SmD1, a conserved component of the small nuclear ribonucleoprotein complex, participates in splicing and in the partitioning of aberrant RNAs between RNA quality control and RNA silencing pathways.
Abstract
RNA quality control (RQC) eliminates aberrant RNAs based on their atypical structure, whereas posttranscriptional gene silencing (PTGS) eliminates both aberrant and functional RNAs through the sequence-specific action of short interfering RNAs (siRNAs). The Arabidopsis thaliana mutant smd1b was identified in a genetic screen for PTGS deficiency, revealing the involvement of SmD1, a component of the Smith (Sm) complex, in PTGS. The smd1a and smd1b single mutants are viable, but the smd1a smd1b double mutant is embryo-lethal, indicating that SmD1 function is essential. SmD1b resides in nucleoli and nucleoplasmic speckles, colocalizing with the splicing-related factor SR34. Consistent with this, the smd1b mutant exhibits intron retention at certain endogenous mRNAs. SmD1 binds to RNAs transcribed from silenced transgenes but not nonsilenced ones, indicating a direct role in PTGS. Yet, mutations in the RQC factors UPFRAMESHIFT3, EXORIBONUCLEASE2 (XRN2), XRN3, and XRN4 restore PTGS in smd1b, indicating that SmD1 is not essential for but rather facilitates PTGS. Moreover, the smd1b mtr4 double mutant is embryo-lethal, suggesting that SmD1 is essential for mRNA TRANSPORT REGULATOR4-dependent RQC. These results indicate that SmD1 interplays with splicing, RQC, and PTGS. We propose that SmD1 facilitates PTGS by protecting transgene-derived aberrant RNAs from degradation by RQC in the nucleus, allowing sufficient amounts to enter cytoplasmic siRNA bodies to activate PTGS.
INTRODUCTION
Posttranscriptional gene silencing (PTGS) controls a wide diversity of processes in eukaryotes through mRNA degradation mediated by small 21- to 22-nucleotide short interfering RNAs (siRNAs) (Baulcombe, 2004; Voinnet, 2009; Martínez de Alba et al., 2013). PTGS starts with the production of double-stranded RNAs (dsRNAs) and their processing into siRNAs. These small siRNAs trigger the sequence-specific cleavage of mRNAs containing complementary sequences. When PTGS is induced by viral or transgenic RNAs, siRNAs target the degradation of the invading RNAs but also of homologous endogenous mRNAs, if any. A forward genetic screen based on the transgenic Arabidopsis thaliana line L1, which carries a posttranscriptionally silent p35S:GUS sense transgene, identified ∼50 PTGS-deficient mutants that defined 12 independent SUPPRESSOR OF GENE SILENCING (SGS) loci. Mutations in these 12 SGS loci also impair PTGS in line 2a3, which carries a p35S:NIA2 sense transgene that triggers cosuppression of the endogenous genes NIA1 and NIA2. A forward genetic screen directly based on line 2a3 identified three additional loci (SGS13, SGS14, and SGS15) required for 2a3 but not L1 silencing (Jauvion et al., 2010). So far, SGS2/RDR6, SGS3, SGS4/AGO1, SGS5/HEN1, SGS6/MET1, SGS7/SDE5, SGS8/JMJ14, SGS9/HPR1, and SGS13/SDE3 have been characterized (Elmayan et al., 1998; Fagard et al., 2000; Mourrain et al., 2000; Morel et al., 2002; Boutet et al., 2003; Jauvion et al., 2010; Le Masson et al., 2012). During PTGS triggered by sense transgenes (S-PTGS), primary siRNAs are produced from an aberrant RNA (Parent et al., 2015), methylated at their 3′ end by the methyltransferase HEN1 (HUA ENCHANCER1) (Boutet et al., 2003; Li et al., 2005) before loading into AGO1 (ARGONAUTE1), which cleaves complementary target RNAs (Morel et al., 2002; Baumberger and Baulcombe, 2005). AGO1-mediated cleavage generates RNA fragments that escape degradation due to the protective activity of SGS3 and are transformed into dsRNA by RDR6 (RNA-DEPENDENT-RNA-POLYMERASE6; Mourrain et al., 2000). These dsRNA are processed into siRNA duplexes by DICER-LIKE4 to produce secondary siRNAs. These secondary siRNAs are also loaded onto AGO1, which cleaves complementary transgene mRNAs, resulting in an amplification loop that reinforces silencing. MET1 and JMJ14 encode a DNA methyltransferase and a histone demethylase, respectively, which likely play a role in remodeling chromatin to allow the transcription of transgene-derived aberrant RNAs that induce PTGS (Le Masson et al., 2012). Also, SDE5 and HPR1 encode RNA trafficking proteins, which likely play a role in bringing RNA molecules at the right place during PTGS (Hernandez-Pinzon et al., 2007; Jauvion et al., 2010; Yelina et al., 2010).
Components of RNA processing complexes that counteract PTGS also have been identified. Known endogenous PTGS suppressors include 5′→3′ EXORIBONUCLEASE2 (XRN2), XRN3, XRN4, and their regulator FIERY1 (Gazzani et al., 2004; Gy et al., 2007); exosome components HEN2, mRNA TRANSPORT REGULATOR4 (MTR4), RIBOSOMAL RNA PROCESSING4 (RRP4), RRP6L1, RRP41, RRP44a, and SUPERKILLER3 (Moreno et al., 2013; Lange et al., 2014; Yu et al., 2015); decapping components DECAPPING1 (DCP1), DCP2, and VARICOSE (Thran et al., 2012; Martínez de Alba et al., 2015); nonsense-mediated decay components UPFRAMESHIFT1 (UPF1) and UPF3 (Moreno et al., 2013); and 3′ end processing factors ENHANCED SILENCING PHENOTYPE1 (ESP1), ESP4, ESP5, CARBONE CATABOLITE REPRESSOR4a, and 3′→5′ POLY(A)-SPECIFIC RIBONUCLEASE (Herr et al., 2006; Moreno et al., 2013). This revealed the diversity of RNA regulation processes intertwined with siRNA-mediated PTGS in all types of compartments (nucleolus, nucleoplasm, and cytoplasm). Likely, a tug of war between RNA quality control and RNA silencing contributes to determine the final transcriptome of the cell by addressing aberrant RNAs to one or the other degradation pathway. However, cellular factors that influence the partitioning of aberrant RNAs to one or the other pathway remain unknown.
Here, we show that the PTGS-defective mutant sgs14 recovered from a genetic screen based on the p35S:NIA2 sense transgene carries a deletion of the SmD1b gene, which encodes one of the two orthologs of the yeast Sm domain-containing protein SmD1, a small nuclear ribonucleoprotein of the conserved Smith (Sm) complex (Wang and Brendel, 2004). The Sm group of proteins was named after Stephanie Smith, the first patient in which the systemic lupus erythematosus-associated anti-Sm autoimmune antibodies were identified. Sm proteins are highly conserved among protists, fungi, animals, and plants and can be classified in several groups. A first group comprises the canonical proteins SmB, SmD1, SmD2, SmD3, SmE, SmF, and SmG; a second group comprises related LSM proteins LSM1 to LSM8. SmB/D1/D2/D3/E/F/G form the core particles of the U1, U2, U4, and U5 spliceosomal ribonucleoproteins, while LSM2-8 is part of the U6 small nuclear ribonucleoprotein also involved in pre-mRNA splicing. LSM1-7 proteins form a different complex, which participates in mRNA decapping in cytoplasmic processing bodies (P-bodies). Additional components (up to LSM16) play various roles, including maturation of U3 small nucleolar RNA, participation in the U7 ribonucleoprotein involved in the maturation of histone mRNA, degradation of mRNA precursors in the nucleus, mRNA translational control, and formation of P-bodies (Golisz et al., 2013, and references therein). The Arabidopsis genome contains 42 Sm and LSM genes (Cao et al., 2011), among which very few have been characterized (Perea-Resa et al., 2012; Golisz et al., 2013). In particular, the role of Arabidopsis SmD1 identified here is not known, although its implication in splicing could be suspected based on the function of yeast SmD1 in this process (Zhang et al., 2001). As we report here, localization studies revealed that Arabidopsis SmD1b colocalizes with the splicing-related factor SR34 in nuclear speckles. Consistent with this, the smd1b mutation affects the splicing of several endogenous mRNAs. Arabidopsis SmD1b also binds to RNAs transcribed from silenced transgenes but not nonsilenced ones, indicating a connection between splicing and PTGS. Nevertheless, PTGS is restored in smd1b upf3, smd1b xrn2, smd1b xrn3, and smd1b xrn4 double mutants, indicating that SmD1b is not essential for PTGS. Moreover, smd1b mtr4 mutants are not viable, indicating that SmD1b also participates to RQC, at least MTR4-dependent RQC. Together, these results indicate that SmD1 influences splicing and the partitioning of aberrant RNAs between RNA quality control and RNA silencing pathways, revealing a broad role of SmD1 in the regulation of gene expression.
RESULTS
SGS14 Encodes an Ortholog of Yeast SmD1
The SGS14 locus is defined by a unique mutant allele, which was identified in a screen for PTGS-deficient mutants using the 2a3 line, which carries a transgene consisting of the NIA2 gene under the control of the 35S promoter (Elmayan et al., 1998). In addition to PTGS deficiency, this mutant exhibits developmental defects, including reduced stature, leaf serration, and early flowering (Figures 1A and 1B). The sgs14 mutation was mapped to a 164-kb interval on chromosome 4. Whole-genome sequencing revealed that fast-neutron mutagenesis had induced a deletion in this interval, removing six protein-coding genes (At4g02800, At4g02810, At4g02820, At4g02830, At4g02840, and At4g02850). Mutant lines harboring T-DNA insertions in the open reading frames of At4g02800, At4g02810, At4g02820, At4g02830, and At4g02850 did not exhibit developmental defects, suggesting that deletion of At4g02840 was responsible for the developmental defects of the sgs14 mutants. However, the only available mutant in this gene had an insertion upstream of the open reading frame and did not exhibit developmental defects. Therefore, we transformed the sgs14 mutant with a 6-kb genomic fragment carrying the At4g02840 gene. Because At4g02840 is one of the two Arabidopsis genes encoding a protein homologous to yeast SmD1 (Wang and Brendel, 2004), this 6-kb genomic fragment is referred to as pSmD1b:SmD1b in Figure 1C. At first, we transformed sgs14 mutant plants from which the 2a3 locus has been segregated away. Among 40 sgs14/pSmD1b:SmD1b transformants, 39 developed like wild-type plants with regards to stature, leaf shape, and flowering time (Figure 1C), indicating that the deletion of At4g02840 is responsible for the developmental defects of the sgs14 deletion mutant. However, because the deletion removed six adjacent genes, PTGS deficiency could be due to the deletion of any of the six genes and not related to the developmental defect observed. Thus, we also transformed sgs14 mutant plants carrying the 2a3 locus. Among 86 2a3/sgs14/pSmD1b:SmD1b transformants, 84 exhibited NIA2 cosuppression, i.e., they died within the first 2 weeks of growth, indicating that the deletion of At4g02840 is responsible for both developmental defects and PTGS-deficiency in the sgs14 mutant, hereafter referred to as smd1b.
Figure 1.
Developmental Defects of smd1 Mutants.
(A) Photographs of 20-d-old plants of wild-type Col, smd1a and smd1b single mutants, and plants homozygous for smd1a and heterozygous for smd1b. Note that plants homozygous for smd1b and heterozygous for smd1a, or homozygous for both smd1a and smd1b are not viable.
(B) Photographs of 30-d-old plants of the same genotype as in (A).
(C) Photographs of 20-d-old plants of wild-type Col, smd1b mutant, and smd1b/pSmD1b:SmD1b transformants.
(D) Photographs of 20-d-old plants of wild-type Col, smd1b mutant, and smd1b/pUBQ10:SmD1a and smd1b/pUBQ10:SmD1b transformants.
SmD1a and SmD1b Encode Redundant Proteins That Are Essential for Plant Viability
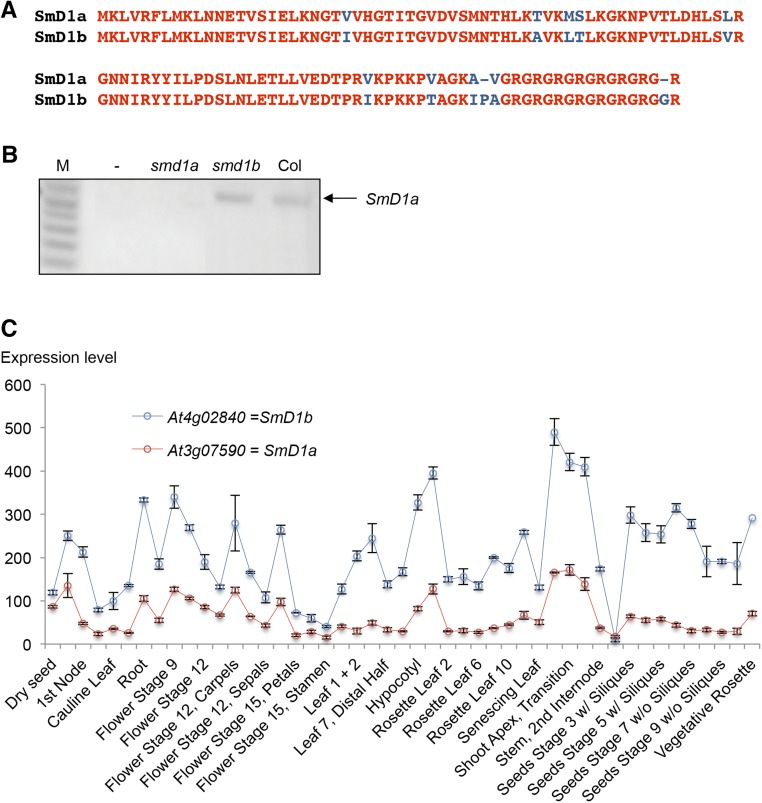
Yeast SmD1 has two orthologs in Arabidopsis, At3g07590 and At4g02840, which are referred to as SmD1a and SmD1b, respectively (Wang and Brendel, 2004). These two proteins only differ by 11 amino acids (Figure 2 A), suggesting that they could play redundant roles. Many T-DNA insertion lines exist around the SmD1a gene, but only SALK_024397 corresponds to an smd1a null allele (Figure 2B). Homozygous smd1a plants did not show any developmental defects when grown under standard laboratory conditions, raising questions about the functionality of this protein. However, both pUBQ10:SmD1a-GFP and pUBQ10:SmD1b-GFP constructs restored wild-type development and NIA2 PTGS when introduced in smd1b or 2a3/smd1b plants, respectively (Figure 1D), indicating that these two proteins have similar function. Analysis of expression arrays revealed that SmD1b mRNA accumulates at a higher level than SmD1a mRNA in wild-type plants (Figure 2C). Therefore, the absence of obvious developmental defects in the smd1a single mutant is likely due to the minor contribution of the SmD1a gene to the total amount of the SmD1 protein present in the cell. Supporting this hypothesis, smd1a/smd1a smd1b/SmD1b plants identified in the F2 progeny deriving from a cross between smd1a and smd1b single mutants were viable and exhibited developmental defects milder than those of the smd1b mutant (Figures 1A and 1B), whereas smd1a/SmD1a smd1b/smd1b or smd1a/smd1a smd1b/smd1b plants could not be identified, suggesting that SmD1 function is essential and that the minimum level of protein needed for development requires either one copy of SmD1b or two copies of SmD1a. Accordingly, siliques formed on smd1a/SmD1a smd1b/smd1b plants lacked 25% of the seeds, indicating that the smd1a/smd1a smd1b/smd1b double mutant is embryo-lethal.
Figure 2.
SmD1 Protein Sequence and SmD1 Expression Patterns.
(A) Alignment of SmD1a and SmD1b proteins. Conserved amino acids are indicated in red.
(B) RT-PCR detection of SmD1a mRNA in wild-type (Col), smd1b (sgs14), and smd1a (SALK_024397) plants. M, molecular markers.
(C) ATH1 array expression profiles of SmD1a and SmD1b genes. Expression data were retrieved from the Arabidopsis eFP Browser. The expression of each SmD1 gene is shown at various developmental stages and in different tissues. Normalization methods, the tissue, and the developmental stages of each sample as well as additional information can be found at http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi?.
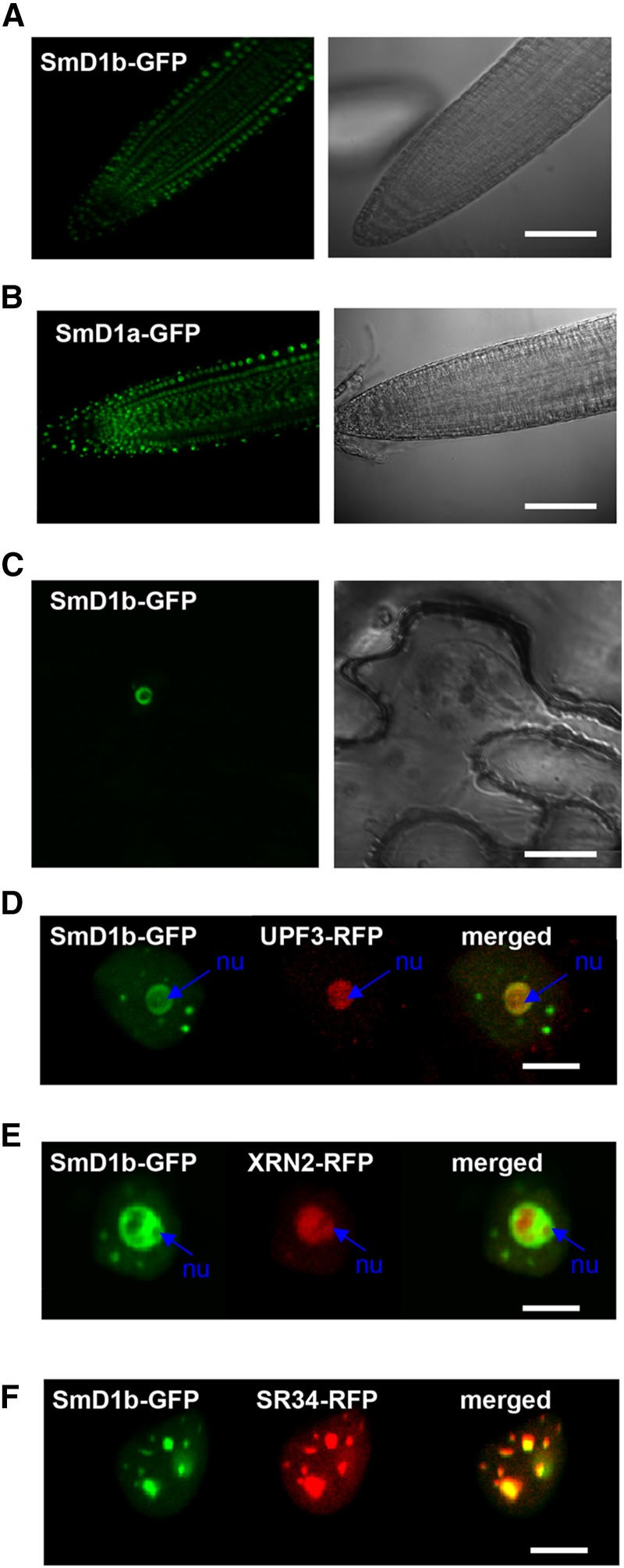
SmD1b Has Dual Localization in Nucleoli and Nuclear Speckles
We explored where SmD1a and SmD1b are expressed in the cell using transgenic smd1b mutants complemented with either the pUBQ10:SmD1a-GFP or pUBQ10:SmD1b-GFP construct. Confocal analysis revealed an exclusive nuclear localization of both fusion proteins (Figures 3A and 3B). Interestingly, two specific subnuclear localizations were observed. Indeed, SmD1b-GFP localized in both nucleoli and nucleoplasmic dots (Figure 3C). Nucleolar localization was confirmed by colocalization experiments using the nucleolar RQC factors UPF3 and XRN2 (Figures 3D and 3E). The nature of the nucleoplasmic dots was further analyzed using the splicing-related factor SR34, which resides in nucleoplasmic speckles (Lorković et al., 2008). Coinfiltration of pUBQ10:SmD1b-GFP and p35S:SR34-RFP indicated that SmD1b and SR34 colocalize in these speckles (Figure 3F).
Figure 3.
Subcellular Localization of SmD1 Proteins.
(A) and (B) Confocal images of Arabidopsis root expressing pUBQ10:SmD1b-GFP (A) or pUBQ10:SmD1a-GFP (B) reveal a nuclear localization.
(C) Confocal image of Nicotiana benthamiana leaf infiltrated with pUBQ10:SmD1b-GFP reveal a localization in nucleoli and nucleoplasmic speckles.
(D) and (E) Confocal images of N. benthamiana leaves infiltrated with pUBQ10:SmD1b-GFP and pUBQ10:UPF3-RFP (D) or pUBQ10:XRN2-RFP (E) reveal colocalization of the two proteins in the nucleolus (nu).
(F) Confocal image of N. benthamiana leaf infiltrated with pUBQ10:SmD1b-GFP and pUBQ10:SR34-RFP reveal a localization in nucleoplasmic speckles.
Bars = 100 µm in (A) and (B), 10 µm in (C), and 5 µm in (D) to (F).
The Splicing of Endogenous mRNAs Is Affected in smd1b Mutants
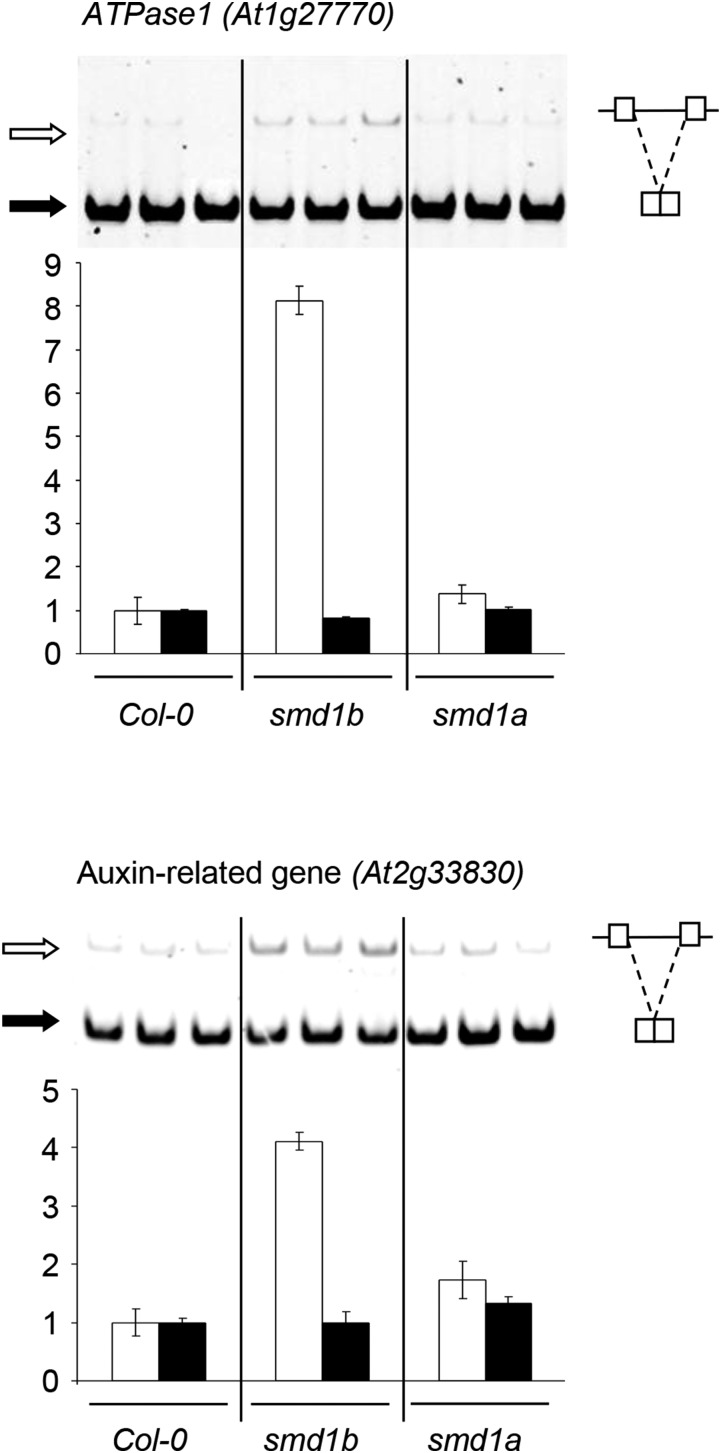
In yeast, the Sm proteins SmB, SmD1, and SmD3 make direct contact with the 5′ splice sites of pre-mRNA substrates and act by stabilizing RNA-RNA interactions between the 5′ end of the U1 small nuclear RNA and the 5′-splice sites (Zhang et al., 2001). The role of yeast SmD1 in pre-mRNA splicing and the colocalization of Arabidopsis SmD1b with the splicing-related factor SR34 in nuclear speckles therefore suggest that Arabidopsis SmD1b could play a role in splicing. To test this hypothesis, the expression of genes known to produce alternatively spliced transcripts (Simpson et al., 2008) was analyzed in the smd1a and smd1b mutants. The auxin-related gene At2g33830 and the ATPase-encoding gene At1g27770 each transcribe a major isoform resulting from full splicing and a minor isoform resulting from intron retention. A higher accumulation of the intron-containing isoform of these two genes was observed in the smd1b mutant compared with Col (Figure 4). No change in the isoform ratio was observed in the smd1a mutant, confirming that SmD1b plays a more important role in splicing regulation than SmD1a.
Figure 4.
Endogenous RNA Accumulation in smd1 Mutants.
RT-PCR and quantification of RNA isoforms of the ATPase1 gene At1g27770 and auxin-related gene At2g33830 on PAGE gels. Black and white arrows indicate spliced and unspliced RNA, respectively. Increased intron retention is observed in smd1b but not smd1a.
SmD1 Does Not Participate in the Endogenous Small RNA Repertoire
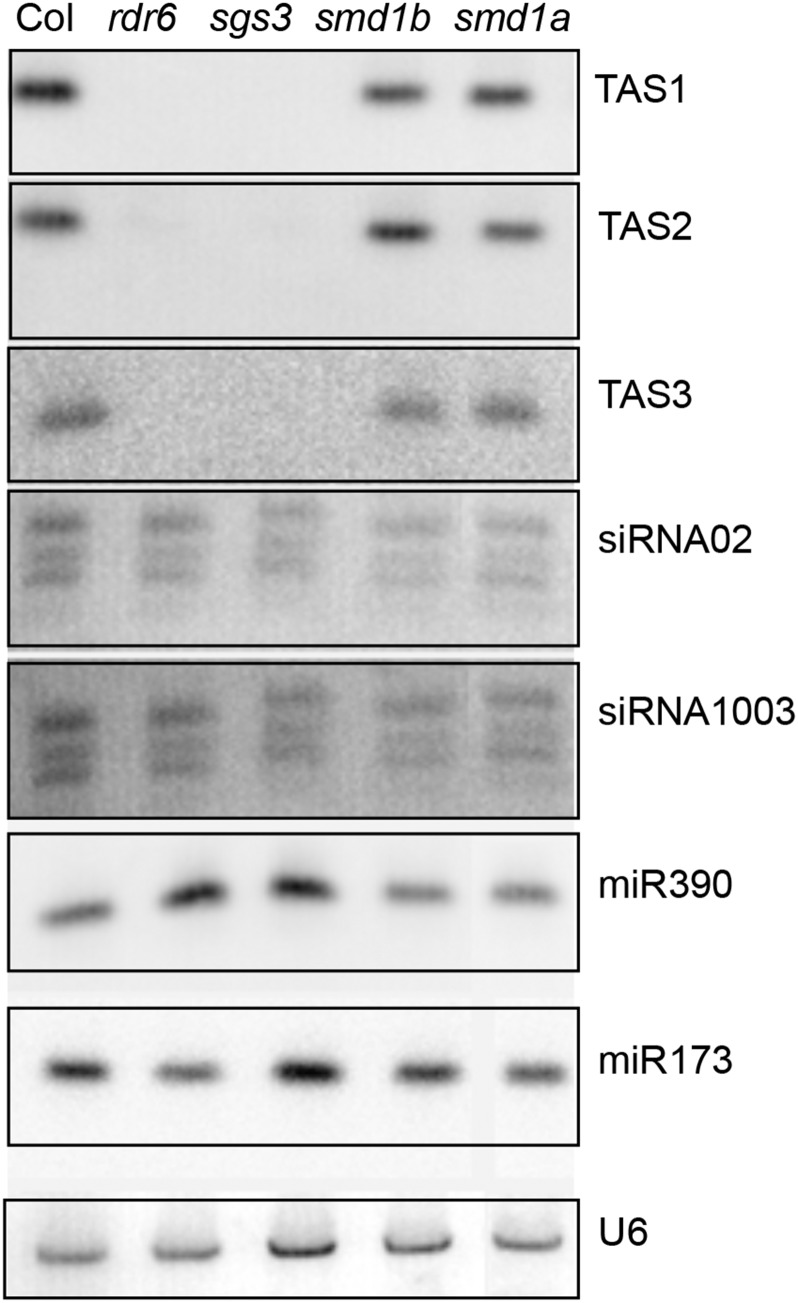
Because many of the PTGS-deficient mutants previously identified in our screen are impaired in components of the cellular machinery producing endogenous siRNAs (Elmayan et al., 1998; Fagard et al., 2000; Mourrain et al., 2000; Morel et al., 2002; Boutet et al., 2003; Jauvion et al., 2010; Le Masson et al., 2012), we examined the accumulation of representative endogenous small RNAs in smd1 mutants. The accumulation of microRNAs (miR173 and miR390), trans-acting siRNAs (TAS1, TAS2, and TAS3), and p4-siRNAs (siRNA02 and siRNA1003) was not affected in the smd1a or smd1b mutants (Figure 5), suggesting that SmD1 does not generally participate in the production of small RNAs but likely affects transgene PTGS at a different step.
Figure 5.
Endogenous Small RNA Accumulation in smd1 Mutants.
RNA gel blot analysis of representative endogenous small RNAs in smd1a and smd1b mutants. Wild-type Col and rdr6 and sgs3 mutants are used as controls. U6 snRNA hybridization served as loading controls for low molecular weight RNA gel blots.
The Effect of smd1b on Transgene PTGS Is Not Specific to Intron-Containing Transgenes but Rather Depends on the Strength of the Silencing Locus
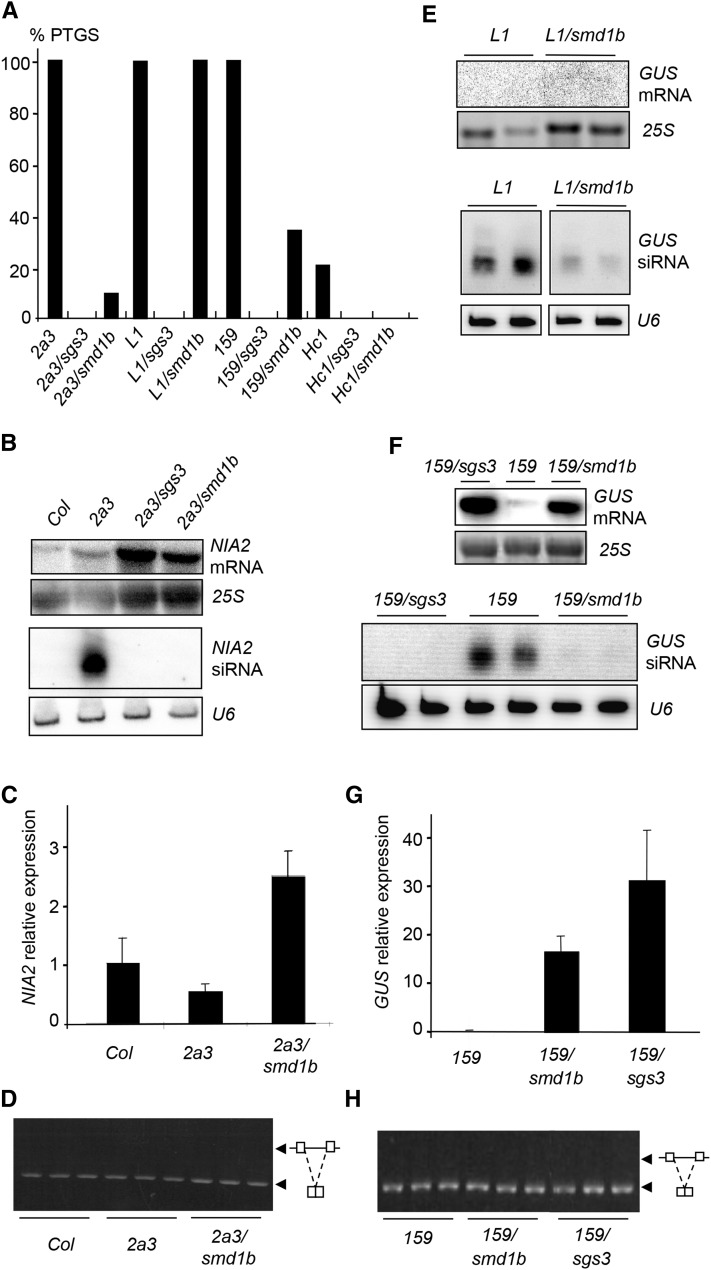
The genetic screen that allowed recovering the smd1b mutant is based on the 2a3 line, which carries a silenced 35S:NIA2 transgene. At each generation, 10% of 2a3/smd1b plants eventually trigger cosuppression of NIA2 (Figure 6A), indicating that the smd1b mutation does not protect against NIA2 cosuppression with 100% efficiency. To further characterize the effect of the smd1b mutation at the molecular level, high and low molecular weight RNAs were extracted from wild-type Col, 2a3 silenced plants, and 2a3/smd1b and 2a3/sgs3 mutants and hybridized with a NIA2 probe. 2a3/sgs3 mutants were used as a control because the sgs3 mutation completely abolishes NIA2 cosuppression triggered by the 2a3 locus (Mourrain et al., 2000). Whereas 2a3 plants accumulated NIA2 siRNAs, 2a3/smd1b mutants lacked NIA2 siRNAs, similar to 2a3/sgs3 mutants (Figure 6B). However, 2a3/smd1b mutants accumulated NIA2 mRNA at a lower level than 2a3/sgs3 mutants (Figures 6B and 6C), consistent with the incomplete erasure of PTGS by the smd1b mutation (Figure 6A).
Figure 6.
Transgene PTGS in smd1b Mutants.
(A) Percentages of silenced 2a3, L1, Hc1, and 159 plants in the indicated genotypes determined by quantitative GUS activity measurements (n = 96 plants for each genotype).
(B) RNA gel blot analyses of NIA mRNA and siRNAs in the indicated genotypes.
(C) RT-qPCR quantification of mature NIA2 mRNA in the indicated genotypes.
(D) Analysis of NIA2 RNA splicing by RT-PCR using primers spanning an intron.
(E) RNA gel blot analyses of L1 GUS mRNA and siRNAs in the indicated genotypes.
(F) RNA gel blot analyses of 159 GUS mRNA and siRNAs in the indicated genotypes.
(G) RT-qPCR quantification of mature GUS mRNA in the indicated genotypes.
(H) Analysis of GUS RNA splicing by RT-PCR using primers spanning the intron.
25S rRNA hybridization or ethidium bromide staining served as loading controls for high molecular weight RNA gel blots. U6 snRNA hybridization served as loading controls for low molecular weight RNA gel blots.
The smd1b mutant was not recovered in the PTGS genetic screen based on the L1 line, which carries a silenced p35S:GUS transgene, although the L1-based screen identified a much larger number of PTGS-deficient mutants than the 2a3-based screen. To test if smd1b has an effect on L1 PTGS, the L1 locus was introduced into smd1b by crossing. Unlike 2a3/smd1b plants, which escaped NIA2 cosuppression with 90% efficiency (Figure 6A), none of the L1/smd1b plants escaped GUS PTGS (Figure 6A). Indeed, L1/smd1b plants lacked GUS mRNA, similar to L1 controls (Figure 6E). Nevertheless, L1/smd1b plants accumulated GUS siRNAs at a lower level than L1 plants (Figure 6E), indicating that the smd1b mutation has an effect on GUS PTGS, although weaker than its effect on NIA2 PTGS. Whether the smd1a smd1b double mutation could abolish GUS PTGS in L1 could not be tested because of the lethality of this double mutant.
The L1 and 2a3 loci differ by many aspects (genomic insertion site of the transgene, sequence of the mature mRNA, and presence or absence of an intron within the pre-mRNA). To determine the basis of the different behavior of the smd1b mutation toward L1 and 2a3, additional transgene loci were tested, including intron-containing and intron-free p35S:GUS loci. At first, the 159 locus was introduced into the smd1b mutant by crossing. The 159 locus carries the same p35S:GUS transgene as L1 except for the presence of a plant intron within the GUS sequence (Vancanneyt et al., 1990). Like L1, line 159 exhibits high GUS activity at early stages of development, low GUS activity at later stages in 100% of the population (Figure 6A), and accumulates high levels of GUS siRNAs when PTGS is triggered (Figure 6F). However, the timing of silencing in line 159 is delayed compared with L1, indicating that the 159 locus is a weaker silencing inducer than the L1 locus. Sixty-nine percent of 159/smd1b plants escaped GUS PTGS and lacked GUS siRNAs, whereas 100% of 159/sgs3 plants escaped GUS PTGS (Figure 6A). Consistent with this, GUS activity and GUS mRNA levels in 159/smd1b plants were high compared with 159 controls, although lower than in a 159/sgs3 plants (Figures 6F and 6G), confirming that smd1b incompletely suppresses PTGS.
Because the 2a3 and 159 loci carry intron-containing transgenes, whereas the L1 locus carries an intron-free transgene, the processing of the 2a3 and 159 pre-mRNAs was further analyzed to determine if the smd1b mutation causes transgene splicing defects that could affect PTGS. Unlike endogenous genes that exhibit intron retention in smd1b (Figure 4), the 2a3 and 159 transgenes did not show detectable changes in their splicing patterns (Figures 6D and 6H), suggesting that the smd1b does not compromise transgene splicing. Thus, PTGS impairment in smd1b does not appear to result from perturbed transgene splicing, suggesting that SmD1 acts in PTGS independent of its role in splicing.
To test this hypothesis, we attempted to determine if smd1b could affect PTGS of an intron-free transgene. To this end, we used the Hc1 locus, which carries the very same p35S:GUS transgene as L1, but triggers GUS PTGS in only 20% of the population at each generation, whereas L1 triggers GUS PTGS with 100% efficiency (Elmayan et al., 1998; Gy et al., 2007; Martínez de Alba et al., 2011, 2015). None of the Hc1/smd1b plants triggered GUS PTGS (Figure 6A), indicating that the smd1b mutation affects both GUS and NIA2 PTGS. Altogether, these results indicate that the smd1b mutation abolishes PTGS of weak silencing lines (Hc1), reduces PTGS efficiency of medium-strength silencing lines (2a3 and 159), and only causes a reduction of siRNA accumulation in strong silencing lines (L1), which is insufficient to prevent PTGS.
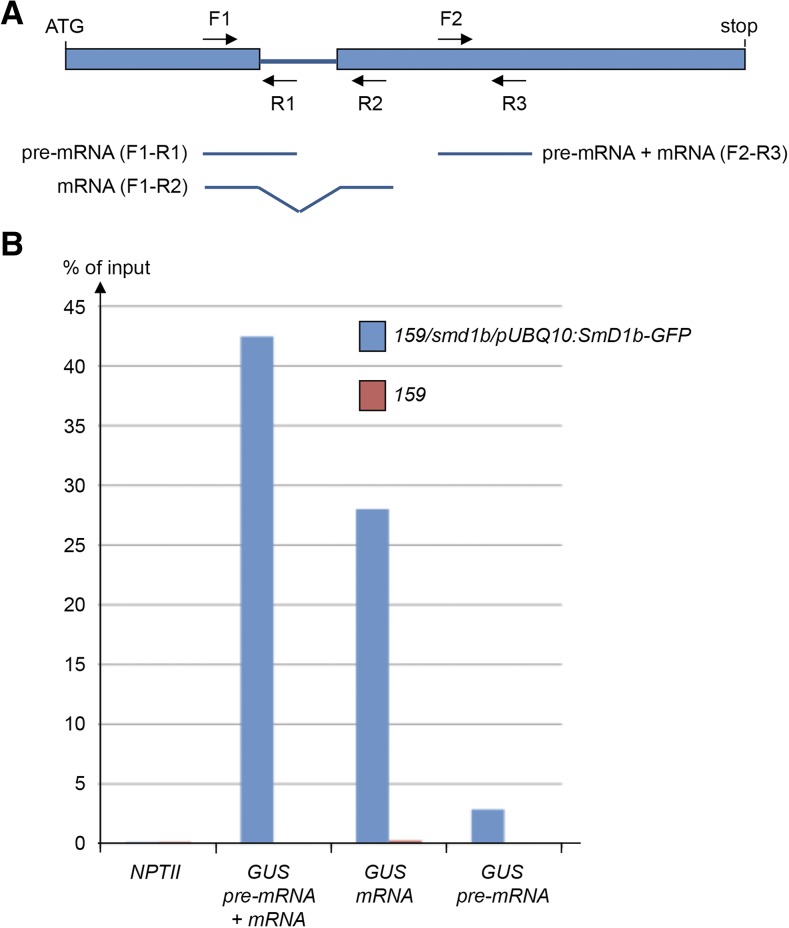
SmD1b Binds to Pre-mRNA and mRNA Produced from Silenced Transgenes
The absence of a detectable effect of smd1b on the splicing of transgene RNA suggests that SmD1 facilitates PTGS independently of its role in splicing. To determine if SmD1 directly interacts with transgene RNA, the 159/smd1b line was transformed with the pUBQ10:SmD1b-GFP construct, and complemented transformants that developed like wild-type plants and lacked GUS activity were selected. The nuclei extract (input) of the 159 line and one 159/smd1b/pUBQ10:Smd1b-GFP transgenic line that triggered GUS PTGS as efficiently as the 159 line were used for RNA immunoprecipitation using anti-GFP antibodies to detect transgene RNAs bound to SMD1-GFP. Specific pairs of primers that amplify GUS pre-mRNA, GUS mRNA, or both forms (Figure 7A) were used to perform reverse transcription followed by quantitative real-time PCR on the RNA immunoprecipitation and input samples. The NptII gene that is adjacent to the GUS gene on the T-DNA was used as a nonsilenced control. Results indicate a strong enrichment of both GUS pre-mRNA and GUS mRNA, but not NptII mRNA (Figure 7B), indicating that SmD1 binds to RNAs produced by silenced transgenes but not from neighboring nonsilenced transgenes, thus supporting a direct role for SmD1 in PTGS.
Figure 7.
Transgene RNA Immunoprecipitation in smd1b/pUBQ10:SmD1b-GFP Plants.
(A) Schematic representation of the GUS RNA. Primers F1 + R1 specifically amplify GUS pre-mRNA. Primers F1+ R2 specifically amplify GUS mRNA. Primers F2 + R3 amplify both GUS pre-mRNA and mRNA.
(B) RNA immunoprecipitation using GFP antibodies, followed by RT and PCR using GUS and NPTII primers.
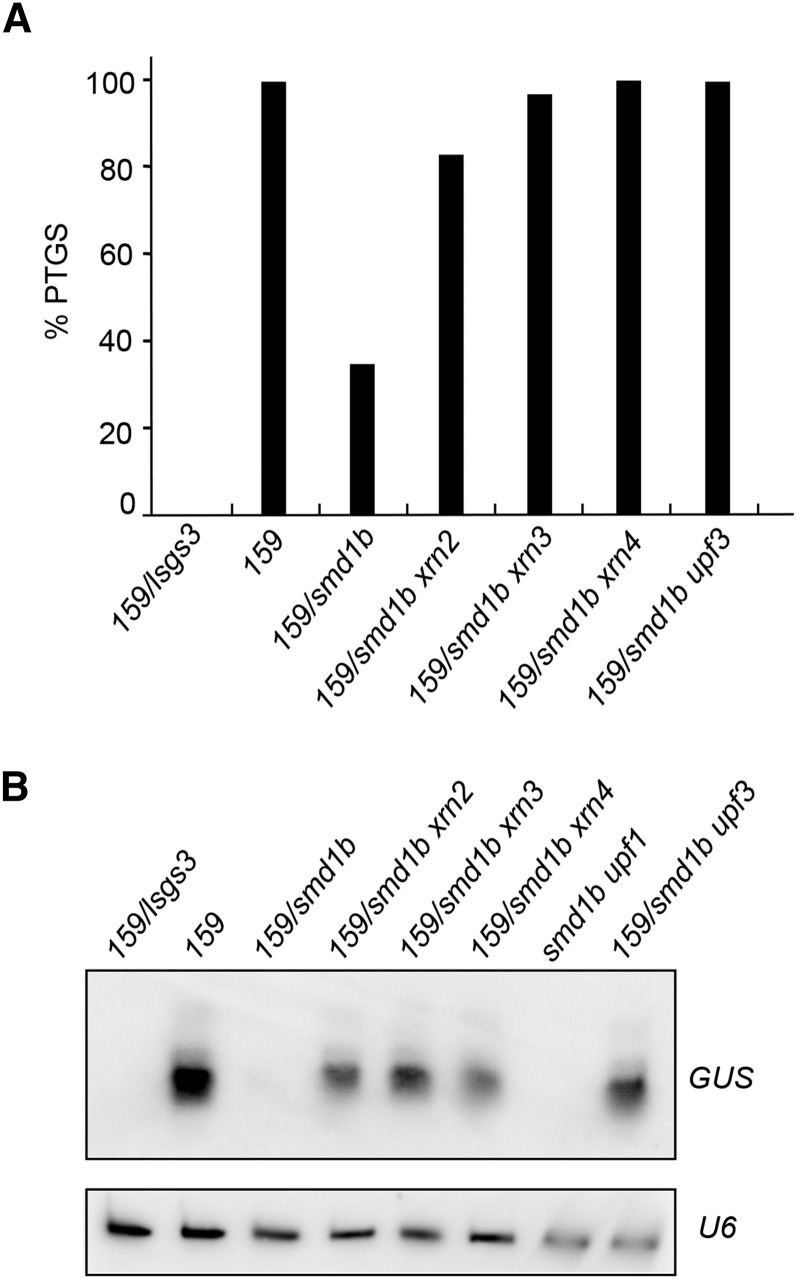
Mutations in UPF3, XRN2, XRN3, or XRN4 RQC Factors Restore PTGS in smd1b Mutants
The results presented above suggest that SmD1 is not essential for PTGS but rather facilitates PTGS, in particular at weak silencing loci. Because RQC limits PTGS by degrading part of the transgene aberrant RNAs that provoke the activation of PTGS (Gy et al., 2007; Moreno et al., 2013; Lange et al., 2014; Martínez de Alba et al., 2015; Parent et al., 2015), we propose that SmD1 facilitates PTGS by limiting the degradation of transgene aberrant RNAs by the RQC machinery, thus favoring their entry into cytoplasmic siRNA bodies where they can trigger PTGS. To test this hypothesis, we crossed 159/smd1b plants with various RQC-deficient mutants, including mtr4, upf1, upf3, xrn2, xrn3, and xrn4, to generate the corresponding double mutants. Remarkably, it was not possible to obtain viable smd1b mtr4 double mutants. The siliques that formed on smd1b/SmD1b mtr4/mtr4 or smd1b/smd1b mtr4/MTR4 plants lacked 25% of the seeds, indicating that the smd1b/smd1b mtr4/mtr4 double mutant is embryo-lethal. This result therefore suggests that SmD1 functions in the MTR4-dependent RQC pathway. In contrast, smd1b upf1, smd1b upf3, smd1b xrn2, smd1b xrn3, and smd1b xrn4 double mutants were viable. However, the upf1 and 159 loci were too close for obtaining plants carrying the 159 locus in a smd1b upf1 background, thus limiting PTGS analysis to smd1b upf3, smd1b xrn2, smd1b xrn3, and smd1b xrn4 double mutants. Whereas GUS PTGS occurred in 100% of 159 control plants and was reduced to 31% in 159/smd1b mutant plants, it was restored to 100, 80, 90, and 100% in smd1b upf3, smd1b xrn2, smd1b xrn3, and smd1b xrn4 plants, respectively (Figure 8A). Indeed, these plants accumulated GUS siRNA, whereas 159/smd1b lacked GUS siRNA (Figure 8B), indicating that PTGS degrades GUS mRNA in these double mutants. These results demonstrate that PTGS can efficiently occur in the smd1b mutant, thus confirming that SmD1 is not part of the core PTGS machinery. Rather, they support the hypothesis that SmD1 facilitates PTGS by protecting transgene aberrant RNAs from degradation by the RQC machinery in the nucleus, thus increasing the amount of transgene aberrant RNAs that succeed to reach the cytoplasm. There aberrant RNAs still have to escape from cytoplasmic RQC (including XRN4) to enter siRNA bodies where they are transformed into dsRNA by RDR6, which eventually activates PTGS (Gy et al., 2007; Moreno et al., 2013; Lange et al., 2014; Martínez de Alba et al., 2015; Parent et al., 2015).
Figure 8.
PTGS and siRNA Accumulation in Double Mutants Involving smd1b.
(A) Percentages of plants silenced by PTGS in the indicated genotypes determined by quantitative GUS activity measurements (n = 96 plants for each genotype).
(B) RNA gel blot analyses of GUS siRNAs in the indicated genotypes. U6 snRNA hybridization served as a loading control. Note that the sm1d upf1 double mutant does not contain the 159 locus because upf1 and 159 loci are very close.
DISCUSSION
It has been hypothesized that transgene loci that produce large amounts of aberrant RNAs activate the PTGS pathway because these RNAs exceed the degradation capacity of the RQC pathways (Gy et al., 2007; Moreno et al., 2013; Lange et al., 2014; Martínez de Alba et al., 2015; Parent et al., 2015). Although this hypothesis probably holds true, it is possible that, in addition, cellular components protect aberrant RNAs from degradation by RQC, thus contributing to addressing larger amounts of these RNAs to the cytoplasm where they can activate PTGS. Our results suggest that the Arabidopsis nuclear ribonucleoprotein SmD1 facilitates PTGS by protecting transgene aberrant RNAs from degradation by the RQC machinery in the nucleus, thus increasing the amount of transgene aberrant RNAs that succeed to enter siRNA bodies in the cytoplasm to eventually activate PTGS.
SmD1 is encoded by two closely related although differentially expressed genes. Both smd1a and smd1b single mutants are viable, but the double mutant cannot be obtained, indicating that SmD1 function is essential for the plant. Consistent with their expression level, SmD1b is more important than SmD1a because smd1b but not smd1a mutants exhibit developmental defects. Moreover, smd1a/smd1a smd1b/SmD1B plants exhibit developmental defects milder than those of the smd1b homozygous mutant, whereas smd1a/SmD1a smd1b/smd1b cannot be identified, indicating that either one copy of SmD1b or two copies of SmD1a is necessary for the plant to survive. Lastly, both pUBQ10:SmD1b and pUBQ10:SmD1a constructs restored wild-type development when introduced in smd1b, indicating that SmD1a and SmD1b proteins have redundant activity. In yeast, SmD1 participates in stabilizing RNA-RNA interactions between the 5′ end of the U1 small nuclear RNA and the 5′splice sites of pre-mRNA substrates (Zhang et al., 2001). We found that Arabidopsis SmD1 colocalizes with the splicing factor SR34 in nucleoplasmic speckles, suggesting that SmD1 also participates in splicing in plants. Even though no global defects in mRNA splicing were found, intron retention at certain endogenous genes was observed in the smd1b mutant. Given the redundant function of SmD1a and SmD1b, it is possible that the smd1a smd1b double mutant is lethal because of splicing defects in essential genes.
SmD1 also localizes in the nucleolus, suggesting additional functions besides splicing. Attempts to obtain double mutants between smd1b and mtr4, upf3, or xrn2 mutants impaired in RQC components that also reside in the nucleolus revealed that the smd1b mtr4 double mutant is embryo-lethal, whereas smd1b upf3 and smd1b xrn2 double mutants are viable. MTR4 acts in the nucleolar exosome (Lange et al., 2014). Whether SmD1 associates with the nucleolar exosome remains to be determined, but this result suggests that, in addition to splicing, SmD1 could play a role in MTR4-dependent RQC function in the nucleolus.
How does SmD1 affect PTGS, and is the effect of SmD1 on PTGS related to its splicing function? Although mutations in the putative splicing factor ESP3/PRP2 were identified in a screen for enhanced PTGS (Herr et al., 2006), very little is known about possible links between splicing and PTGS. Intron-free transgenes appear more prone to trigger PTGS than intron-containing transgenes (Christie et al., 2011), suggesting that the splicing process and/or the splicing machinery somehow protects RNA from entering into the PTGS pathway. The fact that the splicing factor SmD1 facilitates PTGS is somehow inconsistent with these two reports, asking whether SmD1 could affect PTGS indirectly through the deregulation of components of the PTGS machinery. However, PTGS occurs efficiently in smd1b upf3, smd1b xrn2, smd1b xrn3, and smd1b xrn4 double mutants, indicating that the smd1b mutation does not compromise the splicing of a component of the PTGS machinery. Moreover, no detectable effect of the smd1b mutation on the splicing of transgene RNA was observed, suggesting that SmD1 facilitates PTGS through a mechanism that is independent of its role in splicing. Supporting this hypothesis, we found that the smd1b mutation affects PTGS triggered by both intron-containing and intron-free transgenes. Finally, we found that SmD1b binds to both pre-mRNA and mRNA produced by silenced transgenes but not by nonsilenced transgenes, strongly suggesting a direct role in facilitating PTGS independent of splicing.
A facilitating role rather than an essential role in PTGS is supported by the fact that the smd1b mutation does not prevent transgene PTGS triggered by the strong inducing line L1 (which triggers PTGS with 100% efficiency), reduces PTGS triggered by lines 159 and 2a3 (which also trigger PTGS with 100% efficiency but at a slower rate than L1), and abolishes PTGS triggered by the weak inducing line Hc1 (which only triggers PTGS with 20% efficiency). Although we cannot exclude that the smd1a smd1b double mutation could completely abolish PTGS in L1, 159, and 2a3 lines, these results suggest that SmD1 facilitates PTGS triggered by weak inducers, but is dispensable for PTGS triggered by strong inducers. We propose that SmD1 participates in the PTGS of weak inducers by limiting the degradation of transgene aberrant RNAs by RQC in the nucleus, thus facilitating their addressing to the cytoplasm where they need to enter into siRNA bodies to activate PTGS. If this hypothesis is correct, SmD1 should not be part of the core PTGS machinery, and PTGS should still operate in smd1b mutants if large amounts of transgene aberrant RNAs are produced or if RQC is compromised. This is exactly what happens in the strong silencing line L1. PTGS likely occurs in L1/smd1b plants because L1 produces very high amounts of transgene aberrant RNAs, which exceed the capacity of the RQC pathways. Even if part of the transgene aberrant RNAs produced by L1 is degraded by RQC pathways, the amount that remains is probably sufficient to enter the PTGS pathway without the requirement of SmD1. Also consistent with our hypothesis, we observed that PTGS of line 159, which was strongly reduced in smd1b, was restored to wild-type levels in smd1b upf3, smd1b xrn2, smd1b xrn3, and smd1b xrn4 double mutants, confirming that the smd1b mutation does not impair the functioning of the PTGS machinery. Rather, transgene aberrant RNAs, which are less abundant in 159 than in L1, likely are more efficiently degraded by nuclear RQC in the absence of SmD1, thus limiting the triggering of PTGS. Only in double mutants between smd1b and either upf3, xrn2, xrn3, or xrn4, the impairment of one or the other RQC component limits the degradation of transgene aberrant RNAs, allowing a sufficient amount to reach siRNA bodies in the cytoplasm to trigger PTGS.
To summarize, the impairment of PTGS in smd1b and the reestablishment of PTGS in double mutants between smd1b and several RQC-deficient mutants suggest that, in addition to its role in splicing, SmD1 facilitates PTGS by limiting the degradation of transgene aberrant RNAs by nuclear RQC, revealing new roles for the SmD1 splicing regulator in RQC and PTGS. Because the role of SmD1 in splicing involves stabilizing weak RNA-RNA interactions between U1 small nuclear RNA and pre-mRNA splicing substrates (Zhang et al., 2001), it is possible that the role of SmD1 in PTGS also involves the stabilization of weak RNA-RNA interactions. In the case of PTGS, RNA-RNA interactions that need to be stabilized could involve secondary structures within aberrant RNAs, which need to be protected to prevent degradation by RQC components and favor RNA export to the cytoplasm.
METHODS
Plant Material and Growth Conditions
All Arabidopsis thaliana plants are in the Columbia accession. Transgenic lines 2a3, L1, and Hc1 and mutants mtr4-2, sgs3-1, upf1-6, upf3-3, xrn2-2, xrn3-3, and xrn4-5 were previously described (Elmayan et al., 1998; Mourrain et al., 2000; Gy et al., 2007; Moreno et al., 2013; Lange et al., 2014). The smd1a T-DNA insertion mutant SALK_024397 was obtained from NASC (Alonso et al., 2003). Line 159 was produced during this study by screening lines undergoing PTGS among the homozygous progeny of Arabidopsis transformants carrying the same p35S:GUS transgene as in lines L1 and Hc1 except for the addition of a plant intron (Vancanneyt et al., 1990). Plants were grown on Bouturage media (Duchefa) in standard long-day conditions (16 h light, 8 h dark at 20 to 22°C), transferred to soil after 2 weeks, and grown in controlled growth chambers in standard long-day conditions.
Plasmid Constructs
The pSmD1b:SmD1b construct was generated as follows: a 2943-bp genomic fragment starting 1 kb upstream the ATG of At4g02840 and ending 500 bp downstream its stop codon was amplified with Phusion High-fidelity DNA polymerase (Thermo) using primers SmD1b-1 and SmD1b-2, which carry HindIII and EcoRI sites at their ends, respectively (Supplemental Table 1). After cloning in TOPO blunt-end vector (Life Technologies) and verification by sequencing, the HindIII-EcoRI insert was subcloned in the binary pBINplus vector (van Engelen et al., 1995).
The pUBQ10:SmD1a, pUBQ10:SmD1b, pUBQ10:SmD1a-GFP, and pUBQ10:SmD1b-GFP constructs were made using Gateway technology (Invitrogen) as follows. For SmD1b, a 1621-bp genomic fragment starting 96 bp upstream the ATG of At4g02840 and ending at the stop codon was amplified with Phusion High-Fidelity DNA polymerase (Thermo) using primers SmD1b-3 and SmD1b-4 (Supplemental Table 1). After recombination into pENTR/D vector through the Gateway BP recombinase reaction (Invitrogen) and verification by sequencing, final recombination into pUB-DEST and pUBC-GFP through the Gateway LR recombinase reaction (Invitrogen) created pUBQ10:SmD1b and pUBQ10:SmD1b-GFP, respectively. For SmD1a, a 863-bp genomic fragment starting 53 bp upstream the ATG of At3g07590 and ending at the stop codon was amplified with Phusion High-Fidelity DNA polymerase (Thermo) using primers SmD1a-1 and SmD1a-2 (Supplemental Table 1). After recombination into pDONR-207 vector through the gateway BP recombinase reaction (Invitrogen) and verification by sequencing, final recombination into pUB-DEST and pUBC-GFP through the Gateway LR recombinase reaction (Invitrogen) created pUBQ10:SmD1a and pUBQ10:SmD1a-GFP, respectively.
The p35S:SR34-RFP, p35S:UPF3-RFP, and p35S:XRN2-RFP constructs have been described previously (Lorković et al., 2008; Moreno et al., 2013).
Arabidopsis Transformation and Nicotiana benthamiana Agroinfiltration
Agrobacterium strains carrying plasmids of interest were grown overnight at 28°C in 3 mL Luria-Bertani medium containing the appropriate antibiotics to a final OD600 between 1 and 2. For Arabidopsis transformation, the bacteria were pelleted and resuspended in 300 mL of infiltration medium (5% sucrose, 10 mM MgCl2, and 0.015% Silwet L-77) to a final OD600 of 1, which was used for floral dipping. For N. benthamiana agroinfiltration, the bacteria were pelleted and resuspended in 1 mL of infiltration medium (10 mM MgCl2, 10 mM MES, pH 5.2, and 150 mM acetosyringone) to a final OD600 of 0.1. The solution containing the bacteria was injected into the abaxial side leaves using a 1-mL syringe and samples were observed in a confocal microscope 3 d after infiltration.
Imaging and Image Analysis
After agroinfiltration, fluorescent cells were imaged by confocal microscopy (Leica TCS SP2; Leica Microsystems) with excitation at 488 nm and fluorescence emission signal between 495 and 530 nm for GFP fusions, and excitation at 543 nm and emission signal between 555 and 620 nm for DsRed or RFP fusions. The Leica confocal software was used for image acquisition and for the quantification of fluorescence profiles. Sequential scans were performed when necessary. Spectral profiles were calculated for five cells. Data processing was performed using ImageJ (http://rsbweb.nih.gov/ij/).
RNA Extraction and RNA Gel Blot Analysis
For RNA gel blot analyses, frozen tissue was homogenized in a buffer containing 0.1 M NaCl, 2% SDS, 50 mM Tris-HCl, pH 9.0, 10 mM EDTA, pH 8.0, and 20 mM β-mercaptoethanol and RNAs were extracted two times with phenol and recovered by ethanol precipitation. To obtain high molecular weight RNA fraction, resuspended RNAs were precipitated overnight in 2 M LiCl at 4°C and recovered by centrifugation. For low molecular weight RNA analysis, total RNA was separated on a 15% denaturing PAGE gel, stained with ethidium bromide, and transferred to nylon membrane (HybondNX; Amersham). Low molecular weight RNA and U6 hybridizations were at 50°C with hybridization buffer containing 5× SSC, 20 mM Na2HPO4, pH 7.2, 7% SDS, 2× Denhardt’s solution, and denatured sheared salmon sperm DNA (Invitrogen). High molecular weight RNA hybridization was at 37°C in PerfectHyb Plus buffer (Sigma-Aldrich). Blots were hybridized with a radioactively labeled random-primed DNA probes for GUS mRNA and GUS siRNAs and an end-labeled oligonucleotide probe for U6 detection.
RT-PCR Analysis
Total RNA was prepared from roots and plantlets at different developmental stages using the Qiagen RNeasy plant mini kit. The DNase treatment was performed according to the manufacturer’s protocols. For reverse transcription with SuperScript II (Invitrogen), 2.5 μg of total DNase-treated RNA was used. One microliter of the resulting cDNA solution was used for RT-PCR or RT-qPCR analyses. The latter was done using standard protocols and a complete list of RT-qPCR primers is available in Supplemental Table 1. Each cDNA sample was precisely calibrated and verified for two constitutive genes, AT1G13320 and AT4G26410 (Czechowski et al., 2005). For RT-PCR, the amplification was performed as follows: one cycle of 4 min at 98°C, 26 cycles of 30 s at 98°C, 30 s at 59°C, and 1 min at 72°C. The products were separated on a 7.5% polyacrylamide gel stained with SyBr green (Invitrogen) and revealed by Pharos Imager (Bio-Rad). Band profiles were quantified using ImageJ (http://rsbweb.nih.gov/ij/). RT-qPCR was performed using a Roche Light Cycler 480 standard protocol (40 cycles, 60°C annealing).
RNA Immunoprecipitation
Eleven-day-old plants grown in Petri dishes were irradiated three times with UV using a CL-508 cross-linker (Uvitec) at 0.400 J/cm2. Briefly, fixed material was ground in liquid nitrogen and homogenized and nuclei isolated and lysed according to Gendrel et al. (2005).RNA immunoprecipitation was basically performed as described by Carlotto et al. (2016). The nuclei extract (input) was used for the immunoprecipitation performed by the Direct ChIP Protocol of the Diagenode IP-Star SX-86 Compact robot, using 50 μL of Dynabeads-Protein A (Novex 10008D; Life Technologies) and anti-GFP antibodies (632381; Clontech). Beads were washed twice for 5 min at 4°C with wash buffer 1 (150 mM NaCl, 1% Triton, 0.5% Nonidet P-40, 1 mM EDTA, and 20 mM Tris-HCl, pH7.5) and twice with wash buffer 2 (20 mM Tris-HCl, pH 8) instead of the Diagenode assigned buffers and finally resuspended in 100 μL Proteinase K buffer (100 mM Tris-HCl, pH 7.4, 50 mM NaCl, and 10 mM EDTA). After Proteinase K (AM2546; Ambion) treatment, beads were removed with a magneto, and the supernatants were transferred to a 2-mL tube. Each RNA sample was extracted from 800 μL (8 IP-Star tubes) of RNA immunoprecipitation product using 1 mL of TriReagent (Sigma-Aldrich T9424) as indicated by the manufacturer. Eighty microliters of nuclei extracts was used for input RNA extraction. The immunoprecipitation and input samples were treated with DNase, and random hexamers were used for subsequent RT. Quantitative real-time PCR reactions were performed using specific primers. Results were expressed as a percentage of cDNA detected after immunoprecipitation, taking the input sample as 100%.
GUS Extraction and Activity Quantification
GUS protein was extracted and GUS activity was quantified as described before (Gy et al., 2007) from cauline leaves of flowering plants by measuring the quantity of 4-methylumbelliferone product generated from the substrate 4-methylumbelliferyl-β-d-glucuronide (Duchefa) on a fluorometer (Fluoroscan II; Thermo Scientific).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: SmD1b (At4g02840) and SmD1a (At3g07590).
Supplemental Data
Supplemental Figure 1. Original blot for Figure 6E.
Supplemental Table 1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank T. Elmayan for helpful discussions and P. Grillot, H. Ferry, and P. Marechal for plant care. This work was supported by the Agence Nationale de la Recherche ANR-10-BLAN-1707 (to H.V.) and ANR-10-LABX-40 (to M.D.C. and H.V.).
AUTHOR CONTRIBUTIONS
H.V. and M.D.C. designed the experiments. All authors contributed to the production and analysis of the results. H.V. wrote the article with contribution from all the other authors.
Glossary
- PTGS
posttranscriptional gene silencing
- siRNA
short interfering RNA
- dsRNA
double-stranded RNA
- RQC
RNA quality control
Footnotes
Articles can be viewed online without a subscription.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. (2004). RNA silencing in plants. Nature 431: 356–363. [DOI] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102: 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet S., Vazquez F., Liu J., Béclin C., Fagard M., Gratias A., Morel J.B., Crété P., Chen X., Vaucheret H. (2003). Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13: 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Shi F., Liu X., Jia J., Zeng J., Huang G. (2011). Genome-wide identification and evolutionary analysis of Arabidopsis sm genes family. J. Biomol. Struct. Dyn. 28: 535–544. [DOI] [PubMed] [Google Scholar]
- Carlotto N., Wirth S., Furman N., Ferreyra Solari N., Ariel F., Crespi M., Kobayashi K. (2016). The chloroplastic DEVH-box RNA helicase INCREASED SIZE EXCLUSION LIMIT 2 involved in plasmodesmata regulation is required for group II intron splicing. Plant Cell Environ. 39: 165–173. [DOI] [PubMed] [Google Scholar]
- Christie M., Croft L.J., Carroll B.J. (2011). Intron splicing suppresses RNA silencing in Arabidopsis. Plant J. 68: 159–167. [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan T., Balzergue S., Béon F., Bourdon V., Daubremet J., Guénet Y., Mourrain P., Palauqui J.C., Vernhettes S., Vialle T., Wostrikoff K., Vaucheret H. (1998). Arabidopsis mutants impaired in cosuppression. Plant Cell 10: 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M., Boutet S., Morel J.B., Bellini C., Vaucheret H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97: 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani S., Lawrenson T., Woodward C., Headon D., Sablowski R. (2004). A link between mRNA turnover and RNA interference in Arabidopsis. Science 306: 1046–1048. [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Golisz A., Sikorski P.J., Kruszka K., Kufel J. (2013). Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res. 41: 6232–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gy I., Gasciolli V., Lauressergues D., Morel J.B., Gombert J., Proux F., Proux C., Vaucheret H., Mallory A.C. (2007). Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19: 3451–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Pinzon I., Yelina N.E., Schwach F., Studholme D.J., Baulcombe D., Dalmay T. (2007). SDE5, the putative homologue of a human mRNA export factor, is required for transgene silencing and accumulation of trans-acting endogenous siRNA. Plant J. 50: 140–148. [DOI] [PubMed] [Google Scholar]
- Herr A.J., Molnàr A., Jones A., Baulcombe D.C. (2006). Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 14994–15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauvion V., Elmayan T., Vaucheret H. (2010). The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell 22: 2697–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H., et al. (2014). The RNA helicases AtMTR4 and HEN2 target specific subsets of nuclear transcripts for degradation by the nuclear exosome in Arabidopsis thaliana. PLoS Genet. 10: e1004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Masson I., Jauvion V., Bouteiller N., Rivard M., Elmayan T., Vaucheret H. (2012). Mutations in the Arabidopsis H3K4me2/3 demethylase JMJ14 suppress posttranscriptional gene silencing by decreasing transgene transcription. Plant Cell 24: 3603–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang Z., Yu B., Liu J., Chen X. (2005). Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15: 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković Z.J., Hilscher J., Barta A. (2008). Co-localisation studies of Arabidopsis SR splicing factors reveal different types of speckles in plant cell nuclei. Exp. Cell Res. 314: 3175–3186. [DOI] [PubMed] [Google Scholar]
- Martínez de Alba A.E., Elvira-Matelot E., Vaucheret H. (2013). Gene silencing in plants: a diversity of pathways. Biochim. Biophys. Acta 1829: 1300–1308. [DOI] [PubMed] [Google Scholar]
- Martínez de Alba A.E., Jauvion V., Mallory A.C., Bouteiller N., Vaucheret H. (2011). The miRNA pathway limits AGO1 availability during siRNA-mediated PTGS defense against exogenous RNA. Nucleic Acids Res. 39: 9339–9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez de Alba A.E., Moreno A.B., Gabriel M., Mallory A.C., Christ A., Bounon R., Balzergue S., Aubourg S., Gautheret D., Crespi M.D., Vaucheret H., Maizel A. (2015). In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 43: 2902–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel J.B., Godon C., Mourrain P., Béclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno A.B., Martínez de Alba A.E., Bardou F., Crespi M.D., Vaucheret H., Maizel A., Mallory A.C. (2013). Cytoplasmic and nuclear quality control and turnover of single-stranded RNA modulate post-transcriptional gene silencing in plants. Nucleic Acids Res. 41: 4699–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542. [DOI] [PubMed] [Google Scholar]
- Parent J.S., Jauvion V., Bouche N., Beclin C., Hachet M., Zytnicki M., Vaucheret H. (2015). Post-transcriptional gene silencing triggered by sense transgenes involves uncapped antisense RNA and differs from silencing intentionally triggered by antisense transgenes. Nucleic Acids Res. 43: 8464–8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Resa C., Hernández-Verdeja T., López-Cobollo R., del Mar Castellano M., Salinas J. (2012). LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell 24: 4930–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C.G., Fuller J., Maronova M., Kalyna M., Davidson D., McNicol J., Barta A., Brown J.W. (2008). Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant J. 53: 1035–1048. [DOI] [PubMed] [Google Scholar]
- Thran M., Link K., Sonnewald U. (2012). The Arabidopsis DCP2 gene is required for proper mRNA turnover and prevents transgene silencing in Arabidopsis. Plant J. 72: 368–377. [DOI] [PubMed] [Google Scholar]
- Vancanneyt G., Schmidt R., O’Connor-Sanchez A., Willmitzer L., Rocha-Sosa M. (1990). Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol. Gen. Genet. 220: 245–250. [DOI] [PubMed] [Google Scholar]
- van Engelen F.A., Molthoff J.W., Conner A.J., Nap J.P., Pereira A., Stiekema W.J. (1995). pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res. 4: 288–290. [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687. [DOI] [PubMed] [Google Scholar]
- Wang B.B., Brendel V. (2004). The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 5: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina N.E., Smith L.M., Jones A.M., Patel K., Kelly K.A., Baulcombe D.C. (2010). Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc. Natl. Acad. Sci. USA 107: 13948–13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A., Saudemont B., Bouteiller N., Elvira-Matelot E., Lepère G., Parent J.S., Morel J.B., Cao J., Elmayan T., Vaucheret H. (2015). Second-site mutagenesis of a hypomorphic argonaute1 allele identifies SUPERKILLER3 as an endogenous suppressor of transgene posttranscriptional gene silencing. Plant Physiol. 169: 1266–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Abovich N., Rosbash M. (2001). A biochemical function for the Sm complex. Mol. Cell 7: 319–329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.