Abstract
Flowering plants have strikingly distinct genomes, although they contain a similar suite of expressed genes. The diversity of genome structures and organization is largely due to variation in transposable elements (TEs) and whole-genome duplication (WGD) events. We review evidence that chromatin modifications and epigenetic regulation are intimately associated with TEs and likely play a role in mediating the effects of WGDs. We hypothesize that the current structure of a genome is the result of various TE bursts and WGDs and it is likely that the silencing mechanisms and the chromatin structure of a genome have been shaped by these events. This suggests that the specific mechanisms targeting chromatin modifications and epigenomic patterns may vary among different species. Many crop species have likely evolved chromatin-based mechanisms to tolerate silenced TEs near actively expressed genes. These interactions of heterochromatin and euchromatin are likely to have important roles in modulating gene expression and variability within species.
INTRODUCTION
Chromatin modifications play critical roles in the regulation of gene expression, three-dimensional organization of genomic DNA, recombination, and DNA repair. There are a number of distinct chromatin modifications and they vary substantially in their effects and stability. Some chromatin modifications, such as DNA methylation, are relatively stable and heritable. Other chromatin modifications, such as histone acetylation or phosphorylation, are quite labile and may provide important transient functions. The potential for chromatin modifications to provide information beyond the primary sequence of DNA has led to the use of the term “epigenome” to describe the genome-wide pattern of particular chromatin modifications despite the fact that specific modifications have varying stability.
Chromatin modifications influence gene expression during development and in response to environmental cues. In some cases, chromatin modifiers are likely to be recruited to achieve and stabilize alterations in transcription. Several recent reviews have provided excellent summaries of these activities and point toward the conservation of these pathways among flowering plants (Baulcombe and Dean, 2014; Bond and Baulcombe, 2014; Pikaard and Mittelsten Scheid, 2014; Probst and Mittelsten Scheid, 2015; Vriet et al., 2015; Xiao and Wagner, 2015). In this Review, we focus on aspects of chromatin regulation that might be variable or drive variation among closely related species. In particular, we focus on the interaction of chromatin modifications with transposable elements (TEs) and the changes in chromatin following polyploidy events in plants with complex genomes.
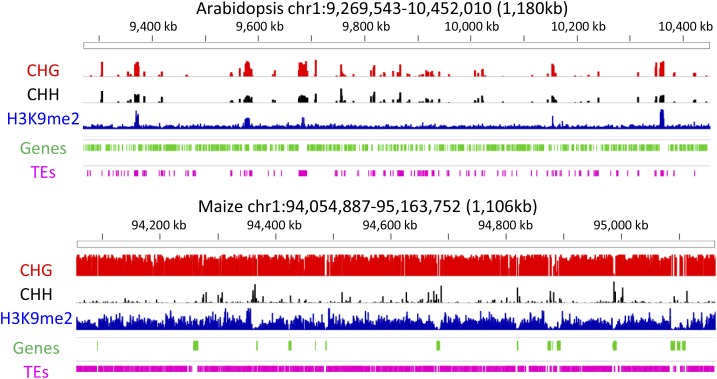
Much of what is known about the role of chromatin in plants is based on studies of the model plant Arabidopsis thaliana, which has a relatively compact genome. However, as we attempt to translate this knowledge to crop species, we are faced with genomes that are much larger and more complex. A visual examination of one megabase from Arabidopsis and maize (Zea mays) reveals striking difference in genome organization and epigenome patterns (Figure 1). Maize has much lower gene density and there is an abundance of transposons surrounding genes (Schnable et al., 2009). The presence of CHG (H = A, C, or T) methylation and H3K9me2 are often associated with TE-associated heterochromatin, and these marks are prevalent in maize. In contrast, these marks are restricted in Arabidopsis (Figure 1). In addition, the profile of CHH methylation, often associated with RNA-directed DNA methylation, is quite different in the two species. Most regions with CHG methylation also contain high CHH methylation in Arabidopsis, but in maize, CHH methylation is only observed at a small number of regions and many of these are located near genes (Gent et al., 2013, 2014). The above example of maize and Arabidopsis simply exhibits differences in two species. A growing set of chromatin profiles in crop species reveals more interesting patterns of heterochromatin and a variety of ways that the epigenome is used to organize complex genome structures (Wang et al., 2009; He et al., 2010; Gent et al., 2013; Makarevitch et al., 2013; Regulski et al., 2013; Schmitz et al., 2013; Zhong et al., 2013; Baker et al., 2015; Kim et al., 2015). Much of the variation among species in genome size and complexity is attributable to two factors: bursts of TEs and/or whole-genome duplications (WGDs). These changes in genome structure create the potential for varying roles of chromatin to organize the information in the genome and allow continued expression of required genes.
Figure 1.
Striking Differences in Genome and Epigenome Organization in Different Plant Species.
The organization of genes (green) and TEs (pink) is shown for portions of the maize and Arabidopsis genomes (annotations from TAIR10 [ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/] and maize RefGen2.0 [ftp://ftp.gramene.org/pub/gramene/maizesequence.org/release-5b/]). The relative abundance of three chromatin modifications, CHG DNA methylation (red), CHH DNA methylation (black), and H3K9me2 methylation (blue), are also shown (primary maize data are from West et al. [2014], Arabidopsis DNA methylation is from Schmitz et al. [2011], and H3K9me2 data are from Stroud et al. [2014]). Whereas Arabidopsis is quite gene rich and only has limited regions with CHG or H3K9me2, maize has fewer genes and the majority of the genome is decorated with CHG methylation and H3K9me2. While CHG and CHH often occur together in Arabidopsis, CHG and H3K9me2 are closely related in maize, and there are limited regions with high CHH in maize, most of which is close to genes.
TRANSPOSON-GENOME INTERACTIONS
Transposon Bursts and Genome Turnover
Even though there is marked conservation of gene order and sequences in related plant species (Devos, 2005), the portion of the genome derived from transposons is often completely distinct. For example, maize and oat (Avena sativa) are related closely enough to allow for introgressions of maize chromosomes following interspecies hybridization (Riera-Lizarazu et al., 1996). However, fluorescence in situ hybridization with maize repetitive elements provides evidence that oat TEs are entirely distinct from maize TEs. Similarly, analyses of Arabidopsis lyrata and Arabidopsis alpina, two close relatives of A. thaliana, demonstrate that TE families, TE abundance, and local TE contents are dramatically different (Willing et al., 2015). This suggests that extant plant genomes are survivors of independent “blooms” of TE activity followed by epigenetic silencing and/or deletion (Lim et al., 2007). Plant genomes are, then, remarkably dynamic structures that manage to maintain their essential functions over hundreds of millions of years. In this Review, we will discuss in more detail the ways in which these contrasting forces may have influenced the evolution of gene function in plants.
Initiating TE Activity
Although there is clear evidence for periodic blooms of TE activity, we actually know very little about what initiates them in natural populations. Horizontal transfer plays an important role in this process in animals (Daniels et al., 1990; Gilbert et al., 2012; Syvanen, 2012) and possibly in plants (Diao et al., 2006; Schaack et al., 2010; El Baidouri et al., 2014). However, it also appears that TE amplification can occur in the absence of horizontal transfer (Piegu et al., 2006; Ungerer et al., 2006; Deininger, 2011; Estep et al., 2013) and horizontal transfer can occur without subsequent TE amplification (Diao et al., 2006). It has also been suggested that wide hybridization and subsequent WGD may play a role in TE amplification (Ungerer et al., 2006; Petit et al., 2010; Madlung and Wendel, 2013; Senerchia et al., 2015). Other potentially important factors include abiotic stresses (Kalendar et al., 2000; Ito et al., 2011; Grandbastien, 2015), viral infection (Wieczorek and Obrępalska-Stęplowska, 2015), and chromosomal breaks (McClintock, 1950). Since tissue culture is also frequently associated with TE activation, natural conditions that mimic that condition might also be expected to be a factor (Hirochika et al., 1996; Huang et al., 2009; Zhang et al., 2014). However, with respect to all of these influences, it should be emphasized that TE blooms can last for hundreds of thousands of years, suggesting that factors contributing to the persistence of those blooms represent chronic rather than acute triggers for activity.
Mutants that compromise the efficiency of silencing pathways can result in global activation of TEs (Lippman et al., 2003; Kato et al., 2004). Interestingly, in at least one case it appears that long-term amplification of TEs is associated with a naturally occurring reduction in the efficiency of symmetric DNA methylation (Willing et al., 2015). Similarly, gymnosperms such as Norway spruce (Picea abies) show evidence for both long-term and gradual increases in TE copy number and a reduced quantity of small RNAs associated with TE silencing (Nystedt et al., 2013).
Selection at the level of individual TEs may also be an important variable (Lisch and Slotkin, 2011). For example, the ONSEN retrotransposon promoter lacks CG and CHG sites, which may limit the ability of the host to stably silence this element via DNA methylation (Cavrak et al., 2014). Similarly, many MITE transposons are exceptionally AT-rich and sometimes prefer to insert into AT-rich regions (Le et al., 2000; Naito et al., 2009). Furthermore, at least some TEs are competent to combat silencing by inactivating systems that have evolved to recognize them (Fu et al., 2013; McCue et al., 2013).
Finally, it is worth considering variation in selection against TE proliferation at the level of the host. TE proliferation represents a balance between selection at the level of the gene and selection at the level of the host (Le Rouzic et al., 2007). Effective population size, mating systems, drift, and cell cycle rates all play a role in the degree to which selection acts (or fails to act) to prevent TE amplification (Whitney et al., 2010).
Arresting TE Activity
The daunting task faced by any host is to recognize and heritably silence TEs while avoiding “off-target” effects on legitimate genes. TEs are remarkably diverse, and it is unlikely that any single pathway can account for all instances of silencing. However, the available evidence suggests that most active TEs are eventually recognized and successfully silenced, and the key variable appears to be the presence of aberrant transcripts. Some active TEs are likely to produce transcripts that are intrinsically distinct from those produced by host genes. This appears to be case for the retrotransposon Evade (Marí-Ordóñez et al., 2013). In this case, although low levels of element transcripts are insufficient to trigger silencing, higher levels of expression associated with high copy number eventually cause RDR6-dependent production of small RNAs that can trigger heritable silencing of the elements. Similar observations have been made for reactivated Athila retroelements, whose resilencing requires RDR6 as well as AGO6 and is associated with 21- to 22-nucleotide small RNAs derived from transcripts produced by the active elements (McCue et al., 2015). Finally, rearranged versions of TEs can also trigger silencing, as is the case for Mu killer, a variant of the MuDR transposon that expresses a hairpin transcript that triggers stable and heritable silencing of one or many MuDR elements (Slotkin et al., 2003; Slotkin et al., 2005; Li et al., 2010). As the copy number of any given element increases, it would seem increasingly likely that that this TE will be recognized and heritably silenced, either due to increases in intrinsically produced triggers (Marí-Ordóñez et al., 2013) or due to the production of a single variant that triggers silencing of all of the active elements (Slotkin et al., 2005). Once silenced, it is hypothesized that these elements are maintained in that state via heritably propagated DNA methylation and histone modifications, as well as tissue-specific reinforcement via trans-acting small RNAs (Slotkin et al., 2009; Creasey et al., 2014; Kim and Zilberman, 2014; Sigman and Slotkin, 2016).
Due to the complexity of the population of TEs in any given genome as well as an ongoing competition between TEs and their hosts, it is quite possible that the initiation of TE silencing in any given plant will be the result of a wide variety of triggers. These could include small quantities of small RNAs produced by elements themselves whose titer reaches some threshold as copy numbers increase (Marí-Ordóñez et al., 2013), preexisting pools of trans-acting small RNAs derived from previously silenced elements (Bousios et al., 2016), novel sense-antisense transcript combinations due to nested insertions (Lisch and Slotkin, 2011), or novel rearrangements such as is observed with Mu killer. There is also evidence for variation in chromatin marks at TEs of differing lengths (Zemach et al., 2013) or differing families (Eichten et al., 2012; Wang et al., 2015).
Living with a New Genome Structure
The proliferation and subsequent silencing of a TE family will result in a reshaped genome. There could be a number of new heterochromatin-euchromatin boundaries potentially changing the chromatin “neighborhood” for many genes and creating the opportunity for TE-associated chromatin to influence nearby genes (Figure 2). Even within species there is evidence for major differences in TE content among distinct haplotypes (Fu and Dooner, 2002; Wang and Dooner, 2006). The complex patchwork of euchromatin and heterochromatin (visualized in Figure 1) must be relatively stable to allow for normal gene function and plant species have likely evolved mechanisms to preserve gene function even when being closely associated with TEs. In this section, we will consider the interactions of genes and TEs both in terms of how chromatin variation at these TEs may influence gene expression and how gene expression may influence the TE chromatin.
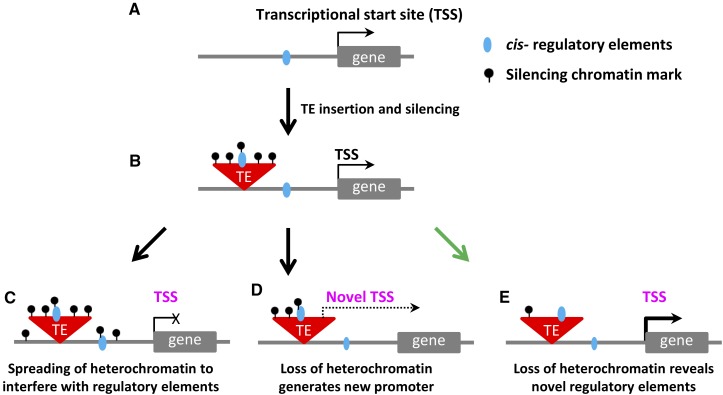
Figure 2.
Potential Interactions between TE Chromatin State and Expression of Nearby Genes.
(A) and (B) A simple model in which a new TE insertion occurs near a gene. The new TE insertion accumulates chromatin modifications associated with silencing.
(C) to (E) Various scenarios of altered TE chromatin and gene expression are illustrated.
(C) The silencing chromatin marks present within the TE could spread to surrounding sequences and alter the accessibility of cis-regulatory elements resulting in reduced gene expression.
(D) Loss of heterochromatic chromatin modifications near the edge of the transposon could result in activation of an outward reading promoter that would influence gene expression.
(E) The loss of heterochromatic modification within the TE could expose cis-regulatory that would then alter expression of a nearby gene. The loss of heterochromatic chromatin modifications in the TE could be a stochastic effect or could be due to spreading of euchromatin from a highly active nearby gene.
Immediately following the proliferation of a TE family, it is expected to generate substantial allelic diversity for specific insertions among different individuals in a population. The specific signals that trigger TE silencing will also likely segregate in the population; therefore, some TE insertions will be exposed to this signal in trans while others will not. This will result in potential epiallelic variation where TEs are silenced in some individuals but remain active in other individuals, which will create the potential for TEs near genes to have varying effects on the expression of nearby genes. Selection upon both genetic variation (TE presence/absence) and epigenetic variation (TE chromatin state) at each site is expected to shape the allele frequency and chromatin state at TEs in subsequent populations. TEs can influence genes through a variety of mechanisms (Lisch, 2013; Figure 2). Oliver et al. (2013) reviewed a number of examples in which TEs influence genes and plant phenotypes. In many cases these may have relatively simple genetic mechanisms of creating loss-of-function alleles or interfering with cis-regulatory elements. However, there are also examples in which the chromatin state at the transposon influences the expression level of the nearby genes. The expression of the FWA gene in Arabidopsis is influenced by the DNA methylation level for a SINE element located around the transcriptional start site (Soppe et al., 2000; Kinoshita et al., 2004). A recent study documented that tissue culture in oil palm (Elaeis guineensis) induces alterations in the DNA methylation level of a retrotransposon inserted within an intron of the DEFICIENS gene, which were associated with proper splicing and termination patterns for this gene (Ong-Abdullah et al., 2015). The BNS locus in Arabidopsis (Saze and Kakutani, 2007), the FAE1 locus in Sinapis alba (Zeng and Cheng, 2014), and the CmWIP1 locus in melon (Cucumis melo; Martin et al., 2009) provide additional examples in which the chromatin state at a TE is linked to expression of a nearby gene. It is likely that the influence of TE chromatin state on nearby genes has been underestimated due to the difficulties in analyzing chromatin state at highly repetitive sequences. The handful of known examples was identified by careful genetic analysis of phenotypic variation followed by locus-specific examination of chromatin. There are additional examples in which TE insertions have been shown to influence gene expression and are heavily methylated (Xiao et al., 2008; Studer et al., 2011; Butelli et al., 2012; Castelletti et al., 2014), but in these examples, it is not clear whether the chromatin state itself is important for the altered phenotype.
One mechanism by which TE chromatin could influence nearby genes would be through the spreading of silencing marks from the TE to flanking low-copy or genic sequences (Figure 2). Genomic analyses of Arabidopsis provided evidence for an evolutionary trade-off between silencing of transposons and maintaining expression for nearby genes (Hollister and Gaut, 2009; X. Wang et al., 2013; Diez et al., 2014). There is evidence for spreading of DNA methylation from transposons to adjacent sequences in Arabidopsis (Ahmed et al., 2011) and maize (Eichten et al., 2012). This effect has not been observed for all transposon families, and the spreading distance seems to have limited range (∼500 bp to 1 kb). The spreading of heterochromatin from TEs to adjacent sequences could interfere with cis-regulatory elements and/or promoters and result in altered expression. The analysis of DNA methylation in several genotypes of maize identified a number of differentially methylated regions that occur at the edge of methylated-unmethylated boundaries, providing evidence for the variable spreading among haplotypes (Li et al., 2015a). An alternative mechanism by which TE chromatin might influence gene expression could be through alterations in the TE chromatin itself. Epiallelic variation for chromatin within a transposon could result in changes in outward reading promoters within the transposon or alter access to cis-regulatory information within the TE itself (Barkan and Martienssen, 1991; Figure 2). There are examples of tissue-specific or stress-responsive expression patterns for alleles containing TE insertions providing evidence for cis-regulatory information (Selinger and Chandler, 2001; Naito et al., 2009; Ito et al., 2011; Butelli et al., 2012; Makarevitch et al., 2015)
While many of the analyses on TE-gene relationships have focused on how TE chromatin influences genes, there is recent evidence suggesting that chromatin plays important roles in mediating the influence of genes on nearby TEs as well. The total portion of methylated DNA seems to be associated with genome size (Mirouze and Vitte, 2014), largely due to the fact that TEs driving genome size increases are highly methylated. Interestingly, the level of CHH methylation does not seem to scale with genome size. For example, maize has lower total levels of CHH methylation than Arabidopsis despite having a genome that is >15-fold larger (West et al., 2014). Gent et al. (2013) noted that regions of high CHH methylation, termed “CHH islands” were often found near highly expressed genes. The majority of the “heterochromatic” portion of the maize genome, defined by H3K9me2 and CHG methylation, contains very little CHH methylation (Figure 1). The CHH methylation region is more often found at the borders between heterochromatin and euchromatin that occur near genes or conserved noncoding sequences (Gent et al., 2014; Li et al., 2015c). The analyses of mutants that affect CHH methylation suggest that the function of these CHH islands is to protect/ensure the silencing of TEs from the open chromatin of nearby genes. Similar findings have been reported for changes in DNA methylation in response to phosphorous stress in rice (Oryza sativa; Secco et al., 2015). A limited number of loci (n = 175) exhibit altered methylation in plants grown in low-phosphorous stress conditions (Secco et al., 2015). Many of these are due to increases in CHH methylation at TEs located near genes that are upregulated under low phosphorous. By assessing the timing of transcriptional and methylation changes under stress and recovery conditions, the authors provided evidence that CHH methylation is targeted to these regions after gene expression has been increased and often is no longer maintained if the gene expression returns to low levels (Secco et al., 2015). Interestingly, similar experiments in Arabidopsis found very few changes in DNA methylation, likely due to the lack of TEs located near genes that undergo transcriptional activation under phosphorous stress. This suggests that DNA methylation and possibly other chromatin modifications play important roles in maintaining TE-gene boundaries in plant with complex genomes and that the primary function of these boundaries may be to keep the gene activity from activating the TE.
CHROMATIN DYNAMICS FOLLOWING WGDs
WGD events are expected to result in significant genetic redundancy. Autopolyploid events will result in full doubling of chromosomes along with their TEs and are expected to provide full redundancy. In contrast, allopolyploid fusions will occur when the full genome complement from two related species is brought together. In these examples it is expected that quite similar complements of genes and regulatory networks will be brought together resulting in high levels of redundancy. Over longer periods of time, much of the redundancy generated by WGD is expected to be resolved through loss of function in one of the copies, subfunctionalization of the duplicates, or neofunctionalization of one gene. Temporal aspects of chromatin dynamics in WGD events worth considering are the perturbation of chromatin and regulatory mechanisms immediately following a WGD event and the role of chromatin in the longer-term loss of function or subfunctionalization of duplicate genes.
Chromatin Perturbation by WGD in Early Generations
Newly formed allopolyploids often exhibit a number of changes relative to the parental lines including genomic sequence changes, gene expression changes, and altered chromatin (reviewed in Renny-Byfield and Wendel, 2014; Song and Chen, 2015). Numerous studies have found evidence for altered DNA methylation patterns in newly synthesized allopolyploids (Lee and Chen, 2001; Shaked et al., 2001; Kashkush et al., 2002; Madlung et al., 2002; Lukens et al., 2006; Gaeta et al., 2007; Parisod et al., 2009; Xu et al., 2009; Yaakov and Kashkush, 2012; Madlung and Wendel, 2013). In some of these studies, the authors focused on assessing the DNA methylation levels for genes with altered expression, and it is difficult to ascertain whether the altered DNA methylation levels are a cause or an effect of the gene expression change. In other cases, studies have focused on changes at transposable elements in newly synthesized polyploids, including changes in DNA methylation, transcription, and transposition (Kashkush et al., 2003; Madlung et al., 2005; Parisod et al., 2009). Studies of transgene expression in diploid and autotetraploid Arabidopsis plants provide evidence for unexpected epigenetic shifts in autopolyploids as well (Mittelsten Scheid et al., 2003; Baubec et al., 2010).
The reactivation of TEs in new polyploids may provide a clue for many of the alterations of gene expression observed in polyploids. A number of studies have found that transposons can be activated in newly formed polyploids (Kashkush et al., 2002, 2003; Madlung et al., 2005). There is evidence for novel or reduced expression of small interfering RNAs (siRNA) in some of these newly formed polyploids as well (Ha et al., 2009; Kenan-Eichler et al., 2011; Ghani et al., 2014; Li et al., 2014; Shen et al., 2014). There are several possible explanations for the presence of novel small RNAs in polyploids (Figure 3). One simple explanation could be that the two parental genomes are expected to have accumulated small RNA pools to allow for recognition and silencing of their unique TE complement. However, when these two genomes are brought together, they may either trigger aberrant novel silencing patterns or they may fail to generate the full small RNA complement. This raises the question, why would the early generations of polyploids not simply contain the full siRNA complement of both parents? Recent findings in Arabidopsis suggest that accessory cells in the male and female gametophytes may play very important roles in generating siRNA pools and reinforcing silencing of TEs (Slotkin et al., 2009; Ibarra et al., 2012; Kawashima and Berger, 2014). Studies also suggest that some small RNAs are maternally inherited/expressed (Brennecke et al., 2008; Mosher et al., 2009), which could potentially contribute to directional genome changes observed in polyploids (Song et al., 1995), and the observation that successful generation of an allopolyploid can only be achieved with one species as maternal parent (Comai et al., 2000). The initial formation of a polyploid will have siRNA pools differing from both parents, and there are some examples in which there are differences in silencing of TEs in newly synthesized polyploids based on the direction of the cross (Parisod et al., 2009). In addition to this potential for parent-of-origin requirement for proper inheritance of siRNAs, there is also evidence that F1 hybrids within plant species have unexpected inheritance of small RNAs (Groszmann et al., 2011; Barber et al., 2012; Li et al., 2012). The hybridization of two species may have even more drastic consequence for the level of expression of small RNAs, which could perturb epigenetic regulation. These observations of perturbations in the small RNA content and in DNA methylation patterns in wide crosses within species have led to the suggestion that hybrid vigor in the F1 and inbreeding depression in subsequent generations could involve alterations to small RNAs and chromatin (He et al., 2010, 2013; Groszmann et al., 2011; Barber et al., 2012; Chodavarapu et al., 2012; Greaves et al., 2012; Shen et al., 2012; Li et al., 2012, 2015b).
Figure 3.
Illustration of Potential for Altered siRNA, TE, and Gene Expression in Newly Formed Allopolyploids.
Two parental species with varying TE content and homologous siRNA populations are diagrammed. These are brought together in the same nucleus in the allopolyploid, and in this case we diagram novel (pink) or reduced/loss of (blue) siRNA levels for specific TEs. This could lead to altered chromatin at the TEs as well as increases or decreases in the expression of adjacent changes.
The alterations in chromatin state at TEs could affect expression of numerous genes (Kashkush et al., 2003). Many of the parental species of polyploids contain complex genome organizations with interspersed TEs and genes. As discussed above, altering the chromatin state at TEs could result in expression changes at nearby genes. In these cases, a large set of gene expression changes observed in new polyploids would be the result of local chromatin changes at nearby TEs, and the ability to protect genic chromatin from spreading of nearby TEs chromatin would stabilize the polyploidy event. An intriguing aspect of this model is that the local chromatin states could show some instability, which could provide abundant opportunities for variation and selection of gene expression states of many genes in newly synthesized polyploids.
Chromatin-Based Long-Term Contributions to Regulatory Diversity of Duplicate Genes
Beyond the early stages of polyploids, chromatin modifications are also expected to play important roles in shaping the eventual genome and transcriptome of stabilized polyploids. Many plant genomes contain evidence of multiple WGD events (Adams and Wendel, 2005; Schmutz et al., 2010) that are supported by colinearity of retained paralogs within a genome, but there is also strong evidence for fractionation (Schnable et al., 2011). Fractionation describes the loss of one member of a duplicate pair such that two subgenomes arising from a WGD event will retain some pairs of duplicate genes but in many other cases there will only be a single gene retained. In some cases, there is evidence of subgenome dominance such that one of the two subgenomes is more fractionated resulting in a higher degree of gene loss (Thomas et al., 2006; Woodhouse et al., 2010; Wang et al., 2011). Ancient WGD events tend to retain relatively few pairs of duplicate genes, while more recent WGD events will often have many examples of retained duplicate pairs. The evolutionary fate of duplicate genes (Prince and Pickett, 2002; Conant and Wolfe, 2008) likely involves gene balance, nonfunctionalization through silencing or fractionation (if they continue to have redundant function), subfunctionalization, or neofunctionalization. Chromatin modifications may play important roles in genome dominance and generation of diversity among retained duplicates in crop genomes.
As researchers have utilized complete genome sequences and high-quality transcriptomes that allow resolution of duplicate gene pairs, it has become clear that many allopolyploids exhibit genome dominance such that one of the two subgenomes plays a greater functional role than the other (Thomas et al., 2006; Wang et al., 2006; Flagel and Wendel, 2010; Woodhouse et al., 2010, 2014; Schnable et al., 2011; Wang et al., 2011; Freeling et al., 2012). This was initially observed through the analysis of the retention of genes in the two subgenomes, but subsequent work has found evidence for preferential transcription and functional impact of genes in one subgenome (Schnable and Freeling, 2011; Schnable et al., 2011). While subgenome dominance has been observed in many plant polyploids, the underlying mechanisms are not understood (Freeling et al., 2012). One suggestion is that chromatin modifications could differentiate the two subgenomes. While there are many examples of genes with differences in DNA methylation or histone modifications between the two subgenomes of maize, the meta-profile of DNA methylation or H3K27me3 for maize genes in subgenomes 1 and 2 does not find global differences that would explain the dominance of subgenome 1 (Eichten et al., 2011; Makarevitch et al., 2013; West et al., 2014). This suggests that while the genic portion of the two subgenomes is affected by genome dominance, it may not be the direct target. Instead, there is growing evidence that differences in TE content and chromatin may provide the differentiation between the subgenomes. A recent analysis of subgenome dominance in Brassica rapa found differences in 24-nucleotide small RNA coverage of transposons located near genes in the two subgenomes (Woodhouse et al., 2014). Woodhouse et al. (2014) suggest that subgenome dominance may arise from differences in TE silencing trade-offs in the two parental genomes. It seems that chromatin modifications and siRNAs may play an important role in genome dominance that is observed in polyploids, but these effects may be mediated by flanking sequences rather than chromatin differences within the coding regions of genes.
Despite the process of fractionation, many crop genomes are replete with examples of retained gene duplicates. In many cases, these duplicates genes could have redundant function and the process of fractionation may still be occurring (Schnable et al., 2011). In other cases, these duplicate pairs may have undergone subfunctionalization or neofunctionalization. These processes could involve alterations to the function of the gene product or changes in the expression pattern (Blanc and Wolfe, 2004). In the context of this Review, we are interested in the potential contributions of chromatin modifications to divergence in expression of the duplicate gene pairs. While it is possible that gene expression patterns had already diverged in the two parents of a polyploid, it is likely that most duplicate gene pairs had similar expression patterns prior to the polyploid fusion event. The analysis of expression levels for retained duplicates in extant crop species finds widespread divergence in expression of the homoeologs (Pont et al., 2011; Schnable et al., 2011; Roulin et al., 2013; Schmitz et al., 2013; Renny-Byfield et al., 2014; Woodhouse et al., 2014). A recent investigation found that 98% of retained duplicate pairs in maize have subfunctionalized tissue-specific expression patterns or have diverged consistently in expression level (Pophaly and Tellier, 2015). Soybean (Glycine max) provides a good system for studying the divergence of expression following a recent (∼13 million years ago) WGD event (Lin et al., 2010; Schmutz et al., 2010; Roulin et al., 2013). The analysis of gene expression in seven tissues suggests that approximately half of the retained duplicates from the recent WGD event exhibit divergent expression patterns while the others have similar expression (Roulin et al., 2013). The factors driving the divergence of expression for the retained duplicates are largely unknown. Analysis of the soybean methylome reveals that many duplicates have similar levels of DNA methylation, but ∼6% of the pairs have divergent non-CG methylation levels that are associated with altered expression (Schmitz et al., 2013). Many of the methylation differences in the duplicate soybean genes can be attributed to transposon insertions that are present in one member of the pair (Schmitz et al., 2013; Kim et al., 2015). These studies along with data from specific wheat (Triticum aestivum) loci (Shitsukawa et al., 2007; Hu et al., 2013; Zhang et al., 2015) and maize (West et al., 2014) suggest that DNA methylation does not play a major global role in differentiating subgenomes, but it can contribute to locus-specific divergence of retained duplicates.
While the majority of this section has focused on the potential role for chromatin and small RNAs in gene regulation following WGD events, there is also a likely role for chromatin in the regulation of smaller duplication events. Gene duplication can occur via unequal crossing-over or transposition and has been classified into different types based on duplication mode (Wang et al., 2012). It was further shown that duplicated genes arising from different modes have distinct patterns in gene body DNA methylation (Y. Wang et al., 2013). Gene copies that are located in nonsyntenic genomic positions relative to other species often have significantly elevated levels of DNA methylation in maize (Eichten et al., 2011; West et al., 2014). It is not known whether this is due to many of these “genes” being transposons or due to the plant host recognizing these insertions as transposons and targeting silencing chromatin modifications. Tandemly duplicated genes arising from unequal crossover can also exhibit substantial epigenetic diversity. The large family of F-box proteins in Arabidopsis provides evidence for diversity of chromatin states within complex loci containing these genes (Hua et al., 2013).
CONCLUSION
Chromatin modifications play important roles in many plant processes. While chromatin states are a critical part of transcriptional responses during development (reviewed in He et al., 2011; Holec and Berger, 2012; Grimanelli and Roudier, 2013; Pikaard and Mittelsten Scheid, 2014; Xiao and Wagner, 2015) or in response to the environment (reviewed in Baulcombe and Dean, 2014; Bond and Baulcombe, 2014; Pikaard and Mittelsten Scheid, 2014; Probst and Mittelsten Scheid, 2015; Vriet et al., 2015), they also play critical roles in creating order in the genome, allowing for the maintenance of proper transcription and genome stability in the face of varying TE landscapes and polyploid changes. We suggest that diverse plant species have evolved variations in the specific mechanisms of chromatin regulation that have allowed them to survive the repeated transposon bursts or polyploidy fusions. Understanding the diversity of chromatin regulation details in plants with complex genomes will be critical to understanding how to engineer traits in crop plants. We also suggest that chromatin-based regulation will play critical roles in creating diversity within many crop species. Transcriptional variation of different alleles due to the chromatin modifications of nearby TEs is likely to result in allelic or epiallelic diversity. The perturbation of chromatin following polyploidy fusion events may also create ample opportunity for selection to act upon chromatin and identify optimal new epialleles. Increasing our understanding of the sources of chromatin diversity is likely to improve our ability to select or engineer ideal crop varieties and to stabilize the performance in these lines.
Acknowledgments
We thank Robert J. Schmitz, Steven R. Eichten, and Cory D. Hirsch for providing important comments and feedback on this Review. This work was supported by the National Science Foundation (Grant DBI-1237931 to N.M.S. and D.L.).
AUTHOR CONTRIBUTIONS
All authors contributed to writing the article.
References
- Adams K.L., Wendel J.F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141. [DOI] [PubMed] [Google Scholar]
- Ahmed I., Sarazin A., Bowler C., Colot V., Quesneville H. (2011). Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res. 39: 6919–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K., Dhillon T., Colas I., Cook N., Milne I., Milne L., Bayer M., Flavell A.J. (2015). Chromatin state analysis of the barley epigenome reveals a higher-order structure defined by H3K27me1 and H3K27me3 abundance. Plant J. 84: 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber W.T., Zhang W., Win H., Varala K.K., Dorweiler J.E., Hudson M.E., Moose S.P. (2012). Repeat associated small RNAs vary among parents and following hybridization in maize. Proc. Natl. Acad. Sci. USA 109: 10444–10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Martienssen R.A. (1991). Inactivation of maize transposon Mu suppresses a mutant phenotype by activating an outward-reading promoter near the end of Mu1. Proc. Natl. Acad. Sci. USA 88: 3502–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T., Dinh H.Q., Pecinka A., Rakic B., Rozhon W., Wohlrab B., von Haeseler A., Mittelsten Scheid O. (2010). Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic states in Arabidopsis. Plant Cell 22: 34–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D.C., Dean C. (2014). Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 6: a019471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond D.M., Baulcombe D.C. (2014). Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol. 24: 100–107. [DOI] [PubMed] [Google Scholar]
- Bousios A., Diez C.M., Takuno S., Bystry V., Darzentas N., Gaut B.S. (2016). A role for palindromic structures in the cis-region of maize Sirevirus LTRs in transposable element evolution and host epigenetic response. Genome Res. 26: 226–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Malone C.D., Aravin A.A., Sachidanandam R., Stark A., Hannon G.J. (2008). An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E., Licciardello C., Zhang Y., Liu J., Mackay S., Bailey P., Reforgiato-Recupero G., Martin C. (2012). Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24: 1242–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelletti S., Tuberosa R., Pindo M., Salvi S. (2014). A MITE transposon insertion is associated with differential methylation at the maize flowering time QTL Vgt1. G3 (Bethesda) 4: 805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavrak V.V., Lettner N., Jamge S., Kosarewicz A., Bayer L.M., Mittelsten Scheid O. (2014). How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet. 10: e1004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu R.K., Feng S., Ding B., Simon S.A., Lopez D., Jia Y., Wang G.L., Meyers B.C., Jacobsen S.E., Pellegrini M. (2012). Transcriptome and methylome interactions in rice hybrids. Proc. Natl. Acad. Sci. USA 109: 12040–12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Tyagi A.P., Winter K., Holmes-Davis R., Reynolds S.H., Stevens Y., Byers B. (2000). Phenotypic instability and rapid gene silencing in newly formed arabidopsis allotetraploids. Plant Cell 12: 1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant G.C., Wolfe K.H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9: 938–950. [DOI] [PubMed] [Google Scholar]
- Creasey K.M., Zhai J., Borges F., Van Ex F., Regulski M., Meyers B.C., Martienssen R.A. (2014). miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels S.B., Peterson K.R., Strausbaugh L.D., Kidwell M.G., Chovnick A. (1990). Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 124: 339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. (2011). Alu elements: know the SINEs. Genome Biol. 12: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos K.M. (2005). Updating the ‘crop circle’. Curr. Opin. Plant Biol. 8: 155–162. [DOI] [PubMed] [Google Scholar]
- Diao X., Freeling M., Lisch D. (2006). Horizontal transfer of a plant transposon. PLoS Biol. 4: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez C.M., Roessler K., Gaut B.S. (2014). Epigenetics and plant genome evolution. Curr. Opin. Plant Biol. 18: 1–8. [DOI] [PubMed] [Google Scholar]
- Eichten S.R., et al. (2011). Heritable epigenetic variation among maize inbreds. PLoS Genet. 7: e1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S.R., Ellis N.A., Makarevitch I., Yeh C.T., Gent J.I., Guo L., McGinnis K.M., Zhang X., Schnable P.S., Vaughn M.W., Dawe R.K., Springer N.M. (2012). Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet. 8: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baidouri M., Carpentier M.C., Cooke R., Gao D., Lasserre E., Llauro C., Mirouze M., Picault N., Jackson S.A., Panaud O. (2014). Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res. 24: 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estep M.C., DeBarry J.D., Bennetzen J.L. (2013). The dynamics of LTR retrotransposon accumulation across 25 million years of panicoid grass evolution. Heredity (Edinb) 110: 194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L.E., Wendel J.F. (2010). Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 186: 184–193. [DOI] [PubMed] [Google Scholar]
- Freeling M., Woodhouse M.R., Subramaniam S., Turco G., Lisch D., Schnable J.C. (2012). Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr. Opin. Plant Biol. 15: 131–139. [DOI] [PubMed] [Google Scholar]
- Fu H., Dooner H.K. (2002). Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99: 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Kawabe A., Etcheverry M., Ito T., Toyoda A., Fujiyama A., Colot V., Tarutani Y., Kakutani T. (2013). Mobilization of a plant transposon by expression of the transposon-encoded anti-silencing factor. EMBO J. 32: 2407–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta R.T., Pires J.C., Iniguez-Luy F., Leon E., Osborn T.C. (2007). Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Ellis N.A., Guo L., Harkess A.E., Yao Y., Zhang X., Dawe R.K. (2013). CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23: 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Madzima T.F., Bader R., Kent M.R., Zhang X., Stam M., McGinnis K.M., Dawe R.K. (2014). Accessible DNA and relative depletion of H3K9me2 at maize loci undergoing RNA-directed DNA methylation. Plant Cell 26: 4903–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghani M.A., Li J., Rao L., Raza M.A., Cao L., Yu N., Zou X., Chen L. (2014). The role of small RNAs in wide hybridisation and allopolyploidisation between Brassica rapa and Brassica nigra BMC Plant Biol. 14: 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C., Hernandez S.S., Flores-Benabib J., Smith E.N., Feschotte C. (2012). Rampant horizontal transfer of SPIN transposons in squamate reptiles. Mol. Biol. Evol. 29: 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien M. (2015). LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochim. Biophys. Acta 1849: 403–416. [DOI] [PubMed] [Google Scholar]
- Greaves I.K., Groszmann M., Ying H., Taylor J.M., Peacock W.J., Dennis E.S. (2012). Trans chromosomal methylation in Arabidopsis hybrids. Proc. Natl. Acad. Sci. USA 109: 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimanelli D., Roudier F. (2013). Epigenetics and development in plants: green light to convergent innovations. Curr. Top. Dev. Biol. 104: 189–222. [DOI] [PubMed] [Google Scholar]
- Groszmann M., Greaves I.K., Albertyn Z.I., Scofield G.N., Peacock W.J., Dennis E.S. (2011). Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc. Natl. Acad. Sci. USA 108: 2617–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., Lu J., Tian L., Ramachandran V., Kasschau K.D., Chapman E.J., Carrington J.C., Chen X., Wang X.J., Chen Z.J. (2009). Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. USA 106: 17835–17840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., Chen B., Wang X., Li X., Li J., He H., Yang M., Lu L., Qi Y., Wang X., Deng X.W. (2013). Conservation and divergence of transcriptomic and epigenomic variation in maize hybrids. Genome Biol. 14: R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., Elling A.A., Deng X.W. (2011). The epigenome and plant development. Annu. Rev. Plant Biol. 62: 411–435. [DOI] [PubMed] [Google Scholar]
- He G., et al. (2010). Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H., Sugimoto K., Otsuki Y., Tsugawa H., Kanda M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93: 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holec S., Berger F. (2012). Polycomb group complexes mediate developmental transitions in plants. Plant Physiol. 158: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister J.D., Gaut B.S. (2009). Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 19: 1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Han Z., Song N., Chai L., Yao Y., Peng H., Ni Z., Sun Q. (2013). Epigenetic modification contributes to the expression divergence of three TaEXPA1 homoeologs in hexaploid wheat (Triticum aestivum). New Phytol. 197: 1344–1352. [DOI] [PubMed] [Google Scholar]
- Hua Z., Pool J.E., Schmitz R.J., Schultz M.D., Shiu S.H., Ecker J.R., Vierstra R.D. (2013). Epigenomic programming contributes to the genomic drift evolution of the F-box protein superfamily in Arabidopsis. Proc. Natl. Acad. Sci. USA 110: 16927–16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhang K., Shen Y., Huang Z., Li M., Tang D., Gu M., Cheng Z. (2009). Identification of a high frequency transposon induced by tissue culture, nDaiZ, a member of the hAT family in rice. Genomics 93: 274–281. [DOI] [PubMed] [Google Scholar]
- Ibarra C.A., et al. (2012). Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Gaubert H., Bucher E., Mirouze M., Vaillant I., Paszkowski J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472: 115–119. [DOI] [PubMed] [Google Scholar]
- Kalendar R., Tanskanen J., Immonen S., Nevo E., Schulman A.H. (2000). Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc. Natl. Acad. Sci. USA 97: 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K., Feldman M., Levy A.A. (2002). Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K., Feldman M., Levy A.A. (2003). Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33: 102–106. [DOI] [PubMed] [Google Scholar]
- Kato M., Takashima K., Kakutani T. (2004). Epigenetic control of CACTA transposon mobility in Arabidopsis thaliana. Genetics 168: 961–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T., Berger F. (2014). Epigenetic reprogramming in plant sexual reproduction. Nat. Rev. Genet. 15: 613–624. [DOI] [PubMed] [Google Scholar]
- Kenan-Eichler M., Leshkowitz D., Tal L., Noor E., Melamed-Bessudo C., Feldman M., Levy A.A. (2011). Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188: 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.D., El Baidouri M., Abernathy B., Iwata-Otsubo A., Chavarro C., Gonzales M., Libault M., Grimwood J., Jackson S.A. (2015). A comparative epigenomic analysis of polyploidy-derived genes in soybean and common bean. Plant Physiol. 168: 1433–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.Y., Zilberman D. (2014). DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 19: 320–326. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Miura A., Choi Y., Kinoshita Y., Cao X., Jacobsen S.E., Fischer R.L., Kakutani T. (2004). One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523. [DOI] [PubMed] [Google Scholar]
- Le Q.H., Wright S., Yu Z., Bureau T. (2000). Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97: 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.S., Chen Z.J. (2001). Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc. Natl. Acad. Sci. USA 98: 6753–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic A., Boutin T.S., Capy P. (2007). Long-term evolution of transposable elements. Proc. Natl. Acad. Sci. USA 104: 19375–19380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., et al. (2014). mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26: 1878–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Freeling M., Lisch D. (2010). Epigenetic reprogramming during vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 107: 22184–22189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., et al. (2015c). RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc. Natl. Acad. Sci. USA 112: 14728–14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Li Y., Moose S.P., Hudson M.E. (2015b). Transposable elements, mRNA expression level and strand-specificity of small RNAs are associated with non-additive inheritance of gene expression in hybrid plants. BMC Plant Biol. 15: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Song J., West P.T., Zynda G., Eichten S.R., Vaughn M.W., Springer N.M. (2015a). Examining the causes and consequences of context-specific differential DNA methylation in maize. Plant Physiol. 168: 1262–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Varala K., Moose S.P., Hudson M.E. (2012). The inheritance pattern of 24 nt siRNA clusters in arabidopsis hybrids is influenced by proximity to transposable elements. PLoS One 7: e47043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.Y., Kovarik A., Matyasek R., Chase M.W., Clarkson J.J., Grandbastien M.A., Leitch A.R. (2007). Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol. 175: 756–763. [DOI] [PubMed] [Google Scholar]
- Lin J.Y., Stupar R.M., Hans C., Hyten D.L., Jackson S.A. (2010). Structural and functional divergence of a 1-Mb duplicated region in the soybean (Glycine max) genome and comparison to an orthologous region from Phaseolus vulgaris. Plant Cell 22: 2545–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., May B., Yordan C., Singer T., Martienssen R. (2003). Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1: E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. (2013). How important are transposons for plant evolution? Nat. Rev. Genet. 14: 49–61. [DOI] [PubMed] [Google Scholar]
- Lisch D., Slotkin R.K. (2011). Strategies for silencing and escape: the ancient struggle between transposable elements and their hosts. Int. Rev. Cell Mol. Biol. 292: 119–152. [DOI] [PubMed] [Google Scholar]
- Lukens L.N., Pires J.C., Leon E., Vogelzang R., Oslach L., Osborn T. (2006). Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 140: 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A., Masuelli R.W., Watson B., Reynolds S.H., Davison J., Comai L. (2002). Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129: 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A., Tyagi A.P., Watson B., Jiang H., Kagochi T., Doerge R.W., Martienssen R., Comai L. (2005). Genomic changes in synthetic Arabidopsis polyploids. Plant J. 41: 221–230. [DOI] [PubMed] [Google Scholar]
- Madlung A., Wendel J.F. (2013). Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet. Genome Res. 140: 270–285. [DOI] [PubMed] [Google Scholar]
- Makarevitch I., Eichten S.R., Briskine R., Waters A.J., Danilevskaya O.N., Meeley R.B., Myers C.L., Vaughn M.W., Springer N.M. (2013). Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27. Plant Cell 25: 780–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I., Waters A.J., West P.T., Stitzer M., Hirsch C.N., Ross-Ibarra J., Springer N.M. (2015). Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 11: e1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí-Ordóñez A., Marchais A., Etcheverry M., Martin A., Colot V., Voinnet O. (2013). Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 45: 1029–1039. [DOI] [PubMed] [Google Scholar]
- Martin A., Troadec C., Boualem A., Rajab M., Fernandez R., Morin H., Pitrat M., Dogimont C., Bendahmane A. (2009). A transposon-induced epigenetic change leads to sex determination in melon. Nature 461: 1135–1138. [DOI] [PubMed] [Google Scholar]
- McClintock B. (1950). The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 36: 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue A.D., Nuthikattu S., Slotkin R.K. (2013). Genome-wide identification of genes regulated in trans by transposable element small interfering RNAs. RNA Biol. 10: 1379–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue A.D., Panda K., Nuthikattu S., Choudury S.G., Thomas E.N., Slotkin R.K. (2015). ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 34: 20–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M., Vitte C. (2014). Transposable elements, a treasure trove to decipher epigenetic variation: insights from Arabidopsis and crop epigenomes. J. Exp. Bot. 65: 2801–2812. [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid O., Afsar K., Paszkowski J. (2003). Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat. Genet. 34: 450–454. [DOI] [PubMed] [Google Scholar]
- Mosher R.A., Melnyk C.W., Kelly K.A., Dunn R.M., Studholme D.J., Baulcombe D.C. (2009). Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286. [DOI] [PubMed] [Google Scholar]
- Naito K., Zhang F., Tsukiyama T., Saito H., Hancock C.N., Richardson A.O., Okumoto Y., Tanisaka T., Wessler S.R. (2009). Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130–1134. [DOI] [PubMed] [Google Scholar]
- Nystedt B., et al. (2013). The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584. [DOI] [PubMed] [Google Scholar]
- Oliver K.R., McComb J.A., Greene W.K. (2013). Transposable elements: powerful contributors to angiosperm evolution and diversity. Genome Biol. Evol. 5: 1886–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong-Abdullah M., et al. (2015). Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 525: 533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisod C., Salmon A., Zerjal T., Tenaillon M., Grandbastien M.A., Ainouche M. (2009). Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol. 184: 1003–1015. [DOI] [PubMed] [Google Scholar]
- Petit M., Guidat C., Daniel J., Denis E., Montoriol E., Bui Q.T., Lim K.Y., Kovarik A., Leitch A.R., Grandbastien M.A., Mhiri C. (2010). Mobilization of retrotransposons in synthetic allotetraploid tobacco. New Phytol. 186: 135–147. [DOI] [PubMed] [Google Scholar]
- Piegu B., Guyot R., Picault N., Roulin A., Sanyal A., Kim H., Collura K., Brar D.S., Jackson S., Wing R.A., Panaud O. (2006). Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 16: 1262–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard C.S., Mittelsten Scheid O. (2014). Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 6: a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pont C., Murat F., Confolent C., Balzergue S., Salse J. (2011). RNA-seq in grain unveils fate of neo- and paleopolyploidization events in bread wheat (Triticum aestivum L.). Genome Biol. 12: R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pophaly S.D., Tellier A. (2015). Population level purifying selection and gene expression shape subgenome evolution in maize. Mol. Biol. Evol. 32: 3226–3235. [DOI] [PubMed] [Google Scholar]
- Prince V.E., Pickett F.B. (2002). Splitting pairs: the diverging fates of duplicated genes. Nat. Rev. Genet. 3: 827–837. [DOI] [PubMed] [Google Scholar]
- Probst A.V., Mittelsten Scheid O. (2015). Stress-induced structural changes in plant chromatin. Curr. Opin. Plant Biol. 27: 8–16. [DOI] [PubMed] [Google Scholar]
- Regulski M., et al. (2013). The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renny-Byfield S., Gallagher J.P., Grover C.E., Szadkowski E., Page J.T., Udall J.A., Wang X., Paterson A.H., Wendel J.F. (2014). Ancient gene duplicates in Gossypium (cotton) exhibit near-complete expression divergence. Genome Biol. Evol. 6: 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renny-Byfield S., Wendel J.F. (2014). Doubling down on genomes: polyploidy and crop plants. Am. J. Bot. 101: 1711–1725. [DOI] [PubMed] [Google Scholar]
- Riera-Lizarazu O., Rines H.W., Phillips R.L. (1996). Cytological and molecular characterization of oat x maize partial hybrids. Theor. Appl. Genet. 93: 123–135. [DOI] [PubMed] [Google Scholar]
- Roulin A., Auer P.L., Libault M., Schlueter J., Farmer A., May G., Stacey G., Doerge R.W., Jackson S.A. (2013). The fate of duplicated genes in a polyploid plant genome. Plant J. 73: 143–153. [DOI] [PubMed] [Google Scholar]
- Saze H., Kakutani T. (2007). Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 26: 3641–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack S., Gilbert C., Feschotte C. (2010). Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol. Evol. (Amst.) 25: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R.J., He Y., Valdés-López O., Khan S.M., Joshi T., Urich M.A., Nery J.R., Diers B., Xu D., Stacey G., Ecker J.R. (2013). Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 23: 1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R.J., Schultz M.D., Lewsey M.G., O’Malley R.C., Urich M.A., Libiger O., Schork N.J., Ecker J.R. (2011). Transgenerational epigenetic instability is a source of novel methylation variants. Science 334: 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Schnable J.C., Freeling M. (2011). Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS One 6: e17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J.C., Springer N.M., Freeling M. (2011). Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Secco D., Wang C., Shou H., Schultz M.D., Chiarenza S., Nussaume L., Ecker J.R., Whelan J., Lister R. (2015). Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 4: e09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger D.A., Chandler V.L. (2001). B-Bolivia, an allele of the maize b1 gene with variable expression, contains a high copy retrotransposon-related sequence immediately upstream. Plant Physiol. 125: 1363–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senerchia N., Felber F., Parisod C. (2015). Genome reorganization in F1 hybrids uncovers the role of retrotransposons in reproductive isolation. Proc. Biol. Sci. 282: 20142874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H., Kashkush K., Ozkan H., Feldman M., Levy A.A. (2001). Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., He H., Li J., Chen W., Wang X., Guo L., Peng Z., He G., Zhong S., Qi Y., Terzaghi W., Deng X.W. (2012). Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 24: 875–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Zhao Q., Zou J., Wang W., Gao Y., Meng J., Wang J. (2014). Characterization and expression patterns of small RNAs in synthesized Brassica hexaploids. Plant Mol. Biol. 85: 287–299. [DOI] [PubMed] [Google Scholar]
- Shitsukawa N., Tahira C., Kassai K., Hirabayashi C., Shimizu T., Takumi S., Mochida K., Kawaura K., Ogihara Y., Murai K. (2007). Genetic and epigenetic alteration among three homoeologous genes of a class E MADS box gene in hexaploid wheat. Plant Cell 19: 1723–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigman M.J., Slotkin R.K. (2016). The first rule of plant transposable element silencing: location, location, location. Plant Cell 28: 304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Freeling M., Lisch D. (2005). Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat. Genet. 37: 641–644. [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Freeling M., Lisch D. (2003). Mu killer causes the heritable inactivation of the Mutator family of transposable elements in Zea mays. Genetics 165: 781–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Lu P., Tang K., Osborn T.C. (1995). Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92: 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q., Chen Z.J. (2015). Epigenetic and developmental regulation in plant polyploids. Curr. Opin. Plant Biol. 24: 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe W.J., Jacobsen S.E., Alonso-Blanco C., Jackson J.P., Kakutani T., Koornneef M., Peeters A.J. (2000). The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell 6: 791–802. [DOI] [PubMed] [Google Scholar]
- Stroud H., Do T., Du J., Zhong X., Feng S., Johnson L., Patel D.J., Jacobsen S.E. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A., Zhao Q., Ross-Ibarra J., Doebley J. (2011). Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43: 1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syvanen M. (2012). Evolutionary implications of horizontal gene transfer. Annu. Rev. Genet. 46: 341–358. [DOI] [PubMed] [Google Scholar]
- Thomas B.C., Pedersen B., Freeling M. (2006). Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16: 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer M.C., Strakosh S.C., Zhen Y. (2006). Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr. Biol. 16: R872–R873. [DOI] [PubMed] [Google Scholar]
- Vriet C., Hennig L., Laloi C. (2015). Stress-induced chromatin changes in plants: of memories, metabolites and crop improvement. Cell. Mol. Life Sci. 72: 1261–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Beyene G., Zhai J., Feng S., Fahlgren N., Taylor N.J., Bart R., Carrington J.C., Jacobsen S.E., Ausin I. (2015). CG gene body DNA methylation changes and evolution of duplicated genes in cassava. Proc. Natl. Acad. Sci. USA 112: 13729–13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H.S., Wei N.E., Jiang H., Watson B., Madlung A., Osborn T.C., Doerge R.W., Comai L., Chen Z.J. (2006). Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Dooner H.K. (2006). Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc. Natl. Acad. Sci. USA 103: 17644–17649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Elling A.A., Li X., Li N., Peng Z., He G., Sun H., Qi Y., Liu X.S., Deng X.W. (2009). Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Weigel D., Smith L.M. (2013). Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet. 9: e1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. ; Brassica rapa Genome Sequencing Project Consortium (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang X., Lee T.H., Mansoor S., Paterson A.H. (2013). Gene body methylation shows distinct patterns associated with different gene origins and duplication modes and has a heterogeneous relationship with gene expression in Oryza sativa (rice). New Phytol. 198: 274–283. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang X., Paterson A.H. (2012). Genome and gene duplications and gene expression divergence: a view from plants. Ann. N. Y. Acad. Sci. 1256: 1–14. [DOI] [PubMed] [Google Scholar]
- West P.T., Li Q., Ji L., Eichten S.R., Song J., Vaughn M.W., Schmitz R.J., Springer N.M. (2014). Genomic distribution of H3K9me2 and DNA methylation in a maize genome. PLoS One 9: e105267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney K.D., Baack E.J., Hamrick J.L., Godt M.J.W., Barringer B.C., Bennett M.D., Eckert C.G., Goodwillie C., Kalisz S., Leitch I.J., Ross-Ibarra J. (2010). A role for nonadaptive processes in plant genome size evolution? Evolution 64: 2097–2109. [DOI] [PubMed] [Google Scholar]
- Wieczorek P., Obrępalska-Stęplowska A. (2015). Suppress to survive—implication of plant viruses in PTGS. Plant Mol. Biol. Rep. 33: 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing E., Rawat V., Mandáková T., Maumus F., James G.V., Nordström K.J., Becker C., Warthmann N., Chica C., Szarzynska B. (2015). Genome expansion of arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat. Plants 1: 14023. [DOI] [PubMed] [Google Scholar]
- Woodhouse M.R., Cheng F., Pires J.C., Lisch D., Freeling M., Wang X. (2014). Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc. Natl. Acad. Sci. USA 111: 5283–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse M.R., Schnable J.C., Pedersen B.S., Lyons E., Lisch D., Subramaniam S., Freeling M. (2010). Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS Biol. 8: e1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Jiang N., Schaffner E., Stockinger E.J., van der Knaap E. (2008). A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319: 1527–1530. [DOI] [PubMed] [Google Scholar]
- Xiao J., Wagner D. (2015). Polycomb repression in the regulation of growth and development in Arabidopsis. Curr. Opin. Plant Biol. 23: 15–24. [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhong L., Wu X., Fang X., Wang J. (2009). Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta 229: 471–483. [DOI] [PubMed] [Google Scholar]
- Yaakov B., Kashkush K. (2012). Mobilization of Stowaway-like MITEs in newly formed allohexaploid wheat species. Plant Mol. Biol. 80: 419–427. [DOI] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Hsieh P.H., Coleman-Derr D., Eshed-Williams L., Thao K., Harmer S.L., Zilberman D. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F., Cheng B. (2014). Transposable element insertion and epigenetic modification cause the multiallelic variation in the expression of FAE1 in Sinapis alba. Plant Cell 26: 2648–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Wang B., Zhao J., Zhao X., Zhang L., Liu D., Dong L., Wang D., Mao L., Li A. (2015). Divergence in homoeolog expression of the grain length-associated gene GASR7 during wheat allohexaploidization. The Crop Journal 3: 1–9. [Google Scholar]
- Zhang D., et al. (2014). Tissue culture-induced heritable genomic variation in rice, and their phenotypic implications. PLoS One 9: e96879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Fei Z., Chen Y.R., Zheng Y., Huang M., Vrebalov J., McQuinn R., Gapper N., Liu B., Xiang J., Shao Y., Giovannoni J.J. (2013). Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31: 154–159. [DOI] [PubMed] [Google Scholar]