A decapping activator complex ensures the adequacy and specificity of the response of Arabidopsis to different abiotic stresses by promoting selective mRNA degradation.
Abstract
In eukaryotes, the decapping machinery is highly conserved and plays an essential role in controlling mRNA stability, a key step in the regulation of gene expression. Yet, the role of mRNA decapping in shaping gene expression profiles in response to environmental cues and the operating molecular mechanisms are poorly understood. Here, we provide genetic and molecular evidence that a component of the decapping machinery, the LSM1-7 complex, plays a critical role in plant tolerance to abiotic stresses. Our results demonstrate that, depending on the stress, the complex from Arabidopsis thaliana interacts with different selected stress-inducible transcripts targeting them for decapping and subsequent degradation. This interaction ensures the correct turnover of the target transcripts and, consequently, the appropriate patterns of downstream stress-responsive gene expression that are required for plant adaptation. Remarkably, among the selected target transcripts of the LSM1-7 complex are those encoding NCED3 and NCED5, two key enzymes in abscisic acid (ABA) biosynthesis. We demonstrate that the complex modulates ABA levels in Arabidopsis exposed to cold and high salt by differentially controlling NCED3 and NCED5 mRNA turnover, which represents a new layer of regulation in ABA biosynthesis in response to abiotic stress. Our findings uncover an unanticipated functional plasticity of the mRNA decapping machinery to modulate the relationship between plants and their environment.
INTRODUCTION
Plants have evolved sophisticated mechanisms to perceive and rapidly respond to the numerous abiotic stresses to which they are continuously challenged. A decisive component of all stress responses is the ability to reprogram transcriptomes. Transcriptome reprogramming in response to adverse environments has been generally considered as a complex process mainly based on the modulation of transcriptional activity of stress-related genes (Yamaguchi-Shinozaki and Shinozaki, 2006; Gujjar et al., 2014; Nakashima et al., 2014). However, recent data indicate that posttranscriptional mechanisms also influential to make fine and timely adjustments of the plant transcriptomes to unfavorable environments (Guerra et al., 2015). The control of mRNA stability, in particular, is critical for the regulation of gene expression in response to abiotic stress. Stability determinants for intrinsically unstable eukaryotic mRNAs include the 3′ poly(A) tail and the 5′ cap. Transcript abundance is then fine-tuned by a major pathway of cytoplasmic mRNA degradation involving deadenylation, decapping, and subsequent exoribonuclease 5′-3′ activity (Parker, 2012). mRNA decapping is achieved in distinct cytoplasmic foci named processing bodies (P-bodies) by the action of the decapping complex, a protein conglomerate highly conserved among eukaryotes (Coller and Parker, 2004; Fillman and Lykke-Andersen, 2005; Xu et al., 2006; Goeres et al., 2007). This complex includes the decapping enzyme DCP2 and its activators DCP1, DCP5, VCS, DHH1, PAT1, and the LSM1-7 complex (Coller and Parker, 2004; Fillman and Lykke-Andersen, 2005; Xu et al., 2006; Goeres et al., 2007; Xu and Chua, 2009). mRNA decapping has been shown to play important roles in development (Xu et al., 2006; Goeres et al., 2007; Xu and Chua, 2009). By contrast, the role of the decapping complex in abiotic stress signaling is barely documented. To date, only DCP1 and DCP5 have been described to associate under drought conditions to promote drought tolerance in Arabidopsis thaliana (Xu and Chua, 2012). Whether mRNA decapping is involved in regulating eukaryotic responses to other abiotic stresses and the molecular mechanisms operating in mRNA decapping during such responses is largely unknown.
The SM-like proteins (LSMs) are implicated in numerous aspects of RNA metabolism in eukaryotes. The Arabidopsis genome contains 11 LSM genes encoding eight central, highly evolutionarily conserved LSM proteins (LSM1-LSM8) (Perea-Resa et al., 2012). LSM1, LSM3, and LSM6 are each duplicated and code for pairs of functionally redundant proteins (LSM1A, B; LSM3A, B; LSM6A, B). In Arabidopsis, as in yeast and animals, the eight conserved LSM proteins are organized in two heteroheptameric ring complexes, LSM1-7 and LSM2-8, specifically localized in the cytoplasm and nucleus and defined by the subunits LSM1 and LSM8, respectively. The LSM2-8 complex functions in pre-mRNA splicing through U6 small nuclear RNA stabilization and ensures normal Arabidopsis development (Perea-Resa et al., 2012). Moreover, alternative splicing analysis in lsm4 and sad1/lsm5 mutants uncovered that this complex acts as a positive regulator of salt tolerance in Arabidopsis (Zhang et al., 2011; Cui et al., 2014). The LSM1-7 complex is involved in accurate mRNA turnover by promoting decapping and subsequent 5′-3′ degradation and is required for the formation of P-bodies (Perea-Resa et al., 2012). In the yeast Saccharomyces cerevisiae, the LSM1-7 complex has been described to operate through the interaction of LSM1 with oligoadenylated mRNAs that are then targeted for degradation (Chowdhury et al., 2007). The analysis of an Arabidopsis lsm1a lsm1b double mutant defective in LSM1A and LSM1B expression unveiled that LSM1 proteins are essential for the assembly of the LSM cytoplasmic complex and that this complex is needed for correct plant development (Perea-Resa et al., 2012). However, the participation of the LSM1-7 decapping activator complex in abiotic stress responses has not yet been established in any organism. Here, we demonstrate that this complex regulates Arabidopsis tolerance to freezing, drought, and high salt by modulating the transcriptome reprogramming that takes place in response to these adverse conditions. More important, RNA immunoprecipitation (RIP) assays revealed that, depending on the abiotic stress to which the plant is subjected, the LSM cytoplasmic complex targets selected stress-inducible transcripts for decapping and degradation, thus controlling their levels and, therefore, ensuring the adequate transcriptomic response. Interestingly, among the selected mRNAs that are differentially targeted for decapping in response to low temperature, water deficiency, and high salt are those encoding NCED3 and NCED5, two key enzymes in abscisic acid (ABA) biosynthesis. We show that, as a consequence, the LSM1-7 complex determines the appropriate levels of ABA in Arabidopsis plants exposed to different abiotic stresses. Our findings thus reveal a previously unrecognized layer of regulation in the biosynthesis of this phytohormone and uncover unexpected functional plasticity of the mRNA decapping machinery in influencing the responses of plants to their environment.
RESULTS
Arabidopsis LSM1 Proteins Localize to P-Bodies in Response to Abiotic Stress
The molecular characterization of Arabidopsis LSM1 genes uncovered that they were responsive to low temperature. qPCR assays showed that LSM1A and LSM1B transcripts accumulated in response to 4°C, reaching a peak of accumulation after 1 d of treatment (Figure 1A). This accumulation was detected broadly through the adult Arabidopsis plant (Supplemental Figure 1A). Transcripts corresponding to LSM2-7 proteins exhibited similar cold-induced accumulation as LSM1 mRNAs (Supplemental Figure 1B). However, LSM transcripts did not accumulate in plants exposed to other related abiotic stresses, such as drought (55% polyethylene glycol [PEG]) or high salt (150 mM NaCl) (Supplemental Figure 1C). We concluded that the expression of genes encoding the Arabidopsis cytoplasmic LSM complex is positively regulated by low temperature.
Figure 1.
Arabidopsis LSM1 Proteins Accumulate in Response to Low Temperature and Localize to P-Bodies.
(A) Expression of LSM1A and LSM1B in 2-week-old Col-0 plants exposed for the indicated hours (h) or days (d) to 4°C. Levels, determined by qPCR, are represented as relative to their corresponding values at 0 h. Error bars indicate the sd of the mean (n ≥ 3). In all cases, differences between cold-treated and control (0 h) plants were significant (P ≤ 0.0001), as determined by ANOVA test.
(B) Immunoblots showing levels of LSM1A-GFP and LSM1B-GFP in 2-week-old transgenic Arabidopsis plants exposed for the indicated times to 4°C. A lane with Col-0 plants was added in the immunoblot as a negative control. Coomassie staining of the large subunit of Rubisco was used as a loading control.
(C) Subcellular localization of LSM1A-GFP and LSM1B-GFP in root tip cells from 6-d-old transgenic Arabidopsis seedlings grown under control conditions (20°C) or exposed for 24 h to 4°C in the presence or absence of cycloheximide (CHX). Bars = 20 µm.
(D) Colocalization of LSM1A-GFP and LSM1B-GFP with RFP-DCP1 in root tip cells from 6-d-old transgenic Arabidopsis seedlings exposed for 24 h to 4°C. Bars = 20 µm.
(E) Subcellular localization of GFP-DCP2 and GFP-VCS in root tip cells from 6-d-old Col-0 and lsm1a lsm1b Arabidopsis seedlings grown under control conditions and subsequently exposed for 24 h to 4°C. Bars = 20 µm.
Since LSM1A and LSM1B transcripts accumulated in response to low temperature, we assessed whether this accumulation was followed by an increase of the corresponding proteins. Immunoblot experiments using Arabidopsis plants containing genomic LSM1-GFP fusions driven by the corresponding LSM1 promoters (LSM1PRO) (Perea-Resa et al., 2012) showed that LSM1A-GFP and LSM1B-GFP proteins were notably more abundant after some days of cold exposure (Figure 1B). Consistent with the expression results, water and salt stresses did not alter the levels of LSM1 proteins (Supplemental Figure 2A). Therefore, concomitantly with the accumulation of their transcripts, the levels of LSM1 proteins also increase in response to low temperature.
Previous data revealed that heat treatment promotes the localization of LSM1A and LSM1B to P-bodies (Perea-Resa et al., 2012). We decided to investigate if LSM1 proteins also localize to these foci under cold, drought, or high salt conditions by examining the distribution of green fluorescence in root cells from the LSM1APRO-LSM1A-GFP and LSM1BPRO-LSM1B-GFP plants. In according with earlier results (Perea-Resa et al., 2012), confocal microscopy analysis indicated that, at 20°C, GFP activity was in both cases broadly distributed in the cytoplasm (Figure 1C). However, when transgenic plants were exposed to 4°C, water deficiency, or salt stress, LSM1A-GFP and LSM1B-GFP fusion proteins were found aggregated in discrete cytoplasmic spots that resembled P-bodies. After cycloheximide treatment, which causes loss of P-bodies (Sheth and Parker, 2003), no cytoplasmic foci were observed in any case, suggesting that the detected LSM1-GFP spots indeed corresponded to P-bodies (Figures 1C; Supplemental Figure 2B). The identity of these foci was confirmed by colocalization studies with DCP1, a protein that accumulates in P-bodies (Xu et al., 2006; Weber et al., 2008; Motomura et al., 2015). The examination of LSM1APRO-LSM1A-GFP and LSM1BPRO-LSM1B-GFP plants cotransformed with a 35S-RFP-DCP1 fusion (Perea-Resa et al., 2012) demonstrated that, in fact, LSM1A-GFP and LSM1B-GFP colocalized with RFP-DCP1 in root cells exposed to 4°C (Figure 1D). Identical results were obtained when LSM1APRO-LSM1A-GFP and LSM1BPRO-LSM1B-GFP transgenic plants were subjected to drought or high salt (Supplemental Figure 2C), providing evidence that LSM1 proteins localize to P-bodies in response to abiotic stress.
LSM1 proteins have been reported to be crucial for P-body integrity under heat stress conditions (Perea-Resa et al., 2012). We examined whether they were also required for P-body formation in response to low temperature. With this aim, the subcellular localization of DCP2 and VCS, two P-body markers (Sheth and Parker, 2003; Xu et al., 2006), was compared in Col-0 (wild type) and lsm1a lsm1b double mutant seedlings containing 35S-GFP-DCP2 or 35S-GFP-VCS fusions (Perea-Resa et al., 2012) exposed to 4°C. While in wild-type plants both proteins accumulated in P-bodies, a mostly dispersed cytosolic signal was observed in lsm1a lsm1b mutants (Figure 1E), showing that, in addition to accumulating in P-bodies, LSM1 proteins are also essential for the formation of these cytoplasmic foci in response to low temperature. Similar results were obtained when plants were subjected to water and salt stresses (Supplemental Figure 2D), demonstrating that LSM1 proteins also localize to P-bodies under these abiotic stresses and are necessary for the assembly of these foci in response to water deficiency and high salt.
The LSM1-7 Complex Regulates Arabidopsis Tolerance to Abiotic Stress
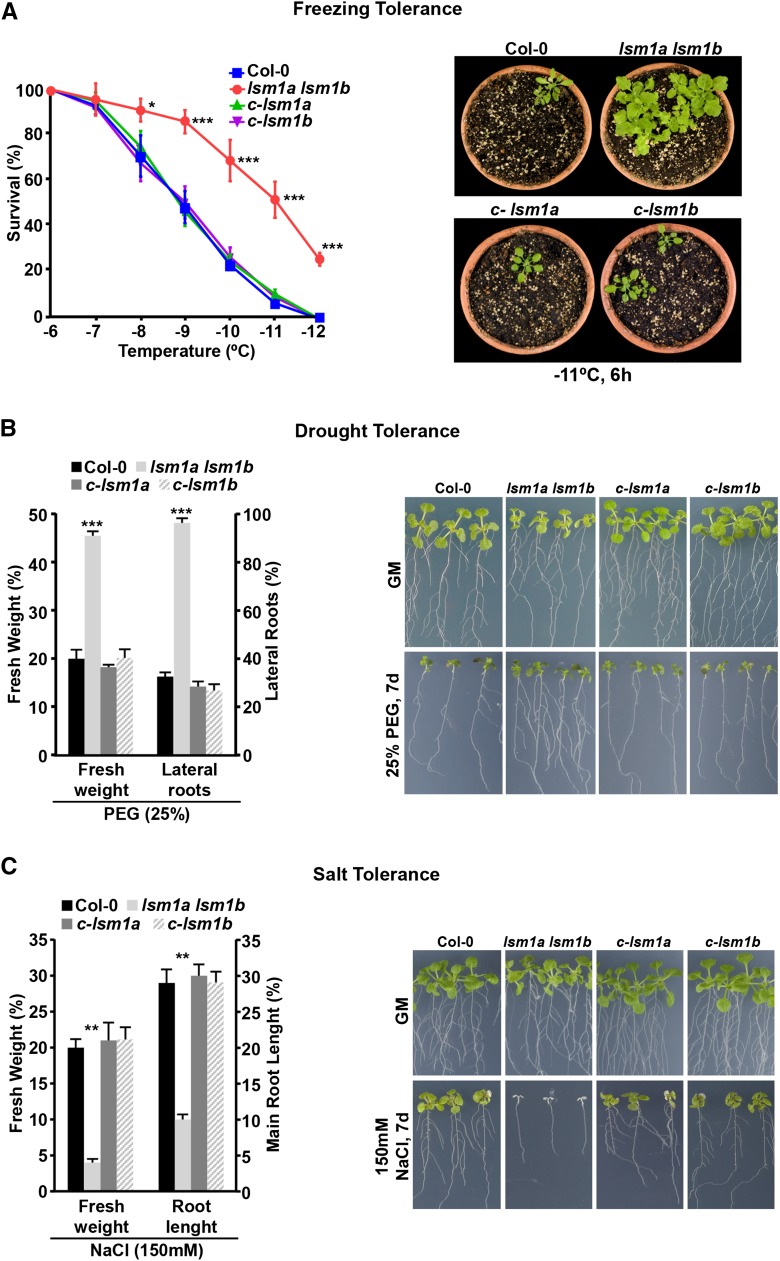
To further characterize the role of LSM1 proteins and, therefore, of the LSM cytoplasmic complex in plant adaptation to abiotic stress, we evaluated their possible implication in Arabidopsis tolerance to freezing, drought, and salinity. Freezing tolerance was analyzed in nonacclimated and cold-acclimated (7 d, 4°C) lsm1a lsm1b mutants exposed for 6 h to different freezing temperatures (Catalá et al., 2014). Nonacclimated mutants presented a similar capacity to tolerate freezing as the wild type, the LT50 (temperature that causes 50% lethality) values being in both cases around −4.5°C (Supplemental Figure 3A). By contrast, the freezing tolerance of cold-acclimated lsm1a lsm1b mutants was significantly higher than that of wild-type plants. In this case, the determined LT50 values were −11°C and −9°C, respectively (Figure 2A).
Figure 2.
The LSM1-7 Complex Differentially Regulates Abiotic Stress Tolerance in Arabidopsis.
(A) Freezing tolerance of cold-acclimated 2-week-old Col-0, lsm1a lsm1b, c-lsm1a, and c-lsm1b plants (left). Representative cold-acclimated plants 7 d after being exposed to −11°C for 6 h (right).
(B) Drought tolerance of 7-d-old Col-0, lsm1a lsm1b, c-lsm1a, and c-lsm1b seedlings (left). Representative seedlings grown on GM or exposed 7 d to 25% PEG (right).
(C) Salt tolerance of 7-d-old Col-0, lsm1a lsm1b, c-lsm1a, and c-lsm1b seedlings (left). Representative seedlings grown on GM or exposed 7 d to 150 mM NaCl (right).
In all graphs, error bars indicate the sd of the mean (n ≥ 3). Asterisks indicate significant differences (*P ≤ 0.01, **P ≤ 0.001, and ***P ≤ 0.0001) between lsm1a lsm1b and the other plants, as determined by ANOVA test. No significant differences between Col-0 and c-lsm1a or c-lsm1b plants were observed in any case.
Drought tolerance was examined in lsm1a lsm1b seedlings transferred to plates containing 25% PEG (Verslues et al., 2006). After 1 week, they exhibited significantly enhanced tolerance compared with wild-type seedlings as revealed by the quantification of their fresh weights and lateral roots (Figure 2B). Tolerance to high salt was assayed in lsm1a lsm1b seedlings grown one further week on plates containing 150 mM NaCl (Verslues et al., 2006). Mutants displayed lower fresh weights and shorter main roots than wild-type seedlings (Figure 2C), manifesting more sensitivity to salt stress. It is worth noting that lsm1a lsm1b plants grown on soil showed identical drought-tolerant and salt-sensitive tolerance phenotypes (Supplemental Figures 3B and 3C).
In all cases, lsm1a lsm1b mutants complemented with either LSM1APRO-LSM1A-GFP (c-lsm1a) or LSM1BPRO-LSM1B-GFP (c-lsm1b) fusions (Perea-Resa et al., 2012) had wild-type capacity to cold acclimate and to tolerate water deficiency and high salt (Figure 2; Supplemental Figure 3), establishing that the stress tolerance phenotypes of lsm1a lsm1b were a direct consequence of the absence of LSM1A and LSM1B expression. Together, these data provided genetic evidence that the LSM1-7 complex is differentially involved in plant tolerance to abiotic stresses. It negatively regulates the ability of Arabidopsis to cold acclimate and tolerate drought but functions as a positive regulator of salt tolerance.
The Arabidopsis LSM Cytoplasmic Complex Differentially Regulates Gene Expression in Response to Abiotic Stress
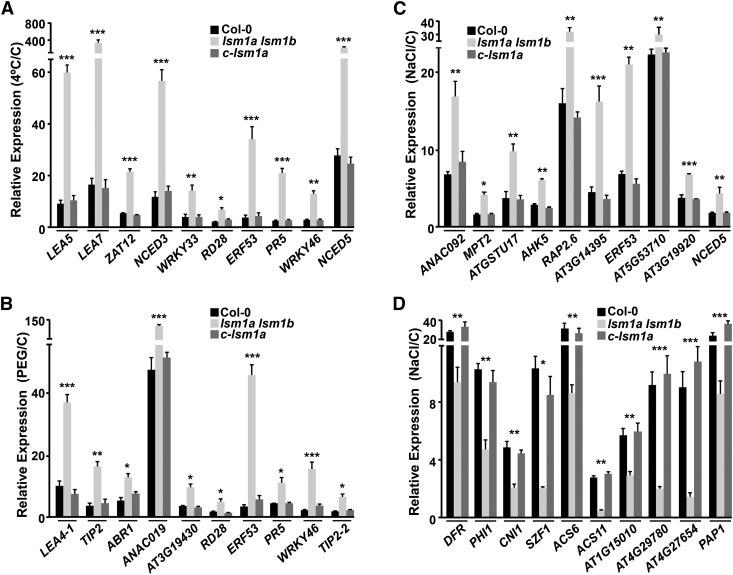
Since the Arabidopsis LSM1-7 complex influences gene expression by promoting RNA decapping and decay (Perea-Resa et al., 2012), we examined whether it might affect gene expression in response to abiotic stresses as a first step to understanding how it differentially regulates plant tolerance to environmental challenges. High-throughput RNA sequencing (RNA-seq) was used to estimate the effect of lsm1a and lsm1b mutations on the transcriptomes of Arabidopsis plants subjected to cold, drought, or high-salt conditions. For this, we sequenced cDNA libraries prepared from stress-treated lsm1a lsm1b and wild-type plants. The resulting reads (±12 Mb/sample) were mapped to the Arabidopsis genome (TAIR10 version) and gene expression changes in the double mutant evaluated. In each treatment, the top 1000 upregulated and top 1000 downregulated genes in lsm1a lsm1b, based on fold change ratios with respect to their corresponding controls, were considered for analysis. The expression levels of the top 1000 upregulated genes in mutant plants exposed for 24 h to 4°C were increased at least 3.4-fold compared with the wild type (Supplemental Data Set 1). Remarkably, 53.3% of these genes (533) have been reported to be induced (≥2-fold) in response to cold (Kilian et al., 2007) (Supplemental Data Set 2). Many of them, moreover, have been associated with the development of cold acclimation (Vogel et al., 2005; Cuevas et al., 2008; Miura and Furumoto, 2013; Shi et al., 2015) and therefore could account for the tolerance phenotype of the double mutant. The upregulation of some of these genes, including LEA5, LEA7, ZAT12, NCED3, WRKY33, RD28, ERF53, PR5, WRKY46, and NCED5, in lsm1a lsm1b was verified by qPCR, validating the RNA-seq results (Figure 3A). Compared with the wild type, the top 1000 downregulated genes in cold-treated mutants displayed a decreased expression of at least 2.1-fold (Supplemental Data Set 3). Of them, 311 (31.1%) have been described to be cold induced (Kilian et al., 2007; Supplemental Data Set 4) but none as functioning as a negative regulator of cold acclimation. Representative cold-inducible genes that were downregulated in lsm1a lsm1b were also validated by qPCR (Supplemental Figure 4A).
Figure 3.
The Arabidopsis LSM1-7 Complex Differentially Regulates Gene Expression in Response to Abiotic Stresses.
(A) Expression of different cold-inducible genes upregulated in lsm1a lsm1b. Levels, determined by qPCR, in 2-week-old Col-0, lsm1a lsm1b, and c-lsm1a plants exposed for 24 h to 4°C are represented relative to their corresponding values in control plants (C).
(B) Expression of different drought-inducible genes upregulated in lsm1a lsm1b. Levels, determined by qPCR, in 2-week-old Col-0, lsm1a lsm1b, and c-lsm1a plants exposed for 10 h to 55% PEG are represented relative to their corresponding values in control plants (C).
(C) and (D) Expression of different salt-inducible genes upregulated (C) or downregulated (D) in lsm1a lsm1b. Levels, determined by qPCR, in 2-week-old Col-0, lsm1a lsm1b and c-lsm1a plants exposed for 10 h to 150 mM NaCl are represented relative to their corresponding values in control plants (C).
In all graphs, error bars indicate the sd of the mean (n ≥ 3). Differences between lsm1a lsm1b and Col-0 or c-lsm1a plants were always significant (*P ≤ 0.01, **P ≤ 0.001, and ***P ≤ 0.0001), as determined by ANOVA test. No significant differences between Col-0 and c-lsm1a plants were observed in any case.
Under conditions of water deficiency (10 h, 55% PEG), the transcript levels of the top 1000 upregulated genes in lsm1a lsm1b plants were found to be higher, by at least 2-fold, than in the wild type (Supplemental Data Set 5). Interestingly, 372 of these genes (37.2%) have been shown to be induced (≥2-fold) in response to drought (Kilian et al., 2007) (Supplemental Data Set 6) and some of them, such as ABR1, ANAC019, or ERF53, to have a positive role in Arabidopsis tolerance to drought (Tran et al., 2004; Pandey et al., 2005; Cheng et al., 2012). qPCR experiments confirmed the upregulation in lsm1a lsm1b plants of these and other genes that promote drought tolerance (Figure 3B), which would account for the drought tolerance phenotype of the mutants and validate the RNA-seq data. Otherwise, the top 1000 downregulated genes in mutant plants subjected to drought exhibited at least 1.9-fold lower expression than in wild-type plants (Supplemental Data Set 7). Among them, 32.6% (326) have been previously shown to be induced by water deficiency (Kilian et al., 2007) (Supplemental Data Set 8), although none acting as negative regulator of drought tolerance. These results were also validated by analyzing representative drought-inducible genes that were downregulated in lsm1a lsm1b using qPCR (Supplemental Figure 4B).
When comparing the transcriptome profiles of mutant and wild-type plants exposed to salt stress (10 h, 150 mM NaCl), the expression levels of the top 1000 upregulated genes in lsm1a lsm1b were at least 2.4-fold higher than in the wild type (Supplemental Data Set 9). In this case, 47% of the genes (470) have been reported to be salt induced (≥2-fold) (Kilian et al., 2007) (Supplemental Data Set 10), and several of them, such as ANAC092, ATGSTU17, or AHK5, to act as negative regulators of salt tolerance in Arabidopsis (Balazadeh et al., 2010; Chen et al., 2012; Pham et al., 2012). The upregulation of these and other salt-responsive genes was verified by qPCR, validating the RNA-seq data (Figure 3C). The top 1000 downregulated genes in mutant plants showed at least 2.1-fold lower expression levels than in the wild type (Supplemental Data Set 11). Strikingly, 457 (45.7%) of these genes were salt induced (Kilian et al., 2007) (Supplemental Data Set 12), and some of them, including DFR, PHI1, or CNI1, have been described to be positive regulators of Arabidopsis tolerance to high salt (Cui et al., 2014; Peng et al., 2014). qPCR experiments confirmed that the expression of these and other salt-regulated genes was lower in lsm1a lsm1b than in wild-type plants after salt treatment, validating once again the RNA-seq data (Figure 3D). The altered expression of the negative and positive regulators of salt tolerance described above was fully consistent with the salt-sensitive phenotype exhibited by the double mutant.
In all cases, when RNA-seq experiments were validated by qPCR, we found a strong, statistically significant correlation (Pearson r ≥ 0.909; P value ≤ 0.0001) between the fold change results obtained from both kinds of analysis (Supplemental Figure 5). Consistent with the close relationship existing between plant responses to low temperature, water deficiency, and high salt (Kilian et al., 2007), several genes were regulated by the LSM1-7 complex in response to more than one stress condition. Nonetheless, most LSM1-7-regulated genes were stress specific (Supplemental Figure 6 and Supplemental Data Sets 1 to 12). c-lsm1a plants presented wild-type expression patterns for all validated genes (Figure 3). Overall, these results indicated that the Arabidopsis LSM cytoplasmic complex regulates Arabidopsis tolerance to abiotic stresses by differentially modulating stress-responsive gene expression.
The Arabidopsis LSM1-7 Complex Regulates the Turnover of Selected Stress-Dependent Transcripts in Response to Abiotic Stresses
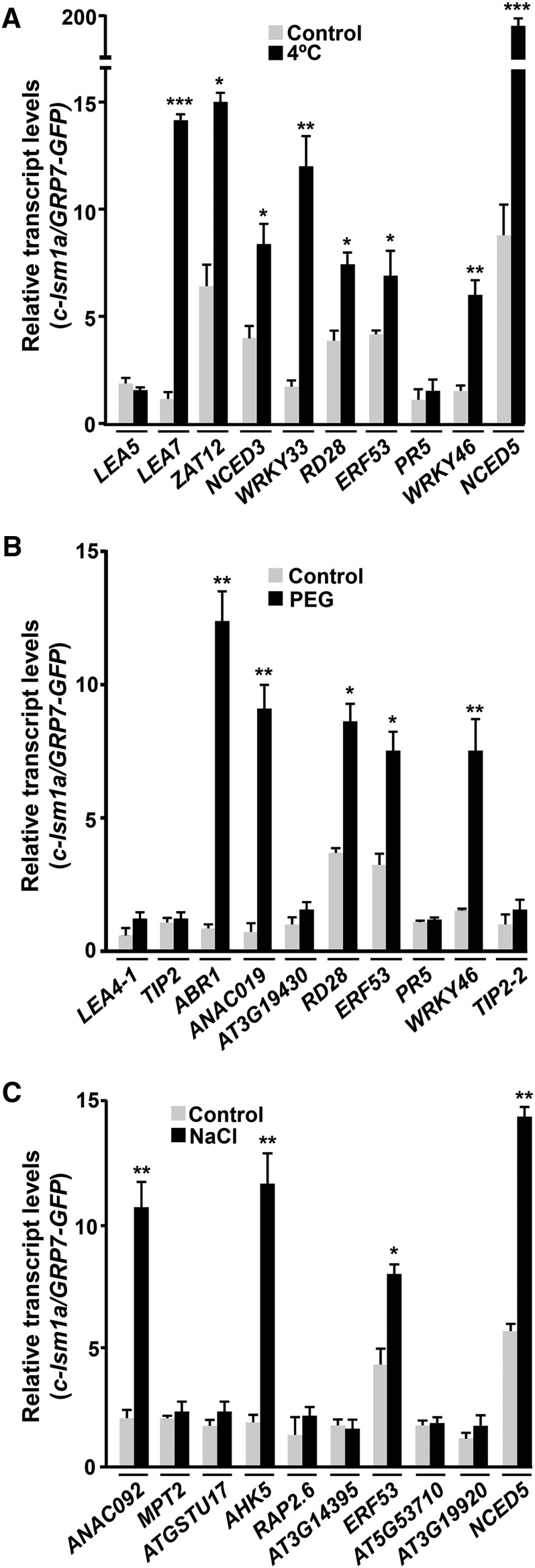
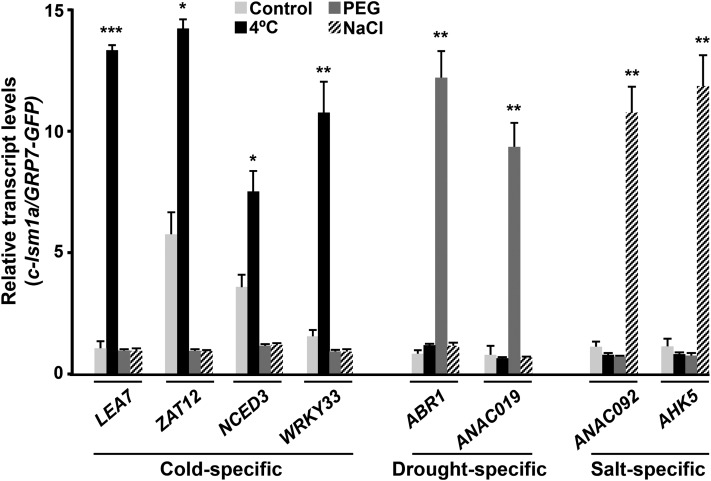
Considering the capacity of the LSM cytoplasmic complex to control transcript turnover through the interaction of LSM1 with target mRNAs, promoting their decapping and subsequent degradation (Chowdhury et al., 2007), we reasoned that it could differentially regulate gene expression in response to abiotic stresses by inducing the decay of selected transcripts in a stress-dependent manner. To investigate this idea, we first identified direct targets of the complex under low temperature, drought, and high salt conditions by means of RIP assays. These assays were performed with c-lsm1a plants grown under control conditions or exposed to 4°C, 55% PEG, or 150 mM NaCl. Transcripts interacting with LSM1A-GFP were coimmunoprecipitated (co-IP) with anti-GFP antibody and then identified by qPCR. Arabidopsis transgenic plants expressing a GFP-tagged GRP7 protein (Streitner et al., 2012) were also used in RIP experiments as a control for the specificity of the detected interactions. Given the function of the LSM1-7 complex, we expected some of its direct targets to be among the mRNAs upregulated in the stress-treated lsm1a lsm1b mutants. Therefore, targets were searched for among the cold-, drought-, and salt-inducible transcripts whose levels we had confirmed were upregulated in lsm1a lsm1b mutants in response to low temperature, water stress, and high salt, respectively (Figures 3A to 3C). As anticipated, several cold-induced transcripts, including LEA7, ZAT12, NCED3, WRKY33, RD28, ERF53, WRKY46, and NCED5, were significantly enriched (≥2-fold) in co-IP RNA samples from cold-treated c-lsm1a plants but not in the RNA samples from GRP7-GFP plants (Figure 4A), providing evidence that they were direct targets of the Arabidopsis LSM cytoplasmic complex in response to low temperature. Some mRNAs, such as ZAT12, NCED3, RD28, ERF53, and NCED5, were already significantly detected, although at lower levels, in co-IP RNA samples recovered from unstressed c-lsm1a plants (Figure 4A), suggesting that they were also targets of the complex under control conditions.
Figure 4.
The Arabidopsis LSM1-7 Complex Interacts with Selected Target Transcripts in Response to Abiotic Stresses.
RIP assays of 2-week-old c-lsm1a plants grown under control conditions: control exposed for 24 h to 4°C (A), 10 h to 55% PEG (B), or 10 h to 150 mM NaCl (C), using an anti-GFP antibody. RIP assays of Arabidopsis containing a GRP7PRO-GRP7-GFP fusion grown under control and stress conditions were also performed as interaction specificity controls. Co-IP RNA samples corresponding to different cold- (A), drought- (B), and salt-inducible (C) genes were quantified by qPCR. Transcript levels in c-lsm1a plants were corrected with respect to their corresponding input values and are represented relative to the levels obtained from RIP control assays. In all graphs, error bars indicate the sd of the mean (n ≥ 3). Asterisks indicate significant differences (*P ≤ 0.01, **P ≤ 0.001, and ***P ≤ 0.0001) in transcript levels between RIP assays from stressed and control plants, as determined by t test.
Compared with those obtained from GRP7-GFP plants, co-IP RNA samples obtained from c-lsm1a plants exposed to water deficiency revealed a specific and significant enrichment in ABR1, ANAC019, RD28, ERF53, and WRKY46 transcripts (Figure 4B). Consistent with the results obtained when looking for targets of the complex under cold conditions (Figure 4A), RD28 and ERF53 mRNAs were also found to be significantly detected in co-IP RNA samples from control c-lsm1a plants (Figure 4B). In the case of RNA samples collected from c-lsm1a and GRP7-GFP plants subjected to high salt, RIP assays uncovered a specific and significant enrichment of ANAC092, AHK5, ERF53, and NCED5 mRNAs in c-lsm1a samples (Figure 4C). ERF53 and NCED5 transcripts were also significantly detected in co-IP RNA samples from nonstressed c-lsm1a plants (Figure 4C).
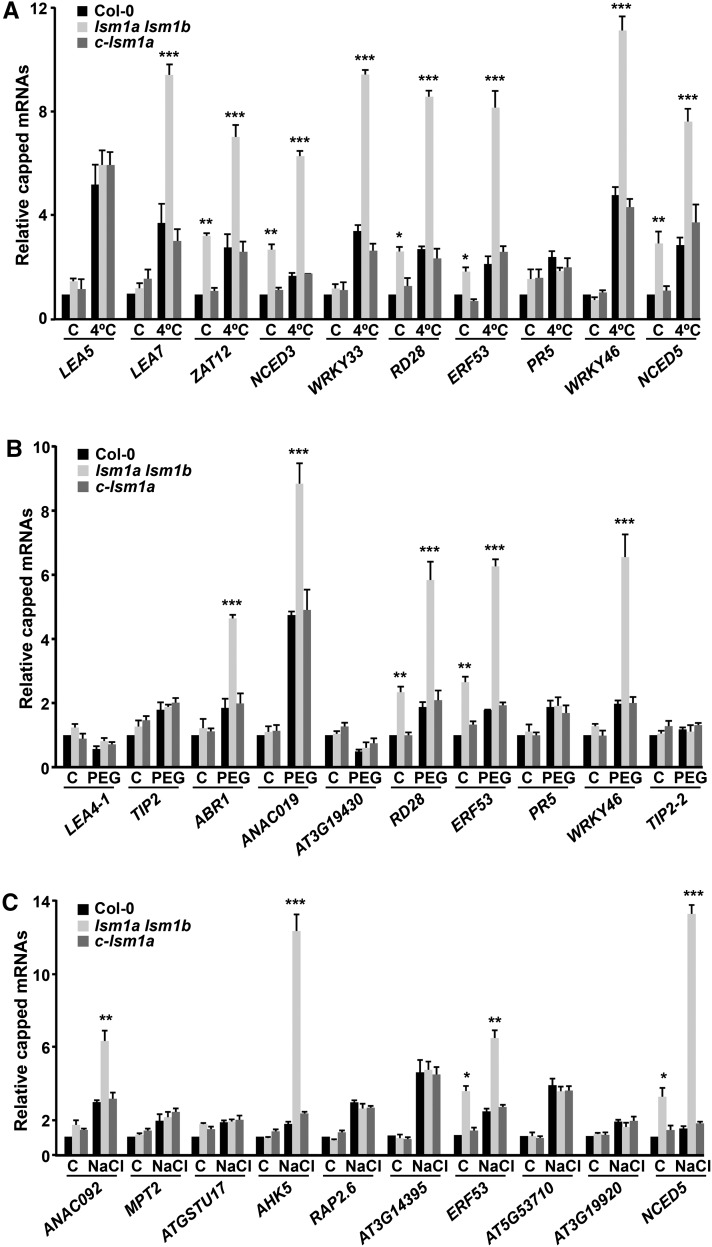
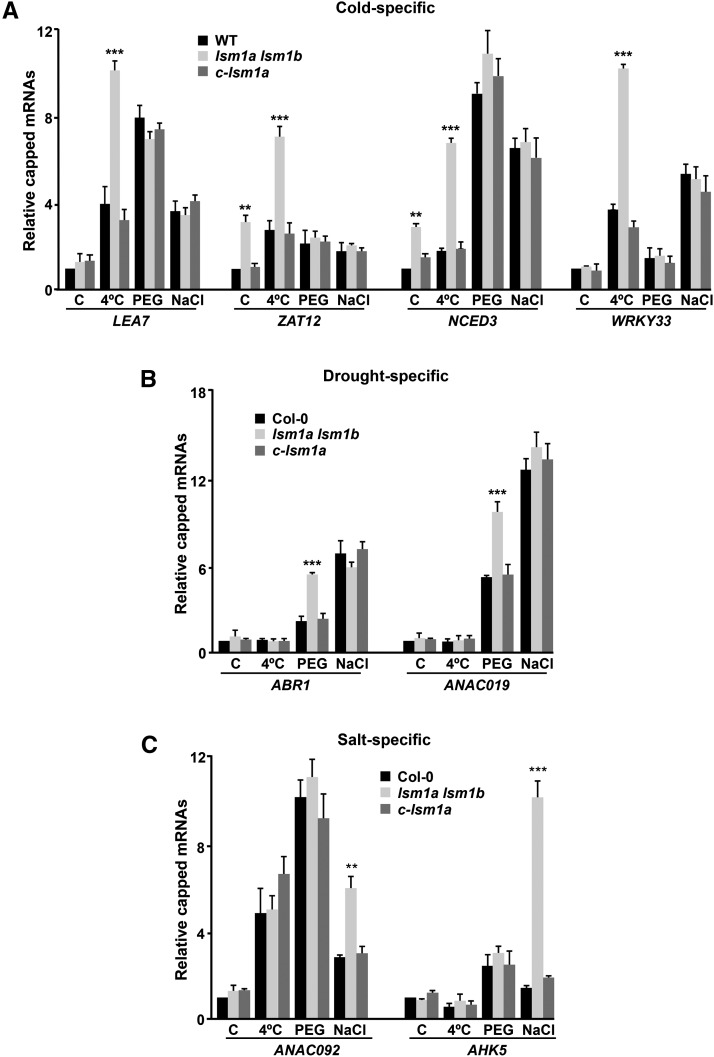
As anticipated from the function of the LSM cytoplasmic complex in mRNA degradation by promoting the decapping of its targets (Perea-Resa et al., 2012), RACE-qPCR experiments revealed that LEA7, ZAT12, NCED3, WRKY33, RD28, ERF53, WRKY46, and NCED5 messengers accumulated in their capped forms in lsm1a lsm1b mutants compared with wild-type plants exposed to low temperature (Figure 5A). Additionally, the levels of capped ZAT12, NCED3, RD28, ERF53, and NCED5 transcripts were higher in mutant than in wild-type plants grown under control conditions (Figure 5A). RACE-qPCR experiments with RNAs from water-stressed lsm1a lsm1b and wild-type plants showed that capped ABR1, ANAC019, RD28, ERF53, and WRKY46 messengers were significantly increased in the mutants. Moreover, as expected, RD28 and ERF53 transcripts were also augmented in their capped forms in unstressed mutants (Figure 5B). The levels of capped ANAC092, AHK5, ERF53, and NCED5 mRNAs were clearly higher in lsm1a lsm1b than in wild-type plants exposed to NaCl (Figure 5C). Furthermore, capped ERF53 and NCED5 transcripts also accumulated in mutants grown under standard conditions (Figure 5C). The capped forms of all mRNAs analyzed were unaltered in c-lsm1a plants (Figure 5).
Figure 5.
The Arabidopsis LSM1-7 Complex Promotes the Decapping of Selected Target Transcripts in Response to Abiotic Stresses.
Capped transcripts in 2-week-old Col-0, lsm1a lsm1b, and c-lsm1a plants grown under control conditions (C), exposed for 24 h to 4°C (A), 10 h to 55% PEG (B), or 10 h to 150 mM NaCl (C). The levels of capped transcripts corresponding to different cold- (A), drought- (B), and salt-inducible (C) genes were corrected with respect to the levels of their corresponding total transcripts and are represented relative to control Col-0 plants. In all graphs, error bars indicate the sd of the mean (n ≥ 3). Asterisks indicate significant differences (*P ≤ 0.01, **P ≤ 0.001, and ***P ≤ 0.0001) between lsm1a lsm1b and Col-0 or c-lsm1a plants, as determined by ANOVA tests. No significant differences between Col-0 and c-lsm1a plants were observed in any case.
All of these results strongly suggested that the LSM1-7 complex regulates the turnover of different selected stress-responsive target transcripts, including both specific and nonspecific ones, in response to different abiotic stresses. The existence of nonspecific target mRNAs, such as RD28, ERF53, WRKY46, and NCED5, has been already evidenced (Figures 4 and 5). The existence of specific targets was established by assessing the affinity of the complex for LEA7, ZAT12, NCED3, WRKY33, ABR1, ANAC019, ANAC092, and AHK5 transcripts under different unfavorable conditions. Thus, RIP experiments showed that LEA7, ZAT12, NCED3, and WRKY33, direct targets of the LSM cytoplasmic complex in response to low temperature (Figures 4A and 5A), were not enriched in co-IP RNA samples from c-lsm1a plants exposed to water deficiency and high salt (Figure 6). In turn, ABR1 and ANAC019, direct targets of the complex in response to drought (Figures 4B and 5B), were not recovered with LSM1A-GFP in response to cold and salt stresses (Figure 6). Similarly, ANAC092 and AHK5, direct targets of the complex in response to high salt (Figures 4C and 5C), were not enriched in co-IP RNA samples from c-lsm1a plants subjected to 4°C and drought (Figure 6). In addition, as expected from the RIP assays, the levels of capped LEA7, ZAT12, NCED3, and WRKY33 messengers did not increase in lsm1a lsm1b mutants under drought and high salt conditions (Figure 7A), and ABR1 and ANAC019 mRNAs did not accumulate in their capped forms in mutant plants in response to low temperature and salt stress (Figure 7B). Moreover, capped ANAC092 and AHK5 transcripts did not increase in lsm1a lsm1b in response to cold and water deficiency (Figure 7C). In all cases, c-lsm1a plants presented equivalent levels of capped mRNAs as wild-type plants (Figure 7).
Figure 6.
The Interaction between the Arabidopsis LSM1-7 Complex and Some of Its Target Transcripts Depends on the Abiotic Stress Conditions.
RIP assays of 2-week-old c-lsm1a plants grown under control conditions, exposed for 24 h to 4°C, 10 h to 55% PEG, or 10 h to 150 mM NaCl, using an anti-GFP antibody. RIP assays of 2-week-old Arabidopsis plants containing a GRP7PRO-GRP7-GFP fusion grown under control and stressed conditions were also performed as interaction specificity controls. Co-IP RNA samples corresponding to LEA7, ZAT12, NCED3, WRKY33, ABR1, ANAC019, ANAC092, and AHK5 genes were quantified by qPCR. Transcript levels in c-lsm1a plants were corrected with respect to their corresponding input values and are represented relative to the levels obtained from RIP control assays. In all graphs, error bars indicate the sd of the mean (n ≥ 3). Asterisks indicate significant differences (*P ≤ 0.01, **P ≤ 0.001, and ***P ≤ 0.0001) in transcript levels between RIP assays from stressed and control plants, as determined by ANOVA tests.
Figure 7.
Decapping of Some Target Transcripts by the Arabidopsis LSM1-7 Complex Depends on the Abiotic Stress Conditions.
Capped transcripts in 2-week-old Col-0, lsm1a lsm1b, and c-lsm1a plants grown under control conditions (C), exposed for 24 h to 4°C, 10 h to 55% PEG, or 10 h to 150 mM NaCl. The levels of capped transcripts corresponding to LEA7, ZAT12, NCED3, and WRKY33 (A), ABR1 and ANAC019 (B), and ANAC092 and AHK5 (C) were corrected with respect to the levels of their corresponding total transcripts and are represented relative to control Col-0 plants. In all graphs, error bars indicate the sd of the mean (n ≥ 3). Asterisks indicate significant differences (**P ≤ 0.001 and ***P ≤ 0.0001) between lsm1a lsm1b and Col-0 or c-lsm1a plants, as determined by ANOVA tests. No significant differences between lsm1a lsm1b and the other plants were found in any case.
Together, our data indicated that the Arabidopsis LSM1-7 complex differentially regulates gene expression in response to abiotic stresses, and consequently plant tolerance to these challenging situations, by modulating in each case the decay of selected, both specific and non-specific, stress-responsive transcripts. The fact that some of these mRNAs are, furthermore, direct targets of the complex at 20°C, suggests that it also regulates stress-responsive gene expression under control conditions.
The Arabidopsis LSM Cytoplasmic Complex Regulates ABA Biosynthesis in Response to Abiotic Stress
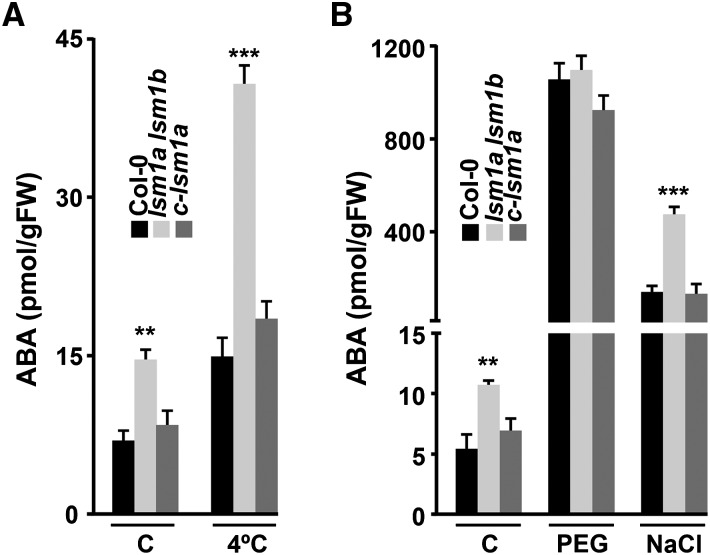
Interestingly, among the mRNAs whose decay was differentially regulated by the Arabidopsis LSM1-7 complex in response to abiotic stresses were those encoding NCED3 and NCED5, two key enzymes in the biosynthesis of the phytohormone ABA that plays a critical role in plant responses to abiotic stresses (Jia et al., 2002; Cuevas et al., 2008; Wu et al., 2009; Frey et al., 2012). In fact, while NCED3 transcripts were direct targets of the complex specifically after cold treatment (Figures 6 and 7A), those of NCED5 were direct targets under both cold and high-salt conditions (Figures 4A, 4C, 5A, and 5C) but not following drought (Supplemental Figure 7). These data strongly suggested that the LSM1-7 complex might differentially regulate ABA biosynthesis in Arabidopsis in response to abiotic stresses. To test this prediction, we measured the ABA content of wild-type and lsm1a lsm1b mutant plants grown under control conditions or exposed to low temperature (4°C), water deficiency (55% PEG), or high salt (150 mM NaCl). When exposed to low temperature, ABA levels increased in both the wild type and lsm1a lsm1b mutants but the increase was significantly higher in mutants than in wild-type plants (Figure 8A). Unstressed lsm1a lsm1b mutants also showed higher levels of ABA than wild-type plants (Figure 8A). However, these levels were much lower than those detected after cold treatment, paralleling the lower binding affinity of the LSM cytoplasmic complex for NCED3 and NCED5 mRNAs at 20°C compared with 4°C (Figures 4A and 5A). c-lsm1a plants exhibited similar ABA content as wild-type plants (Figure 8A), providing evidence that the Arabidopsis LSM1-7 complex attenuates ABA biosynthesis in response to low temperature and, to a lesser extent, under standard conditions by promoting the decay of NCED3 and NCED5 messengers.
Figure 8.
ABA Biosynthesis Is Differentially Regulated by the Arabidopsis LSM1-7 Complex in Response to Abiotic Stresses.
ABA levels in 2-week-old Col-0, lsm1a lsm1b and c-lsm1a plants grown under control conditions (C), exposed for 24 h to 4°C (A), 10 h to 55% PEG (B) or 10 h to 150 mM NaCl (B). In all graphs, error bars indicate the sd of the mean (n = 4). Asterisks indicate significant differences (**P ≤ 0.001 and ***P ≤ 0.0001) between lsm1a lsm1b and Col-0 or c-lsm1a plants, as determined by ANOVA tests. No significant differences between Col-0 and c-lsm1a plants were observed in any case.
ABA levels also increased very prominently in Arabidopsis wild-type plants exposed to water stress or high salt (Figure 8B). In the case of lsm1a lsm1b mutants, water deficiency caused an accumulation of ABA similar to that of wild-type plants. By contrast, salt stress provoked an accumulation of ABA that was significantly higher than in wild-type plants. c-lsm1a plants exhibited similar ABA content as the wild type in response to both drought and salt stresses (Figure 8B). Therefore, as predicted from the expression and RIP analyses, the LSM cytoplasmic complex also mediates the biosynthesis of ABA in Arabidopsis plants exposed to salt stress, but it is not involved in ABA biosynthesis in response to water stress. Collectively, these findings demonstrated that the Arabidopsis LSM1-7 complex guarantees adequate levels of ABA in Arabidopsis plants exposed to different abiotic stresses by differentially regulating the decay of NCED3 and NCED5 transcripts.
DISCUSSION
The control of mRNA stability is a key step in the regulation of gene expression. However, the effect of mRNA decapping on plant transcriptome reprogramming in response to abiotic stress and its importance to stress tolerance are largely unknown. Here, we show that the LSM1-7 decapping activator complex serves as an integration node of regulatory pathways mediating plant tolerance to abiotic stresses. It regulates Arabidopsis tolerance to freezing, drought, and high salt by interacting with selected, specific, and nonspecific stress-inducible transcripts under each stress condition to promote their decapping and subsequent degradation, which ultimately ensures the appropriate patterns of downstream stress-responsive gene expression. Interestingly, one of the regulatory pathways mediated by the LSM1-7 complex is the one leading to ABA biosynthesis. We demonstrate that this complex modulates the levels of ABA in response to adverse environmental situations through the differential regulation of NCED3 and NCED5 mRNA turnover.
The expression analysis described in this work revealed that LSM1-7 genes are differentially regulated in response to abiotic stresses. LSM1-7 mRNAs accumulate in response to cold but not in response to drought or high salt. The mechanisms underlying this regulation are still under investigation. Consistent with the expression results, an increase in LSM1A and LSM1B protein levels was observed only in plants exposed to low temperature. Remarkably, however, Arabidopsis LSM1 proteins were found to localize to P-bodies in response to the different stresses assayed, indicating that the molecular mechanisms mediating LSM1 localization to these foci do not depend on protein levels. In this way, DCP1 has also been reported to localize to P-bodies under stress conditions even though its levels do not increase (Motomura et al., 2015). These findings suggest that the localization of LSM1 to P-bodies, like that of DCP1 and other Arabidopsis RNA decay-related proteins (Merret et al., 2013; Motomura et al., 2015), is induced in plants exposed to abiotic stresses. Moreover, our results show that LSM1 proteins are required for P-body formation under challenging situations, indicating that the assembly of these foci is also promoted by adverse environmental conditions. How abiotic stresses regulate the cytoplasmic dynamics of proteins involved in RNA degradation and, ultimately, P-body formation is poorly understood. Phosphorylation of different components of human and Arabidopsis mRNA decapping machineries by mitogen-activated protein kinases (MPKs) during stress responses seems to be necessary for their cytoplasmic localization and for P-body assembly (Rzeczkowski et al., 2011; Roux et al., 2015). The sequence of LSM1 proteins (Perea-Resa et al., 2012) contains a consensus motif S/T-P for phosphorylation by MPK. Therefore, it is tempting to speculate that the subcellular localization of LSM1 proteins could be regulated by MPKs in response to abiotic stresses.
Our results demonstrate that the LSM1-7 complex differentially regulates Arabidopsis tolerance to challenging environmental situations. In fact, the complex restrains the plant’s capacity to cold acclimate and tolerate drought while promoting its tolerance to high salt. The implication of a single component of the mRNA decay apparatus in a range of abiotic stress responses has not been shown in any system and uncovers the enormous potential of this apparatus to precisely modulate the adaptation of a given organism to its surroundings. Therefore, the LSM cytoplasmic complex seems to serve as a regulatory node where pathways mediating abiotic stress responses converge and integrate to guarantee the precise development of Arabidopsis tolerance to freezing, drought, and salinity. Furthermore, our results provide evidence that the LSM1-7 complex operates in Arabidopsis tolerance to abiotic stresses by modulating stress-responsive gene expression. Hundreds of genes are specifically regulated by the complex under low temperature, water deficiency, or high salt. Moreover, the complex also regulates the expression of many genes in response to more than one abiotic stress, which indicates that it mediates gene expression under different unfavorable situations via specific and shared signaling pathways. Interestingly, numerous specific and nonspecific genes that are regulated by the LSM cytoplasmic complex under each stress condition have been described to have a role in plant tolerance to the relevant condition, substantiating a major and differential function for the complex in plant tolerance to abiotic stresses by ensuring appropriate patterns of stress-responsive gene expression. DCP5 has also been shown to act in abiotic stress tolerance through the regulation of stress-related gene expression. It enhances Arabidopsis tolerance to dehydration by modulating numerous dehydration-responsive genes (Xu and Chua, 2012). Whether DCP5 and/or other components of the Arabidopsis mRNA decay apparatus have the ability, similar to the LSM1-7 complex, to differentially regulate gene expression in a stress-dependent manner remains unknown.
The role of the LSM cytoplasmic complex in the transcriptome reprogramming that takes place in Arabidopsis when subjected to abiotic stresses is a consequence of its capacity to modulate the turnover of stress-selected target transcripts. In fact, by analyzing the interaction between mRNAs whose levels are attenuated by the complex in response to abiotic stresses and the LSM1 protein, we have been able to identify transcripts that are direct targets of the complex when Arabidopsis plants are exposed to low temperature, drought, or high salt and may act as positive or negative regulators of gene expression. Stress-selected targets are then decapped and subsequently degraded. It is noteworthy that most of them are specific for each stress, while others are selected by the complex in response to various adverse conditions. Therefore, modulating the differential accumulation of the selected specific and nonspecific target transcripts ensures adequate stress-responsive gene expression under each abiotic stress and ultimately contributes to the capacity of the LSM1-7 complex to regulate Arabidopsis tolerance to freezing, drought, and salinity. The Arabidopsis decapping activator DCP5 has also been reported to select target transcripts in response to an individual abiotic stress, namely, water deficiency (Xu and Chua, 2012). Our results nonetheless unveil a more intricate scenario when considering the regulation of gene expression and plant adaptation by the mRNA decay apparatus under unfavorable circumstances. They provide evidence that one component can select different targets depending on the stress situations, highlighting a previously unknown regulatory mechanism of mRNA turnover during stress responses.
Remarkably, among the selected stress-dependent transcripts that are triggered by the Arabidopsis LSM cytoplasmic complex for decapping in response to abiotic stresses, we found those encoding NCED3 and NCED5, two key enzymes in ABA biosynthesis (Tan et al., 2003; Frey et al., 2012). NCED3 and NCED5 messengers are direct targets of the complex under low temperature but not under drought conditions. Under salt stress, only NCED5 is targeted by the LSM1-7 complex. ABA is biosynthesized de novo in plants subjected to cold, drought, or salinity and plays a pivotal role in plant responses to abiotic stresses (Jia et al., 2002; Cuevas et al., 2008; Wu et al., 2009). In Arabidopsis, the accumulation of ABA after abiotic stress exposure seems to be the result of NCED3 and NCED5 expression (Tan et al., 2003; Frey et al., 2012). So far, available data had indicated that this expression is regulated at the transcriptional level (Barrero et al., 2006; Cuevas et al., 2008; Jiang et al., 2012). The results reported here show that the mRNA decay machinery, and in particular the decapping apparatus, has a crucial function in establishing the levels of NCED3 and NCED5 mRNAs when plants are exposed to stressful circumstances, indicating that these levels are also regulated by posttranscriptional mechanisms. Therefore, our findings reveal a layer of regulation of ABA biosynthesis in response to abiotic stresses. Consistent with its role in differentially modulating the turnover of NCED3 and NCED5 transcripts under adverse environmental circumstances, our data also demonstrate that the LSM cytoplasmic complex contributes to establish the appropriate levels of ABA under cold and high salt conditions. However, it is not involved in modulating the accumulation of ABA caused by water deficiency.
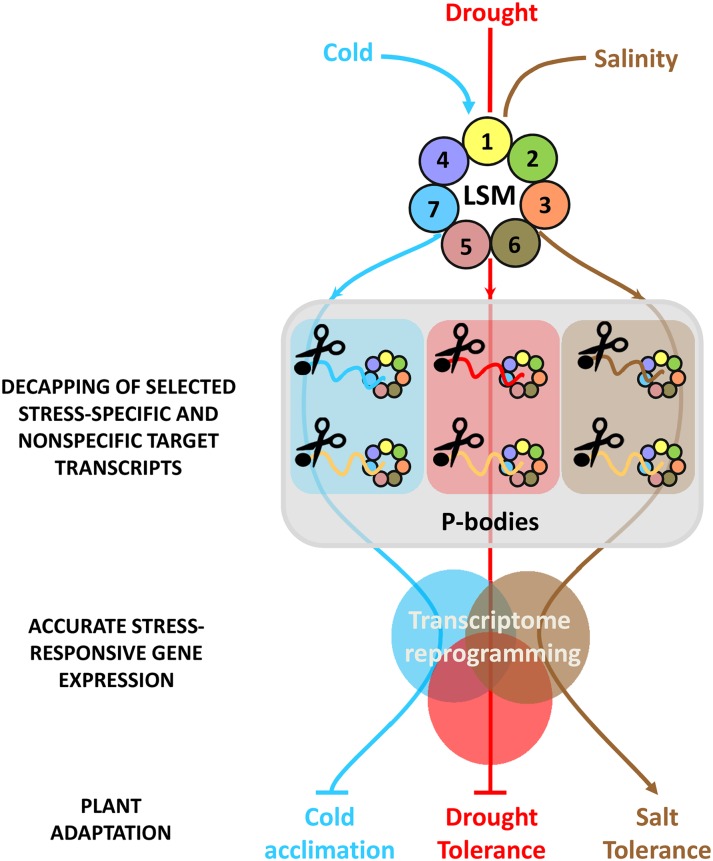
Based on the results described in this work, a hypothetical model for the function of the Arabidopsis LSM1-7 complex in plant adaptation to abiotic stresses is presented in Figure 9. In response to low temperature, drought, and salinity, the complex would preferentially localize to P-bodies. There, depending on the stress, the complex would interact with selected transcripts promoting their decapping and subsequent 5′-3′ degradation. The vast majority of selected transcripts are stress specific; others, however, interact with the complex under more than one stress condition. In most cases, target transcripts would correspond to stress-inducible genes, and some of them encode proteins involved in regulating, positively or negatively, Arabidopsis tolerance to abiotic stresses. The degradation of the LSM1-7 target transcripts selected in response to a given abiotic stress would have, in turn, a substantial effect on the downstream stress-regulated gene expression that would contribute to shape the transcriptome reprogramming required for plant adaptation to that stress. In conclusion, the results presented here reveal that the LSM1-7 decapping activator complex plays a critical role in plant adaptation to abiotic stresses by modulating the turnover of selected stress-specific and nonspecific target transcripts and, consequently, stress-regulated gene expression. Identifying the molecular mechanisms whereby the LSM cytoplasmic complex selects different targets depending on the stress conditions is a clear goal of future studies.
Figure 9.
Proposed Model for the Function of LSM1-7 Complex in Plant Responses to Abiotic Stresses.
In response to abiotic stress conditions, the LSM1-7 complex would localize to P-bodies where it would interact with selected, specific, and nonspecific mRNA targets to promote their decapping and subsequent decay. This function would contribute to shape the transcriptome reprogramming required for plant adaptation to abiotic stresses. Arrowheads and end lines indicate positive and negative regulation, respectively.
METHODS
Plant Materials, Growth Conditions, Treatments, and Tolerance Assays
Arabidopsis thaliana Columbia (Col-0) ecotype was used in all experiments. The lsm1a lsm1b double mutants, transgenic lines LSM1APRO-LSM1A-GFP (c-lsm1a) and LSM1BPRO-LSM1B-GFP (c-lsm1b), lines LSM1APRO-LSM1A-GFP and LSM1BPRO-LSM1B-GFP expressing 35S-DCP1-RFP construct, and lsm1a lsm1b lines expressing the 35S-GFP-DCP2 or 35S-GFP-VCS constructs were previously described (Perea-Resa et al., 2012). The Arabidopsis transgenic line expressing the GRP7PRO-GRP7-GFP fusion (Streitner et al., 2012) was kindly provided by Dorothee Staiger (University of Bielefeld, Germany). Plants were grown at 20°C under a long-day photoperiod (16 h of cool-white fluorescent light, photon flux of 90 µmol m−2 s−1) in pots containing a mixture of organic substrate and vermiculite (3:1 [v/v]) or in Petri dishes containing Murashige and Skoog medium supplemented with 1% sucrose (GM) and solidified with 0.8% (w/v) agar. Low-temperature treatment for gene expression and immunoblot analyses was performed by transferring plants growing in Petri dishes or on soil under control conditions to a growth chamber set to 4°C for different periods of time under long-day photoperiods with a photon flux of 40 mmol m−2 s−1. For the rest of experiments, plants were always exposed to 4°C for 24 h. In all cases, water and salt stress treatments were accomplished by transferring plants growing in Petri dishes under standard conditions to plates containing GM medium supplemented with 55% PEG or 150 mM NaCl, respectively, for 10 h. For immunoblot analysis, water and salt stress treatments were also performed for 24 h. Tolerance to freezing temperatures was determined on 2-week-old plants grown on soil as described (Catalá et al., 2014). In vitro tolerance to water (25% PEG) and salt (150 mM NaCl) stresses was assayed on 7-d-old seedlings grown on GM medium as reported (Verslues et al., 2006). The tolerance of 2-week-old plants growing on soil to these stresses was estimated as the number of surviving individuals after 10 d of water deprivation and 5 d of rewatering or after watering with 250 mM NaCl for 10 d. In all cases, data reported are expressed as means of three independent experiments with 50 plants each.
Microscopy Analysis
Subcellular localization of LSM1A-GFP, LSM1B-GFP, RFP-DCP1, GFP-DCP2, and GFP-VCS fusion proteins was performed by confocal microscopy in roots from 6-d-old transgenic seedlings grown under control or subjected to stress conditions as described above. Treatments with cycloheximide were performed by incubating stressed seedlings in GM liquid medium supplemented with 200 µg/mL cycloheximide for 2 h at 20°C or 4°C. Microscopy images were collected using a TCS SP5 confocal laser spectral microscope (Leica Microsystems). The excitation lines for imaging GFP and RFP fusions were 488 and 561 nm, respectively.
ABA Measurements
ABA levels were determined in 20 mg of 2-week-old wild-type, lsm1a lsm1b, and c-lsm1a plants grown under control or stress conditions as described (Turečková et al., 2009). All experiments were repeated as four biological replicates employing 10 pmol stable isotope-labeled standard to validate the liquid chromatography-mass spectrometry method.
Immunoblot Analysis
Total protein was extracted from 2-week-old Arabidopsis LSM1APRO-LSM1A-GFP, LSM1BPRO-LSM1B-GFP, and wild-type plants grown under control or stress conditions as reported (Catalá et al., 2014). Monoclonal anti-GFP (ab290; GR158277-1; Abcam) was used as primary antibody and horseradish peroxidase-conjugated anti-rabbit as secondary antibody. Coomassie Brilliant Blue staining of the large subunit of Rubisco was used as a loading control. All assays were performed in triplicate employing three independent protein samples.
Gene Expression Analysis and RNA-Seq Experiments
For gene expression, qPCR experiments were performed as described (Catalá et al., 2014). In all cases, the relative expression values were determined using AT4G24610 as a control (Czechowski et al., 2005). Primers used are listed in Supplemental Data Set 13. All reactions were performed in triplicate employing three independent RNA samples.
For RNA-seq experiments, total RNA was extracted from 2-week-old wild-type and lsm1a lsm1b plants subjected to stress conditions (see above) using TRIzol reagent (Life Technologies) and cleaned with the RNeasy Plant Mini Kit (Qiagen). cDNA libraries were generated from three independent RNA preparations each. RNA quality, library preparation, and subsequent sequencing were performed by the staff of the Beijing Genome Institute. RNA-seq reads were aligned to the TAIR10 Col-0 reference genome using TopHat2 (Kim et al., 2013) with default parameters. Uniquely mapped reads (Supplemental Data Set 14) were counted per representative gene model (excluding introns) according to the TAIR10 annotation using custom R scripts. Only genes with reads per kilobase per million > 1 in at least one sample were used for differential expression analysis between wild-type and lsm1a lsm1b plants in each condition using DEseq2 (Love et al., 2014). This package internally estimates size factors for each sample, calculates dispersion for each gene, and then fits a negative binomial GLM to detect differentially expressed genes taking into account the size factors and dispersion values.
Capped mRNA Analysis and RIP Experiments
Capped transcript levels were determined essentially as reported (Perea-Resa et al., 2012). Total RNA from 2-week-old plants grown on GM plates under control or stress conditions (see above) was employed. Capped mRNAs were quantified by qRT-PCR using reverse specific-gene primers (Supplemental Data Set 13) and the 5′RACE Inner Primer included in the First Choice RLM-RACE kit (Ambion). The levels of capped transcripts were corrected with respect to the levels of their corresponding total transcripts, determined by qPCR using gene-specific primers (Supplemental Data Set 13), and represented relative to the levels in wild-type plants under control conditions for each gene. In all cases, reactions were performed in triplicate employing three independent RNA samples.
In vivo RIP experiments were performed essentially as described (Streitner et al., 2012). Extracts from 2-week-old c-lsm1a and GRP7-GFP plants grown on GM plates under control or stress conditions (see above) were incubated overnight at 4°C with GFP-Trap beads (Chromotek). Beads were subsequently washed three times prior to RNA extraction using TRIzol reagent (Life Technologies). RNAs resulting from inputs, prepared in parallel, and from coprecipitated samples, were subsequently quantified by qPCR (see above) using gene-specific primers (Supplemental Data Set 13). Transcript levels in coprecipitated samples from c-lsm1a plants were corrected with respect to their corresponding input values and represented relative to their levels in coprecipitated samples from GRP7-GFP plants for each gene. All assays were performed in triplicate employing three independent RNA samples.
Statistical Analysis
Data sets were analyzed using Prism 6 software (GraphPad Software). Comparisons between two groups were made using Student's t test. Comparisons between multiple groups were made using one‐way or two-way ANOVA tests depending whether one or two different variables were considered, respectively. Correlation plots were computed using Prism 6 software (GraphPad Software) from log2-transformed values and shown the relationship between qRT-PCR results (x axis) and the corresponding data from RNA-seq (y axis).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Supplemental Data Set. The full names of the genes mentioned in this article are also included in Supplemental Data Set 15. The complete genome-wide data from this publication have been submitted to the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) and assigned the identifier accession GSE70491.
Supplemental Data
Supplemental Figure 1. Expression analysis of Arabidopsis LSM1-7 genes in response to abiotic stresses.
Supplemental Figure 2. Arabidopsis LSM1 proteins do not accumulate in response to drought and salinity but localize to P-bodies under these abiotic stresses.
Supplemental Figure 3. Constitutive freezing tolerance and tolerance of drought and salinity of lsm1a lsm1b plants grown on soil.
Supplemental Figure 4. Expression levels of genes downregulated in lsm1a lsm1b plants in response to cold and drought.
Supplemental Figure 5. Correlation between RNA-seq and qPCR results.
Supplemental Figure 6. Venn diagrams of deregulated transcripts in lsm1a lsm1b plants under cold, drought, and salt stresses.
Supplemental Figure 7. The Arabidopsis LSM1-7 complex differentially interacts with and promotes the decapping of NCED5 transcripts in response to drought and high salt.
Supplemental Data Set 1. Top 1000 upregulated genes in lsm1a lsm1b mutants in response to low temperature.
Supplemental Data Set 2. Cold-inducible genes upregulated in lsm1a lsm1b mutants in response to low temperature.
Supplemental Data Set 3. Top 1000 downregulated genes in lsm1a lsm1b mutants in response to low temperature.
Supplemental Data Set 4. Cold-inducible genes downregulated in lsm1a lsm1b mutants in response to low temperature.
Supplemental Data Set 5. Top 1000 upregulated genes in lsm1a lsm1b mutants in response to drought.
Supplemental Data Set 6. Drought-inducible genes upregulated in lsm1a lsm1b mutants in response to drought.
Supplemental Data Set 7. Top 1000 downregulated genes in lsm1a lsm1b mutants in response to drought.
Supplemental Data Set 8. Drought-inducible genes downregulated in lsm1a lsm1b mutants in response to drought.
Supplemental Data Set 9. Top 1000 upregulated genes in lsm1a lsm1b mutants in response to salt stress.
Supplemental Data Set 10. Salt-inducible genes upregulated in lsm1a lsm1b mutants in response to salt stress.
Supplemental Data Set 11. Top 1000 downregulated genes in lsm1a lsm1b mutants in response to salt stress.
Supplemental Data Set 12. Salt-inducible genes downregulated in lsm1a lsm1b mutants in response to salt stress.
Supplemental Data Set 13. Oligonucleotides used in this work.
Supplemental Data Set 14. Total mapped and unmapped reads obtained for each sample by RNA-seq experiments.
Supplemental Data Set 15. List of accession numbers appearing in this study.
Supplementary Material
Acknowledgments
We thank D. Staiger for the LSMGRP7PRO-GRP7-GFP Arabidopsis transgenic line and J.J. Sanchez-Serrano, J.A. Jarillo, and R. Solano for discussions and comments. This work was supported by Grants BIO2010-17545 and BIO2013-47788-R from MINECO to J.S., GA14-34792S from CSF to O.N., and MCB-1022435 from the National Science Foundation to L.S. R.C. is supported by a JAE-DOC contract from the CSIC, and C.C.-L. is a recipient of a FPI fellowship from MINECO.
AUTHOR CONTRIBUTIONS
C.P.-R., C.C.-L., O.N., L.S., and J.S. conceived and designed the experiments. C.P.-R., C.C.-L., R.C., V.T., and W.Z. performed the experiments. C.P.-R., C.C.-L., R.C., O.N., L.S., J.M.J.-G., and J.S. analyzed the data. C.P.-R. and J.S. wrote the article.
Glossary
- RIP
RNA immunoprecipitation
- ABA
abscisic acid
- PEG
polyethylene glycol
- co-IP
coimmunoprecipitated
- MPK
mitogen-activated protein kinase
References
- Balazadeh S., Siddiqui H., Allu A.D., Matallana-Ramirez L.P., Caldana C., Mehrnia M., Zanor M.I., Köhler B., Mueller-Roeber B. (2010). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62: 250–264. [DOI] [PubMed] [Google Scholar]
- Barrero J.M., Rodríguez P.L., Quesada V., Piqueras P., Ponce M.R., Micol J.L. (2006). Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 29: 2000–2008. [DOI] [PubMed] [Google Scholar]
- Catalá R., López-Cobollo R., Mar Castellano M., Angosto T., Alonso J.M., Ecker J.R., Salinas J. (2014). The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 26: 3326–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.H., Jiang H.W., Hsieh E.J., Chen H.Y., Chien C.T., Hsieh H.L., Lin T.P. (2012). Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol. 158: 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.C., Hsieh E.J., Chen J.H., Chen H.Y., Lin T.P. (2012). Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 158: 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A., Mukhopadhyay J., Tharun S. (2007). The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13: 998–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J., Parker R. (2004). Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73: 861–890. [DOI] [PubMed] [Google Scholar]
- Cuevas J.C., López-Cobollo R., Alcázar R., Zarza X., Koncz C., Altabella T., Salinas J., Tiburcio A.F., Ferrando A. (2008). Putrescine is involved in Arabidopsis freezing tolerance and cold acclimation by regulating abscisic acid levels in response to low temperature. Plant Physiol. 148: 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P., Zhang S., Ding F., Ali S., Xiong L. (2014). Dynamic regulation of genome-wide pre-mRNA splicing and stress tolerance by the Sm-like protein LSm5 in Arabidopsis. Genome Biol. 15: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillman C., Lykke-Andersen J. (2005). RNA decapping inside and outside of processing bodies. Curr. Opin. Cell Biol. 17: 326–331. [DOI] [PubMed] [Google Scholar]
- Frey A., Effroy D., Lefebvre V., Seo M., Perreau F., Berger A., Sechet J., To A., North H.M., Marion-Poll A. (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 70: 501–512. [DOI] [PubMed] [Google Scholar]
- Goeres D.C., Van Norman J.M., Zhang W., Fauver N.A., Spencer M.L., Sieburth L.E. (2007). Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19: 1549–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra D., Crosatti C., Khoshro H.H., Mastrangelo A.M., Mica E., Mazzucotelli E. (2015). Post-transcriptional and post-translational regulations of drought and heat response in plants: a spider’s web of mechanisms. Front. Plant Sci. 6: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujjar R.S., Akhtar M., Singh M. (2014). Transcription factors in abiotic stress tolerance. Indian J. Plant. Physiol. 19: 306–316. [Google Scholar]
- Jia W., Wang Y., Zhang S., Zhang J. (2002). Salt-stress-induced ABA accumulation is more sensitively triggered in roots than in shoots. J. Exp. Bot. 53: 2201–2206. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Liang G., Yu D. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 5: 1375–1388. [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merret R., Descombin J., Juan Y.T., Favory J.J., Carpentier M.C., Chaparro C., Charng Y.Y., Deragon J.M., Bousquet-Antonelli C. (2013). XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Reports 5: 1279–1293. [DOI] [PubMed] [Google Scholar]
- Miura K., Furumoto T. (2013). Cold signaling and cold response in plants. Int. J. Mol. Sci. 14: 5312–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomura K., Le Q.T.N., Hamada T., Kutsuna N., Mano S., Nishimura M., Watanabe Y. (2015). Diffuse decapping enzyme DCP2 accumulates in DCP1 foci under heat stress in Arabidopsis thaliana. Plant Cell Physiol. 56: 107–115. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yamaguchi-Shinozaki K., Shinozaki K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G.K., Grant J.J., Cheong Y.H., Kim B.G., Li L., Luan S. (2005). ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol. 139: 1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. (2012). RNA degradation in Saccharomyces cerevisae. Genetics 191: 671–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Li Z., Wen X., Li W., Shi H., Yang L., Zhu H., Guo H. (2014). Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 10: e1004664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Resa C., Hernández-Verdeja T., López-Cobollo R., del Mar Castellano M., Salinas J. (2012). LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell 24: 4930–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham J., Liu J., Bennett M.H., Mansfield J.W., Desikan R. (2012). Arabidopsis histidine kinase 5 regulates salt sensitivity and resistance against bacterial and fungal infection. New Phytol. 194: 168–180. [DOI] [PubMed] [Google Scholar]
- Roux M.E., Rasmussen M.W., Palma K., Lolle S., Regué À.M., Bethke G., Glazebrook J., Zhang W., Sieburth L., Larsen M.R., Mundy J., Petersen M. (2015). The mRNA decay factor PAT1 functions in a pathway including MAP kinase 4 and immune receptor SUMM2. EMBO J. 34: 593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzeczkowski K., Beuerlein K., Müller H., Dittrich-Breiholz O., Schneider H., Kettner-Buhrow D., Holtmann H., Kracht M. (2011). c-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J. Cell Biol. 194: 581–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Ding Y., Yang S. (2015). Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 56: 7–15. [DOI] [PubMed] [Google Scholar]
- Streitner C., Köster T., Simpson C.G., Shaw P., Danisman S., Brown J.W.S., Staiger D. (2012). An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana. Nucleic Acids Res. 40: 11240–11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B.C., Joseph L.M., Deng W.T., Liu L., Li Q.B., Cline K., McCarty D.R. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35: 44–56. [DOI] [PubMed] [Google Scholar]
- Tran L.S.P., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turečková V., Novák O., Strnad M. (2009). Profiling ABA metabolites in Nicotiana tabacum L. leaves by ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Talanta 80: 390–399. [DOI] [PubMed] [Google Scholar]
- Verslues P.E., Agarwal M., Katiyar-Agarwal S., Zhu J., Zhu J.K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45: 523–539. [DOI] [PubMed] [Google Scholar]
- Vogel J.T., Zarka D.G., Van Buskirk H.A., Fowler S.G., Thomashow M.F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41: 195–211. [DOI] [PubMed] [Google Scholar]
- Weber C., Nover L., Fauth M. (2008). Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 56: 517–530. [DOI] [PubMed] [Google Scholar]
- Wu Y., Deng Z., Lai J., Zhang Y., Yang C., Yin B., Zhao Q., Zhang L., Li Y., Yang C., Xie Q. (2009). Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19: 1279–1290. [DOI] [PubMed] [Google Scholar]
- Xu J., Chua N.H. (2009). Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 21: 3270–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Chua N.H. (2012). Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J. 31: 1975–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Yang J.Y., Niu Q.W., Chua N.H. (2006). Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18: 3386–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57: 781–803. [DOI] [PubMed] [Google Scholar]
- Zhang Z., et al. (2011). Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 23: 396–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.