-1484. [DOI] [PubMed] [Google Scholar]
- 8.Cowley, J. A., C. M. Dimmock, K. M. Spann, and P. J. Walker. 2000. Detection of Australian gill-associated virus (GAV) and lymphoid organ virus (LOV) of Penaeus monodon by RT-nested PCR. Dis. Aquat. Org. 39:159-167. [DOI] [PubMed] [Google Scholar]
- 9.Cowley, J. A., C. M. Dimmock, and P. J. Walker. 2002. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic mRNAs that do not possess 5′-leader sequences. J. Gen. Virol. 83:929-937. [DOI] [PubMed] [Google Scholar]
- 10.den Boon, J. A., E. J. Snijder, E. D. Chirnside, A. A. F. de Vries, M. C. Horzinek, and W. J. M. Spaan. 1991. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 65:2910-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duckmanton, L. M., R. Tellier, P. Liu, and M. Petric. 1998. Bovine torovirus: sequencing of the structural genes and expression of the nucleocapsid protein of Breda virus. Virus Res. 58:83-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godeny, E. K., L. Chen, S. N. Kumar, S. L. Methven, E. V. Koonin, and M. A. Brinton. 1993. Complete genomic sequence and phylogenetic analysis of the lactate dehydrogenase-elevating virus (LDV). Virology 194:585-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 14.Harlow, E., and D. P. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Horzinek, M. C., J. Ederveen, and M. Weiss. 1985. The nucleocapsid of Berne virus. J. Gen. Virol. 66:1287-1296. [DOI] [PubMed] [Google Scholar]
- 16.Jitrapakdee, S., S. Unajak, N. Sittidilokratna, R. A. J. Hodgson, J. A. Cowley, P. J. Walker, S. Panyim, and V. Boonsaeng. 2002. Identification and analysis of gp116 and gp64 structural glycoproteins of the yellow head nidovirus of Penaeus monodon shrimp. J. Gen. Virol. 84:863-873. [DOI] [PubMed] [Google Scholar]
- 17.Khanobdee, K., C. Soowannayan, T. W. Flegel, S. Ubol, and B. Withyachumnarnkul. 2002. Evidence for apoptosis correlated with mortality in the giant black tiger shrimp Penaeus monodon infected with yellow head virus. Dis. Aquat. Org. 48:279-290. [DOI] [PubMed] [Google Scholar]
- 18.Kroneman, A., L. A. H. M. Cornelissen, M. C. Horzinek, R. J. de Groot, and H. F. Egberink. 1998. Identification and characterization of a porcine torovirus. J. Virol. 72:3507-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laude, H., and P. S. Masters. 1995. The coronavirus nucleocapsid protein, p. 141-163. In S. G. Siddell (ed.), The coronaviridae. Plenum Press, New York, N.Y.
- 20.Limsuwan, C. 1991. Handbook for cultivation of black tiger prawns. Tansetakit Co. Ltd., Bangkok, Thailand.
- 21.Martin, G. G., M. Quintero, M. Quigley, and H. Khosrovian. 2000. Elimination of sequestered material from the gills of decapod crustaceans. J. Crustac. Biol. 20:209-217. [Google Scholar]
- 22.Mayo, M. A. 2002. A summary of taxonomic changes recently approved by the ICTV. Arch. Virol. 147:1655-1656. [DOI] [PubMed] [Google Scholar]
- 23.Meulenberg, J. J., M. M. Hulst, E. J. de Meijer, P. L. Moonen, A. den Besten, E. P. de Kluyver, G. Wensvoort, and R. J. Moormann. 1993. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology 192:62-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadala, E. C. B., L. M. Tapay, and P. C. Loh. 1997. Yellow-head virus: a rhabdovirus-like pathogen of penaeid shrimp. Dis. Aquat. Org. 31:141-146. [Google Scholar]
- 25.Pearson, R., S. Muharsini, G. Wijffels, and T. Vuocolo. 2000. Purification of recombinant peritrophic membrane proteins of the old world screwworm fly, Chrysomya bezziana. Ilmu Ternak Dan Veteriner 5:185-191. [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-219. [DOI] [PubMed] [Google Scholar]
- 28.Sittidilokratna, N., R. A. J. Hodgson, J. A. Cowley, S. Jitrapakdee, V. Boonsaeng, S. Panyim, and P. J. Walker. 2002. Complete ORF1b-gene sequence indicates yellow head virus is an invertebrate nidovirus. Dis. Aquat. Org. 50:87-93. [DOI] [PubMed] [Google Scholar]
- 29.Sittidilokratna, N. 2003. Ph.D. thesis. Mahidol University, Bangkok, Thailand.
- 30.Snijder, E. J., J. A. den Boon, W. J. M. Spaan, G. M. G. M. Verjans, and M. Horzinek. 1989. Identification and primary structure of the gene encoding the Berne virus nucleocapsid protein. J. Gen. Virol. 70:3363-3370. [DOI] [PubMed] [Google Scholar]
- 31.Snijder, E. J., M. C. Horzinek, and W. J. M. Spaan. 1990. A 3′-coterminal nested set of independently transcribed mRNAs is generated during Berne virus replication. J. Virol. 64:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soowannayan, C., T. W. Flegel, P. Sithigorngul, J. Slater, A. Hyatt, T. Wise, M. J. Crane, J. A. Cowley, R. J. McCulloch, and P. J. Walker. 2003. Detection and differentiation of yellow head complex viruses using monoclonal antibodies. Dis. Aquat. Org. 57:193-200. [DOI] [PubMed] [Google Scholar]
- 33.Spann, K. M., J. E. Vickers, and R. J. G. Lester. 1995. Lymphoid organ virus of Penaeus monodon from Australia. Dis. Aquat. Org. 23:127-134. [Google Scholar]
- 34.Spann, K. M., J. A. Cowley, P. J. Walker, and R. J. G. Lester. 1997. Gill-associated virus (GAV), a yellow head-like virus from Penaeus monodon cultured in Australia. Dis. Aquat. Org. 31:169-179. [Google Scholar]
- 35.Spann, K. M., R. J. McCulloch, J. A. Cowley, I. J. East, and P. J. Walker. 2003. Detection of gill-associated virus (GAV) by in situ hybridization during acute and chronic infections of Penaeus monodon and P. esculentus shrimp. Dis. Aquat. Org. 56:1-10. [DOI] [PubMed] [Google Scholar]
- 36.Tang, K. F.-J., and D. V. Lightner. 1999. A yellow head virus gene probe: application to in situ hybridization and determination of its nucleotide sequence. Dis. Aquat. Org. 35:165-173. [DOI] [PubMed] [Google Scholar]
- 37.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. M. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Vliet, A. L., S. L. Smits, P. J. Rottier, and R. J. de Groot. 2002. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 21:6571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker, P. J., J. A. Cowley, K. M. Spann, R. A. J. Hodgson, M. R. Hall, and B. Withyachumnarnkul. 2001. Yellow head complex viruses: transmission cycles and topographical distribution in the Asia-Pacific region, p. 227-237. In C. L. Browdy and D. E. Jory (ed.), The new wave: proceedings of the special session on sustainable shrimp culture, Aquaculture 2001. The World Aquaculture Society, Baton Rouge, La.
- 40.Wang, Y.-C., and P.-S. Chang. 2000. Yellow head virus infection in the giant tiger prawn Penaeus monodon cultured in Taiwan. Fish Pathol. 35:1-10. [Google Scholar]
- 41.Weiss, M., and M. C. Horzinek. 1987. The proposed family Toroviridae: agents of enteric infections. Arch. Virol. 92:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss, M., F. Steck, and M. C. Horzinek. 1983. Purification and partial characterization of a new enveloped RNA virus (Berne virus). J. Gen. Virol. 64:1849-1858. [DOI] [PubMed] [Google Scholar]
- 43.Wongteerasupaya, C., S. Sriurairatana, J. E. Vickers, A. Akrajamorn, V. Boonsaeng, S. Panyim, A. Tassanakajon, B. Withyachumnarnjul, and T. W. Flegel. 1995. Yellow-head virus of Penaeus monodon is an RNA virus. Dis. Aquat. Org. 22:45-50. [Google Scholar]
- 44.Ziebuhr, J., S. Bayer, J. A. Cowley, and A. E. Gorbalenya. 2003. The 3C-like proteinase of an invertebrate nidovirus links coronavirus and potyvirus homologs. J. Virol. 77:1415-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]