Abstract
Nitric oxide (NO) may affect the genomes of various pathogens, and this mutagenesis is of particular interest for viral pathogenesis and evolution. Here, we investigated the effect of NO on viral replication and mutation. Exogenous or endogenous NO had no apparent antiviral effect on influenza A virus and Sendai virus. The mutagenic potential of NO was analyzed with Sendai virus fused to a green fluorescent protein (GFP) gene (GFP-SeV). GFP-SeV was cultured in SW480 cells transfected with a vector expressing inducible NO synthase (iNOS). The mutation frequency of GFP-SeV was examined by measuring loss of GFP fluorescence of the viral plaques. GFP-SeV mutation frequency in iNOS-SW480 cells was much higher than that in parent SW480 cells and was reduced to the level of mutation frequency in the parent cells by treatment with an NO synthase (NOS) inhibitor. Immunocytochemistry showed generation of more 8-nitroguanosine in iNOS-SW480 cells than in SW480 cells without iNOS transfection. Authentic 8-nitroguanosine added exogenously to GFP-SeV-infected CV-1 cells increased the viral mutation frequency. Profiles of the GFP gene mutations induced by 8-nitroguanosine appeared to resemble those of mutations occurring in mouse lungs in vivo. A base substitution that was characteristic of both mutants (those induced by 8-nitroguanosine and those occurring in vivo) was a C-to-U transition. NO-dependent oxidative stress in iNOS-SW480 cells was also evident. Together, the results indicate unambiguously that NO has mutagenic potential for RNA viruses such as Sendai virus without affecting viral replication, possibly via 8-nitroguanosine formation and cellular oxidative stress.
Nitric oxide (NO) is an endogenous inorganic radical exhibiting a diverse array of biological functions (32). It has been implicated in various conditions, including inflammation, neurodegenerative disease, cardiovascular disease, and neoplasia. Production of NO by the inducible isoform of NO synthase (iNOS) as a host defense has been observed in most types of infections caused by pathogens, such as bacteria, protozoa, fungi, and viruses (2, 27, 34, 37, 38, 52). This NO production plays a critical role in microbial clearance, especially for bacteria (9, 18, 19, 58). However, the contribution of NO to antiviral defense and to viral pathogenesis varies among species of virus. The role of NO in viral infection is thus not fully understood.
We have been working for some time on oxygen radical- and NO-induced viral pathogenesis with several animal models of infections caused by neurotropic and pneumotropic viruses such as herpes simplex virus and influenza and Sendai viruses (2, 3, 7, 40). NO and its reactive derivatives peroxynitrite (ONOO−) and nitrogen dioxide (NO2) have cytotoxic and proinflammatory effects (14, 24, 43, 45, 46); in addition, our recent work has indicated that NO has a unique biological effect on the genome of both pathogen and host via chemical modification of nucleic acids (4, 8). This effect was most evident in our earlier study with a recombinant Sendai virus (SeV) constructed with a green fluorescent protein (GFP) gene, GFP-SeV, the gene providing an indication of endogenous mutagenesis of the viral genome. We thus determined that NO could be a potent mutagen for the RNA virus, such that the mutation frequency of GFP-SeV was significantly elevated as a result of high NO production by iNOS in vivo (4). More important, our recent work illustrated a unique nucleic acid modification, i.e., formation of 8-nitroguanosine, brought about in vivo by NO generated from iNOS, as evidenced by intense immunostaining in airway epithelial cells in virus-infected tissue (8). In addition, we found that 8-nitroguanosine has a potent redox-active property involving superoxide anion radical (O2·−) generation catalyzed by NADPH-cytochrome P450 reductase (P450 reductase) and various isoforms of NO synthases (NOSs) (8, 47). These results thus indicate that NO may affect genomic structure and function via chemical modification and may cause mutagenesis of various pathogens and host cells as well.
In the present study, we sought to clarify the role of NO in viral pathogenesis, with a focus on the mutagenic potential of NO through 8-nitroguanosine formation. Viral mutation was examined with the use of GFP-SeV propagated in cells cultured with or without NO. The mutation was quantified on the basis of phenotypic alteration (loss of GFP fluorescence), as reported previously (4). To further explore the molecular mechanisms of NO-induced mutagenesis, the formation and mutagenic potential of 8-nitroguanosine were examined with an antibody specific for 8-nitroguanosine. We thus clearly demonstrated that NO has potent mutagenic activity, without apparent antiviral effect, possibly via formation of 8-nitroguanosine.
MATERIALS AND METHODS
Viruses and virus assay.
Influenza virus A/Kumamoto/Y5/67(H2N2) and SeV strain Z were used to examine the effect of NO on viral replication in Madin-Darby canine kidney (MDCK) cells in culture. Virus yield for each cell culture was quantified by means of the plaque-forming assay; MDCK cells were used for influenza virus, and CV-1 cells were used for SeV cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen Corp., Carlsbad, Calif.) plus 0.2% bovine serum albumin (BSA; Sigma-Aldrich Fine Chemicals, St. Louis, Mo.), as described previously (6). GFP-SeV prepared as described earlier (4) was used for the viral mutation assay.
Treatment of cells with NO donors.
MDCK cell monolayers were inoculated with either influenza virus or SeV at a multiplicity of infection of 3.0 PFU per cell. At 1, 12, 24, and 36 h after starting the culture of virus-infected cells in DMEM containing 0.2% BSA, various concentrations of the NO donors S-nitrosoglutathione (GS-NO) and S-nitroso-N-acetyl-d,l-penicillamine (SNAP) were added to the culture. At different time points after infection, the yield of virus in the culture supernatant was assessed by use of the plaque-forming assay just mentioned. In some experiments, the MDCK cells were infected with influenza virus or SeV at a multiplicity of infection of 0.01 or 0.1 PFU per cell, followed by incubation in DMEM plus 0.2% BSA containing 0.5 μg of trypsin (Sigma-Aldrich) per ml for 2 days. At 1, 12, 24, and 36 h after virus inoculation, SNAP was added to the cultures at a final concentration of 0.1 mM. Under these conditions, the half-life of the NO donors added to cultures of infected cells was 2.5 h and 4.0 h for GS-NO and SNAP, respectively, as assessed by high-performance liquid chromatography coupled with flow reactor analysis, which we reported earlier (5).
Virus culture in iNOS-transfected cells.
The growth of virus was also examined with a cell line expressing iNOS. Cells stably expressing iNOS were obtained with a human adenocarcinoma cell line, SW480, after transfection with plasmid pSK(+)/CMV containing a rat iNOS gene as reported earlier (1). Briefly, SW480 cells were transfected with the iNOS expression vector by the calcium phosphate coprecipitation method, and selection was accomplished by culturing the cells with 800 μg of G418 (Invitrogen) per ml for more than 2 weeks. A clone of SW480 cells expressing iNOS (iNOS-SW480 cells), which showed positive staining for NADPH diaphorase activity, was used for virus culture. In addition, cellular expression of iNOS was verified by Western blotting for iNOS protein, and the activity of the enzyme was confirmed by measuring nitrite and nitrate (NO2− and NO3−) in the culture supernatant of the cells, as reported previously (5).
Peritoneal exudate cells (PEC) (macrophages) served as the positive control for iNOS expression. PEC were obtained by peritoneal lavage of mice 4 days after administration of OK-432 as described previously (41). Cell lysates (25 μg of protein for SW480 cells and iNOS-SW480 cells and 10 μg of protein for PEC) were used for Western blotting. The iNOS-SW480 cells were infected with influenza virus or SeV at a multiplicity of infection of 3.0 PFU per cell, and virus yield was quantified at various time points after virus inoculation as described above. The effect of NO produced from iNOS on viral replication in the cells was examined by inhibiting cellular NOS activity with the NOS inhibitor Nω-monomethyl-l-arginine (l-NMMA). Specifically, after virus inoculation, infected cells were cultured in DMEM plus 0.2% BSA with or without 1.0 or 10 mM l-NMMA, and the production of virus was assessed as just mentioned. The effect of iNOS expression on growth of virus was also investigated by comparing growth of virus in iNOS-SW480 cells and their parent SW480 cells that had no iNOS expression after viral infection, as described above.
Viral mutation assay.
The mutagenic potential of NO for SeV was analyzed by assessing mutation of GFP-SeV according to a method described earlier (4). Monolayers of iNOS-SW480 cells or parent SW480 cells were inoculated with recombinant GFP-SeV at a multiplicity of infection of 3.0 PFU per cell, followed by culture in DMEM plus 0.2% BSA with or without l-NMMA at a concentration of 1.0 or 10 mM. The culture supernatant obtained 48 h after infection was then subjected to the viral mutation assay. Similarly, mutation of GFP-SeV was examined after multiple-cycle replications of GFP-SeV in iNOS-SW480 and parent SW480 cells. Monolayers of each cell line inoculated with GFP-SeV at a multiplicity of infection of 0.1 PFU per cell were cultured in the presence of 0.5 μg of trypsin per ml for 72 h, and the mutation frequency of virus produced in the culture was analyzed.
In these assays, the mutation frequency of GFP-SeV was determined by counting the number of fluorescence-negative viral plaques among the fluorescence-positive plaques formed after GFP-SeV replication. The plaques were formed on monolayers of CV-1 cells overlaid with 0.4% agarose containing 0.5 μg of trypsin per ml in DMEM plus 0.2% BSA. Mutations were identified and quantified via hemadsorption and fluorescence. More than 1,000 plaques were assessed for each determination obtained by this mutation assay.
In some experiments, GFP-SeV was cultured in CV-1 cells in the same manner as for culture in iNOS-SW480 or SW480 cells as just described except that cells were grown in various concentrations of authentic 8-nitroguanosine, which was synthesized and purified as described elsewhere (8).
Sequence analysis for GFP-SeV mutants found in vitro and in vivo.
GFP-SeV mutants were isolated and cloned by obtaining a single plaque that had formed on the CV-1 cell monolayer, as reported previously (4). Briefly, fluorescence-negative plaques formed with culture supernatant of GFP-SeV-infected CV-1 cells with or without 8-nitroguanosine (500 μM) were isolated, and suspensions of virus derived from these plaques were then inoculated onto other cultures of CV-1 cells. After 48 h of culture in DMEM plus 0.2% BSA with 0.5 μg of trypsin per ml, total RNA extracted from the infected cells was subjected to reverse transcription-PCR (RT-PCR) with the following oligonucleotide primers: sense 22-mer, 5′-TGAGCAAGGGCGAGGAGCTGTT-3′; and antisense 23-mer, 5′-TACAGCTCGTCCATGCCGAGAGT-3′. The 712-bp GFP cDNA fragments thus obtained were ligated to the PCR4-TOPO cloning vector (Invitrogen).
Initial transformation was carried out with Escherichia coli DH5α, and the cDNA sequence was determined by a dideoxy method with a DNA sequencer (model 373A; Applied Biosystems, Foster City, Calif.) with a BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems). To investigate GFP mutation generated in vivo, C57BL/6 mice (4 weeks old; male) were infected with GFP-SeV by inhalation of a viral suspension at twice the 50% lethal dose, as described elsewhere (4). On day 7 after the infection, GFP-SeV in infected lungs were isolated by the plaque-forming assay with CV-1 cells. Fluorescence-negative plaques were analyzed further for the nucleotide sequence of GFP mutants as just described.
Analysis of mutation frequency of both the endogenous viral gene and the GFP insert gene.
To confirm that NO affects not only the GFP gene exogenously inserted but also the endogenous genes in SeV, we compared the frequencies of NO-induced GFP mutation and mutation of the gene encoding the F protein, which is critical for internalization of SeV into host cells. F protein fusion activity occurs via proteolytic processing by trypsin-like proteases (31). Because the susceptibility of the F protein to proteases is determined by the amino acid sequence at the cleavage site of the protein (49), alteration of coding for bases around the cleavage site, which results in a change in the sensitivity of the F protein to these enzymes, allowed us to select different protease-sensitive mutants.
For example, an amino acid change at the P1 site of the F protein (Arg-166-Ile) occurs via point mutation of the nucleotide at position 347 (C to A [negative sense]), the result being a chymotrypsin-sensitive SeV mutant. Thus, the mutation frequency of the SeV F protein gene can be estimated by selecting chymotrypsin-sensitive SeV mutants after culture of virus-infected cells in the presence of chymotrypsin. To do this, wild-type SeV that was produced by reverse genetics without GFP insertion and GFP-SeV were treated for 2.5 min with peroxynitrite via a constant flux system to maintain an effective concentration of peroxynitrite of 0.8 μM in the reaction mixture, as described earlier (4). Both SeV strains (each at 107 PFU/ml) were treated with peroxynitrite under the same constant flux reaction conditions.
We chose peroxynitrite because it is the most important NO derivative that seems to be a major contributor to NO-induced mutagenesis, as reported earlier (2, 8, 42), and we can directly determine its effect on viral genes, including those encoding GFP and F protein, under consistent and well-defined reaction conditions.
After peroxynitrite treatment of GFP-SeV, aliquots of the reaction mixture were inoculated onto CV-1 cells. The viral plaques that formed were analyzed, and the mutation frequency of GFP was quantified as described above. Similarly, SeV treated with peroxynitrite was inoculated onto a CV-1 cell monolayer, followed by plaque formation in an agarose-overlaid culture containing 2.0 μg of chymotrypsin per ml, as reported earlier (4). The mutation frequency of F protein was then determined by counting the number of chymotrypsin-sensitive SeV plaques. We also determined the number of trypsin-sensitive plaques that had formed on a CV-1 cell monolayer overlaid with trypsin-containing agarose in the same manner as for the GFP-SeV plaque-forming assay.
The genetic basis of the F protein mutation was verified by sequence analysis of F protein cDNA obtained by RT-PCR, as was done for the GFP cDNA sequence analysis mentioned above. Briefly, after chymotrypsin-sensitive SeV mutants were cloned by use of a single plaque, they were cultured again in CV-1 cells in DMEM plus 0.2% BSA with 2.0 μg of chymotrypsin per ml. The total RNA extracted from the infected cells was then subjected to RT-PCR with the following oligonucleotide primers: sense 24-mer, 5′-AAGATAGCTGGATCCCACGAATCG-3′; and antisense 30-mer, 5′-AGGCTTTGATGAGCGCTATGTCTCTTTTGG-3′. A 334-bp F protein cDNA fragment including the coding region for the amino acid sequence of the protease cleavage site was thereby obtained. The sequence of the F protein cDNA fragment was determined in the same manner as for the GFP cDNA.
Identification of NO-induced nucleoside modification.
The effect of NO on nucleic acid was examined by identifying a chemical modification of guanosine, nitration, induced by NO. We recently developed a mouse monoclonal antibody for 8-nitroguanosine according to the standard protocol described earlier (57). BDF1 mice (SLC, Inc., Shizuoka, Japan) were immunized by intraperitoneal injection of an 8-nitroguanosine-BSA conjugate, which was prepared as described elsewhere (8), plus Freund's complete adjuvant, followed by injection of antigen with Freund's incomplete adjuvant. After the antibody titer in the blood increased, the spleen was harvested for preparation of splenocytes, which were then fused with murine myeloma cells (SP2/0) for hybridoma production. Cell fusion was performed with polyethylene glycol, and hybridomas were selected by incubation in hypoxanthine, aminopterin, and thymidine medium. Hybridomas were screened with an enzyme-linked immunosorbent assay (ELISA) and subsequently cloned by limited dilution. The specificity of the monoclonal antibody finally obtained was confirmed by a competitive ELISA, which showed that the antibody reacted with 8-nitroguanosine and 8-nitroguanine, but not with other endogenous nucleosides and nucleotides or with related compounds, including guanine, guanosine, 8-oxoguanine, 8-oxoguanosine, 8-bromoguanine, 8-bromoguanosine, 8-chloroguanine, xanthine, adenine, adenosine, thymine, deoxythymidine, uracil, uridine, 3-nitrotyrosine, nitroimidazole, and cytosine.
8-Nitroguanosine formation in iNOS-SW480 cells was thus detected by immunocytochemical analysis with the monoclonal 8-nitroguanosine antibody. Briefly, iNOS-SW480 cells and their parent SW480 cells cultured in DMEM plus 10% fetal bovine serum were fixed by the method of Zamboni and colleagues (59). After cells were treated with a blocking reagent (BlockAce; Dainippon Pharmaceuticals Co., Ltd., Osaka, Japan), they were reacted overnight with 10 μg of anti-8-nitroguanosine antibody per ml, followed by reaction with Cy3-labeled secondary antibody (Amersham Biosciences Corp., Piscataway, N.J.). Bound antibody was visualized by fluorescence microscopy.
Analysis for oxidative stress caused by NO and 8-nitroguanosine.
Oxidative stress occurring in iNOS-SW480 cells was analyzed by means of a flow cytometer (FACSCalibur; Becton Dickinson Immunocytometry System, San Jose, Calif.), with dihydrorhodamine 123 as a fluorescent indicator of the intracellular peroxidation reaction as described previously (25). iNOS-SW480 cells were incubated with 2.5 μM dihydrorhodamine 123 (Sigma-Aldrich) in the presence or absence of 1 mM l-NMMA in Krebs-Ringer phosphate buffer (KRP, pH 7.4) containing 0.2% BSA and 1 mM l-arginine for 6 h at 37°C. After the cells were washed twice with KRP, they were analyzed by flow cytometry for intracellular oxidation of dihydrorhodamine 123. Similarly, intracellular peroxidation of dihydrorhodamine was examined with CV-1 cells after treatment with 8-nitroguanosine in KRP containing 0.2% BSA for 12 h.
Statistical analysis.
All data are expressed as means ± standard error. Statistical differences were determined by the unpaired t test.
RESULTS
No inhibitory effect of NO on viral replication in cultured cells.
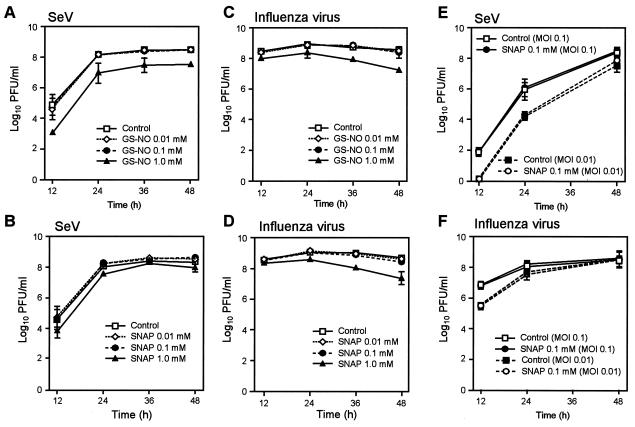
To see whether NO affects viral replication, SeV (Z strain) and influenza virus were grown in MDCK cells in culture in the presence and absence of the NO donors SNAP and GS-NO. No appreciable effect was observed for yields of SeV and influenza virus throughout the course of infection when infected cells were treated with repeated additions, at 12-h intervals, of 0.01 or 0.1 mM GS-NO or SNAP (Fig. 1A to D). Physiological concentrations of NO were released, as assessed by the presence of oxidized metabolites of NO nitrite and nitrate (data not shown). Although viral propagation was slightly attenuated by the 1.0 mM concentration of the NO donors, such a high concentration is not biologically relevant, and in fact microscopic observation of treated cells revealed nonspecific cytotoxicity. We also examined the effects of GS-NO and SNAP (each at 0.01 and 0.1 mM) when added to the cultures at 1, 6, 12, and 24 h (6-h intervals) after viral inoculation on growth of SeV and influenza virus (data not shown). The replication of the viruses was not affected by treatment with these NO donors. Moreover, no appreciable suppressive effects of SNAP were observed in multicycle replications of SeV and influenza virus (Fig. 1E and F).
FIG. 1.
Effects of NO formed from GS-NO and SNAP on propagation of SeV and influenza virus. (A to D) After MDCK monolayers were inoculated with influenza virus or SeV at a multiplicity of infection of 3.0 PFU per cell, infected cells were incubated in DMEM containing 0.2% BSA and various concentrations of the NO donors GS-NO and SNAP. (E and F) MDCK cells were infected with SeV or influenza virus at a multiplicity of infection (MOI) of 0.1 or 0.01 PFU per cell, followed by culture with 0.1 mM SNAP as just described. At different time points after infection, the yield of virus in culture supernatants was assessed by use of the plaque-forming assay. Data are means ± standard error (n = 3). See text for details.
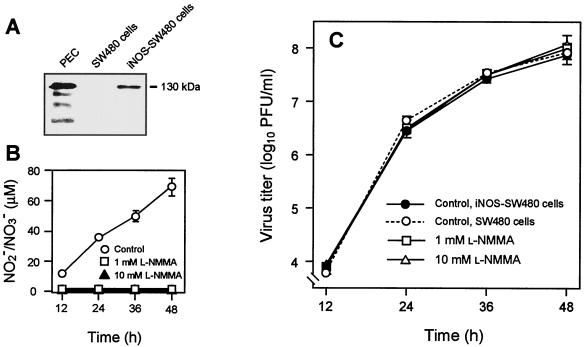
The effect of NO on viral replication was also investigated with NO formed endogenously by iNOS-SW480 cells, a transfected cell line stably expressing iNOS, in which a significant amount of NO is constantly produced by the cells (Fig. 2A and B). After the iNOS-SW480 cells were infected with SeV, the time profile of viral production was monitored in the presence or absence of l-NMMA. No appreciable difference was found in virus yield between the groups cultured with l-NMMA (1 and 10 mM) and the controls (Fig. 2C). l-NMMA almost completely inhibited NO synthesis in virus-infected cells, however (Fig. 2B). Similar results were observed with influenza virus propagated in iNOS-SW480 cells endogenously forming NO. As shown in Fig. 2C, the same level of SeV growth was observed in iNOS-SW480 cells and their parent cells after infection. Because parent SW480 cells did not express a measurable level of iNOS, as determined by Western blotting (Fig. 2A), neither NO formation nor iNOS expression suppressed viral replication in these cells. These data thus indicate that replication of SeV and influenza virus in the cells is not affected by NO regardless of the origin of NO.
FIG. 2.
Effect of endogenous NO on SeV replication in cultured cells. A cell line stably expressing iNOS (iNOS-SW480 cells) was established. iNOS protein expression and NO overproduction were confirmed by Western blotting (A) and by assay for nitrite (NO2−) and nitrate (NO3−) formed in the supernatant of the cell culture (B). PEC, mouse peritoneal exudate cells, which served as a positive control for iNOS expression. (C) The effect of an NOS inhibitor (l-NMMA) on SeV replication in iNOS-SW480 cells is shown. iNOS-SW480 cells and parent SW480 cells served as controls for iNOS- and NO-producing and nonproducing cells, respectively. iNOS-SW480 and SW480 cells were infected with SeV at a multiplicity of infection of 3.0 PFU per cell, and the yield of virus in the culture supernatant was quantified by means of the plaque-forming assay. Data are means ± standard error (n = 3). See text for details.
Viral mutation enhanced by NO.
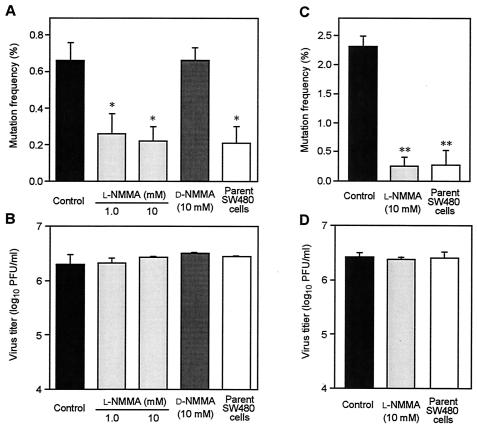
We further explored the mutagenic potential of NO for SeV by use of the mutagenesis assay with GFP-SeV. After GFP-SeV was cultured in iNOS-SW480 cells with or without l-NMMA for 48 h, the virus in the culture supernatant was subjected to analysis for mutation of the GFP gene. As illustrated in Fig. 3A, the mutation frequency in iNOS-SW480 cells was markedly attenuated by treatment with 1 and 10 mM l-NMMA without affecting viral growth in the cells (Fig. 3B). In fact, this treatment reduced the mutation frequency of GFP-SeV to a level similar to that found for the parent SW480 cells. When the GFP-SeV-infected cells were incubated with d-NMMA, which is an enantiomer of l-NMMA without NOS-inhibitory activity, no significant change in the mutation frequency of GFP-SeV compared with the control was obtained.
FIG. 3.
Mutation frequency (A and C) and virus yield (B and D) in iNOS-SW480 cells and parent SW480 cells with or without l-NMMA and d-NMMA. The mutagenic potential of NO for SeV was determined by analysis of the mutation of GFP-SeV. (A and B) Monolayers of iNOS-SW480 cells were inoculated with GFP-SeV at a multiplicity of infection of 3.0 PFU per cell, followed by culture with or without l-NMMA or d-NMMA. The culture supernatant obtained 48 h after infection was used for determination of viral mutation by the mutation assay (A). The virus yield in the same samples used for the mutation assay is shown in B. (C and D) Monolayers of iNOS-SW480 cells and SW480 cells were inoculated with GFP-SeV at a multiplicity of infection of 0.1 PFU per cell (multicycle replications). After 72 h of culture of the infected cells with or without l-NMMA, the mutation frequency (C) and virus yield (D) were assessed in the same manner as in A and B. Control, iNOS-SW480 cells without l-NMMA; l-NMMA (1.0 or 10 mM), iNOS-SW480 cells treated with 1.0 or 10 mM l-NMMA; d-NMMA, iNOS-SW480 cells treated with 10 mM d-NMMA. Data are means ± standard error of four independent experiments (>1,000 plaques counted/assay); *, P < 0.05, and **, P < 0.01 versus the control and the d-NMMA-treated cells. See text for details.
An important finding was a clearly enhanced mutation frequency of GFP-SeV for virus propagated after multicycle replication in iNOS-SW480 cells (Fig. 3C). The increase in mutation frequency of GFP-SeV was threefold (0.6% in iNOS-SW480 cells versus 0.2% in SW480 cells or l-NMMA-treated iNOS-SW480 cells) after single-step replication (Fig. 3A). The mutation frequency increase was even greater (increased almost 10-fold) after multicycle replication: 2.3% in iNOS-SW480 cells versus 0.25 and 0.27% in SW480 cells and l-NMMA-treated iNOS-SW480 cells, respectively (Fig. 3C). Viral propagation in these multicycle replications in iNOS-SW480 cells with or without l-NMMA and in parent SW480 cells did not differ (Fig. 3D). These findings strongly suggest that SeV mutation is greatly enhanced by the presence of NO during viral replication.
It was quite important to determine whether NO could affect viral function via induction of mutation in endogenous RNA genes of the virus as well as mutation of the GFP insert. Alteration of the phenotype of the RNA viruses would serve as evidence of this effect. Thus, we compared the frequency of peroxynitrite-induced GFP mutation with the mutation frequency of the gene encoding the F protein, which is critical for internalization of SeV into host cells. The mutation frequency of the SeV F protein gene was estimated by selecting a chymotrypsin-sensitive SeV mutant. In this mutant, F protein can be proteolytically activated by chymotrypsin so as to obtain membrane fusion activity, which confers viral infectivity. Furthermore, to confirm the genetic basis of this chymotrypsin-sensitive mutation of F protein, the sequence of the F protein gene of the virus clone selected was analyzed, and only the clone having a point mutation at nucleotide 347 was considered to be the chymotrypsin-sensitive mutant.
As shown in Table 1, when SeV was treated in vitro with 0.8 μM peroxynitrite (the concentration maintained in a constant-flux system) for 2.5 min, the frequency of generation of chymotrypsin-sensitive SeV mutants was 8.3 × 10−5. The background value for the same mutation of SeV without peroxynitrite treatment was 4.7 × 10−7. Thus, the mutation rate at a single nucleotide position of F protein (C347A), which increased after peroxynitrite treatment of GFP-SeV, was 8.2 × 10−5 (8.3 × 10−5 − 4.7 × 10−7). To compare the nucleotide-based mutations of the GFP and F protein genes, the mutation rate per nucleotide for the GFP gene was determined: if it is assumed that nucleotide substitution occurs at a similar rate for each nucleotide of the GFP gene, the mutation frequency per nucleotide of the GFP gene is given by the formula GFP gene mutation frequency × [(number of nucleotide substitutions in mutant GFP genes)/(number of virus clones sequenced) × (nucleotide number of GFP gene)].
TABLE 1.
Comparison of mutation rates of GFP and F protein genes in SeVa
| Gene | Frequency of base substitutions
|
Mutation rate per nucleotide generated by peroxynitrite treatment (B − A) | |
|---|---|---|---|
| Control, background (A) | Peroxynitrite-induced mutants (B) | ||
| F protein (C347A) | 4.7 × 10−7 | 8.3 × 10−5 | 8.2 × 10−5 |
| GFP | 8.6 × 10−6 | 8.5 × 10−5 | 7.6 × 10−5 |
Values for F protein gene are means of three or four different determinations. The mutation frequency of the GFP gene was obtained from three different determinations in which more than 105 plaques were formed and used for cloning fluorescence- negative GFP-SeV mutants. The frequency per nucleotide was then obtained by the formula described in the text.
For example, the GFP gene (712 bases) in GFP-SeV was sequenced for 32 clones of GFP-SeV mutants that had occurred spontaneously and existed before peroxynitrite treatment, and 75 nucleotide substitutions were found. A similar sequence analysis performed for the GFP gene in GFP-SeV mutants generated by peroxynitrite treatment showed 94 nucleotide substitutions among 25 clones. The background value of GFP gene mutations in GFP-SeV, as evidenced by loss of fluorescence, was 2.6 × 10−3, which increased to 1.6 × 10−2 after peroxynitrite treatment. The mutation frequencies of the GFP gene per nucleotide for the spontaneous (background) and peroxynitrite-induced mutations were thus calculated to be 8.6 × 10−6 and 8.5 × 10−5, respectively (Table 1). For comparison of mutation rates per nucleotide of the GFP and F protein genes that were obtained after peroxynitrite treatment, the value of the background mutation frequency per nucleotide for the GFP gene was subtracted from the value of the mutation frequency induced by peroxynitrite. A similar mutation rate for the F protein and GFP genes was obtained, 8.2 × 10−5 for the F protein gene versus 7.6 × 10−5 for the GFP gene. This finding supports the proposal that the RNA mutation occurred at the same rate in both the GFP and endogenous viral genes. In addition, it confirmed the reliability and validity of our GFP-related SeV mutation assay for general analysis of NO-induced mutagenesis in RNA viruses.
NO-induced 8-nitroguanosine formation and mutagenic potential.
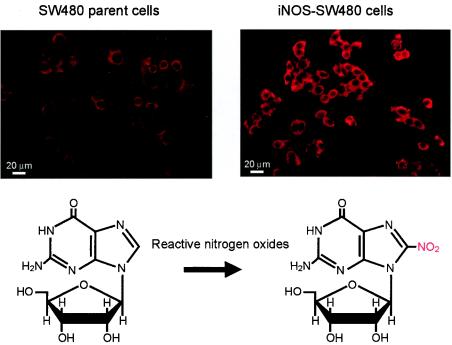
To analyze the molecular mechanism of NO-dependent mutagenesis of SeV, chemical modification of nucleic acids was examined with a focus on guanosine nitration, i.e., formation of 8-nitroguanosine (Fig. 4). We recently reported that guanosine nitration occurs in vivo in influenza virus- and SeV-induced pneumonia in mice via excessive formation of NO (8), which was clearly identified by use of a specific polyclonal antibody against 8-nitroguanosine. Here, we used an immunocytochemical method with a monoclonal 8-nitroguanosine antibody to investigate NO-dependent 8-nitroguanosine formation in iNOS-SW480 cells. As shown in Fig. 4, iNOS-SW480 cells showed much more intense immunostaining than the parent cells. This result indicates that greater nucleic acid nitration occurred in the iNOS-SW480 cells than in SW480 cells, which correlates well with the increased viral mutation frequency seen with iNOS-SW480 cells (Fig. 3).
FIG. 4.
Increased formation of 8-nitroguanosine in iNOS-SW480 cells. 8-Nitroguanosine formation in iNOS-SW480 cells was identified by immunocytochemical analysis with the monoclonal 8-nitroguanosine antibody. After iNOS-SW480 cells and their parent SW480 cells cultured in DMEM plus 10% fetal bovine serum were fixed and blocked, they were reacted overnight with 10 μg of anti-8-nitroguanosine antibody per ml, followed by reaction with Cy3-labeled secondary antibody. The antibody bound on the cells was visualized by fluorescence microscopy. The reaction for generation of 8-nitroguanosine appears below the fluorescent images.
We also found, in a separate analysis by confocal laser scanning microscopy, that the 8-nitroguanosine was localized mainly in the cytosol of iNOS-SW480 cells (data not shown). This result is consistent with our earlier observation showing a similar intracellular localization in bronchial epithelial cells of influenza virus-infected mouse lungs (8). This cytoplasmic localization of 8-nitroguanosine may be explained by the following: the 8-nitroguanine moiety in deoxyguanosine, which is formed in DNA, is unstable and undergoes spontaneous depurination to produce an apurinic site in DNA (56). Thus, 8-nitroguanine (8-nitrodeoxyguanine) does not accumulate in DNA in an amount sufficient to be detected by our immunohistochemical analysis; cytosol 8-nitroguanosine, however, is stably formed in the nucleotide pool and RNA in the cytosol of the cells, as reported earlier (8).
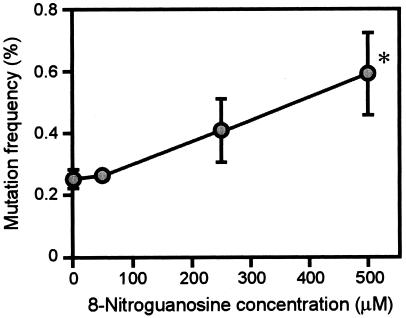
More important, we now verified that 8-nitroguanosine is mutagenic for GFP-SeV (Fig. 5). When GFP-SeV was cultured in CV-1 cells in the presence of various concentrations of 8-nitroguanosine, the mutation frequency of the progeny virus was markedly elevated in a concentration-dependent fashion. The increased frequency mutation became apparent at a concentration of 250 μM and was further augmented at 500 μM 8-nitroguanosine. In a separate experiment with a murine macrophage cell line expressing iNOS, the intracellular concentration of 8-nitroguanosine, which was quantified by a competitive ELISA, was found to be higher than 100 μM (unpublished observation). No cytotoxic effect on GFP-SeV-infected cells was observed, at least with the concentrations of 8-nitroguanosine used in this study. Also, the virus yield among the experimental groups of virus-infected cells was not significantly different with or without 8-nitroguanosine. It is therefore conceivable that physiologically relevant concentrations of 8-nitroguanosine enhanced the mutation of GFP-SeV in the cells.
FIG. 5.
Effect of 8-nitroguanosine on mutation frequency of GFP-SeV replicated in CV-1 cells. GFP-SeV was cultured in CV-1 cells in the same manner as for the culture with iNOS-SW480 cells described for Fig. 3 except that GFP-SeV was allowed to propagate in the cells in the presence of various concentrations of authentic 8-nitroguanosine. Data are means ± standard error of four independent experiments (>1,000 plaques counted/assay); *, P < 0.05 versus the control without 8-nitroguanosine. See text for details.
Mutation spectra of GFP-SeV mutants obtained from cells treated with 8-nitroguanosine or untreated and of mutants obtained from virus-infected lungs.
The GFP genes inserted into various GFP-SeV mutants were sequenced, and mutation spectra of different groups were compared: GFP-SeV mutants occurring spontaneously in CV-1 cells without the use of 8-nitroguanosine; GFP-SeV mutants obtained with 8-nitroguanosine-treated CV-1 cells; and GFP-SeV mutants obtained from mouse lungs in vivo. No deletions or insertions were found in the mutant GFP gene, and all mutants contained point mutations. As summarized in Table 2, the A-to-G transition was frequently observed in all groups of GFP-SeV mutants and was predominant in the spontaneously occurring GFP-SeV mutants. This finding is consistent with our earlier results (4). The present analysis found another predominant point mutation, i.e., a C-to-U transition, especially in GFP-SeV mutants generated in CV-1 cells treated with 8-nitroguanosine and in mouse lung tissues infected with GFP-SeV. Because the frequency of C-to-U substitution was much lower in mutants occurring spontaneously in CV-1 cells than in other groups, this point mutation seems to be relatively characteristic of GFP gene mutation induced by 8-nitroguanosine and that occurring in vivo. G-specific mutation was not evident in any group of GFP-SeV mutants, although a relatively high incidence of G-to-A substitutions was observed in GFP-SeV mutants recovered from infected mouse lungs in vivo.
TABLE 2.
Mutation of GFP genes in various GFP-SeV mutants
| Change | No. and frequency (%) of base substitutions
|
||
|---|---|---|---|
| Mutants from 8-nitroguanosine- treated cells (35 clones) | Mutants from lungs in vivo (29 clones) | Spontaneous mutants (32 clones) | |
| Transition | |||
| A→G | 27 (40.9) | 9 (17.0) | 45 (60.0) |
| G→A | 1 (1.5) | 10 (18.9) | 8 (10.7) |
| U→C | 12 (18.2) | 9 (17.0) | 8 (10.7) |
| C→U | 18 (27.3) | 16 (30.2) | 3 (4.0) |
| Subtotal | 58 (87.9) | 44 (83.0) | 64 (85.3) |
| Transversion | |||
| A→U | 7 (10.6) | 1 (1.9) | 5 (6.7) |
| A→C | 0 | 0 | 2 (2.7) |
| G→U | 0 | 1 (1.9) | 0 |
| G→C | 0 | 1 (1.9) | 0 |
| U→A | 1 (1.5) | 1 (1.9) | 2 (2.7) |
| U→G | 0 | 2 (3.8) | 2 (2.7) |
| C→A | 0 | 2 (3.8) | 0 |
| C→G | 0 | 1 (1.9) | 0 |
| Subtotal | 8 (12.1) | 9 (17.0) | 11 (14.7) |
| Total | 66 (100) | 53 (100) | 75 (100) |
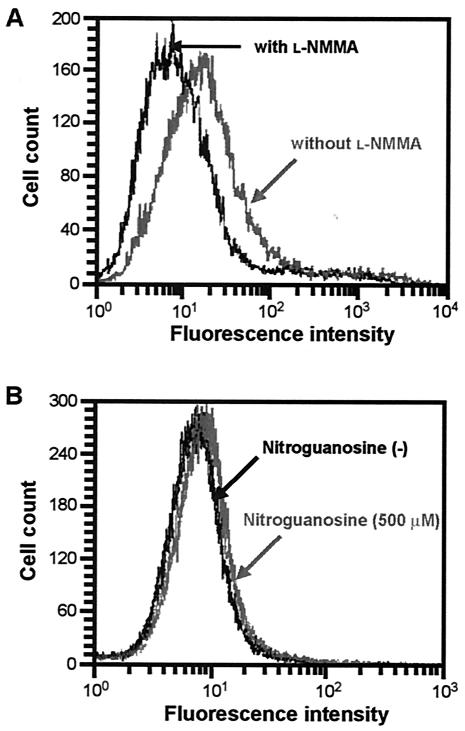
Cellular oxidative stress induced by NO and 8-nitroguanosine.
It is now well documented that excessive NO production causes oxidative stress via the formation of reactive nitrogen oxides such as peroxynitrite and nitrogen dioxide. Because oxidative stress is thought to be involved in mutagenesis occurring during infection and inflammation, the oxidative stress induced by NO in iNOS-SW480 cells was analyzed by flow cytometric measurement of the intensity of fluorescence caused by oxidation of dihydrorhodamine 123. As demonstrated in Fig. 6A, the level of fluorescence intensity was reduced significantly by treatment with l-NMMA; the mean fluorescence intensity for iNOS-SW480 cells without l-NMMA treatment was 26.7 ± 0.42, whereas that with l-NMMA treatment was 12.5 ± 0.16 (n = 3, P < 0.001). This result suggests that excessive production of NO from iNOS induced oxidative stress in the iNOS-SW480 cells.
FIG. 6.
Oxidative stress induced in cells by NO and 8-nitroguanosine. Intensity of fluorescence related to oxidation of dihydrorhodamine 123, as measured by flow cytometry, is shown for iNOS-SW480 cells treated with l-NMMA (A) and for CV-1 cells treated with 8-nitroguanosine (B). (A) iNOS-SW480 cells were incubated with dihydrorhodamine 123 in the presence or absence of 1 mM l-NMMA in KRP (pH 7.4) containing 0.2% BSA and 1 mM l-arginine for 6 h at 37°C. Rhodamine fluorescence was measured as an indication of intracellular peroxidation. (B) Oxidative stress was examined with CV-1 cells after treatment with 500 μM 8-nitroguanosine in KRP containing 0.2% BSA for 12 h. Each panel represents data for three different flow cytometric measurements.
Although the change in CV-1 cells was not as marked as the change in iNOS-SW480 cells, the intensity of fluorescence related to dihydrorhodamine oxidation occurring in CV-1 cells was significantly elevated by 8-nitroguanosine added exogenously to the cell culture (Fig. 6B). Mean fluorescence intensities for control CV-1 cells and for cells treated with 8-nitroguanosine were 8.4 ± 0.06 and 10.2 ± 0.17, respectively (n = 3, P < 0.001). This result, as well as the immunocytochemistry result shown in Fig. 4, indicates that enhanced oxidative stress caused by NO production in iNOS-SW480 cells may be attributable, at least in part, to 8-nitroguanosine formation.
DISCUSSION
In the present study, we verified that NO possesses potent mutagenic activity for SeV, which is consistent with our previous in vivo experiment showing that GFP-SeV propagated in iNOS-deficient mice had a much lower level of mutation frequency than GFP-SeV grown in wild-type mice (4). No appreciable antiviral effect of NO was observed on influenza virus and SeV replicated in cells in culture. This finding again supports our earlier data obtained with mouse models of influenza virus- and SeV-induced pneumonia (4, 7, 8). Furthermore, it is of great importance that our present work provides the first demonstration that 8-nitroguanosine, which is produced endogenously by NO or its reactive derivatives, as evidenced here and in our earlier report (8), induced mutagenesis of GFP-SeV in cultured cells.
The mutation frequency of GFP-SeV increased almost 10-fold during multicycle replication of GFP-SeV in iNOS-SW480 cells compared with the GFP-SeV mutation frequency in l-NMMA-treated iNOS-SW480 cells and in parent SW480 cells that had no appreciable iNOS expression (Fig. 3). A similar magnitude of increase in NO-dependent viral mutation was observed with GFP-SeV replicated in vivo in mouse lungs, which was reported previously (4). The mutation rates of GFP and F protein genes were found to be similar (Table 1), which suggests that mutation of viral genes other than the GFP and F genes may also be accelerated by NO to the same extent during viral replication. This NO-induced viral mutagenesis may explain the heterogeneity and increased repertories of variants from which a particular genotype can evolve rapidly under selective pressure. Therefore, GFP-SeV mutation induced by NO is considered biologically relevant and may have important implications for viral pathogenesis and evolution, particularly when a virus is replicating in vivo in the presence of NO.
In our recent work with influenza virus- and SeV-induced pneumonia in mice, formation of 8-nitroguanosine was found to be localized mainly in bronchial and bronchiolar epithelial cells of the lung (8), where viral replication primarily occurs. The same study found an appreciable amount of 8-nitroguanosine in the total RNA isolated from iNOS-expressing cells in culture. It is therefore highly plausible that NO-dependent viral mutagenesis was brought about at least in part by NO-generated 8-nitroguanosine, which is in turn incorporated into the viral genome during replication and thus accelerates viral mutation (Fig. 7).
FIG. 7.
Schematic drawing of hypothetical mechanisms for NO-induced viral mutagenesis proposed by the present work. NO may accelerate viral mutation via formation of 8-nitroguanosine (8-nitroGuo), which may be a substantial contributor to erroneous RNA replication of the virus. NO-generated 8-nitroguanosine may cause viral mutation via two different mechanisms: directly, through incorporation into template RNAs for viral replication (pathway shown on the right), and indirectly, by enhanced oxidative stress because of its potent redox-active property (pathway shown on the left).
Peroxynitrite formed via superoxide and NO generation during infections possesses the potential for potent nitrating and oxidizing effects on many biomolecules, including nucleic acids (14, 43, 45-48). Peroxynitrite has mutagenic effects on prokaryotic DNA, possibly via nitration of guanine residues of DNA (35). Wogan's group documented NO-induced mutation of an endogenous hypoxanthine-guanine phosphoribosyltransferase (hprt) gene in murine macrophages expressing iNOS (60). The same group showed that mutagenicity was enhanced by NO overproduction in vivo, as evidenced by mutation of an exogenously expressed lacZ with lacZ-containing pUR288 transgenic mice (26). Also important, Ohshima's group reported that p53 was inactivated by peroxynitrite, which may indirectly increase genetic mutation related to oxidative damage of DNA (15). Therefore, excess production of NO by iNOS induced by proinflammatory cytokines, possibly through reactive nitrogen intermediates, may cause nucleic acid modifications and thus mutagenesis in various pathogens as well as hosts. This process may occur during infections in biological systems as a result of host defense.
In addition, oxidative stress caused by NO and 8-nitroguanosine may have a great impact in terms of mutagenic potential (2, 33, 42). Our other studies have revealed that 8-nitroguanosine has strong redox activity, which stimulates superoxide generation from NADPH-cytochrome P450 reductase and various isoforms of NOS (8, 47). It has been known for a long time that many naturally occurring mutagens and carcinogens may act as generators of free radicals (10). Moreover, oxygen radicals and reactive oxygen species, as endogenous initiators of DNA damage and mutation, are involved in multiple stages of carcinogenesis (11, 29, 53, 55). In fact, human leukocytes producing superoxide but not leukocytes lacking superoxide-generating activity from patients with chronic granulomatous disease caused mutation of Salmonella enterica serovar Typhimurium TA100 (54). It is therefore logical that NO-induced viral mutation may be mediated by NO-generated 8-nitroguanosine through two different mechanisms: direct modification of nucleic acid (e.g., via 8-nitroguanosine formation), and indirect augmentation by 8-nitroguanosine of oxidative stress via superoxide generation (Fig. 7).
It is intriguing that the mutation profile of the GFP gene in the GFP-SeV mutants induced by 8-nitroguanosine appeared to resemble that of the mutants occurring in mouse lungs in vivo (with a predominant C-to-U transition), in which NO is produced in excess from iNOS (4). A base substitution that was relatively characteristic of the GFP mutants induced by 8-nitroguanosine and mutants occurring in vivo was the C-to-U transition (Table 2). This finding suggests indirectly that 8-nitroguanosine formed in vivo could indeed contribute to enhanced viral mutation induced by NO.
The mechanism for the C-to-U point mutation may involve incorporation of 8-nitroguanosine into the positive-strand antigenomic RNA, with subsequent G-to-A (positive sense) and C-to-U transitions in the viral genome during RNA replication. However, because a G-to-A substitution did not occur very often in the GFP gene of GFP-SeV mutants, except for mutants produced in vivo, other mechanisms may be involved in the C-to-U mutation. Similarly, the exact mechanism for the frequent A-to-G mutation, which was found in various GFP-SeV mutants, has not yet been identified. In addition, no G-specific alteration was detected in GFP-SeV mutants, which is consistent with our previous analysis (4). These mutation profiles seem to differ from the DNA mutations induced by NO, in which G-to-T transversion was typical in eukaryotic and prokaryotic DNA treated with peroxynitrite (35, 42). In fact, transversions occurred much less frequently than transitions did, and the G-to-U transversion in the GFP-SeV mutants isolated in the present study was rare (Table 2). This result suggests again that NO may cause RNA mutagenesis by a mechanism different from that of NO-elicited DNA mutagenesis. Further analyses in a cell-free replication system are needed to elucidate the molecular mechanism of RNA mutagenesis involving NO and 8-nitroguanosine.
The most striking feature of a virus is its considerable adaptability to various environmental stresses (21, 30). For example, RNA viruses exist as highly heterogeneous populations, called quasispecies, primarily because of the error-prone nature of the replicase of the viruses. In general, RNA viruses have a high mutation rate, ranging from 10−3 to 10−5 misincorporations/nucleotide site/replication, which is more than 104-fold higher than the error rate for DNA viruses (20-23, 30). The low fidelity of RNA replication has been believed to be due to the lack of proofreading and repair functions of RNA polymerase or reverse transcriptase (21, 36). Our earlier and present studies, however, showed that RNA viral mutation was greatly affected by NO and its reactive derivatives and that guanosine nitration (8-nitroguanosine formation) occurred more in RNA than in DNA. Also, the degree of RNA viral mutation was reportedly increased by chemical mutagens, including nitrous acid (16, 28, 50, 51). Thus, the higher incidence of erroneous RNA viral replication may be due to greater susceptibility of RNA than of DNA to NO or oxidative stress.
Several reports have showed a possible association between oxidative stress and viral mutation. For example, oxidative stress augmented the integration of duck hepatitis B virus DNA into genomic DNA in cells through DNA damage and deficient DNA repair (44). Beck et al. documented that the pathogenicity of coxsackievirus B3 is potentiated in vivo in mice fed a selenium-deficient diet, which impairs the antioxidant systems of the host (13). Similar results that were obtained with animals deficient in vitamin E and glutathione peroxidase suggest that oxidative stress facilitates the selection and generation of virulent mutants (13). Impaired immunological clearance of virus that is induced by oxidative stress aids the survival of heterogeneous mutants, which would result in selection of highly pathogenic variants of coxsackievirus (12). In this context, it is of great interest that NO has immunosuppressive and regulatory effects by modulating the T-cell immune response during viral infection (39).
In conclusion, NO-induced mutagenesis may result in greater heterogeneity of variants of RNA viruses, which would lead to rapid viral evolution under selective pressure and to the production of drug-resistant, immunologically tolerant, and cell tropism-altered mutants. It is now accepted that NO is generated during infection caused by any type of pathogen. Therefore, further clarification of the mechanism of NO-induced mutation of viruses is quite important, with particular focus on the role of 8-nitroguanosine in NO-dependent mutagenesis, as suggested by our current work.
Acknowledgments
We thank Judith B. Gandy for excellent editorial work on the manuscript. Thanks are also due to Yoshiyuki Nagai for critical discussions.
This work was supported by a Grant-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan and a grant from the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Adachi, H., S. Iida, S. Oguchi, H. Ohshima, H. Suzuki, K. Nagasaki, H. Kawasaki, T. Sugimura, and H. Esumi. 1993. Molecular cloning of a cDNA encoding an inducible calmodulin-dependent nitric-oxide synthase from rat liver and its expression in COS 1 cells. Eur. J. Biochem. 217:37-43. [DOI] [PubMed] [Google Scholar]
- 2.Akaike, T. 2001. Role of free radicals in viral pathogenesis and mutation. Rev. Med. Virol. 11:87-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akaike, T., M. Ando, T. Oda, T. Doi, S. Ijiri, S. Araki, and H. Maeda. 1990. Dependence on O2− generation by xanthine oxidase of pathogenesis of influenza virus infection in mice. J. Clin. Investig. 85:739-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akaike, T., S. Fujii, A. Kato, J. Yoshitake, Y. Miyamoto, T. Sawa, S. Okamoto, M. Suga, M. Asakawa, Y. Nagai, and H. Maeda. 2000. Viral mutation accelerated by nitric oxide production during infection in vivo. FASEB J. 14:1447-1454. [DOI] [PubMed] [Google Scholar]
- 5.Akaike, T., K. Inoue, T. Okamoto, H. Nishino, M. Otagiri, S. Fujii, and H. Maeda. 1997. Nanomolar quantification and identification of various nitrosothiols by high performance liquid chromatography coupled with flow reactors of metals and Griess reagent. J. Biochem. 122:459-466. [DOI] [PubMed] [Google Scholar]
- 6.Akaike, T., A. Molla, M. Ando, S. Araki, and H. Maeda. 1989. Molecular mechanism of complex infection by bacteria and virus analyzed by a model using serratial protease and influenza virus in mice. J. Virol. 63:2252-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akaike, T., Y. Noguchi, S. Ijiri, K. Setoguchi, M. Suga, Y. M. Zheng, B. Dietzschold, and H. Maeda. 1996. Pathogenesis of influenza virus-induced pneumonia: involvement of both nitric oxide and oxygen radicals. Proc. Natl. Acad. Sci. USA 93:2448-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akaike, T., S. Okamoto, T. Sawa, J. Yoshitake, F. Tamura, K. Ichimori, K. Miyazaki, K. Sasamoto, and H. Maeda. 2003. 8-Nitroguanosine formation in viral pneumonia and its implication for pathogenesis. Proc. Natl. Acad. Sci. USA 100:685-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alam, M. S., T. Akaike, S. Okamoto, T. Kubota, J. Yoshitake, T. Sawa, Y. Miyamoto, F. Tamura, and H. Maeda. 2002. Role of nitric oxide in host defense in murine salmonellosis as a function of its antibacterial and antiapoptotic activities. Infect. Immun. 70:3130-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ames, B. N. 1983. Dietary carcinogens and anticarcinogens. Oxygen radicals and degenerative diseases. Science 221:1256-1264. [DOI] [PubMed] [Google Scholar]
- 11.Ames, B. N., M. K. Shigenaga, and T. M. Hagen. 1993. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. USA 90:7915-7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beck, M. A., R. S. Esworthy, Y.-S. Ho, and F. F. Chu. 1998. Glutathione peroxidase protects mice from viral-induced myocarditis. FASEB J. 12:1143-1149. [DOI] [PubMed] [Google Scholar]
- 13.Beck, M. A., Q. Shi, V. G. Morris, and O. A. Levander. 1995. Rapid genomic evolution of a non-virulent coxsackievirus B3 in selenium-deficient mice results in selection of identical virulent isolates. Nat. Med. 1:433-436. [DOI] [PubMed] [Google Scholar]
- 14.Beckman, J. S., and W. H. Koppenol. 1996. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am. J. Physiol. 271:C1424-1437. [DOI] [PubMed] [Google Scholar]
- 15.Calmels, S., P. Hainaut, and H. Ohshima. 1997. Nitric oxide induces conformational and functional modifications of wild-type p53 tumor suppressor protein. Cancer Res. 57:3365-3369. [PubMed] [Google Scholar]
- 16.Carp, R. I., and H. Koprowski. 1962. Mutation of type 3 poliovirus with nitrous acid. Virology 17:99-109. [DOI] [PubMed] [Google Scholar]
- 17.Cody, C. W., D. C. Prasher, W. M. Westler, F. G. Prendergast, and W. W. Ward. 1993. Chemical structure of the hexapeptide chromophore of the Aequorea green-fluorescent protein. Biochemistry 32:1212-1218. [DOI] [PubMed] [Google Scholar]
- 18.DeGroote, M. A., D. Granger, Y. Xu, G. Campbell, R. Prince, and F. C. Fang. 1995. Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc. Natl. Acad. Sci. USA 92:6399-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doi, T., M. Ando, T. Akaike, M. Suga, K. Sato, and H. Maeda. 1993. Resistance to nitric oxide in Mycobacterium avium complex and its implication in pathogenesis. Infect. Immun. 61:1980-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domingo, E. 1997. Rapid evolution of viral RNA genomes. J. Nutr. 127:958S-961S. [DOI] [PubMed] [Google Scholar]
- 21.Domingo, E., L. Menendez-Arias, and J. J. Holland. 1997. RNA virus fitness. Rev. Med. Virol. 7:87-96. [DOI] [PubMed] [Google Scholar]
- 22.Drake, J. W. 1993. Rates of spontaneous mutation among RNA viruses. Proc. Natl. Acad. Sci. USA 90:4171-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drake, J. W., B. Charlesworth, D. Charlesworth, and J. F. Crow. 1998. Rates of spontaneous mutation. Genetics 148:1667-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eiserich, J. P., M. Hristova, C. E. Cross, A. D. Jones, B. A. Freeman, B. Halliwell, and A. van der Vliet. 1998. Formation of nitric oxide-derived inflammatory oxidants by myeloperoxidase in neutrophils. Nature (London) 391:393-397. [DOI] [PubMed] [Google Scholar]
- 25.Emmendörffer, A., M. Hecht, M. L. Lohmann-Matthes, and J. Roesler. 1990. A fast and easy method to determine the production of reactive oxygen intermediates by human and murine phagocytes using dihydrorhodamine 123. J. Immunol. Methods 131:269-275. [DOI] [PubMed] [Google Scholar]
- 26.Gal, A., and G. N. Wogan. 1996. Mutagenesis associated with nitric oxide production in transgenic SJL mice. Proc. Natl. Acad. Sci. USA 93:15102-15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Granger, D. L., J. B. Hibbs, Jr., J. R. Perfect, and D. T. Durack. 1988. Specific amino acid (l-arginine) requirement for the microbiostatic activity of murine macrophages. J. Clin. Investig. 81:1129-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granoff, A. 1961. Induction of Newcastle disease virus mutants with nitrous acid. Virology 13:402-408. [DOI] [PubMed] [Google Scholar]
- 29.Harris, C. C. 1991. Chemical and physical carcinogenesis: advances and perspectives for the 1990s. Cancer Res. 51:5023s-5044s. [PubMed] [Google Scholar]
- 30.Holland, J., K. Spindler, F. Horodyski, E. Grabau, S. Nichol, and S. VandePol. 1982. Rapid evolution of RNA genomes. Science 215:1577-1585. [DOI] [PubMed] [Google Scholar]
- 31.Homma, M., and M. Ouchi. 1973. Trypsin action on the growth of Sendai virus in tissue culture cells. 3. Structural difference of Sendai viruses grown in eggs and tissue culture cells. J. Virol. 12:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ignarro, L. J. 2000. Introduction and overview, p. 3-19. In L. J. Ignarro (ed.), Nitric oxide: biology and pathobiology. Academic Press, San Diego, Calif.
- 33.Jaiswal, M., LaRusso, N. F., Burgart, L. J., and G. L. Gores. 2000. Inflammatory cytokines induce DNA damage and inhibit DNA repair in cholangiocarcinoma cells by a nitric oxide-dependent mechanism. Cancer Res. 60:184-190. [PubMed] [Google Scholar]
- 34.James, S. L. 1995. Role of nitric oxide in parasitic infections. Microbiol. Rev. 59:533-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juedes, M. J., and G. N. Wogan. 1996. Peroxynitrite-induced mutation spectra of pSP189 following replication in bacteria and in human cells. Mutat. Res. 349:51-61. [DOI] [PubMed] [Google Scholar]
- 36.Leider, J. M., P. Palese, and F. I. Smith. 1988. Determination of the mutation rate of a retrovirus. J. Virol. 62:3084-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nathan, C. 1997. Inducible nitric oxide synthase: what difference does it make? J. Clin. Investig. 100:2417-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nathan, C., and M. U. Shiloh. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 97:8841-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niedbala, W., X. Q. Wei, C. Campbell, D. Thomson, M. Komai-Koma, and F. Y. Liew. 2002. Nitric oxide preferentially induces type 1 T cell differentiation by selectively up-regulating IL-12 receptor β2 expression via cGMP. Proc. Natl. Acad. Sci. USA 99:16186-16191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oda, T., T. Akaike, T. Hamamoto, F. Suzuki, T. Hirano, and H. Maeda. 1989. Oxygen radicals in influenza-induced pathogenesis and treatment with pyran polymer-conjugated SOD. Science 244:974-976. [DOI] [PubMed] [Google Scholar]
- 41.Oda, T., K. Kojima, T. Akaike, S. Ijiri, A. Molla, and H. Maeda. 1990. Inactivation of chemotactic activity of C5a by the serratial 56-kilodalton protease. Infect. Immun. 58:1269-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohshima, H., and H. Bartsch. 1994. Chronic infections and inflammatory processes as cancer risk factors: possible role of nitric oxide in carcinogenesis. Mutat. Res. 305:253-264. [DOI] [PubMed] [Google Scholar]
- 43.Okamoto, T., T. Akaike, T. Nagano, S. Miyajima, M. Suga, M. Ando, K. Ichimori, and H. Maeda. 1997. Activation of human neutrophil procollagenase by nitrogen dioxide and peroxynitrite: a novel mechanism for procollagenase activation involving nitric oxide. Arch. Biochem. Biophys. 342:261-274. [DOI] [PubMed] [Google Scholar]
- 44.Petersen, J., M. Dandri, A. Burkle, L. Zhang, and C. E. Rogler. 1997. Increase in the frequency of hepadnavirus DNA integrations by oxidative DNA damage and inhibition of DNA repair. J. Virol. 71:5455-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radi, R., J. S. Beckman, K. M. Bush, and B. A. Freeman. 1991. Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J. Biol. Chem. 266:4244-4250. [PubMed] [Google Scholar]
- 46.Rubbo, H., V. Darley-Usmar, and B. A. Freeman. 1996. Nitric oxide regulation of tissue free radical injury. Chem. Res. Toxicol. 9:809-820. [DOI] [PubMed] [Google Scholar]
- 47.Sawa, T., T. Akaike, K. Ichimori, T. Akuta, K. Kaneko, H. Nakayama, D. J. Stuehr, and H. Maeda. 2003. Superoxide generation mediated by 8-nitroguanosine, a highly redox-active nucleic acid derivative. Biochem. Biophys. Res. Commun. 311:300-306. [DOI] [PubMed] [Google Scholar]
- 48.Sawa, T., T. Akaike, and H. Maeda. 2000. Tyrosine nitration by peroxynitrite formed from nitric oxide and superoxide generated by xanthine oxidase. J. Biol. Chem. 275:32467-32474. [DOI] [PubMed] [Google Scholar]
- 49.Scheid, A., and P. W. Choppin. 1976. Protease activation mutants of Sendai virus. Activation of biological properties by specific proteases. Virology 69:265-277. [DOI] [PubMed] [Google Scholar]
- 50.Singer, B., and H. Fraenkel-Conrat. 1969. Mutagenicity of alkyl and nitroso-alkyl compounds acting on tobacco mosaic virus and its RNA. Virology 39:395-399. [DOI] [PubMed] [Google Scholar]
- 51.Tsugita, A., and H. Fraenkel-Conrat. 1962. The composition of proteins of chemically evoked mutants of TMV RNA. J. Mol. Biol. 4:73-82. [DOI] [PubMed] [Google Scholar]
- 52.Umezawa, K., T. Akaike, S. Fujii, M. Suga, K. Setoguchi, A. Ozawa, and H. Maeda. 1997. Induction of nitric oxide synthesis and xanthine oxidase and their roles in the antimicrobial mechanism against Salmonella typhimurium infection in mice. Infect. Immun. 65:2932-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vuillaume, M. 1987. Reduced oxygen species, mutation, induction and cancer initiation. Mutat. Res. 186:43-72. [DOI] [PubMed] [Google Scholar]
- 54.Weitzman, S. A., and T. P. Stossel. 1981. Mutation caused by human phagocytes. Science 212:546-547. [DOI] [PubMed] [Google Scholar]
- 55.Witz, G. 1991. Active oxygen species as factors in multistage carcinogenesis. Proc. Soc. Exp. Biol. Med. 198:675-682. [DOI] [PubMed] [Google Scholar]
- 56.Yermilov, V., Rubio, J., and H. Ohshima. 1995. Formation of 8-nitroguanine in DNA treated with peroxynitrite in vitro and its rapid removal from DNA by depurination. FEBS Lett. 376:207-210. [DOI] [PubMed] [Google Scholar]
- 57.Yokoyama, W. M. 1991. Production of monoclonal antibody, p. 2.5.1-2.5.17. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley & Sons, Inc., New York, N.Y.
- 58.Yoshida, K., T. Akaike, T. Doi, K. Sato, S. Ijiri, M. Suga, M. Ando, and H. Maeda. 1993. Pronounced enhancement of NO-dependent antimicrobial action by an NO-oxidizing agent, imidazolineoxyl N-oxide. Infect. Immun. 61:3552-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zamboni, L., D. R. Mishell, Jr., J. H. Bell, and M. Baca. 1966. Fine structure of the human ovum in the pronuclear stage. J. Cell Biol. 30:579-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhuang, J. C., C. Lin, D. Lin, and G. N. Wogan. 1998. Mutagenesis associated with nitric oxide production in macrophages. Proc. Natl. Acad. Sci. USA 95:8286-8291. [DOI] [PMC free article] [PubMed] [Google Scholar]