Abstract
RNA Polymerase II (Pol II) regulatory cascades involving transcription factors (TFs) and their targets orchestrate the genetic circuitry of every eukaryotic organism. In order to understand how these cascades function, they can be dissected into small genetic networks, each containing just a few Pol II transcribed genes, that generate specific signal-processing outcomes. Small RNA regulatory circuits involve direct regulation of a small RNA by a TF and/or direct regulation of a TF by a small RNA and have been shown to play unique roles in many organisms. Here, we will focus on small RNA regulatory circuits containing Pol II transcribed microRNAs (miRNAs). While the role of miRNA-containing regulatory circuits as modular building blocks for the function of complex networks has long been on the forefront of studies in the animal kingdom, plant studies are poised to take a lead role in this area because of their advantages in probing transcriptional and posttranscriptional control of Pol II genes. The relative simplicity of tissue- and cell-type organization, miRNA targeting, and genomic structure make the Arabidopsis thaliana plant model uniquely amenable for small RNA regulatory circuit studies in a multicellular organism. In this Review, we cover analysis, tools, and validation methods for probing the component interactions in miRNA-containing regulatory circuits. We then review the important roles that plant miRNAs are playing in these circuits and summarize methods for the identification of small genetic circuits that strongly influence plant function. We conclude by noting areas of opportunity where new plant studies are imminently needed.
INTRODUCTION
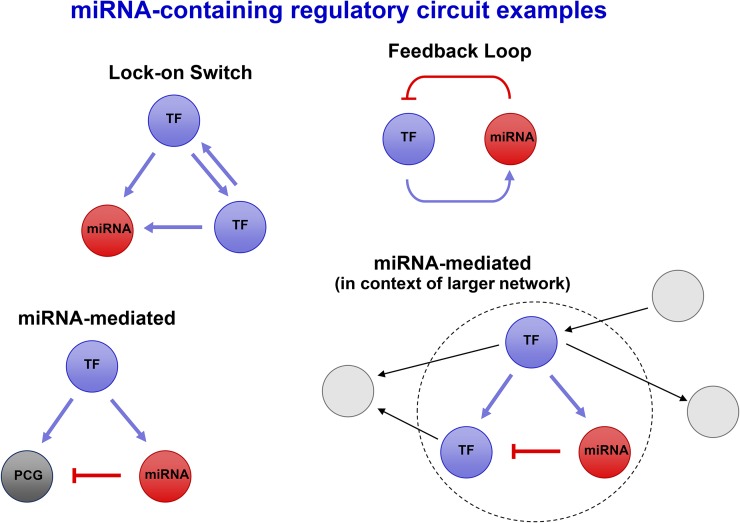
Small genetic circuits are genetic networks that are “small” in the sense that they contain few components, one generally thinks of two to four. Small RNA regulatory circuits are genetic networks involving direct regulation of a small RNA by a transcription factor (TF) and/or direct regulation of a TF by a small RNA. By “circuit” we mean a module that is not a tree-like structure, but rather a network where each component interacts with at least two other components that are not exclusively downstream. The components of a small RNA regulatory circuit or a small genetic circuit as discussed in this article include TFs, small RNAs, and non-TF protein-coding genes (PCGs), which interact with or ultimately influence the activity of RNA polymerase to regulate transcription. This article focuses explicitly on transcriptional regulation by RNA Polymerase II (Pol II) and therefore generally limits discussion of small RNA regulatory circuits to those containing microRNAs (miRNAs). Small RNA regulatory circuits or networks involving one or more miRNAs are often referred to as miRNA-containing. miRNA-mediated regulatory circuits are a special type of miRNA-containing circuit in which both a TF and a miRNA are involved controlling another component (Figure 1).
Figure 1.
Examples of miRNA-Containing Regulatory Circuits.
Several examples of small genetic circuits that contain miRNAs, including “miRNA-mediated” circuits. Lower right: Small genetic circuits usually function in context of larger regulatory cascades and can be thought of as signal processing submodules.
In the first two sections of this Review, we cover analysis, tools, and validation methods for probing the component interactions in small genetic circuits: regulatory interactions between Pol II transcription factors and their target (miRNAs and protein coding gene) promoters and miRNA target interactions. In the final section, we review the important roles that plant miRNAs are playing in genetic networks, along with examples of how small miRNA-containing circuits are central to plant development and environmental adaptation. We conclude with a brief primer on network motif discovery, a method for dissecting a large putative TF-miRNA-gene interaction network into its small two- to four-node component circuits and forming testable hypothesis about the function of the most important subcircuits.
POL II TRANSCRIPTION: IT ALL BEGINS AT THE START SITES
In a very real sense, it all begins at the transcription start sites. The transcription of each component of a genetic circuit (TFs and miRNAs as well as PCGs) ultimately is controlled by Pol II TFs, and the genomic DNA region in the immediate vicinity of each entity’s transcription start site (TSS) encodes cis-regulatory elements (CREs) that enable combinatorial control by upstream TFs. The last decade of research on transcriptional control has uncovered aspects of this process that were entirely unaccounted for in the textbook models of the early 2000s: Each gene has not just one or a few “alternative start sites” but entire contiguous regions of start sites, sometimes several of these regions, and chromatin state may render TF binding sites that are functional in one state entirely irrelevant in another. It is also now recognized that TSS loci may give rise to “bidirectional” or divergent transcripts, and there is ongoing scientific debate over a proposed model that many eukaryotic promoters are inherently bidirectional (Seila et al., 2009), particularly in light of recent evidence in humans that divergent transcription is instead simply an outcome that occurs when both forward and reverse-directed core promoters are present (Duttke et al., 2015). Even the previously drawn lines between a gene’s “core promoter” and “distal promoter” regions blur under the microscope of recent studies in both plants and animals. And while a lack of knowledge about transcriptional control of small RNAs themselves has been a past roadblock, recent transcription start site sequencing technologies adapted from animal systems have enabled progress in this area, opening the field for studies on how small RNAs in small genetic circuits exert system-wide influence on both the timing of specific functions and tissues in which they occur. In the following sections, we cover what is known about the nature of plant promoters and identify gaps calling for further investigation.
The State of the Core: Our Limited Knowledge of Pol II Core Promoter Elements in Plants
Transcriptional regulation is essential for a variety of biological responses ranging from cellular growth, differentiation, and development to rapid responses to biotic and abiotic stimuli. An integral part of this regulation is achieved by TFs binding to CREs, which are short, often degenerate sequences of DNA within the promoter region directly upstream and downstream of the TSSs of genes (Thomas and Chiang, 2006; Heintzman and Ren, 2007). Because of this, knowledge of promoter architecture is critical in understanding how transcription is initiated by RNA polymerases such as Pol II, the polymerase responsible for transcribing protein coding genes (Thomas and Chiang, 2006; Heintzman and Ren, 2007) and miRNAs (Lee et al., 2004b). While promoter lengths are quite variable, typically ranging anywhere from several hundred nucleotides to several kilobases or longer, their architecture has classically been broken down into two regions: a “core” and an “extended” or “distal” promoter region (Kadonaga, 2004; Kumari and Ware, 2013). The core site is the primary docking point of the Pol II preinitiation complex and historically has been described as a site within a region starting around −50 (i.e., 50 bases upstream of the TSS, also known as the +1 site) and ending around position +50 (i.e., 49 bases downstream of the +1 site) (Thomas and Chiang, 2006; Kadonaga, 2012; Kumari and Ware, 2013). Extensive studies in yeast, human, and Drosophila melanogaster early on identified CREs within the core promoter referred to as core promoter elements (CPEs) that are bound by basal or general transcription factors (Kadonaga, 2004, 2012; Thomas and Chiang, 2006; de Boer et al., 2013) including TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, with one of the most well defined and studied CPEs being the TATA box, which is bound by the TATA box binding protein component of TFIID. While these elements were initially thought to be universally present in Pol II gene promoters, it is now apparent that CPEs within the core promoter form a diverse set of CREs with no one CPE being identified universally (Kadonaga, 2004, 2012; Thomas and Chiang, 2006; Kumari and Ware, 2013).
After the discovery of the TATA box, a long list of additional CPEs have come to light that includes, but is not limited to, Initiator (Inr) (Javahery et al., 1994), MTE (Motif ten element) (Lim et al., 2004), TFIIB recognition element (BRE: BREu and BREd, which are the BREs upstream and downstream of the TATA box, respectively) (Lagrange et al., 1998; Deng and Roberts, 2005), DPE (downstream promoter element) (Burke and Kadonaga, 1996), DCE (downstream core element) (Lee et al., 2005), XCPE1 (X core promoter element 1) (Tokusumi et al., 2007), CCAAT box (Dorn et al., 1987), MED-1 (Multiple start site Element Downstream) (Ince and Scotto, 1995), and the GC box (Blake et al., 1990). Additionally, important sequence content stretches such as CpG islands (often found in conjunction with the GC box in mammalian promoters) (Suzuki et al., 2001) and Y Patch (Yamamoto et al., 2007b) (found in plants) also appear to contribute to identification of the core promoter, although CpG islands appear to be absent from plant promoters.
While many of these CPEs have been well studied in animals, studies in plants are not as extensive, leaving many elements that have only been identified computationally (Yamamoto et al., 2007a, 2011; Civán and Švec, 2009; Bernard et al., 2010; Zuo and Li, 2011; Kumari and Ware, 2013). This is especially problematic considering that even some of the most well studied and conserved elements such as TATA and Inr show distinct consensus sequence differences in plants compared with their yeast and mammalian counterparts, making plant-specific consensus sequence identification an important priority in accurate promoter identification in plants (Kumari and Ware, 2013). While CPE identification in yeast and animal systems has been difficult, the situation in plants appears to be even less tractable. For example, recent approaches have used de novo sequence enrichment analyses in an attempt to find new CPEs that would explain transcription in TATA-less promoters (Yamamoto et al., 2007b; Yang et al., 2007), and one study even expanded the core promoter region up to 500 bases to expand the identification of CPEs in plant promoters (Kumari and Ware, 2013).
To highlight the unique difficulty of CPE identification, we used a previously published scanning tool (Megraw and Hatzigeorgiou, 2010) to analyze over 14,000 high-confidence Arabidopsis thaliana TSS promoter sequences provided in a recently published (Cumbie et al., 2015a) data set (Table 1). The scans used the positional weight matrix (PWM) representing each CPE used by Morton et al. (2014), along with additional PWMs from the JASPAR database (Bryne et al., 2008) in order to include TFs analyzed by Kumari and Ware (2013). We found that for any given CPE, very liberal cutoff settings resulted in “identifying” that CPE in 54 to 85% of sequences (Table 1), with only 1% percent of genes having no “identified” CPE. However, when a threshold associated with a low false positive rate is used and one only includes sites located in regions actually associated with their respective CPEs (e.g., −20 to −40 for TATA), then one only identifies such sites in 0 to 10% of the TSS sequences for any given CPE, with the highest percentage identified being TATA (10.19%). Even more striking is that as many as 73% of genes have no strong candidate sites for any of the CPEs listed (Table 1). While this list and analysis are neither exhaustive nor definitive, it is striking how little is really known about an appropriate definition for the “core promoter” region in plants and how much more needs to be done to understand the TSS-proximal regulatory region in plant transcription.
Table 1. Summary of CPE Scans of 14,000 High-Confidence TSSs.
| CPE | 5′ CPE Start Positiona | Percentage of Sequencesb with CPEc | Percentage of Sequencesb Overlapping CPE Startd | Overlapped ROEe |
|---|---|---|---|---|
| GC box | −150 | 8.19,3.41,1.14* | 62.76,39.59,19.4 | No |
| CCAAT box | −75 | 16.1,NA,NA* | 69,NA,NA | No |
| BREu | −38 | 11.06,6.08,1.99* | 65.33,46.05,19.06 | Yes |
| TATA box | −30 | 26.14,20.05,10.19* | 71.88,57.35,29.07 | Yes |
| BREd | −24 | 9.27,5.15,0.47* | 70.1,45.62,3.9 | Yes |
| Y-Patch | −13 | 32.09,24.16,3.98* | 84.92,73.11,18.94 | Yes |
| XCPE1 | −8 | 10.18,4.61,0.89* | 66.15,39.99,9.57 | Yes |
| Inr | −2 | 8.34,7.19,1.6* | 53.92,46.73,14.22 | Yes |
| DCE | +6 | 14.59,8.41,2.15* | 72.57,52.9,18.64 | Yes |
| MTE | +18 | 13.61,8.05,1.23* | 69.86,52.92,12.36 | Yes |
| DPE | +28 | 9.13,4.17,0.81* | 60.55,34.24,7.02 | Yes |
| MED-1 | +88 | 21.01,13.86,6.22* | 74.32,58.04,32.07 | No |
| Percentage of genes with no CPEf | 0.01,0.2,17.35 | |||
| Percentage of genes with no CPE around 5′ CPE startg | 14.65,33.18,72.92 | |||
The asterisk indicates that each percentage was calculated using a threshold that held the false-positive rate at 0.001, 0.0005, and 0.0001 in the same order for each cell identified. This table compares observed positional enrichments for 12 traditionally identified CPEs in the genomic regions surrounding 14,000 high-confidence TSSs in wild-type Arabidopsis whole-root samples. CPE start position is the commonly listed location in the literature for each element with respect to the TSS; for each CPE, a point roughly in the middle of the various ranges reported (Bryne et al., 2008; Kadonaga, 2012; Kumari and Ware, 2013) was used. Region of enrichment (ROE) is the region with respect to TSS where the element was actually cumulatively observed to be enriched using log-likelihood scans over the Arabidopsis promoters. The overlapped ROE column highlights major differences (“No” entries) between expected locations of elements in the general literature and observed enrichment locations in Arabidopsis.
The common midpoint used for each CPE, since most CPEs are reported as having a range of start positions.
14,000 genes were scanned 500 bp upstream and downstream of a high-confidence TSS identified by Cumbie et al. (2015a).
CPEs were found 50 bp up- and downstream of the 5′ start position.
CPEs were found anywhere within 500 bp of the TSS (see footnote b).
ROE identified using an algorithm previously described (Morton et al., 2014) were found to overlap the region surrounding the CPE (see footnote c).
Percentage of genes where no CPE was found within 500 bp of the TSS (see footnote b).
Percentage of genes where no CPE was found around the preferred CPE start site (see footnote c).
Genomic Elements That Matter: TSS Peak Shape, TF Binding Site Position, and the Open-Chromatin Connection
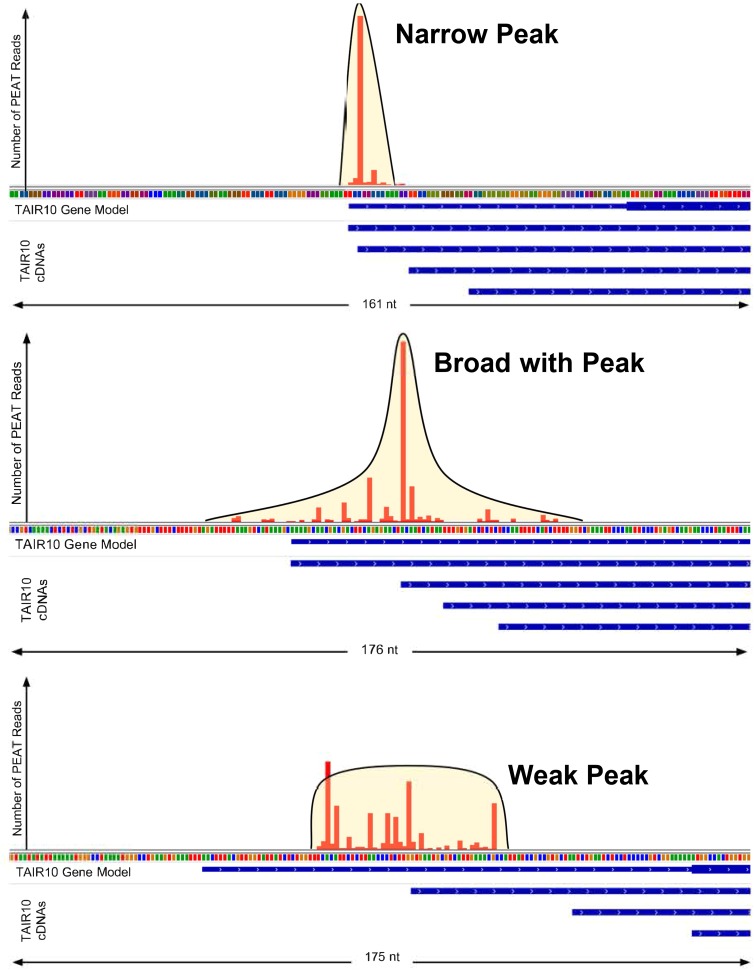
In order to perform precise studies of proximal or distal regulatory regions for miRNAs or PCGs, one must first identify the TSSs associated with Pol II genes. By obtaining TSS sequencing (TSS-Seq) data using recent protocols such as PEAT (Ni et al., 2010), CAGE (Carninci et al., 2005; Takahashi et al., 2012), or variants of nanoCAGE (Salimullah et al., 2011; Cumbie et al., 2015a), it is now possible to examine the nature of the promoters driving highly expressed TSS locations within a sample on a genome-wide scale. TSS-Seq data provide a new window into associations between TSS distribution along the genome, the location of TF binding sites (TFBSs), chromatin state surrounding the TSS regions, and gene function. Studies in both plants and animals conclude that TSSs fall into several general “shape” categories according to the distribution of TSSs along the genome (Carninci et al., 2006; Ni et al., 2010; Morton et al., 2014) (Figure 2). In all of these studies, TSS peak shapes tend to associate with gene functional categories in the following way: Narrow peaks tend to associate with time or tissue-specifically expressed genes, broad/weak peaks with housekeeping and other constitutively and/or ubiquitously expressed genes, and broad with peaks with circadian and other genes that have a mixture of these qualities over space and time. This could be explained by the concept that the availability of CRE sites within a gene’s upstream/downstream region is tuned to a gene’s spatio-temporal expression needs. For example, broad/weak TSS peaks may be present because the nearby sequence offers a large variety of CRE combinations (however weak) at which at least a low level of transcription could begin because the organism needs to ensure that this gene’s protein is produced in a tissue reliably under a wide variety of circumstances (a variety of TFs present at varying concentrations), and similarly for the stringency/specificity of narrow peak expression.
Figure 2.
Tag Cluster Shape Categories.
Examples of narrow peak (top), broad with peak (middle), and broad/weak peak (bottom) TSS-Seq tag clusters. The horizontal axis of each plot displays a region of genomic sequence, with TAIR10 cDNAs in the region displayed below the axis. The vertical axis displays the number of TSS-Seq reads observed at each nucleotide location in the region. (Reproduced from Morton et al. [2014], Figure 1.)
However, most striking and difficult to reconcile with classical views of promoter structure is the position-specific enrichments that TSS-Seq studies have revealed for TFBSs upstream of highly expressed TSS peak locations (Megraw et al., 2009; Morton et al., 2014, 2015). There are many CREs associated with specific TFs that one would not classically consider core elements, but a surprisingly large number of these TF-associated CREs are difficult to categorize purely as enhancer elements because of their strong positional preferences in the proximal promoter region and their apparent importance to defining TSSs. Particularly in plants, these TF-associated CRE enrichment positions can be used together with known CPEs to accurately predict the locations of TSS peaks of all three types (but known plant CPEs alone cannot accurately predict TSS location) (Morton et al., 2014). Interestingly, regions of open chromatin (OC) are proving useful to identify functional CRE sites (Song and Crawford, 2010; Song et al., 2011; Vierstra et al., 2015), yet are not required to accurately predict a highly expressed TSS location (Morton et al., 2015). It may well be the case that knowledge of OC regions are a necessary component to understanding the tissue, time, and/or level at which a given promoter region will express. Adaptations of DNase-I hypersensitive site sequencing protocols have recently yielded studies of OC regions in plants, including Arabidopsis (Zhang et al., 2012a; Sullivan et al., 2014; Cumbie et al., 2015b; Liu et al., 2015) and rice (Oryza sativa; Zhang et al., 2012b; Pajoro et al., 2014), and these data sets offer immense opportunity for further study. These OC data sets suggest that on average the genomic region from the TSS to ∼1 kb upstream tends to be much more open to transcription than other regions.
As a result of TSS-Seq, OC-Seq, and other epigenetic studies in animals, a new paradigm for understanding the structure of promoter regions is starting to emerge in the literature which posits that the classical binary division into core versus distal promoter regions is an oversimplification that no longer aligns well with the totality of current findings, particularly in vertebrates (reviewed in Andersson, 2015; Feuerborn and Cook, 2015; Kim and Shiekhattar, 2015). In particular, it is clear that the presence of one or more previously labeled CPEs within a promoter is not an absolute requirement in order to achieve basal transcription (Kim and Shiekhattar, 2015) and that the structure of regions that function as a “promoter” and the structure of regions that function as an “enhancer” is essentially identical for many human and mouse loci. The most recent plant promoter analysis study based on high-throughput TSS capture (Morton et al., 2014) concurs with this view, showing that up to computational analysis, it appears that strongly expressed TSS peaks result from the presence of a combination of CREs that are specifically located with respect to each other in genomic sequence and that this combination does not necessarily require CPEs. Only when fundamental questions have been answered about the basic structure of plant promoters can we begin to address more complex inquiries, including the fascinating question of whether all instances of bidirectional transcription arise from cognate pairs of reverse-oriented core promoters. Many further investigations into the structure and operation of transcriptional regulatory regions in their entirety are required in order to obtain a detailed functional understanding of plant promoter architecture.
Using Machine Learning to Identify Elements Predictive of Transcription
TSS information for a gene within a sample of interest may already be available from existing gene-by-gene experiments such as 5′-RACE assays (Xie et al. [2005] is the most extensive such study in Arabidopsis, and Zhang et al. [2009] provides a more recent study in maize), or one may have the resources to obtain TSS-Seq data directly in the lab. But what if this data is absent—where should one look for potential regulatory sites? What about the case of miRNAs, where even if TAIR annotation is present for the mature sequence or precursor, the TSS is unannotated and the primary transcript may be of considerably variable length? What are the combinations of cis-regulatory motifs that would lead one to believe that a TSS is present, and which CREs are the most likely contributors to transcription in that case? Looking at the presence of one or several CREs in regions near TSSs, or at simple aggregate statistics such as TF binding site enrichments or general sequence enrichments, certainly is helpful (Yamamoto et al., 2009, 2011; Kumari and Ware, 2013), but many important questions of this nature are very difficult to address “by eye.” This is where statistical pattern recognition methods can play a uniquely helpful role in scientific problem solving, and these methods are well developed for scientific application (reviewed in Jordan and Mitchell, 2015). A potential barrier to their application is the requirement for field-specific knowledge and experience with these methods, along with some facility with general computing. Another requirement is generally a fairly large number of “training examples” (on the order of at least hundreds of examples) to work from when attempting to “learn” a pattern within a data set. Yet understanding when and how these methods can address a scientific problem is immensely useful, even if one may need to seek collaboration with colleagues in order to employ a pattern recognition method. Machine learning methods in general lend themselves well to the problem of identifying TSSs and the patterns of sequence elements that lead to transcription; there has been a long history of progress in this direction largely in animal studies (reviewed in Kapranov, 2009), but recent availability of precise large-scale TSS-Seq data sets have created a unique opportunity for progress particularly in plants (Morton et al., 2014; Cumbie et al., 2015a). Here, we will use an example to explain how machine learning methods can be applied to answer the questions presented above; in particular, where is transcription most likely to begin for a miRNA or gene of interest, and which CREs are most likely to be contributing to transcription at these locations?
Examples of machine learning studies that specifically address the search for miRNA primary transcript TSSs in animals include Zhou et al. (2007), Megraw et al. (2009), and Marsico et al. (2013). Morton et al. (2014) provide a machine-learning based plant study addressing miRNA TSSs as a subcategory of Pol II TSSs. The most recent whole-genome prediction efforts in animals (Bhattacharyya et al., 2012; Georgakilas et al., 2014) focus predominantly on data such as histone modifications, although one can train a remarkably successful TSS prediction model based on sequence alone for Pol II TSSs (Morton et al., 2015); sequence-only based models are particularly useful in organisms where epigenetic data sets are still sparse. The overall machine learning paradigm is to first identify a training set composed of representative TSS locations (in case of miRNAs this may come from other Pol II genes) and non-TSS locations. Second, a “feature set” is identified; this is a set of numerical characteristics of every location in the training set, whether it is a TSS or a non-TSS. For example, features could include TF binding site score or the presence/absence of a histone mark at a certain genomic position relative to the location under examination. A model is “trained” to discriminate between locations that are TSSs and not TSSs within the training set. The model is simply a mathematical function that for any given location under examination takes the features for that location as inputs and provides either a yes/no answer, a score, or a probability for that genomic location to be a TSS as an output. The model is then tested on a previously unseen data set to determine whether its performance generalizes well to cases beyond the training set. A perfectly successful model will correctly identify all TSSs in the test set as TSSs and all non-TSSs as non-TSSs. Finally, if a highly successful model is able to be trained, one can then examine the model to determine which features have contributed most heavily to successful prediction. One can also examine successfully predicted TSS locations to determine which features were present that lead to the determination of this site as a TSS. In doing so, if the feature set consists of CREs, for example, one can determine the combination of CREs that were predictive of transcription happening at this particular genomic location and, therefore, a likely combination of regulators for transcripts that begin at this TSS.
There is much to learn from this paradigm and countless questions remain to be addressed. How well does a model of promoter structure that is identified in one plant species, for example, a dicot such as Arabidopsis, generalize to other dicots with a similar set of TFs and sequence composition characteristics? Does phylogenetic distance matter or has promoter structure been deeply conserved at least within a specific plant tissue from the time of an ancient common ancestor? To what degree has evolution transformed elements used to promote transcription at particular start sites under conditions of stress or in different stages of development? Would information about OC regions allow one to determine not only the genomic locations of strongly expressed TSS locations but their level of expression as well or are we still missing critical causal information, perhaps DNA methylation status or histone marks? Studies (Zhang et al., 2012a; Morton et al., 2014; Pajoro et al., 2014; Sullivan et al., 2014; Cumbie et al., 2015b) are just beginning to provide data to scratch the surface of these types of questions, though many new data sets and machine learning studies will be required in years to come in order to make steady progress in this area.
TARGETING: TOOLS AND TECHNIQUES
In order to identify small functional genetic circuits within a regulatory network, identifying and confirming TF and miRNA targets is just as important as identifying their upstream regulators. Even if we have precisely identified the most highly expressed start site for a given gene within a sample of interest and have a notion of a binding motif for a TF of interest, this motif is often short and sometimes degenerate; there may be dozens of such sites in the vicinity of a single gene and an equal density of matches or partial matches in random intergenic sequence (Siebert and Söding, 2014). What to do? And what about finding the targets of a miRNA of interest—how do we find a path through the array of computational tools and validation techniques available? Here, we provide a review of the most straightforward methods from the recent literature and note current tradeoffs between accuracy and ease-of-use.
Where's My TF Binding Site? Computational TFBS Discovery in a Nutshell
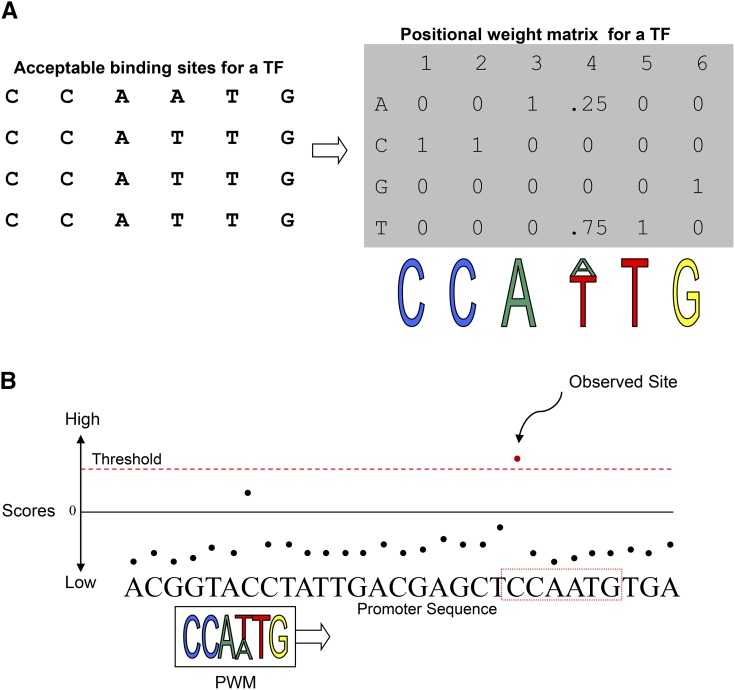
Suppose one has a TF of interest and wishes to identify direct targets of this TF within a gene set of interest. There are two general cases: either the TF:DNA binding motif is currently known in some form, or it is entirely unknown. We will not cover the latter case in extensive detail, but will comment briefly here on several common laboratory approaches to elucidate a TF’s binding profile when entirely unknown. A binding profile can be visualized as a “sequence logo,” that is, a summary of all of the possible DNA sequences of a small fixed size (generally 6 to 10 nucleotides) to which a TF’s binding domain can bind (Figure 3). Traditionally, to obtain a full profile of these possible sequences, one would take an in vitro approach such as SELEX (Kinzler and Vogelstein, 1990; Tuerk and Gold, 1990) where a large number of oligos of a given size are mixed with purified TF protein and a tabulation of bound oligos is used to form the profile. A high-throughput version of this assay called HT-SELEX (Jolma et al., 2010) is now available. This same general concept can also effectively be applied on a microarray chip using protein binding microarrays (Mukherjee et al., 2004), and a large protein binding microarray data set has recently become available in Arabidopsis (Weirauch et al., 2014). An alternate postprocessing strategy for producing binding profiles known as the Seed-and-Wobble algorithm was described previously (Berger and Bulyk, 2009; Gordân et al., 2011). If an antibody to the desired TF is available, chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-Seq) can be performed on a sample of interest; this does not result directly in the bound sequence locations, but the binding signal from a large number of longer sequences must be processed computationally using de novo motif finding ((Kharchenko et al., 2008). As a result of the considerable uncertainty in this process, it is often desirable to combine ChIP-Seq with supporting data from other sources; the DeepBind program (Alipanahi et al., 2015) is an example of a recently published software package for this purpose. Recently, approaches such as SpecSeq (Stormo et al., 2015) have been published, in which bound and unbound fractions in a standard binding reaction are sequenced directly.
Figure 3.
TFBS Log-Likelihood Scanning with a PWM.
(A) Given a collection of sequences that represent observed binding sites for a TF, a PWM “counts up” the number of As, Cs, Gs, and Ts in each position to describe the chance of finding each nucleotide in this position. The PWM can be visualized as a “logo” that describes how often the TF is expected to bind certain types of sites.
(B) An illustration to visualize scanning for binding sites: Only those sites in a promoter sequence that exceed the PWM-specific threshold score are “observed” as putative binding sites.
(Adapted from Megraw and Hatzigeorgiou [2010], Figures 1 and 4, for [A] and [B], respectively. Springer Plant MicroRNAs, Methods in Molecular Biology, Chapter 11, Vol. 592, 2010, pp. 149–161, Megraw and Hatzigeorgiou, © Humana Press a part of Springer Science + Business Media, LLC 2009, with permission of Springer.)
Studies using these experimental methods, along with individual ChIP and other studies, have been compiled from the plant literature into two primary database sources: Plant TRANSFAC (Wingender, 2008) and to a lesser degree JASPAR (Bryne et al., 2008). TRANSFAC is the most comprehensive source of TF binding profiles, containing over 100 profiles in plants; unfortunately, it is not public and must be purchased. On the order of 20 profiles have made their way into secondary free sources, and Weirauch et al. (2014) offer a new resource to be mined. Starting with these binding profile representations or PWMs (Stormo, 2000), the time-tested method for computational TF binding site discovery in both the plant and animal literature is log-likelihood scanning (Durbin et al., 1999). This is the process of computing a score at each nucleotide in a putative promoter sequence as the log of a probability ratio: It is the probability that the fixed-size motif under examination matches a PWM divided by the probability that it matches the background sequence composition. One advantage of this method is that for each PWM, one can explicitly compute specific theoretical false positive rate and false negative rate cutoff thresholds for scanning; for example, one can scan a PWM over a length of sequence such that any location matching the PWM with a log-likelihood score above a given value (representing a false-positive rate of say 0.01%) is deemed a “site.” This was the method used to determine sites in Table 1 above. One can obtain promoter sequences to scan from TAIR or ideally obtain more precise locations if TSS-Seq data are available. Log-likelihood scanning for TF binding sites using a dinucleotide background model has worked extremely well in TSS prediction models that use TF binding sites as features (Megraw et al., 2009; Morton et al., 2014). A downside of this method is that available log-likelihood scanning toolsets that perform both scanning and PWM-based thresholding (Cumbie and Megraw, 2015) generally require a considerable level of comfort with command line computing skills. A reasonable alternative is a tool such as STORM (Schones et al., 2007), which can search for modules of sites and takes an empirical approach to estimate cutoffs for site determination as by Kumari and Ware (2013). Statistically well-grounded current options still leave something to be desired in a user-friendly graphically based software suite oriented toward plant TF binding site discovery for noncomputationally trained scientists.
Techniques That Bash Promoter Bashing: TF Binding Validation in 2015
Functional analysis of Pol II promoters is challenging due to the combinatorial nature of gene regulatory control and can be especially difficult for promoters of miRNAs as a result of inadequate primary transcript annotation. Several experimental approaches can be used to dissect promoter elements for miRNAs and coding genes and to identify TFs interacting with regulatory DNA in vitro and in vivo. These methods include “promoter bashing” or site-directed mutagenesis, ChIP, the yeast one-hybrid (Y1H) system, and electrophoresis mobility shift assays (EMSAs). Additionally, the recently described CRISPRi method offers a potential approach for probing direct binding interactions by silencing specific sites and observing the perturbed expression outcome (Qi et al., 2013).
Promoter bashing is a time-tested and valuable approach for identifying cis-regulatory elements and entails systematic mutagenesis of particular sequence regions followed by testing the effect of mutation or deletion on gene expression. To test the activity of mutagenized promoter variants, their sequences are transcriptionally fused to a reporter gene and expressed in vivo. In plants, GUS or GFP are reporters of choice for the analysis of Pol II promoter activity and tissue specificity (Parizotto et al., 2004; Yan et al., 2012). However, bioluminescence reporters such as firefly luciferase (LUC) offer the advantages of high sensitivity and real-time quantification. Importantly, the dual firefly/Renilla luciferase reporter system allows internal normalization of LUC expression. For example, using site-directed mutagenesis and dual LUC assays, Qian et al. (2011) identified TFBSs in the promoters of the Drosophila intergenic miRNAs bantam and miR-276a. Promoter bashing in combination with sensitive reporter systems is still a powerful tool for dissecting functional regions and for empirically determining or confirming predicted TFBSs in miRNA or PCG promoters, but mutagenesis can be laborious and time-consuming in plants, particularly due to the necessity of obtaining transgenics. The relatively recent development of a transient protoplast system with higher throughput potential may significantly improve promoter bashing in vivo. The Arabidopsis mesophyll protoplast system enables facile, cost-effective, and potentially high-throughput transient gene expression analysis in vivo (Yoo et al., 2007).
Due to the recent rapid progress in high-throughput sequencing technologies, the method of ChIP-Seq (reviewed in Park, 2009) is now a mainstream approach for the discovery of novel CREs in promoters of coding and noncoding RNAs. For example, in mammals, histone modifications histone H3 trimethylated at lysine 3 (H3K4me3) and lysine 36 (and H3K36me3) are enriched in the promoters (Guttman et al., 2009) and along the length (Mikkelsen et al., 2007) of actively transcribed regions, respectively. Using ChIP-Seq data, Guttman et al. (2009) mapped locations of H3K4me3 and H3K36me3 histone marks and identified ∼1600 noncoding RNAs and their promoters. With some exceptions, mammals and higher plants have a very similar histone code (reviewed in Liu et al., 2010). Therefore, use of plant-adapted ChIP-Seq protocols (such as the one described by Kaufmann et al., 2010) is likely to greatly facilitate identification of active promoters of noncoding RNAs in plants in the future.
ChIP-Seq data can be complemented by the analysis of nucleosome positioning. This approach utilizes observations that transcriptionally active promoters are associated with nucleosome-free regions (Lee et al., 2004a; Yuan et al., 2005; Sekinger et al., 2005) and also can be marked by several specific histone modifications (Pokholok et al., 2005; Guenther et al., 2007; Heintzman et al., 2007). By analyzing nucleosome positioning and chromatin modifications, Ozsolak et al. (2008) identified putative promoters of a sizable portion of human miRNAs. Moreover, nucleosome positioning and linker sequence mapping allowed predictions not only of miRNA promoters but also TFs targeting these promoters. Nucleosome depleted regions mapped to the miRNA promoters, whereas DNA sequences corresponding to mature miRNAs were preferentially associated with well positioned nucleosomes (Ozsolak et al., 2008). This approach suggested that many intronic miRNAs utilize their own host gene-independent promoters.
A Gateway-compatible Y1H system enables the large-scale discovery of novel protein-DNA (including TF promoter) interactions in vivo (Deplancke et al., 2006; reviewed in Reece-Hoyes and Marian Walhout, 2012). Using the Y1H system, Martinez et al. (2008) showed that 63 putative Caenorhabditis elegans miRNA promoters can interact with 116 TFs, producing a total of 347 interactions. Similarly, Brady et al. (2011) identified numerous TF-miRNA promoter interactions between eight miRNA promoters and 15 TFs expressed in Arabidopsis root. Genome-size libraries of Y1H-compatible clones of Arabidopsis TFs are maintained in several laboratories (Pruneda-Paz et al., 2009; Ou et al., 2011) and can be used for miRNA promoter screening.
In EMSA assays, a short biotin or (32P)-labeled DNA fragment is incubated with purified protein or nuclear (or cellular) extracts and separated in polyacrylamide gel under nondenaturing conditions. Interacting DNA-protein complexes migrate slowly and are observed in the gel as a shifted band. Addition of an antibody against the protein of the interest may also result in a super-shift of the retarded band. EMSA is a method of choice for validation and functional dissection of TF interactions with their target promoters. EMSA has been broadly used for characterization of core promoter elements in both animals (Morachis et al., 2010; reviewed in Roy and Singer, 2015) and plants (Achard et al., 2003), including characterization of miRNA core promoters (reviewed in Xie et al., 2010). Using gel-shift assays, Bhogale et al. (2014) identified binding sites for the SQUAMOSA PROMOTER BINDING-LIKE9 (SPL9) TF in the potato (Solanum tuberosum) miR172 promoter. Predictably, these binding sites contained a GTAC motif resembling SPL9 binding sites in the promoters of protein coding genes.
EMSA was successfully used to demonstrate that transcriptional activation of miRNA-34a in human cancer cells depends on direct binding of NF-kappaB factor to the miR-34a promoter (Li et al., 2012). This binding was further corroborated by ChIP assays and by super-shift of the miRNA-34a promoter fragment in the presence of anti-NF-kappaB protein antibody. This study is an excellent example of using complementary EMSA, ChIP, and promoter deletion assays to dissect active regulatory elements and to discover novel regulatory mechanisms of miRNA transcription. EMSA can be also employed to confirm the binding of one nucleic acid to another (Hellman and Fried, 2007). Gel-shift assays were successfully used to validate binding of animal small RNA (Morita et al., 2012) or miRNA (Solé et al., 2013) to their mRNA targets. Thus, EMSA and ChIP assays are both well vetted and appropriate choices for the analysis and functional dissection of plant miRNA promoters in vitro and in vivo.
Among emerging novel approaches for genome editing, the CRISPR-Cas nuclease system deserves special attention because it acts through stable transcriptional repression or activation including promoter elements. The system is based on bacterial adaptive immune system and involves clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) endonucleases (reviewed in Haimovich et al., 2015). Recently, the CRISPR-Cas system was adapted for mapping and perturbing cis-regulatory elements in the promoters of human and yeast cells (Gilbert et al., 2013). Using this method (termed CRISPR interference or CRISPRi), Gilbert at al. showed that catalytically inactive dCas9 fusion proteins can target and silence proximal promoter elements (presumably via recruiting chromatin modifiers). CRISPRi-mediated transcriptional repression is highly specific and can be used as a modular and flexible DNA binding platform (Gilbert et al., 2013). Future adaptations of the CRISPRi system for the plant cell may provide a powerful tool for probing CREs via targeted and simultaneous silencing of several miRNA promoters.
Small RNA Target Primer: Prediction and Validation
Many plant miRNAs are thought to play important roles in plant physiology and development. Therefore, identifying functionally important miRNA interactions with their targets is essential to the understanding of functionality of cellular regulatory networks. Nonetheless, the in vivo activities of many plant miRNAs remain unknown. Generally, plant miRNA targets are more amenable to computational prediction compared with their animal counterparts because in plants the recognition of target sites often requires higher complementarity between miRNAs and their targets. Plant miRNAs also frequently guide their target mRNAs to cleavage, whereas their animal counterparts seem to preferentially act by translational inhibition of their targets. It must be noted, however, that cleaved mRNA targets may represent only a portion of all plant miRNA targets, and the possibility that some plant mRNAs are targeted for translational inhibition through imperfect miRNA binding has not been tested extensively (Brodersen and Voinnet, 2009). For example, a mechanism different from target cleavage has been reported for the noncoding RNA IPS1 (INDUCED BY PHOSPHATE STARVATION1) by miR399 (Franco-Zorrilla et al., 2007), which like IPS1 is involved in the response to phosphate starvation (Fujii et al., 2005; Chiou et al., 2006). The miR399 target site in IPS1 contains a three-nucleotide insertion, which prevents cleavage of the IPS1 transcript, resulting instead in sequestration of miR399 in a RISC-miR399 complex, leading to a reduction of active miR399 level. This mechanism resembles the so-called miRNA sponges observed in animals, which are represented by RNA transcripts containing multiple miRNA binding sites that compete with endogenous miRNA targets, thereby reducing the efficiency of miRNAs (Ebert et al., 2007). Mechanisms other than those affecting the miRNA level apparently also exist in plants, which affect the interaction between miRNAs and their targets. For example, it has been reported that despite similar miR159 levels throughout Arabidopsis development, its efficiency in targeting GAMYB-like transcripts MYB33 and MYB65 is attenuated in seeds compared with vegetative tissues (Alonso-Peral et al., 2012). Recent reports indicate that besides target cleavage, plant miRNA interactions with their targets include translational inhibition, difference in AGO1 subcellular localization, and regulation of RISC complex assembly and miRNA loading (Brodersen et al., 2008, 2012; Wang et al., 2011; Zhu et al., 2011; Iki et al., 2012). A novel twist in the miRNA functional relationships with their targets is also indicated by the recent discovery that in addition to miRNAs, primary miRNA transcripts (pri-miRNA) also encode short regulatory peptides able to affect pri-miRNA expression level (Lauressergues et al., 2015). The complexity of miRNA-mediated regulation of plant function is also reflected in the ability of miRNAs to diffuse through plasmodesmata, forming concentration gradients, and to act in a non-cell-autonomous morphogen-like manner on their targets (Carlsbecker et al., 2010; Miyashima et al., 2011; Knauer et al., 2013). In addition, miRNAs can move long-distance through the phloem and serve as systemic signals, e.g., for leaf development and phosphate homeostasis (Juarez et al., 2004; Pant et al., 2008). Thus, in some cases, the effects of plant miRNAs are not limited to simple contexts, as they can act in cell-to-cell or organ-to-organ communication. Furthermore, in comparison to animals, plant miRNAs are typically encoded by larger gene families. Interestingly, in Arabidopsis, a negative correlation between the copy numbers of miRNAs and the size of the target families they regulate has been reported (Takuno and Innan, 2008). Indirect targeting is an additional layer of complexity that one must consider when attempting to identify miRNA targets via a method that does not directly validate miRNA:target interactions. As plant miRNAs often target TFs, which are likely to represent hubs in regulatory networks, many downstream signaling components and effector proteins can become the indirect targets of miRNAs.
Currently, variations on two major types of approaches, computational and experimental, are widely used to predict miRNA targets in both plants and animals. Computational prediction algorithms are based on known miRNA-mRNA interaction rules. A number of computational tools have been developed to predict plant miRNA targets in a web server format. For example, Plant Small RNA Target Analysis (psRNATarget) (Dai and Zhao, 2011) implements two analysis functions: (1) reverse complementary matching between miRNAs and target transcripts, and (2) evaluation of target site accessibility on mRNA by calculating unpaired energy required to “open” secondary structure in the target-site region. A strategy to augment miRNA target predictions based on their conservation in different plant species has also been reported (Chorostecki et al., 2012). Several miRNA target discovery tools in plants have been directly compared on an experimentally supported reference data set in terms of sensitivity and specificity by Ding et al. (2012); these include psRNATarget as well as TAPIR (Bonnet et al., 2010), UEA_sRNA (Moxon et al., 2008), and the Web MicroRNA Designer 3 (WMD3; http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) toolset (Schwab et al., 2006; Ossowski et al., 2008, 2009). Among these, WMD3 offers a conservative tool that favors specificity and provides well-supported, time-tested software. In general, however, computational approaches suffer from an incomplete knowledge of all possible types of miRNA-target interactions. Experimental approaches have been also used that rely on measuring the change in target RNA level caused by a change in the corresponding miRNA level. Recently, approaches aimed at the generation of large-scale collections of knockdowns for Arabidopsis miRNA families have been reported using artificial miRNA target mimics and molecular sponges (Todesco et al., 2010; Reichel et al., 2015). The authors reported morphological defects in the aerial parts for 20% of the analyzed families known to be conserved across land plants. Although valuable, experimental approaches also yield false positives by picking up indirect targets whose expression level is modified. A more direct experimental approach for miRNA target prediction recently tested in Arabidopsis relies on the detection of the endonucleolytic cleavage products guided by specific miRNA. Plant AGO proteins, such as Arabidopsis AGO1, cleave the target RNAs between positions 10 and 11 of the miRNA binding site. Based on this mechanism, a “degradome-mapping” approach called parallel analysis of RNA ends uses a modified RACE PCR to identify target fragment ends in vivo (German et al., 2008).
As a rule, both computational and experimental miRNA target prediction strategies require additional experimental validation of the functionality of each miRNA-target interaction in the biological context of interest. Specific miRNA activity can be validated using naturally occurring or engineered gain-of-function alleles of mRNA that alter the miRNA complementary site without changing the protein sequence of the target but render the target resistant to mRNA cleavage (Emery et al., 2003; Tang et al., 2003; Mallory et al., 2004). Other popular approaches use “miRNA sensors,” which represent miRNA-sensitive sequences fused to reporter genes such as GFP, RFP, and luciferase. In Arabidopsis, stable lines (Parizotto et al., 2004; Carlsbecker et al., 2010; Nodine and Bartel, 2010), transient expression in leaf-mesophyll protoplasts (Martinho et al., 2015), and in vitro systems with plant cell extracts (Iwakawa and Tomari, 2013) have been used with miRNA sensors to asses miRNA interactions with their targets.
SMALL CIRCUITS AS BUILDING BLOCKS FOR UNDERSTANDING PLANT FUNCTION
The recent progress in TF-promoter target analysis and miRNA-mRNA target prediction, along with evolving technologies for validating both, are opening the field for new studies on how small RNAs exert a system-wide influence on plant function. While the role of small RNA regulatory circuits as modular building blocks for the function of complex networks has long been on the forefront of studies in bacteria, yeast, and some insects, plant studies are poised for takeoff in this due to the ideal nature of their genomes for investigation and comparison in a multicellular domain.
Small miRNA-Containing Networks Point to Function
In addition to identification of the miRNA targets, research is beginning to focus on the regulation of miRNA genes themselves. This is still an underexplored area despite its importance for the reconstruction of miRNA-containing subnetworks. Experimental identification of TSSs and promoter regions for pri-miRNAs on a global scale is still lacking. Using a 5′-RACE approach, Xie et al. (2005) identified the TSS locations of ∼63 pri-MIR transcripts in Arabidopsis. Many of these sites are supported by a TSS-Seq approach (Morton et al., 2014). Nevertheless, several specific studies have been performed in Arabidopsis and other plant species, unraveling intriguing miRNA-containing networks. For example, mutant characterizations and genetic analyses have revealed an evolutionarily conserved and tightly regulated network that involves miR156 and miR172 and regulates many biological processes during development (reviewed in Rubio-Somoza and Weigel, 2011; Yu et al., 2015). Targets of miR156 are SPL TFs, which have a wide spectrum of activities (Cardon et al., 1999). For example, SPL9 induces flowering through transcriptional activation of MADS box proteins APETALA1 (AP1), FRUITFULL, and SUPPRESSOR OF OVEREXPRESSION OF CO1 (Wang et al., 2009; Yamaguchi et al., 2009) and promotes terpene biosynthesis through activation of terpene synthase TPS21 (Yamaguchi et al., 2009). SPL9 also represses cytokinin responses by interacting with B-type ARABIDOPSIS RESPONSE REGULATORs and anthocyanin production through binding with MYB transcription factors in the anthocyanin biosynthetic pathway (Gou et al., 2011; Zhang et al., 2015). Interestingly, while miR156 level decreases with plant age, that of miR172 gradually increases because the miR156 target, SPL9, is a positive regulator of pri-miR172B transcription (Wu et al., 2009). miR172 targets AP2-like TFs, which act as repressors of flowering (Mathieu et al., 2009). Another miRNA-containing regulatory circuit has been recently linked to the regulation of leaf complexity (Rubio-Somoza et al., 2014). miR319 targets the transcription of TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) TFs that interfere with the function of miR164-regulated and miR164-independent CUP-SHAPED COTYLEDON (CUC) proteins, preventing the formation of leaf serrations in Arabidopsis and Cardamine hirsuta. As plants age and miR156 declines, miR156-repressed SPLs accumulate and act to destabilize TCP-CUC interactions, allowing for gradual increase of leaf complexity. Increasing evidence also indicates that miRNAs regulate key components of hormone signaling pathways and hormone homeostasis. Auxin signaling in particular is tightly regulated by miRNAs in a manner that was found to be conserved among different plant species. For instance, it has been demonstrated that in Arabidopsis, the auxin response factors ARF10, ARF16, and ARF17 are regulated by miR160 (Rhoades et al., 2002; Bartel and Bartel, 2003). miR160-regulated ARF17 acts to repress genes involved in auxin conjugation and thus repress the level of free (active) auxin. Therefore, plants expressing a form of ARF17 that is resistant to miR160 regulation have altered auxin homeostasis associated with numerous growth defects (Mallory et al., 2005).
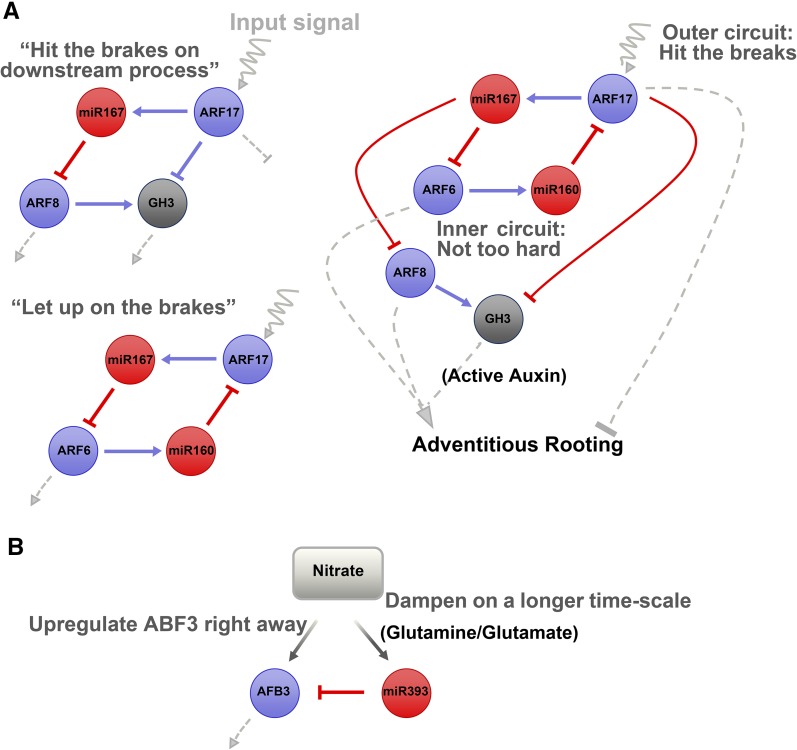
Another auxin-related miRNA network is involved in the regulation of adventitious root formation in Arabidopsis (Figure 4A). As shown in Figure 4, interpreting regulatory cascades as a collection of interlocking small genetic circuits can often aid in understanding how these relatively complex networks might function. In this network, miR160, miR167, ARF6, ARF8, and ARF17 form multiple feedback circuits (Gutierrez et al., 2009; reviewed in Rubio-Somoza and Weigel, 2011). It has also been shown that miR390 is induced during lateral root initiation and triggers the local production of trans-acting siRNAs (tasiRNA). In the lateral root primordium, these tasiRNAs reduce the activity of ARF2, ARF3, and ARF4, thereby promoting lateral root growth. In turn, ARF2, ARF3, and ARF4 are required for proper miR390 expression through feedback transcriptional regulation. Thus, miR390, tasiRNAs, and their ARF targets define a regulatory network for lateral root growth.
Figure 4.
Interpretations of Regulatory Cascades as Interacting Small Genetic Circuits.
(A) The literature-supported network controlling adventitious root formation is complex, and the exact outcome depends on timing and concentrations of each component. By visualizing the two small genetic subcircuits on the left and examining the outcome if one component (in this case ARF17) were suddenly up- or downregulated, one can see that the complex circuit on the right can avoid a sudden “shutoff” (or alternatively, “full throttle”) to a downstream process.
(B) One can extend this thinking to include other input “components”; here, viewing nitrate response as a miRNA-mediated control circuit for AFB3 regulation suggests one way that a plant can respond to continued nitrate input with a damped “pulse” of AFB3.
miRNA-Mediated Circuit Response: Developmental Timing, Tissue Patterning, and Plant Plasticity
In contrast to animals, which establish their body plan during embryonic development, plants continue to elaborate their structures throughout their entire lifespan. Recent evidence indicates that miRNA-mediated networks play crucial regulatory roles in a tissue-specific manner and are able to fine-tune developmental transitions as well as plant response to the surrounding ecosystem. For example, in the Arabidopsis root, expression of pri-miR165a/166b occurs strictly in endodermis, activated by the TFs SHORT ROOT and SCARECROW, whereas mature miR165/166 diffuse into stele forming a gradient that declines toward the stele center; this miR165/166 distribution regulates the radial expression level of the homeodomain leucine zipper III (AD-Zip III) TFs, ATHB14 and ATHB15, resulting ultimately in the characteristic development of xylem, with metaxylem strands more central in stele and protoxylem strands more peripheral in stele (Carlsbecker et al., 2010). In the root tip meristem, miR165a and miR166a,b interact in a similar non-cell-autonomous manner with ATHB14 and ATHB15 in order to establish precise patterning of the root tissue layers (Miyashima et al., 2011). In the mature portion of the root, cleavage of INDOLE-3-ACETIC ACID INDUCIBLE28 mRNA by auxin-inducible miR847 upregulates auxin signaling leading to lateral root formation (Wang and Guo, 2015). In the shoot, the most extensively studied miRNA network is one that involves antagonistic miR156 and miR172 nodes and regulates developmental timing along with progression through different developmental phases in both monocots and dicots (reviewed in Rubio-Somoza and Weigel, 2011). For example, a high level of miR172 in the shoot induces flowering, whereas a high level of miR156 suppresses flowering (reviewed in Yu et al., 2015). The TF AP2, which represses the expression of multiple flowering-promoting transcripts, promotes the expression of miR156e, which in turn represses some of the flowering-promoting TFs of the SPL family, thus stabilizing the delay of flowering time. At the same time, expression of miR172b is downregulated by AP2 and further forms a feedback loop by acting as an AP2 repressor (Yant et al., 2010). In another regulatory module related to flowering, miR172 promotes flowering time by targeting the TF SCHLAFMÜTZE, a repressor of flowering that suppresses the expression of several flowering promoting TFs including the key flowering regulator, FLOWERING LOCUS T known as florigen (Mathieu et al., 2009). miR156 also regulates shoot regenerative capacity: Age-related gradual increase in miR156-targeted SPL TFs leads to a decline in the regenerative capacity of the shoot by attenuating cytokinin response (Zhang et al., 2015). Another interesting example is miRNA regulation of root system architecture in response to nutrient supply. For example, miR167 and its target, the auxin response factor ARF8 mRNA, have been shown to act specifically in the pericycle to control a network of genes, leading to induction of lateral root initiation in response to organic nitrogen (Gifford et al., 2008). Another regulatory module controlling root system architecture in response to nitrogen is represented by a feed-forward loop formed by miR393 and the auxin receptor AUXIN SIGNALING F-BOX3 (AFB3) (Figure 4B), in which AFB3 is induced by nitrate in a concentration-dependent manner and repressed by miR393, whereas miR393 is induced by N metabolites produced by nitrate reduction and assimilation (Vidal et al., 2010). Further evidence shows that AFB3 acts specifically in the context of the nitrate response and regulates a connected network of genes controlled by NAC FAMILY TRANSCRIPTION FACTOR4 (Vidal et al., 2013).
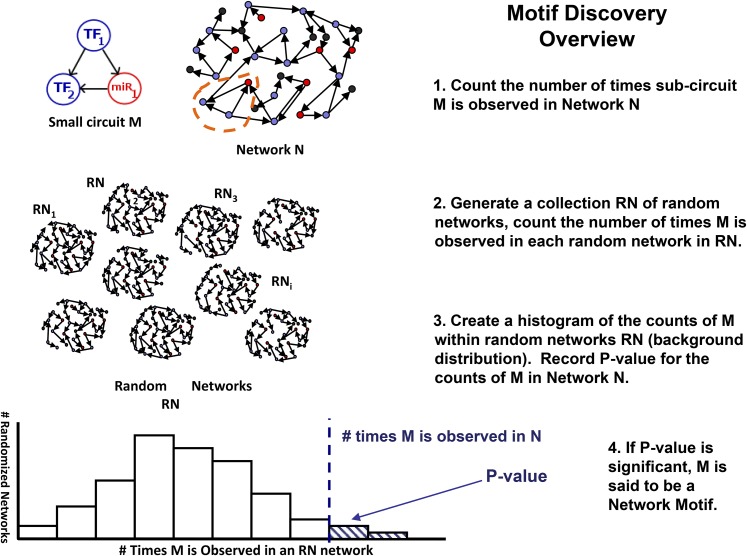
Network Motif Discovery: Distilling Large Complex Networks
All of the examples described above are small circuits of three or four genetic components typically involving TFs, miRNAs, and protein coding genes. Circuits involving miRNAs are unique in that they can elicit particular dynamics that may be difficult to achieve by substituting a transcriptional repressor (Osella et al., 2011); interestingly, miRNAs preferentially target TFs in plants (Jones-Rhoades et al., 2006), creating a variety of these TF-miRNA subcircuits. One of the most well-studied examples among such cases is the miRNA-mediated feed-forward loop (Tsang et al., 2007; Friard et al., 2010) (shown in Figure 5), which is capable of eliciting a controlled genetic pulse. In general, one framework for understanding a large complex genetic network composed of multiple types of genetic interactions is to view it as a composition of smaller subcircuits, each with an identifiable function. Just as one can construct an electronic circuit with complex function from small subcomponents that generate feedback or feed-forward loops, logic gates, or other specific signal-processing outcomes, the composition of genetic circuits can be viewed in a similar way; Alon (2006) provides a thorough overview of this subject. This is a useful viewpoint in the construction of synthetic circuits (reviewed in Medford and Prasad, 2014; Sowa et al., 2015); however, one of the most common needs in the study of endogenous networks is to dissect or “reverse-engineer” a large putative network in order to identify components that are likely to be playing the most critical roles. Network motif discovery is a well-established statistical method for accomplishing this goal in many areas of science (Alon, 2007); the central concept is to compare the frequency of particular subcircuits (such as a three-node feed-forward loop) in a given “real-world” network to its frequency in randomized networks (Figure 5). If the subcircuit appears much more often than in these randomized networks, it is said to be a network motif.
Figure 5.
Concept of Network Motif Discovery.
How network motif discovery works, illustrated using a small circuit of interest M. The method determines whether M is observed a significantly large number of times in an original network N compared with randomized networks RN. If the P value falls below a small predetermined value, the number of times that M is observed in original network N is considered to be significant and therefore M is called a network motif for N. (Adapted from Megraw et al. [2013].)
Historically, the first network motif discovery algorithms used in molecular biology (Milo et al., 2004; Wernicke and Rasche, 2006) focused on networks containing TFs because of their critical importance as master regulators in genetic systems (Jothi et al., 2009). Studies applying these algorithms to TF networks in bacteria (Shen-Orr et al., 2002) and yeast (Milo et al., 2002) met with good success, identifying and validating individual instances of specific motifs that were of strong interest to endogenous function. In these studies, a network of putative interactions was generated, and then the most important and interesting motifs were targeted for further investigation. In this way, motif discovery was shown to be a valuable hypothesis generation tool in predicted networks with a large number of interactions. The general method effectively sorts through a large number of possible circuits, some of which may contain false positive or false negative interactions and identifies statistically highly overrepresented cases as those most likely to be “standing out” above the noise, and thus worthy of further attention. However, a number of qualitative and quantitative challenges arise in the specific application of previous network motif discovery algorithms to very large systems such as Arabidopsis or human regulatory networks. It is also now widely recognized in eukaryotic systems that posttranscriptional gene regulation by small RNAs, including miRNAs, is a critical layer of gene regulation that must often be considered (Alon, 2007; Hobert, 2008). Large eukaryotic biological networks containing TFs share two common properties: a tendency for large hubs (both source hubs and target hubs) and many TF autoregulatory interactions. These two network qualities, in addition to the consideration of unique node types such as miRNAs, which have distinct properties from TFs, pose a significant challenge for network motif discovery background randomization algorithms in terms of efficiency and accuracy (Megraw et al., 2013).
Two recent algorithms address this challenge. The WaRSwap randomization algorithm (Megraw et al., 2013) provides a quick-sampling heuristic that focuses on uniform sampling of biological networks with any combination of interaction types present (TF-only and TF gene as well as those interaction types involving miRNAs); it accounts for TF autoregulation, large source and target hubs, and the possibility that miRNA target rearrangements can occur in evolutionary time. User-friendly software is now available (Ansariola and Megraw, 2015). CoMoFinder (Liang et al., 2015) takes a different approach, focusing on networks with all three component types present (TFs, miRNAs, and genes) where autoregulation and target rearrangements are not present; attention is turned away from uniform sampling and toward a fast method for finding subcircuits that are overrepresented with respect to a subcollection of networks identified by the algorithm as having many differences with the input network. CoMoFinder was applied to human network data, while WaRSwap was computationally validated in Arabidopsis. Astonishingly, the WaRSwap study found that when applied to TF-miRNA-gene networks from both developing Arabidopsis roots and Drosophila embryos, only two motif types were overrepresented and these were identical: an miRNA-mediated feed-forward loop controlling a gene, and a specific three-node signal switch where two mutually regulatory TFs both target a downstream effector (ensuring that the targeted TF, miRNA, or gene stays on or off) (Megraw et al., 2013). Both circuits are instrumental in ensuring reliable body plan formation in complex organisms, with specific circuits that had been previously validated in the literature and many yet to be investigated. Whichever tool one chooses, network motif discovery provides a freshly relevant option that is helpful in distilling the vast number of predicted TF and miRNA regulatory interactions from methods discussed above into a manageable number of hypotheses with specific testable cases, all backed by a form of statistical evidence. As new high-throughput data sets become available in plant samples from a variety of species, tissues, and conditions, motif discovery is a powerful method to quickly sort through many interactions and arrive at genetic circuit components that are playing key roles in plant function.
BACK TO THE BASICS/BACK TO THE FUTURE
This Review highlights several major areas where going “back to the basics” is now necessary for rapid advancements in understanding Pol II transcriptional circuits: These include intensive research on Pol II promoters and their functional control elements in a variety of plant species, in multiple plant tissues and cell types, over different stages of developmental time, and across environmental conditions. Advances that would greatly aid in this endeavor include more comprehensive user-friendly software for examining plant transcriptional control regions including TF binding site analysis and continued resources to hone the accuracy and throughput of validation methods that make improved models possible.
The ability to edit plant genomes at will, particularly in a relatively low-throughput manner as is now possible via the CRISPR-Cas system, only becomes truly powerful when one can predict the edits that will lead to a desired outcome. We are still some distance away from understanding how to edit regulatory regions such that expression in time, tissue, and under varying conditions can be controlled. We are still further from understanding how to construct small genetic circuit components in context, particularly those with powerful miRNA-mediated component behaviors, which play critical roles in organismal function. However, plant studies are poised to take a lead role in this area because of their advantages in probing transcriptional and posttranscriptional control of Pol II genes.
Acknowledgments
This work was supported by National Institutes of Health Grant GM097188 to M.M.
AUTHOR CONTRIBUTIONS
All authors contributed to writing this article.
References
- Achard P., Lagrange T., El-Zanaty A.-F., Mache R. (2003). Architecture and transcriptional activity of the initiator element of the TATA-less RPL21 gene. Plant J. 35: 743–752. [DOI] [PubMed] [Google Scholar]
- Alipanahi B., Delong A., Weirauch M.T., Frey B.J. (2015). Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33: 831–838. [DOI] [PubMed] [Google Scholar]
- Alon U. (2006). An Introduction to Systems Biology: Design Principles of Biological Circuits. (Boca Raton, FL: Chapman and Hall/CRC; ). [Google Scholar]
- Alon U. (2007). Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8: 450–461. [DOI] [PubMed] [Google Scholar]
- Alonso-Peral M.M., Sun C., Millar A.A. (2012). MicroRNA159 can act as a switch or tuning microRNA independently of its abundance in Arabidopsis. PLoS One 7: e34751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R. (2015). Promoter or enhancer, what’s the difference? Deconstruction of established distinctions and presentation of a unifying model. BioEssays 37: 314–323. [DOI] [PubMed] [Google Scholar]
- Ansariola M., Megraw M. (2015). WaRSwap software application. http://megraw.cgrb.oregonstate.edu/software/WaRSwapSoftwareApplication/.
- Bartel B., Bartel D.P. (2003). MicroRNAs: at the root of plant development? Plant Physiol. 132: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M.F., Bulyk M.L. (2009). Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Protoc. 4: 393–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard V., Brunaud V., Lecharny A. (2010). TC-motifs at the TATA-box expected position in plant genes: a novel class of motifs involved in the transcription regulation. BMC Genomics 11: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya M., Feuerbach L., Bhadra T., Lengauer T., Bandyopadhyay S. (2012). MicroRNA transcription start site prediction with multi-objective feature selection. Stat. Appl. Genet. Mol. Biol. 11: Article 6. [DOI] [PubMed] [Google Scholar]
- Bhogale S., Mahajan A.S., Natarajan B., Rajabhoj M., Thulasiram H.V., Banerjee A.K. (2014). MicroRNA156: a potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 164: 1011–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake M.C., Jambou R.C., Swick A.G., Kahn J.W., Azizkhan J.C. (1990). Transcriptional initiation is controlled by upstream GC-box interactions in a TATAA-less promoter. Mol. Cell. Biol. 10: 6632–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer C.G., van Bakel H., Tsui K., Li J., Morris Q.D., Nislow C., Greenblatt J.F., Hughes T.R. (2013). A unified model for yeast transcript definition. Genome Res. 24: 154–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E., He Y., Billiau K., Van de Peer Y. (2010). TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26: 1566–1568. [DOI] [PubMed] [Google Scholar]
- Brady S.M., et al. (2011). A stele-enriched gene regulatory network in the Arabidopsis root. Mol. Syst. Biol. 7: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190. [DOI] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Schaller H., Khafif M., Schott G., Bendahmane A., Voinnet O. (2012). Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 1778–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Voinnet O. (2009). Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 10: 141–148. [DOI] [PubMed] [Google Scholar]
- Bryne J.C., Valen E., Tang M.H., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., Sandelin A. (2008). JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 36: D102–D106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke T.W., Kadonaga J.T. (1996). Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 10: 711–724. [DOI] [PubMed] [Google Scholar]
- Cardon G., Höhmann S., Klein J., Nettesheim K., Saedler H., Huijser P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237: 91–104. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., et al. (2006). Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38: 626–635. [DOI] [PubMed] [Google Scholar]
- Carninci P., et al.; FANTOM Consortium; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) (2005). The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. [DOI] [PubMed] [Google Scholar]
- Chiou T.-J., Aung K., Lin S.-I., Wu C.-C., Chiang S.-F., Su C.-L. (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorostecki U., Crosa V.A., Lodeyro A.F., Bologna N.G., Martin A.P., Carrillo N., Schommer C., Palatnik J.F. (2012). Identification of new microRNA-regulated genes by conserved targeting in plant species. Nucleic Acids Res. 40: 8893–8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civán P., Švec M. (2009). Genome-wide analysis of rice (Oryza sativa L. subsp. japonica) TATA box and Y Patch promoter elements. Genome 52: 294–297. [DOI] [PubMed] [Google Scholar]
- Cumbie J.S., Filichkin S.A., Megraw M. (2015b). Improved DNase-seq protocol facilitates high resolution mapping of DNase I hypersensitive sites in roots in Arabidopsis thaliana. Plant Methods 11: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumbie J.S., Megraw M. (2015). TFBS Scanner Suite. http://megraw.cgrb.oregonstate.edu/software/TFBS_Scanner_Suite.
- Cumbie J.S., Ivanchenko M.G., Megraw M. (2015a). NanoCAGE-XL and CapFilter: an approach to genome wide identification of high confidence transcription start sites. BMC Genomics 16: 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Zhao P.X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39: W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Roberts S.G. (2005). A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 19: 2418–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B., Vermeirssen V., Arda H.E., Martinez N.J., Walhout A.J. (2006). Gateway-compatible yeast one-hybrid screens. CSH Protoc. 2006: pdb.prot4590. [DOI] [PubMed]
- Ding J., Li D., Ohler U., Guan J., Zhou S. (2012). Genome-wide search for miRNA-target interactions in Arabidopsis thaliana with an integrated approach. BMC Genomics 13 (suppl. 3): S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn A., Bollekens J., Staub A., Benoist C., Mathis D. (1987). A multiplicity of CCAAT box-binding proteins. Cell 50: 863–872. [DOI] [PubMed] [Google Scholar]
- Durbin R., Eddy S., Krogh A., Mitchison G. (1999). Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. (Cambridge, UK: Cambridge University Press). [Google Scholar]
- Duttke S.H.C., Lacadie S.A., Ibrahim M.M., Glass C.K., Corcoran D.L., Benner C., Heinz S., Kadonaga J.T., Ohler U. (2015). Human promoters are intrinsically directional. Mol. Cell 57: 674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M.S., Neilson J.R., Sharp P.A. (2007). MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4: 721–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13: 1768–1774. [DOI] [PubMed] [Google Scholar]
- Feuerborn A., Cook P.R. (2015). Why the activity of a gene depends on its neighbors. Trends Genet. 31: 483–490. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Friard O., Re A., Taverna D., De Bortoli M., Corá D. (2010). CircuitsDB: a database of mixed microRNA/transcription factor feed-forward regulatory circuits in human and mouse. BMC Bioinformatics 11: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Chiou T.-J., Lin S.-I., Aung K., Zhu J.-K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15: 2038–2043. [DOI] [PubMed] [Google Scholar]
- Georgakilas G., Vlachos I.S., Paraskevopoulou M.D., Yang P., Zhang Y., Economides A.N., Hatzigeorgiou A.G. (2014). MicroTSS: accurate microRNA transcription start site identification reveals a significant number of divergent pri-miRNAs. Nat. Commun. 5: 5700. [DOI] [PubMed] [Google Scholar]
- German M.A., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26: 941–946. [DOI] [PubMed] [Google Scholar]
- Gifford M.L., Dean A., Gutierrez R.A., Coruzzi G.M., Birnbaum K.D. (2008). Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 105: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L.A., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordân R., Murphy K.F., McCord R.P., Zhu C., Vedenko A., Bulyk M.L. (2011). Curated collection of yeast transcription factor DNA binding specificity data reveals novel structural and gene regulatory insights. Genome Biol. 12: R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J.-Y., Felippes F.F., Liu C.-J., Weigel D., Wang J.-W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. (2007). A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L., Bussell J.D., Păcurar D.I., Schwambach J., Păcurar M., Bellini C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21: 3119–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich A.D., Muir P., Isaacs F.J. (2015). Genomes by design. Nat. Rev. Genet. 16: 501–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N.D., Ren B. (2007). The gateway to transcription: identifying, characterizing and understanding promoters in the eukaryotic genome. Cell. Mol. Life Sci. 64: 386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N.D., et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39: 311–318. [DOI] [PubMed] [Google Scholar]
- Hellman L.M., Fried M.G. (2007). Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2: 1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. (2008). Gene regulation by transcription factors and microRNAs. Science 319: 1785–1786. [DOI] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Meshi T., Ishikawa M. (2012). Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 31: 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince T.A., Scotto K.W. (1995). A conserved downstream element defines a new class of RNA polymerase II promoters. J. Biol. Chem. 270: 30249–30252. [DOI] [PubMed] [Google Scholar]
- Iwakawa H.O., Tomari Y. (2013). Molecular insights into microRNA-mediated translational repression in plants. Mol. Cell 52: 591–601. [DOI] [PubMed] [Google Scholar]
- Javahery R., Khachi A., Lo K., Zenzie-Gregory B., Smale S.T. (1994). DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol. Cell. Biol. 14: 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A., et al. (2010). Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 20: 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. (2006). MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57: 19–53. [DOI] [PubMed] [Google Scholar]
- Jordan M.I., Mitchell T.M. (2015). Machine learning: Trends, perspectives, and prospects. Science 349: 255–260. [DOI] [PubMed] [Google Scholar]
- Jothi R., Balaji S., Wuster A., Grochow J.A., Gsponer J., Przytycka T.M., Aravind L., Babu M.M. (2009). Genomic analysis reveals a tight link between transcription factor dynamics and regulatory network architecture. Mol. Syst. Biol. 5: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez M.T., Kui J.S., Thomas J., Heller B.A., Timmermans M.C.P. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88. [DOI] [PubMed] [Google Scholar]
- Kadonaga J.T. (2004). Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116: 247–257. [DOI] [PubMed] [Google Scholar]
- Kadonaga J.T. (2012). Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip. Rev. Dev. Biol. 1: 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P. (2009). From transcription start site to cell biology. Genome Biol. 10: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472. [DOI] [PubMed] [Google Scholar]
- Kharchenko P.V., Tolstorukov M.Y., Park P.J. (2008). Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 26: 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-K., Shiekhattar R. (2015). Architectural and functional commonalities between enhancers and promoters. Cell 162: 948–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler K.W., Vogelstein B. (1990). The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol. Cell. Biol. 10: 634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer S., Holt A.L., Rubio-Somoza I., Tucker E.J., Hinze A., Pisch M., Javelle M., Timmermans M.C., Tucker M.R., Laux T. (2013). A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 24: 125–132. [DOI] [PubMed] [Google Scholar]
- Kumari S., Ware D. (2013). Genome-wide computational prediction and analysis of core promoter elements across plant monocots and dicots. PLoS One 8: e79011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange T., Kapanidis A.N., Tang H., Reinberg D., Ebright R.H. (1998). New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 12: 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D., Couzigou J.-M., Clemente H.S., Martinez Y., Dunand C., Bécard G., Combier J.-P. (2015). Primary transcripts of microRNAs encode regulatory peptides. Nature 520: 90–93. [DOI] [PubMed] [Google Scholar]
- Lee C.-K., Shibata Y., Rao B., Strahl B.D., Lieb J.D. (2004a). Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36: 900–905. [DOI] [PubMed] [Google Scholar]
- Lee D.-H., Gershenzon N., Gupta M., Ioshikhes I.P., Reinberg D., Lewis B.A. (2005). Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Mol. Cell. Biol. 25: 9674–9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. (2004b). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23: 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang K., Chen X., Meng H., Song M., Wang Y., Xu X., Bai Y. (2012). Transcriptional activation of microRNA-34a by NF-kappa B in human esophageal cancer cells. BMC Mol. Biol. 13: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Li Y., Luo J., Zhang Z. (2015). A novel motif-discovery algorithm to identify co-regulatory motifs in large transcription factor and microRNA co-regulatory networks in human. Bioinformatics 31: 2348–2355. [DOI] [PubMed] [Google Scholar]
- Lim C.Y., Santoso B., Boulay T., Dong E., Ohler U., Kadonaga J.T. (2004). The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 18: 1606–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61: 395–420. [DOI] [PubMed] [Google Scholar]
- Liu M.-J., Seddon A.E., Tsai Z.T.-Y., Major I.T., Floer M., Howe G.A., Shiu S.-H. (2015). Determinants of nucleosome positioning and their influence on plant gene expression. Genome Res. 25: 1182–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Bartel D.P., Bartel B. (2005). MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17: 1360–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Reinhart B.J., Jones-Rhoades M.W., Tang G., Zamore P.D., Barton M.K., Bartel D.P. (2004). MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 23: 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsico A., Huska M.R., Lasserre J., Hu H., Vucicevic D., Musahl A., Orom U., Vingron M. (2013). PROmiRNA: a new miRNA promoter recognition method uncovers the complex regulation of intronic miRNAs. Genome Biol. 14: R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N.J., Ow M.C., Barrasa M.I., Hammell M., Sequerra R., Doucette-Stamm L., Roth F.P., Ambros V.R., Walhout A.J.M. (2008). A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 22: 2535–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinho C., Confraria A., Elias C.A., Crozet P., Rubio-Somoza I., Weigel D., Baena-González E. (2015). Dissection of miRNA pathways using arabidopsis mesophyll protoplasts. Mol. Plant 8: 261–275. [DOI] [PubMed] [Google Scholar]
- Mathieu J., Yant L.J., Mürdter F., Küttner F., Schmid M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medford J.I., Prasad A. (2014). Plant Science. Plant synthetic biology takes root. Science 346: 162–163. [DOI] [PubMed] [Google Scholar]
- Megraw M., Hatzigeorgiou A.G. (2010). MicroRNA promoter analysis. In Methods in Molecular Biology, B.C. Meyers and P.J. Green, eds (Humana Press), pp. 149–161. [DOI] [PubMed] [Google Scholar]
- Megraw M., Mukherjee S., Ohler U. (2013). Sustained-input switches for transcription factors and microRNAs are central building blocks of eukaryotic gene circuits. Genome Biol. 14: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M., Pereira F., Jensen S.T., Ohler U., Hatzigeorgiou A.G. (2009). A transcription factor affinity-based code for mammalian transcription initiation. Genome Res. 19: 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R., Kashtan N., Itzkovitz S., Newman M.E.J., Alon U. (2004). On the uniform generation of random graphs with prescribed degree sequences. http://arxiv.org/abs/cond-mat/0312028.
- Milo R., Shen-Orr S., Itzkovitz S., Kashtan N., Chklovskii D., Alon U. (2002). Network motifs: simple building blocks of complex networks. Science 298: 824–827. [DOI] [PubMed] [Google Scholar]
- Miyashima S., Koi S., Hashimoto T., Nakajima K. (2011). Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138: 2303–2313. [DOI] [PubMed] [Google Scholar]
- Morachis J.M., Murawsky C.M., Emerson B.M. (2010). Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes Dev. 24: 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T., Maki K., Aiba H. (2012). Detection of sRNA-mRNA interactions by electrophoretic mobility shift assay. Methods Mol. Biol. 905: 235–244. [DOI] [PubMed] [Google Scholar]
- Morton T., Petricka J., Corcoran D.L., Li S., Winter C.M., Carda A., Benfey P.N., Ohler U., Megraw M. (2014). Paired-end analysis of transcription start sites in Arabidopsis reveals plant-specific promoter signatures. Plant Cell 26: 2746–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton T., Wong W.-K., Megraw M. (2015). TIPR: transcription initiation pattern recognition on a genome scale. Bioinformatics 31: 3725–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon S., Schwach F., Dalmay T., Maclean D., Studholme D.J., Moulton V. (2008). A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 24: 2252–2253. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Berger M.F., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L. (2004). Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 36: 1331–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T., Corcoran D.L., Rach E.A., Song S., Spana E.P., Gao Y., Ohler U., Zhu J. (2010). A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat. Methods 7: 521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine M.D., Bartel D.P. (2010). MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 24: 2678–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osella M., Bosia C., Corá D., Caselle M. (2011). The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLOS Comput. Biol. 7: e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Fitz J., Schwab R., Riester M., Weigel D. (2009). WMD3 Web microRNA Designer. http://wmd3.weigelworld.org/cgi-bin/webapp.cgi.
- Ossowski S., Schwab R., Weigel D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690. [DOI] [PubMed] [Google Scholar]
- Ou B., Yin K.-Q., Liu S.-N., Yang Y., Gu T., Wing Hui J.M., Zhang L., Miao J., Kondou Y., Matsui M., Gu H.-Y., Qu L.-J. (2011). A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol. Plant 4: 546–555. [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Poling L.L., Wang Z., Liu H., Liu X.S., Roeder R.G., Zhang X., Song J.S., Fisher D.E. (2008). Chromatin structure analyses identify miRNA promoters. Genes Dev. 22: 3172–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajoro A., et al. (2014). Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 15: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant B.D., Buhtz A., Kehr J., Scheible W.-R. (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53: 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parizotto E.A., Dunoyer P., Rahm N., Himber C., Voinnet O. (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18: 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P.J. (2009). ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 10: 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok D.K., et al. (2005). Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527. [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Zhang Z., Liang J., Ge Q., Duan X., Ma F., Li F. (2011). The full-length transcripts and promoter analysis of intergenic microRNAs in Drosophila melanogaster. Genomics 97: 294–303. [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes J.S., Marian Walhout A.J. (2012). Yeast one-hybrid assays: a historical and technical perspective. Methods 57: 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel M., Li Y., Li J., Millar A.A. (2015). Inhibiting plant microRNA activity: molecular SPONGEs, target MIMICs and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol. J. 13: 915–926. [DOI] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. (2002). Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Roy A.L., Singer D.S. (2015). Core promoters in transcription: old problem, new insights. Trends Biochem. Sci. 40: 165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I., Weigel D. (2011). MicroRNA networks and developmental plasticity in plants. Trends Plant Sci. 16: 258–264. [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I., Zhou C.-M., Confraria A., Martinho C., von Born P., Baena-Gonzalez E., Wang J.-W., Weigel D. (2014). Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Biol. 24: 2714–2719. [DOI] [PubMed] [Google Scholar]
- Salimullah M., Sakai M., Plessy C., Carninci P. (2011). NanoCAGE: a high-resolution technique to discover and interrogate cell transcriptomes. CSH Protoc. 2011: pdb.prot5559. [DOI] [PMC free article] [PubMed]
- Schones D.E., Smith A.D., Zhang M.Q. (2007). Statistical significance of cis-regulatory modules. BMC Bioinformatics 8: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila A.C., Core L.J., Lis J.T., Sharp P.A. (2009). Divergent transcription: a new feature of active promoters. Cell Cycle 8: 2557–2564. [DOI] [PubMed] [Google Scholar]
- Sekinger E.A., Moqtaderi Z., Struhl K. (2005). Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell 18: 735–748. [DOI] [PubMed] [Google Scholar]
- Shen-Orr S.S., Milo R., Mangan S., Alon U. (2002). Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31: 64–68. [DOI] [PubMed] [Google Scholar]
- Siebert M., Söding J. (2014). Universality of core promoter elements? Nature 511: E11–E12. [DOI] [PubMed] [Google Scholar]
- Solé A., Mencia N., Villalobos X., Noé V., Ciudad C.J. (2013). Validation of miRNA-mRNA interactions by electrophoretic mobility shift assays. BMC Res. Notes 6: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Crawford G.E. (2010). DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. CSH Protoc. 2010: pdb.prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., et al. (2011). Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 21: 1757–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa S.W., Gelderman G., Contreras L.M. (2015). Advances in synthetic dynamic circuits design: using novel synthetic parts to engineer new generations of gene oscillations. Curr. Opin. Biotechnol. 36: 161–167. [DOI] [PubMed] [Google Scholar]
- Stormo G.D. (2000). DNA binding sites: representation and discovery. Bioinformatics 16: 16–23. [DOI] [PubMed] [Google Scholar]
- Stormo G.D., Zuo Z., Chang Y.K. (2015). Spec-seq: determining protein-DNA-binding specificity by sequencing. Brief. Funct. Genomics 14: 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A.M., et al. (2014). Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Reports 8: 2015–2030. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., et al. (2001). Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 11: 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Kato S., Murata M., Carninci P. (2012). CAGE (cap analysis of gene expression): a protocol for the detection of promoter and transcriptional networks. Methods Mol. Biol. 786: 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S., Innan H. (2008). Evolution of complexity in miRNA-mediated gene regulation systems. Trends Genet. 24: 56–59. [DOI] [PubMed] [Google Scholar]
- Tang G., Reinhart B.J., Bartel D.P., Zamore P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M.C., Chiang C.-M. (2006). The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41: 105–178. [DOI] [PubMed] [Google Scholar]
- Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokusumi Y., Ma Y., Song X., Jacobson R.H., Takada S. (2007). The new core promoter element XCPE1 (X Core Promoter Element 1) directs activator-, mediator-, and TATA-binding protein-dependent but TFIID-independent RNA polymerase II transcription from TATA-less promoters. Mol. Cell. Biol. 27: 1844–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J., Zhu J., van Oudenaarden A. (2007). MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell 26: 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510. [DOI] [PubMed] [Google Scholar]
- Vidal E.A., Araus V., Lu C., Parry G., Green P.J., Coruzzi G.M., Gutiérrez R.A. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E.A., Moyano T.C., Riveras E., Contreras-López O., Gutiérrez R.A. (2013). Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc. Natl. Acad. Sci. USA 110: 12840–12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J., et al. (2015). Functional footprinting of regulatory DNA. Nat. Methods 12: 927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-J., Guo H.-S. (2015). Cleavage of INDOLE-3-ACETIC ACID INDUCIBLE28 mRNA by microRNA847 upregulates auxin signaling to modulate cell proliferation and lateral organ growth in Arabidopsis. Plant Cell 27: 574–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749. [DOI] [PubMed] [Google Scholar]
- Wang W., Ye R., Xin Y., Fang X., Li C., Shi H., Zhou X., Qi Y. (2011). An importin β protein negatively regulates microRNA activity in Arabidopsis. Plant Cell 23: 3565–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch M.T., et al. (2014). Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernicke S., Rasche F. (2006). FANMOD: a tool for fast network motif detection. Bioinformatics 22: 1152–1153. [DOI] [PubMed] [Google Scholar]
- Wingender E. (2008). The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief. Bioinform. 9: 326–332. [DOI] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.-W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138: 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Khanna K., Ruan S. (2010). Expression of microRNAs and its regulation in plants. Semin. Cell Dev. Biol. 21: 790–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Wu M.-F., Yang L., Wu G., Poethig R.S., Wagner D. (2009). The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17: 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y.Y., Ichida H., Abe T., Suzuki Y., Sugano S., Obokata J. (2007a). Differentiation of core promoter architecture between plants and mammals revealed by LDSS analysis. Nucleic Acids Res. 35: 6219–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y.Y., Ichida H., Matsui M., Obokata J., Sakurai T., Satou M., Seki M., Shinozaki K., Abe T. (2007b). Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics 8: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y.Y., Yoshioka Y., Hyakumachi M., Obokata J. (2011). Characteristics of core promoter types with respect to gene structure and expression in Arabidopsis thaliana. DNA Res. 18: 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y.Y., Yoshitsugu T., Sakurai T., Seki M., Shinozaki K., Obokata J. (2009). Heterogeneity of Arabidopsis core promoters revealed by high-density TSS analysis. Plant J. 60: 350–362. [DOI] [PubMed] [Google Scholar]
- Yan K., Liu P., Wu C.-A., Yang G.-D., Xu R., Guo Q.-H., Huang J.-G., Zheng C.-C. (2012). Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell 48: 521–531. [DOI] [PubMed] [Google Scholar]
- Yang C., Bolotin E., Jiang T., Sladek F.M., Martinez E. (2007). Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 389: 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T.T., Ott F., Lanz C., Wollmann H., Chen X., Schmid M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu S., Lian H., Wang J.-W. (2015). Plant developmental transitions: the role of microRNAs and sugars. Curr. Opin. Plant Biol. 27: 1–7. [DOI] [PubMed] [Google Scholar]
- Yuan G.-C., Liu Y.-J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. (2005). Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630. [DOI] [PubMed] [Google Scholar]
- Zhang L., Chia J.-M., Kumari S., Stein J.C., Liu Z., Narechania A., Maher C.A., Guill K., McMullen M.D., Ware D. (2009). A genome-wide characterization of microRNA genes in maize. PLoS Genet. 5: e1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.-Q., Lian H., Tang H., Dolezal K., Zhou C.-M., Yu S., Chen J.-H., Chen Q., Liu H., Ljung K., Wang J.-W. (2015). An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell 27: 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wu Y., Schnable J.C., Zeng Z., Freeling M., Crawford G.E., Jiang J. (2012b). High-resolution mapping of open chromatin in the rice genome. Genome Res. 22: 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhang T., Wu Y., Jiang J. (2012a). Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis. Plant Cell 24: 2719–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Ruan J., Wang G., Zhang W. (2007). Characterization and identification of microRNA core promoters in four model species. PLOS Comput. Biol. 3: e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Hu F., Wang R., Zhou X., Sze S.-H., Liou L.W., Barefoot A., Dickman M., Zhang X. (2011). Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145: 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y.-C., Li Q.-Z. (2011). Identification of TATA and TATA-less promoters in plant genomes by integrating diversity measure, GC-Skew and DNA geometric flexibility. Genomics 97: 112–120. [DOI] [PubMed] [Google Scholar]