Abstract
Although most Salmonella serovars are able to infect a range of animal hosts, some have acquired the ability to cause systemic infections of specific hosts. For example, Salmonella enterica serovar Choleraesuis is primarily associated with systemic infection in swine. Adherence to host epithelial cells is considered a prerequisite for initial infection, and fimbrial appendages on the outer membrane of the bacteria are implicated in this process. Although type 1 fimbriae encoded by the fim gene cluster are commonly found in Salmonella serovars, it is not known whether S. Choleraesuis produces this fimbrial type and if and how fimbriae are involved in pathogenesis. In the present study, we demonstrated that only four out of 120 S. Choleraesuis isolates from pigs with salmonellosis produced type 1 fimbriae as assayed by the yeast agglutination test and electron microscopy. One of the 116 non-type 1 fimbria-producing isolates was transformed with plasmids carrying different fim genes from S. Typhimurium LB5010, a type 1 fimbria-producing strain. Our results indicate that non-type 1 fimbria-producing S. Choleraesuis required only an intact fimH to regain the ability to produce fimbrial appendages. Sequence comparison revealed six amino acid variations between the FimH of the non-type 1 fimbria-producing S. Choleraesuis isolates and those of the type 1 fimbria-producing S. Choleraesuis isolates. S. Choleraesuis that produced type 1 fimbriae contained FimH with an amino acid sequence identical to that of S. Typhimurium LB5010. Site-directed mutagenesis leading to the replacement of the non-conserved residues revealed that a change from glycine to valine at position of 63 (G63V) resulted in a non-type 1 fimbria-producing S. Choleraesuis being able to express type 1 fimbriae on its outer membrane. It is possible that this particular amino acid change prevents this polypeptide from proper interaction with other Fim subunits required for assembly of an intact type 1 fimbrial shaft in S. Choleraesuis; however, it remains to be determined if and how the absence of type 1 fimbriae production is related to the systemic infection of the swine host by S. Choleraesuis.
Introduction
The Salmonella genus is composed of Salmonella enterica and Salmonella bongori species, which comprise six subspecies (I, II, IIa, IIb, IV, and V) subdivided into about 2800 serovars [1]. Most Salmonella serovars that are pathogenic to human and animals belong to the species Salmonella enterica. Many serovars infect a broad range of animal hosts and cause gastroenteritis, but some serovars are highly virulent in specific hosts [2]. For example, Salmonella enterica serovar Choleraesuis is primarily associated with systemic invasive infection in swine, but may sometimes infect other animals, including humans [3].
Fimbriae are hairlike appendages on the outer membrane of many Enterobacteriaceae and are involved in adherence to host epithelial cells, a prerequisite for infection. Although at least 13 gene clusters within Salmonella may have the potential to produce fimbrial appendages, type 1 fimbriae are the most commonly found type and are easily detected in vitro [4, 5]. Some fimbrial proteins can only be detected by flow cytometry from those samples that have been recovered 8 h after infection of bovine ligated ileal loops with S. Typhimurium [6]. The phenotypic expression of type 1 fimbriae is primarily a result of cooperation among several Fim proteins and arginine tRNA encoded by fimU within the fim gene cluster. FimA, FimI, FimH, and FimF are fimbrial subunits that form the shaft of type 1 fimbriae, with FimA as the major fimbrial subunit [7]. Type 1 fimbriae adhere to a variety of cells that possess mannose residues, such as erythrocytes, leukocytes, yeast, and respiratory cells [8]. This binding capacity is conferred by the FimH adhesin [9]. FimC and FimD are a chaperone and usher protein, respectively, whose respective functions are to assist the fimbrial subunit to the outer membrane of the cell and to assemble type 1 fimbriae in the proper order [10]. FimZ, FimY, STM0551, FimW, and fimU are involved in a delicate regulatory circuit for the production of type 1 fimbriae [11–15]. The precise contribution of type 1 fimbriae to virulence is still unclear because several factors, such as the Salmonella strain used, the inoculation route, and the laboratory animal chosen, affect the results [16, 17]. Nonetheless, this type of fimbria may play an important role at some stage of the infectious cycle of Salmonella [18].
Recent genome sequencing studies of host-adapted, invasive Salmonella serovars, including Choleraesuis, have revealed that extensive deletions and truncations occur in these serovars [19, 20]. The majority of the lost genes have functional counterparts in systemically noninvasive serovars; for example, pseudogenes have been found in most fimbrial clusters, including fim in serovar Typhi [21]. Since information regarding the type 1 fimbriae of S. Choleraesuis is limited, the present study investigated the distribution of the expression of fimbrial appendages among S. Choleraesuis isolates from diseased swine in the field. We report that most S. Choleraesuis isolates did not produce type 1 fimbriae and that allelic variation in fimH could be one of the reasons for this non-fimbriate phenotype.
Materials and Methods
Bacterial strains and plasmids
The S. Choleraesuis strains were originally isolated from pigs with a tentative diagnosis of swine salmonellosis in swine farms at different locations of Taiwan. The pigs with clinical signs had either died recently or had been euthanized by electric shock and examined post-mortem by a registered veterinarian. Specimens from liver, spleen, or lung were obtained by the veterinarian and brought back to the laboratory and cultured for Salmonella on Xylose Lysine Desoxycholate Agar (Difco/Becton Dickinson, Franklin Lakes, NJ). Suspected Salmonella colonies were initially screened by Salmonella O antiserum poly A-I, and Vi (Difco/Becton Dickinson). S. Choleraesuis were identified by Salmonella O Group C1 antiserum (Difco/Becton Dickinson) and then confirmed by polymerase chain reaction (PCR) analysis according to the protocol reported by Chiu et al. [22], and stored at -80°C. The major sources of our Salmonella were collected from swine farms located in Taichung, Tainan, Chiayi, Pingtung, and Changhua counties in Taiwan from 2001 to 2015. Each isolate stocked was from one swine. One hundred and twenty strains were selected from our stock (n = 185) by simple random sampling and included in this study. The other specified bacterial strains and plasmids used are listed in Table 1.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Genotype or relevant features | Reference or source |
|---|---|---|
| Salmonella Typhimurium | ||
| LB5010 | a LT2 strain derivative, Fim+ and fimbrial phase variable | [23] |
| Salmonella Choleraesuis | ||
| SC-15, 27, 30, 46, 77 | Clinical isolate, Fim- | This study |
| SC-31, 34, 39 | Clinical isolate, Fim+ and fimbrial phase variable | This study |
| Escherichia coli | ||
| DH5α | [24] | |
| Plasmids | ||
| yT&A | 2.7 kb cloning vector that contains T7 promoter; Amr | Yeastern, Taiwan |
| pACYC184 | 4.2 kb cloning vector; Tetr and Cmr | ATCC |
| pISF101 | 12.8 kb DNA with the complete fim genes of S. Typhimurium cloned into pACYC184 | [25] |
| pfimAICDHF | 8.0 kb fimAICDHF DNA fragment cloned into pACYC184; Cmr | This study |
| pfimZY0551WU | 3.7 kb fimZY0551WU DNA fragment cloned into pACYC184; Cmr | This study |
| pfimAI | 1.4 kb fimAI DNA fragment cloned into pACYC184; Cmr | This study |
| pfimAICD | 6.1 kb fimAICD fragment cloned into pACYC184; Cmr | This study |
| pfimHF | 1.6 kb fimHF fragment cloned into pACYC184; Cmr | This study |
| pfimH | 1.2 kb fimH DNA fragment cloned into pACYC184; Cmr | This study |
| pfimHL57P | A fimH coding sequence with L57P cloned into pACYC184; Cmr | This study |
| pfimHG63V | A fimH coding sequence with G63V cloned into pACYC184; Cmr | This study |
| pfimHR89Q | A fimH coding sequence with R89Q cloned into pACYC184; Cmr | This study |
| pfimHA115T | A fimH coding sequence with A115T cloned into pACYC184; Cmr | This study |
| pfimHS131Y | A fimH coding sequence with S131Y cloned into pACYC184; Cmr | This study |
| pfimHG177S | A fimH coding sequence with G177S cloned into pACYC184; Cmr | This study |
Yeast agglutination test
The expression of type 1 fimbriae on the bacterial surface was analyzed by the yeast agglutination test [26]. Briefly, the bacterial isolate was cultured in 10-mL of Luria-Bertani (LB) broth (Difco/Becton Dickinson, Franklin Lakes, NJ) at 37°C for 48 h statically or grown on LB agar (Difco/ Becton Dickinson) at 37°C for 24 h. Cells were pelleted from broth by centrifugation for 10 min or scraped from agar and resuspended in 1 × phosphate-buffered saline. Aliquots of bacterial suspension and 3% Saccharomyces cerevisiae (Sigma-Aldrich, St. Louis, MO) were mixed on a glass slide and gently agitated for 1 min. Visible agglutination indicated the presence of type 1 fimbriae. Positive samples were mixed again with yeast cells and 3% D-mannose solution (Sigma-Aldrich). Mannose-sensitive agglutination conferred by type 1 fimbriae was indicated by inhibition of agglutination in the presence of mannose.
DNA manipulation
Complementation test
The primers used for amplification are listed in Table 2. The template was genomic DNA extracted from SC-39, a Fim+ strain with a commercial kit (GeneMark, Taipei, Taiwan). Briefly, the PCR mixture contained 1 μg of template DNA, 10 mM of primers, 2.5 mM of dNTP, 5 × Phusion High Fidelity Buffer, and 1 unit of Phusion DNA polymerase (ThermoFisher Scientific, Wltham, Massachusetts, USA). The PCR conditions consisted of initial denaturation at 98°C for 3 min, followed by 35 cycles at 98°C for 30 s, 50°C for 30 s, and 72°C for 30 s/kb, and extended at 72°C for 10 min. The PCR products were cleaned with a GeneJet PCR purification kit (ThermoFisfer Scientific). Amplified DNA fragments were incubated at 37°C for 30 min with 1 unit of Taq DNA polymerase to add an adenine residue before the subsequent TA-cloning step. Insert DNA was cloned into the T&A Cloning Vector by blue-white selection according to the protocol provided by the manufacturer (Yeastern Biotech, Taipei, Taiwan). The recombinant plasmid was isolated with a Plasmid Miniprep Purification kit (GeneMark), and the insert DNA was sequenced (Mission Biotech, Taipei, Taiwan) to confirm that no error had been introduced during PCR. The insert in the plasmid was retrieved after cleavage with BamHI and SphI (FastDigest, ThermoFisher Scientific) and cloned into vector pACYC184. The recombinant plasmid was introduced into the Fim- SC-15 strain by electroporation (ECM 830 Electroporation System, Harvard Apparatus, Holliston, Massachusetts, USA). The transformants were tested for their ability to produce type 1 fimbriae with the yeast agglutination test.
Table 2. Primers used in this study.
| primer | Sequence 5’-3’ |
|---|---|
| fimAICDHF-F | ACGGATGAGGATCCACGTTTGCTTGCGACATAAATCTGTGA |
| fimAICDHF-R | TTCAGCGTGCATGCTCTGGCCAATGAAATGTCTAACAAAGA |
| fimZY0551WU-F | ATTGACTGGGATCCTGATCAATTACAATTAGTGTCCGTTATT |
| fimZY0551WU-R | CTCAAGAGGCATGCCGAAAATAAAAATAGAAGACTTTCGCTT |
| fimAI-R | CTCAAGAGGCATGCGGCGTCTGCGGCAAATT |
| fimAICD-R | CTCAAGAGGCATGCCAAGCCGCATCGATAAAT |
| fimHF-F | AGATGAGGATCCATGAAAATATACTCAGCGCTATTG |
| fimHF-R | TAGCGTGCATGCCTAATTGTAATTGATCAGGAAGTTC |
| fimH-R | TAGCGTGTCGACAATATTCACTTCGCCCAGAGATGAG |
| L57P-F | GTGGTTACGCTGCCGGAAAAATCAGGTTGG |
| L57P-R | CCAACCTGATTTTTCCGGCAGCGTAACCAC |
| G63V-F | AAATCAGGTTGGGTCGGCGTAAACGCG |
| G63V-R | CGCGTTTACGCCGACCCAACCTGATTT |
| R89Q-F | GAATTACGGGTACAAAGCACCGAAGGAAAT |
| R89Q-R | ATTTCCTTCGGTGCTTTGTACCGGTAATTC |
| A115T-F | ACCGATAGTGTCACTGGGGTATTTTAT |
| A115T-R | ATAAAATACGCCAGTCACACTATCGGT |
| S131Y-F | ATGGGCGTCGACTATAACGTGTCGCAGCAA |
| S131Y-R | TTGCTGCGACACGTTATAGTCGACGCCCAT |
| G177S-F | ACGACCTCTACCAGCGACGCGTTGAGCACG |
| G177S-R | CGTGCTCAACGCGTCGCTGGTAGAGGTCGT |
Site-directed mutagenesis of fimH
A mutant allele of fimH was generated by site-directed mutagenesis using an overlapping-extension PCR with SC-15 strain genomic DNA template and mutagenic oligonucleotide primers L57P-F and L57P-R [27]. Briefly, fimH-F and L57P-R were used to amplify the first DNA fragment using Phusion DNA polymerase. The PCR conditions consisted of initial denaturation at 98°C for 30 s, followed by 35 cycles at 98°C for 10 s, 50°C for 30 s, and 72°C for 40 s. The second DNA fragment was amplified using fimH-R and L57P-F primers by the same PCR conditions described above. These two DNA fragments were purified with a GeneJet PCR Extraction Kit. Ligation of these two DNA fragments with two overlapping ends was achieved with fimH-F and fimH-R primers as follows: denaturation at 98°C for 30 s, ligation at 50°C for 30 s, and elongation at 72°C for 45 s, followed by 35 cycles at 98°C for 10 s, 50°C for 30 s, and 72°C for 45 s. The amplified fragments were cloned into the T&A Cloning Vector and sequenced to determine whether the codon encoding leucin at amino acid 57 had been replaced with proline. The other fimH alleles were constructed with the same methods. The insert DNA was cloned into the pACYC184 vector and transformed into a SC-15 strain by electroporation and tested for its ability to produce type 1 fimbriae as described above.
Electron microscopy
The bacterial strains were resuspended in ddH2O, and 20 μL was taken to mix with 20 μL of 2% phosphotungstic acid in a microcentrifuge tube. A drop of this mixture was placed onto a formvar grid and left for 20 s. Excess fluid was removed with filter paper. The grid was observed with a JEOL JEM-1400 transmission electron microscope (JEOL Ltd, Tokyo, Japan).
Results
Among the 120 S. Choleraesuis isolates screened for type 1 fimbrial expression with the yeast agglutination test, only four produced type 1 fimbriae (4/120 = 3.3%). Because many Salmonella isolates exhibited multidrug-resistant phenotypes, which would interfere with our further cloning studies, all of these Fim- isolates were tested for chloramphenicol sensitivity using the Kirby-Bauer method and interpreted according to the CLSI guidelines [28]. We found five S. Choleraesuis isolates that were chloramphenicol sensitive and thus potential recipients of recombinant pACYC184 plasmids. One of these isolates, SC-15, was chosen for complementation tests.
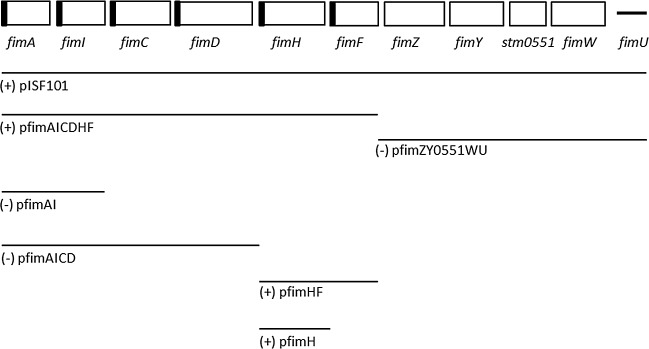
To determine whether the SC-15 type 1 fimbria-negative strain carried defects in the fim gene cluster, pISF101 and different derivatives from the fim of SC-39 were introduced into SC-15. Transformants that carried pACYC184 only did not produce type 1 fimbriae. pISF101, which possessed the entire fim gene cluster, allowed SC-15 to produce type 1 fimbriae and mediate yeast agglutination. To further dissect which structural or regulatory portion of the fim gene cluster was responsible for fimbrial expression in SC-15, pfimAICDHF and pfimZY0551WU were constructed and it was revealed that the SC-15 transformed with pfimAICDHF had fimbriae. On the basis of this finding, pfimAI, pfimAICD, pfimHF, and pfimH were further constructed to test which elements of the structural genes would permit SC-15 to produce type 1 fimbriae. We found that plasmids that carried fimHF or fimH alone were able to produce a fimbriate phenotype in SC-15 (Fig 1).
Fig 1. Summary of the yeast agglutination test results of SC-15 transformed with different recombinant plasmids.
The signal peptide regions of the structural and biosynthetic fim genes are shown as filled black boxes. Solid lines indicate the fim gene(s) retained on the pACYC184. (+): positive result in the yeast agglutination test, (-): negative result in the yeast agglutination test.
Since FimH confers binding specificity for type 1 fimbriae [29], fimH of non-fimbriate isolates SC-15, 27, 30, 46, and 77 and that of fimbriate isolates SC-31, 34, and 39 were sequenced. The FimH of S. Typhimurium LB5010 (Fim+) and S. Choleraesuis SC-B67 (Fim-) [19] were also aligned for comparison. It was interesting to find that the Fim- S. Choleraesuis isolates and SC-B67 strain possessed the same FimH sequences, whereas those of FimH from Fim+ S. Choleraesuis isolates and S. Typhimurium LB5010 were identical. The difference between the Fim+ and Fim- groups resides in only six amino acid residues: Fim+/Fim- (position): P/L (57), V/G (63), Q/R (89), T/A (115), Y/S (131), and S/G (177) (Table 3).
Table 3. Amino acid variations in FimH of Fim+ and Fim- strains.
| Strain\ position | 57 | 63 | 89 | 115 | 131 | 177 |
|---|---|---|---|---|---|---|
| LB5010 (Fim+) | P | V | Q | T | Y | S |
| SC-31(Fim+) | P | V | Q | T | Y | S |
| SC-34 (Fim+) | P | V | Q | T | Y | S |
| SC-39 (Fim+) | P | V | Q | T | Y | S |
| SC-B67 (Fim-) | L | G | R | A | S | G |
| SC-15 (Fim-) | L | G | R | A | S | G |
| SC-27 (Fim-) | L | G | R | A | S | G |
| SC-30 (Fim-) | L | G | R | A | S | G |
| SC-46 (Fim-) | L | G | R | A | S | G |
| SC-77 (Fim-) | L | G | R | A | S | G |
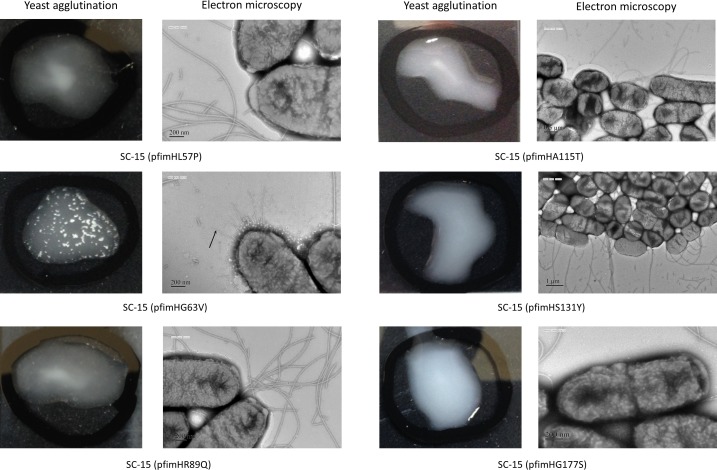
The introduction of plasmids with fimH inserts carrying the various amino acid changes demonstrated that only the SC-15 transformant that carried pfimHG63V changed the phenotype to Fim+ (Fig 2).
Fig 2. Results of the yeast agglutination test and electron microscopy.
SC-15 transformed with pfimHG63V enabled the mediation of yeast agglutination and produced fimbrial appendages (arrow) on the outer membrane of the bacterial cell. Other transformants did not mediate yeast cells to agglutinate, and no fimbrial structures other than flagella were observed.
Discussion
S. Choleraesuis is the most common cause of swine salmonellosis and infection of these animals manifests primarily as septicemia, whereas S. Typhimurium accounts for most cases of enterocolitis in swine [30, 31]. The type 1 fimbriae of S. Typhimurium bind to enterocytes of swine in vivo [32], but little information regarding this type of fimbriae in S. Choleraesuis has been reported. In this study, we found that most S. Choleraesuis isolates from diseased swine (116 of 120) did not mediate yeast agglutination. Appendages like type 1 fimbriae in nontyphoidal Salmonella serovars may play an important role in inducing the inflammatory response during the course of intestinal infection, thus facilitating the survival of fimbriated Salmonella in such a milieu and restricting infection to the gastrointestinal tract [33]. In contrast, some host-restricted Salmonella serovar, such as Gallinarum, are deficient in mannose-sensitive binding activity [34–36], instead, they produce morphologically similar fimbriae called type 2 fimbriae, with a preferable binding ability to avian leukocyte than to mammalian cells [37]. These differences in binding specificity are primarily associated with allelic variations in FimH adhesins [36, 38]. S. Choleraesuis that are deprived of type 1 fimbriae are perhaps unable to colonize the intestinal mucosa. They might cause less of an inflammatory response and are able to enter the bloodstream more rapidly than strains expressing type 1 fimbriae. Most Salmonella serovars isolated from swine carcasses, live swine, or their environments are primarily those with a broad host range, such as Typhimurium, Derby, and Anatum, whereas Choleraesuis has only rarely been identified as predominant serovar [39, 40].
Although proteins other than Fim proteins, such as leucine-responsive regulatory protein, were also found to modulate type 1 fimbrial expression [41], we focused only on the fim gene cluster. Our results demonstrated that fimH from S. Typhimurium was uniquely able to cause a Fim+ phenotype in strain SC-15. In contrast, E. coli which has been reported to have the ability to assemble a fimbrial shaft even in the absence of FimH [42], S. Choleraesuis was unable to produce type 1 fimbriae without FimH. This finding is in line with the report of Zeiner et al. that indicated that FimA, FimF, and FimH are necessary for the assembly of type 1 fimbriae in Salmonella [7].
It has been postulated that the repeated occurrence of phylogenetically unlinked mutations at the same amino acid positions, or hot spot mutations, may represent evidence of adaptive evolution via the molecular convergence of protein variants [43]. A comparison of the amino acid sequences of FimH alleles from Fim+ and Fim- S. Choleraesuis isolates revealed that all six amino acid differences between these two groups were included in the previously described hot spot mutations in the fimH of a large set of Salmonella strains [44, 45]. Fim+/Fim- (position): P/L (57), V/G (63), and Q/R (89) were identified as recent structural mutations, indicating that they have emerged only recently, whereas Y/S (131) is a long-term structural mutation that occurred in protein variants that are persistent in nature over long evolutionary periods [45, 46]. Positions 115 and 177 of Fim- S. Choleraesuis and SC-B67 strain were A and G, respectively; whereas those positions were T and S in Fim+ S. Choleraesuis and S. Typhimurium LB5010. Interestingly, we found that only FimH of S. Schwarzengrund exhibited T and S; all others showed A and G at these positions according to the current Microbial Variome Database provided by Dr. Sokurenko (http://depts.washington.edu/sokurel/variome/). Nevertheless, a switch between these two amino acids in different fimH alleles at position 115 or 177 did not have an impact in terms of fimbrial production in our study.
Site-directed mutagenesis analysis showed that only one amino acid substitution in FimH (G63V) was sufficient to restore the ability of SC-15 to produce type 1 fimbriae. This finding correlated with observations in S. Gallinarum. The FimH of S. Gallinarum and S. Typhimurium only differ by six residues, and one amino acid substitution (I78T) in the hot spot mutation zone was sufficient to restore the ability of serovar Gallinarum to gain mannose-specific binding activity [35, 47]. The ability of Salmonella to colonize or cause diseases in different hosts probably depends not only on the presence of an array of specific genes, but also on the allelic variation within these genes [38]. In our study, the lack of capability to produce type 1 fimbriae is not likely a result of the absence of FimH, but it nonetheless may be attributable to the property of the FimH polypeptides [48]. The amino acid variations in SC-15 are located at the N-terminal lectin-domain of FimH. We can only hypothesize that glycine at residue 63 may interfere with the binding of the chaperone-adhesin FimC/FimH complex to the N-terminal domain of usher protein FimD, which is the first step of type 1 fimbrial biogenesis in the well-studied E. coli model [49].
One limitation of our study was that most of the clinical S. Choleraesuis isolates from diseased swine were multidrug-resistant and could not be used in the same manner as strain SC-15. The fact that many S. Choleraesuis isolates were Fim- could suggest that this phenotype could contribute to the virulence of this serovar in swine.
Conclusions
Although the results from the present study were obtained with only one S. Choleraesuis isolate, it appears likely that allelic variation in fimH is a cause of the widespread nonfimbriate phenotype of S. Choleraesuis shown in this study. This hypothesis is supported by observation that the FimH amino acid sequences of the Fim+ S. Typhimurium LB5010 and Fim- S. Choleraesuis SC-B67 differ only by 6 amino acids. Sequencing of FimH from more Fim- S. Choleraesuis is currently underway in our laboratory.
Acknowledgments
We thank Ms. S.-T. Kuo from the Animal Health Research Institute, Council of Agriculture, Taiwan for assisting electron microscopy.
Data Availability
All relevant data are included in the paper.
Funding Statement
This research was supported by Ministry of Science and Technology, Taiwan, R.O.C. under the contract number MOST 103-2313-B-002-044.
References
- 1.Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemuhl J, Grimont PA, et al. Supplement 2003–2007 (No. 47) to the White-Kauffmann-LeMinor scheme. Res Microbiol. 2010;161:26–9. 10.1016/j.resmic.2009.10.002 [DOI] [PubMed] [Google Scholar]
- 2.Uzzau S, Brown DJ, Wallis T, Rubino S, Leori G, Bernard S, et al. Host adapted serotypes of Salmonella enterica. Epidemiol Infect. 2000;125:229–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones TF, Ingram LA, Cieslak PR, Vugia DJ, Tobin-D'angelo M, Hurd S, et al. Salmonellosis outcomes differ substantially by serotype. J Infect Dis. 2008;198:109–14. 10.1086/588823 [DOI] [PubMed] [Google Scholar]
- 4.Duguid JP, Gillies RR. Fimbriae and adhesive properties in dysentery bacilli. J Pathol Bacteriol. 1957;74:397–411. [Google Scholar]
- 5.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature (London). 2001;413:852–6. [DOI] [PubMed] [Google Scholar]
- 6.Humphries AD, Raffatellu M, Winter S, Weening EH, Kingsley RA, Droleskey R, et al. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol Microbiol. 2003;48:1357–76. [DOI] [PubMed] [Google Scholar]
- 7.Zeiner SA, Dwyer BE, Clegg S. FimA, FimF, and FimH are necessary for assembly of type 1 fimbriae on Salmonella enterica serovar Typhimurium. Infect Immun. 2012;80(9):3289–96. 10.1128/IAI.00331-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duguid JP, Anderson ES, Campbell I. Fimbriae and adhesive properties in Salmonella. J Pathol Bacteriol. 1966;92:107–38. [DOI] [PubMed] [Google Scholar]
- 9.Krogfelt KA, Bergmans H, Klemm P. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect Immun. 1990;5:1995–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol. 2009;7:765–74. 10.1038/nrmicro2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh KS, Hancox LS, Clegg S. Construction and characterization of a fimZ mutant of Salmonella typhimurium. J Bacteriol. 1995;177:6861–5. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tinker JK, Clegg S. Characterization of FimY as a coactivator of type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. Infect Immun. 2000;68:3305–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tinker JK, Hancox LS, Clegg S. FimW is a negative regulator affecting type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2001;183:435–42. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K-C, Hsu Y-H, Huang Y-N, Yeh K-S. A previously uncharacterized gene stm0551 plays a repressive role in the regulation of type 1 fimbriae in Salmonella enterica serotype Typhimurium. BMC Microbiol. 2012;12(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swenson DL, Kim KJ, Six EW, Clegg S. The gene fimU affects expression of Salmonella typhimurium type 1 fimbriae and is related to the Escherichia coli tRNA gene argU. Mol Gen Genet. 1994;244:216–8. . [DOI] [PubMed] [Google Scholar]
- 16.Duguid JP, Darekar MR, Wheater DWF. Fimbriae and infectivity in Salmonella typhimurium. J Med Microbiol. 1976;9:459–73. [DOI] [PubMed] [Google Scholar]
- 17.Lockman HA, Curtiss R III. Virulence of non-type-1-fimbriated and nonfimbriated nonflagellated Salmonella typhimurium mutants in murine typhoid fever. Infect Immun. 1992;60:491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clegg S, Swenson DL. Salmonella fimbriae In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis, and vaccines. Boca Raton, FL: CRC Press; 1994. p. 105–14. [Google Scholar]
- 19.Chiu C-H, Tang P, Chu C, Hu S, Bao Q, Yu J, et al. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 2005;33:1690–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A, et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet. 2004;36:1268–74. [DOI] [PubMed] [Google Scholar]
- 21.Thomson NR, Clayton DJ, Windhorst D, Vernikos G, Davidson S, Churcher C, et al. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 2008;18:1624–37. 10.1101/gr.077404.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu TH, Pang JC, Hwang WZ, Tsen HY. Development of PCR primers for the detection of Salmonella enterica serovar Choleraesuis based on the fliC gene. J Food Protec. 2005;68:1575–80. [DOI] [PubMed] [Google Scholar]
- 23.Bullas LR, Ryu JI. Salmonella typhimurium LT2 strains which are r- m+ for all three chromosomally located systems of DNA restriction and modification. J Bacteriol. 1983;156:471–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–80. [DOI] [PubMed] [Google Scholar]
- 25.Swenson DL, Clegg S, Old DC. The frequency of fim genes among Salmonella serovars. Microb Pathog. 1991;10:487–92. [DOI] [PubMed] [Google Scholar]
- 26.Old DC, Corneil I, Gibson LF, Thomson AD, Duguid JP. Fimbriation, pellicle formation and the amount of growth of salmonellas in broth. J Gen Microbiol. 1968;51:1–16. [DOI] [PubMed] [Google Scholar]
- 27.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–9. [DOI] [PubMed] [Google Scholar]
- 28.CLSI. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-first Informational Supplement M100-21. CLSI, Wayne, PA, USA. 2011.
- 29.Thankavel K, Shah AH, Cohen MS, Ikeda T, Lorenz RG, Curtiss R III, et al. Molecular basis for the enterocyte tropism exhibited by Salmonella typhimurium type 1 fimbriae. J Biol Chem. 1999;274:5797–809. [DOI] [PubMed] [Google Scholar]
- 30.Foley SL, Lynne AM. Food animal-associated Salmonella challenges: Pathogenicity and antimicrobial resistance. J Anim Sci. 2008;86 (E Suppl.):E173–E87. [DOI] [PubMed] [Google Scholar]
- 31.Anderson RC, Nisbet DJ, Buckley SA, Genovese KJ, Harvey RB, Deloach JR, et al. Experimental and natural infection of early weaned pigs with Salmonella choleraesuis. Res Vet Sci. 1998;64:261–2. [DOI] [PubMed] [Google Scholar]
- 32.Althouse C, Patterson S, Fedorka-Cray P, Isaacson RE. Type 1 fimbriae of Salmonella enterica serovar Typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect Immun. 2003;71:6446–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuzminska-Bajor M, Grzymajio K, Ugorski M. Type 1 fimbriae are important factors limiting the dissemination and colonization of mice by Salmonella Enteritidis and contribute to the induction of intestinal inflammation during Salmonella invasion. Front Microbiol. 2015;6:Article 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuzminska-Bajor M, Kuczkowski M, Grzymajio K, Wojciech L, Sabat M, Kisiela D, et al. Decreased colonization of chicks by Salmonella enterica serovar Gallinarum expressing mannose-sensitive FimH adhesin from Salmonella enterica serovar Enteritidis. Vet Microbiol. 2012;158:205–10. 10.1016/j.vetmic.2012.01.029 [DOI] [PubMed] [Google Scholar]
- 35.Hancox LS, Yeh KS, Clegg S. Construction and characterization of type 1 non-fimbriate and non-adhesive mutants of Salmonella typhimurium. FEMS Immun Med Microbiol. 1997;19:289–96. [DOI] [PubMed] [Google Scholar]
- 36.Kisiela DI, Chattopadhyay S, Libby SJ, Karlinsey JE, Fang FC, Tchesnokova V, et al. Evolution of Salmonella enterica virulence via point mutations in the fimbrial adhesin. PLoS Pathogens. 2012;8:e1002733 10.1371/journal.ppat.1002733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo A, Cao S, Tu L, Chen P, Zhang C, Jia A, et al. FimH alleles direct preferential binding of Salmonella to distinct mammalian cells or to avian cells. Microbiology. 2009;155:1623–33. 10.1099/mic.0.026286-0 [DOI] [PubMed] [Google Scholar]
- 38.Yue M, Schifferli DM. Allelic variation in Salmonella: an underappreciated driver of adaptation and virulence. Front Microbiol. 2014;4:Article 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pires AFA, Funk JA, Bolin C. Risk factors associated with persistence of Salmonella shedding in finishing pigs. Prev Vet Med. 2014;116:120–8. 10.1016/j.prevetmed.2014.06.009 [DOI] [PubMed] [Google Scholar]
- 40.Schmidt JW, Brichta-Harhay DM, Kalchayanand N, Bosilevac JM, Shackelford SD, Wheeler TL, et al. Prevalence, enumeration, serotypes, and antimicrobial resistance phenotypes of Salmonella enterica isolates from carcasses at two large United States pork processing plants. Appl Environ Microbiol. 2012;78(8):2716–26. 10.1128/AEM.07015-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McFarland KA, Lucchin S, Hinton JCD, Dorman CJ. The leucine-responsive regulatory protein, Lrp, activates transcription of the fim operon in Salmonella enterica serovar Typhimurium via the fimZ regulatory gene. J Bacteriol. 2008;190:602–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell PW, Orndorff PE. Lesions in 2 Escherichia coli type 1 pilus genes alter pilus number and length without affecting receptor binding. J Bacteriol. 1992;174:5923–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chattopadhyay S, Paul S, Kisiela DI, Linardopoulou EV, Sokurenko EV. Convergent molecular evolution of genomic cores in Salmonella enterica and Escherichia coli. J Bacteriol. 2012;194(18):5002–11. 10.1128/JB.00552-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sokurenko EV, Hasty DL, Dykhuizen DE. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 1999;7:191–5. [DOI] [PubMed] [Google Scholar]
- 45.Chattopadhyay S, Taub F, Paul S, Weissman SJ, Sokurenko EV. Microbial variome database: point mutations, adaptive or not, in bacterial core genomes. Mol Biol Evol. 2013;30(6):1465–70. 10.1093/molbev/mst048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chattopadhyay S, Weissman SJ, Minin VN, Russo TA, Dykhuizen DE, Sokurenko EV. High frequency of hotspot mutations in core genes of Escherichia coli due to short-term positive selection. Proc Natl Acad Sci USA. 2009;106:12412–7. 10.1073/pnas.0906217106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kisiela D, Sapeta A, Kuczkowski M, Stefaniak T, Wieliczko A, Ugorski M. Characterization of FimH adhesin expressed by Salmonella enterica serovar Gallinarum and Pullourm: reconstitution of mannose-binding properties by single amino acid substitution. Infect Immun. 2005;73:6187–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dwyer BE, Newton KL, Kisiela D, Sokurenko EV, Clegg S. Single nucleotide polymorphisms of fimH associated with adherence and biofilm formation by serovars of Salmonella enterica. Microbiology. 2011;157:3162–71. 10.1099/mic.0.051425-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lillington J, Geibel S, Waksman G. Biogenesis and adhesion of type 1 and P pili. Biochim Biophys Acta. 2015;1850:554–64. 10.1016/j.bbagen.2014.07.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are included in the paper.