Abstract
We have previously reported several lines of evidence that support a role for cellular DNA repair systems in completion of the retroviral DNA integration process. Failure to repair an intermediate in the process of integrating viral DNA into host DNA appears to trigger growth arrest or death of a large percentage of infected cells. Cellular proteins involved in the nonhomologous end joining (NHEJ) pathway (DNA-PKCS) and the damage-signaling kinases (ATM and ATR) have been implicated in this process. However, some studies have suggested that NHEJ proteins may not be required for the completion of lentiviral DNA integration. Here we provide additional evidence that NHEJ proteins are required for stable transduction by human immunodeficiency type 1 (HIV-1)-based vectors. Our analyses with two different reporters show that the number of stably transduced DNA-PKCS-deficient scid fibroblasts was reduced by 80 to 90% compared to the number of control cells. Furthermore, transduction efficiency can be restored to wild-type levels in scid cells that are complemented with a functional DNA-PKCS gene. The efficiency of stable transduction by an HIV-1-based vector is also reduced upon infection of Xrcc4 and ligase IV-deficient cells, implying a role for these components of the NHEJ repair pathway. Finally, we show that cells deficient in ligase IV are killed by infection with an integrase-competent but not an integrase-deficient HIV-1 vector. Results presented in this study lend further support to a general role for the NHEJ DNA repair pathway in completion of the retroviral DNA integration process.
Integration of viral DNA into host DNA is an essential step in the replication cycle of retroviruses and retrotransposons (4, 13, 26). The first two steps in this process, viral DNA processing and joining, are catalyzed by the retroviral protein integrase and require specific sequences at the ends of the viral DNA. In the first step, nucleotides (usually two) are removed from the 3′ ends of the viral DNA, and in the second step, these newly created ends are joined to staggered phosphates in the complementary strands of the host cell DNA, creating an integration intermediate with gaps in the DNA sequence. Complete, stable integration of the viral DNA depends on repair of these gaps: filling in the missing nucleotides, removing the short flap on the 5′ ends of the viral DNA, ligation to the newly synthesized 3′ end of host DNA and, likely, reconstitution of appropriate chromatin structure and composition, all by mechanisms not yet fully understood.
At each step, the retroviral DNA integration process may be assisted by host cellular proteins, including those that participate in DNA repair. Mechanistically, retroviral DNA integration resembles RAG1/2-mediated V(D)J recombination, the process by which immunoglobulin genes are assembled (5). Successful completion of V(D)J recombination depends on proteins that make up the nonhomologous end joining (NHEJ) DNA repair pathway, including the DNA-dependent protein kinase (DNA-PK), which consists of a large, phosphatidylinositol 3-kinase-related catalytic subunit (DNA-PKCS) and a DNA binding heterodimer Ku70/Ku80. We have shown that DNA-PKCS-deficient scid mouse pre-B cells undergo apoptotic cell death in response to infection by avian sarcoma virus (ASV) or human immunodeficiency virus type 1 (HIV-1) retroviral vectors and that this cell death requires an active viral integrase (9, 10). We also showed that the ability of an ASV vector to stably transduce selectable markers is reduced by 80 to 90% in scid fibroblasts and other adherent cells that bear mutations in three components of the NHEJ DNA repair pathway: DNA-PKCS, Ku80, and Xrcc4 (9). Therefore, we have proposed that the NHEJ DNA repair pathway is required for efficient completion of the retroviral DNA integration process and that failure in repair of the integration intermediate triggers growth arrest or cell death. Others have shown that yeast Ku80 and Ku70 proteins are required for Ty1 retrotransposition in Saccharomyces cerevisiae (11) and that HIV-1 replication and integration are reduced in Ku80-deficient human cells (16). In addition, we have reported that two DNA-PKCS-related kinases, ATM and ATR, also participate in the retroviral DNA integration process (7, 8). However, whereas ATM is required only for the residual integration observed in the DNA-PK-deficient cells, ATR appears to be required even in the presence of DNA-PKCS (7, 8). These results suggest that integrase-mediated joining of viral to host DNA is sensed as DNA damage by the cell and that the viral DNA integration process has evolved to exploit the cellular DNA damage response and double-strand break DNA repair pathways.
Another group of investigators (1) has reported that DNA-PK is not required for efficient integration by lentiviral vectors. These researchers found that infection by HIV-1-based vectors induced death of DNA-PKCS-deficient mouse embryo fibroblasts (MEFs) and other NHEJ-deficient cell lines; however, they did not observe deficiencies in stable transduction when cells were infected at a low multiplicity. In addition, a second group (20) reported that retroviral infection induces death in a human lymphoid cell line lacking another NHEJ protein, ligase IV. In these experiments, cell death was also induced by a vector carrying an integration-defective integrase; furthermore, an inhibitor of reverse transcriptase suppressed cell death. Therefore, these researchers suggested that the presence of unintegrated viral DNA, rather than an unrepaired integration intermediate, is the causative factor in retrovirus-induced death of NHEJ-deficient cells.
In the experiments presented herein, we measured the transduction efficiency of HIV-1-based vectors with DNA-PKCS-deficient scid MEFs and also examined the viability of a ligase IV-deficient lymphoid cell line after infection with such vectors. Our experiments reveal a striking deficiency in stable transduction of scid MEFs and an Xrcc4-deficient cell line with these lentiviral vectors compared to control cells. We also demonstrate that the transduction deficiency is overcome in scid cells in which a functional DNA-PKCS gene had been introduced. Reduced HIV-1 vector-mediated transduction of the ligase IV-deficient lymphoid cells was also observed, but we found that the vector did not induce cell killing if it carried an inactive integrase. Several possible explanations for the discrepancies noted above are addressed. Data provided in this study lend further support for our proposal that components of the NHEJ DNA repair pathway are required for efficient completion of the retroviral DNA integration process and the survival of cultured cells in which a chromosome has been damaged by integrase-mediated insertion of viral DNA.
MATERIALS AND METHODS
Cells.
scid and isogenic normal MEFs were generously provided by the laboratory of M. Bosma (Fox Chase Cancer Center), and cells were maintained in RPMI 1640 medium in the presence of 10% fetal bovine serum, 5 × 10−6 M 2-mercaptoethanol, and 20 IU of penicillin and 20 μg of streptomycin per ml (PEN/STR). The human lymphoid cell lines Nalm-6 (LIG4+/+) and ligase IV-deficient Nalm-6 (LIG4−/−) were provided by M. Lieber (University of California at Los Angeles) and have been described by Li et al. (20). These cell lines were also maintained in RPMI 1640 medium in the presence of 10% fetal bovine serum, 5 × 10−6 M 2-mercaptoethanol, and PEN/STR. The murine ST.SCID and 100E cell lines were obtained from M. Oettinger (Harvard Medical School). These cell lines were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and PEN/STR. The Chinese hamster ovary CHO-K1 and XR-1 cell lines were obtained from the Fox Chase Cancer Center Cell Culture Facility and D. Roth (Baylor College of Medicine), respectively, and maintained as described by Li et al. (21). Finally, 293T cells were maintained in DMEM with 10% fetal bovine serum and PEN/STR.
Viruses.
The HIV-based vector encoding a lacZ reporter gene (22) was prepared as follows. 293T cells were plated on 100-mm-diameter dishes at a density of approximately 106 cells per dish. The following day, cells were transfected using the three-plasmid system described previously (22). For our preparations, in each dish, we used 25 μg of a lacZ vector genome plasmid, 50 μg of a backbone plasmid that expresses HIV-1 gag and pol genes, and 5 μg of a plasmid that expresses vesicular stomatitis virus (VSV) G protein, and cells were transfected using a Profection kit (calcium chloride) from Promega. On the following day, the medium was replaced, and 2 days after transfection, supernatant from vector-producing 293T cells was harvested, passed through a 0.45-μm-pore-size filter to remove producer cells, and then subjected to centrifugation at 4°C and 27,500 × g for 30 min to concentrate the virus. The virus-containing pellet was dissolved in DMEM. The titer of this virus (number of infectious units per cell) was determined by transduction of the lacZ reporter using HeLa cells.
An HIV-1 vector encoding an enhanced green fluorescent protein (EGFP) reporter was prepared as described by Wu et al. (29) for the experiment shown in Fig. 4 or prepared as described above for the experiments shown in Fig. 7 and Table 1, except that the vector genome plasmid encoded EGFP under control of a cytomegalovirus promoter (3). The titer of this vector was determined with HeLa cells, and scoring transduction was performed by flow cytometry.
FIG. 4.

Transduction efficiencies of normal and scid MEFs infected with a HIV-1 vector encoding an EGFP reporter. Normal and scid MEFs (11th passage) were distributed in 60-mm-diameter dishes at a density of 105 cells per dish. Cells were infected 24 h later with two different dilutions of the HIV-1 EGFP vector (MOI for undiluted cells of ≈0.05) as described in the legend to Fig. 1. Twelve days postinfection, cells were harvested and analyzed by flow cytometry to determine the percentage of EGFP-expressing cells in each sample. The average count of two dishes for each point is shown.
FIG. 7.
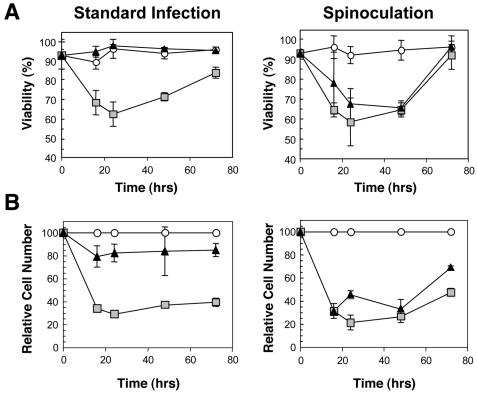
Killing of ligase IV-deficient cells is integrase dependent after infection under standard conditions but not after spinoculation. Nalm-6 LIG4−/− cells were infected at a MOI of ≈2 (titer determined by transduction of HeLa cells) with the HIV-1 lacZ vectors containing either wild-type integrase or integrase with the inactivating E152A substitution. Cells were either infected under standard conditions, as described in Materials and Methods, or by spinoculation (23). In spinoculation, the vector-cell mixtures were subjected to centrifugation at 1,000 × g for 2 h. (A) Viability was determined by trypan blue exclusion. (B) The relative number of viable cells was determined by counting and expressing the value as a percentage of the cells counted in the mock-infected or mock-spinoculated cultures. Results from standard infection methods(left panels) and with spinoculation (right panels) are shown. Two samples were analyzed for each datum point. Symbols: open circles, mock-infected cells; light grey squares, cells infected with wild-type integrase-containing vector; black triangles, cells infected with the E152A integrase-containing vector.
TABLE 1.
Fluorescence-activated cell sorting analyses for expression of an HIV-1-transduced EGFP reporter in ligase IV-null and control cells
| Virus dilution | Nalm-6 cell | % Positive cells |
|---|---|---|
| 10−1 | LIG4+/+ | 37.6 |
| LIG4−/− | 20.3 | |
| 10−2a | LIG4+/+ | 10.0 |
| LIG4−/− | 2.9 | |
| 10−3 | LIG4+/+ | 2.53 |
| LIG4−/− | 0.88 |
MOI of ≈0.1.
For infection of Nalm-6 LIG4+/+ and Nalm-6 LIG4−/− human cell cultures, the HIV-1 vectors were prepared as described above, except that we used backbone plasmids encoding an E152A substitution in the catalytic site of integrase and the corresponding wild-type backbone plasmid, both obtained from F. Bushman, Salk Institute (described by Li et al. [20]). The vector genome plasmids, encoding EGFP or carrying lacZ under control of the cytomegalovirus promoter were as described above. The titer of the EGFP vector that carried wild-type integrase was established by flow cytometry, assuming (from the Poisson distribution) that at a multiplicity of infection (MOI) of 0.1 infectious unit/cell, 10% of the LIG+/+ cells will be transduced. To normalize titers of the integration-competent and integration-deficient (IN, E152A-carrying) vectors, we used an immunological assay that measures the amount of capsid protein, CAp24, in each virus stock, performed according to the manufacturer's instructions (Beckman Coulter). Volumes were then adjusted to obtain the same concentrations of CAp24 in both vector stocks.
The ASV vector-producing RCAN-Eco-(polII-AP) DFJ8 cell line was a gift from E. Barsov (National Institutes of Health). Virus titers were determined by transduction of normal MEFs.
Infections.
For studies with normal and scid MEFs (10th passage), cells were distributed into a 96-well plate at a density of 104 cells per well. Cells were infected 24 h later with the HIV-1 lacZ vector for 2 h in the presence of 10 μg of DEAE dextran per ml. Two days postinfection, cells were stained using a β-galactosidase assay (Stratagene), and all blue cells were counted.
To examine the effects of cell density on transduction, normal and scid MEFs (11th passage) were distributed into a 96-well plate at densities from 5 × 103 to 5 × 104 cells per well. Cells were infected and analyzed as described above.
In the complementation experiment, murine ST.SCID and 100E cells were distributed into a 96-well plate at a density of 104 cells per well. Cells were infected 24 h later as described above with various dilutions of the HIV-1 lacZ vector, and blue cells were counted 2 days later.
For HIV-1 EGFP vector infections of MEFs, wild-type and scid MEFs (11th passage) were plated on 60-mm-diameter dishes at a density of 105 cells per dish. Cells were infected 24 h later with two different dilutions of the vector for 2 h in the presence of 10 μg of DEAE dextran per ml. Cells were passaged every 2 to 3 days, and 12 days postinfection, the cells were harvested and analyzed by flow cytometry to determine the percentage of EGFP-expressing cells in each sample. An average count of cells from two dishes was determined for each point.
ASV alkaline phosphatase vector infections of MEFs were performed as described above for HIV-1. Forty-eight hours postinfection, cells were fixed with 4% paraformaldehyde and 1% glutaraldehyde in phosphate-buffered saline. Endogenous alkaline phosphatase activity was heat inactivated by treating the cells for 30 min at 65°C. The vector-expressed placental alkaline phosphatase was detected by treating the cells with 0.4 mM nitroblue tetrazolium and 0.45 mM bromochloroindolyl phosphate in alkaline phosphatase buffer (100 mM NaCl, 5 mM MgCl2, 100 mM Tris [pH 9.5]) overnight at room temperature.
In the experiments with Chinese hamster ovary (CHO) cells (CHO-K1 and XR-1), cultures were plated at a density of 105 cells per 60-mm-diameter dish. Cells were infected on the following day with the HIV-1 lacZ vector and analyzed as described above.
For analysis of transduction efficiency of Nalm-6 LIG4−/− and Nalm-6 LIG4+/+ control cells, these lymphoid cultures were distributed into the wells of a 24-well plate at a density of 5 × 105 cells per well. The HIV-1 EGFP vector was then added, in a total volume of 1 ml, in the presence of 5 μg of DEAE dextran per ml. The percentage of GFP-positive cells was determined by flow cytometry 3 days after the addition of the virus. For viability analysis, these cultures were distributed into the wells of a 24-well plate at a density of 1.5 × 105 cells per well. HIV-1 lacZ vectors carrying either integrase with an inactivating E152A substitution [IN(E152A)] or wild-type integrase were then added in a total volume of 1 ml in the presence of 5 μg of DEAE dextran per ml. Viability was evaluated using trypan blue dye exclusion.
RESULTS
Efficient transduction of MEFs by an HIV-1 vector requires DNA-PKCS.
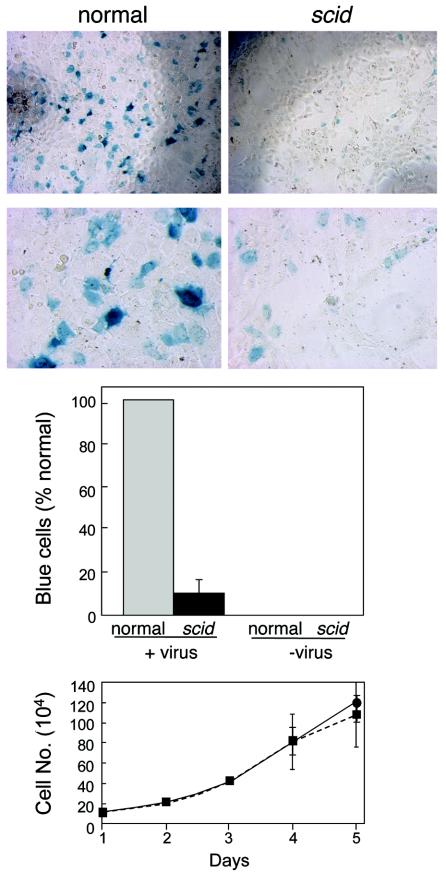
Stable transduction by a retroviral vector is a useful method for monitoring the successful completion of the retroviral DNA integration process and survival of the transduced cells. To determine whether DNA-PKCS-deficient scid MEFs are deficient for stable transduction by lentiviral vectors, we infected the same number of normal and scid MEFs at a low multiplicity (≈0.1 infectious unit/cell) with an HIV-1 lacZ vector, following the conditions reported by Baekelandt et al. (1). As shown in Fig. 1, we detected approximately sixfold-fewer blue scid MEFs than normal MEFs and no blue cells in the uninfected control under these conditions. This experiment was reproduced five times, consistently showing 6- to 10-fold differences in blue-staining scid and control MEFs, confirming the differences shown in Fig. 1. To verify that this result was due primarily to a difference in the efficiency of stable transduction, we infected the scid and normal MEFs under the same conditions, split the cultures, and allowed them to expand for another week. Any potential expression from unintegrated DNA would thus be diluted, and formation of blue colonies should indicate the presence of a stably integrated provirus. Under these conditions, we consistently observed even greater differences in the numbers of blue cells in the infected normal and scid MEF cultures (data not shown). Although we have observed no difference in the growth rate of these scid and control MEFs (Fig. 1, bottom graph), such passaging could introduce an unknown bias. However, the simplest interpretation of these results is that the differences illustrated in Fig. 1 are due to differences in the efficiency of formation or survival of stably transduced scid and normal MEFs.
FIG. 1.
Transduction efficiencies of normal and scid MEFs infected with an HIV-1 vector. Normal (wild-type) and scid MEFs (11th passage) were distributed in a 96-well plate at a density of 104 cells per well. Cells were infected 24 h later with the HIV-1 lacZ vector (22) for 2 h in the presence of 10 μg of DEAE dextran per ml. Two days postinfection, cells were stained using a β-galactosidase assay kit (Stratagene), and digital micrographs were taken at magnifications of ×4 (top micrographs) and ×20 (bottom micrographs). Results are expressed as the percentage of blue cells in scid cultures compared to normal cultures, with the averages and standard deviations (error bars) obtained from six experiments in which cells were stained and counted 2 to 7 days postinfection. The micrograph shows results with cells infected at a MOI of ≈0.1; averaged data in the bar graph are from experiments with MOIs from 0.0001 to 0.1. The growth curves for scid and normal MEFs are shown at the bottom. Cells were plated at a density of 105 cells per 600-mm-diameter dish, and two plates were counted for each datum point. The average values and standard deviations (error bars) are shown.
Cell density affects the total number of transductants but not the relative transduction efficiencies in scid and normal MEFs.
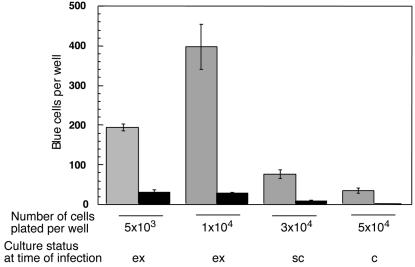
Our previous analyses monitored results after retroviral vector infection of exponentially growing cultures (8, 9). Comparisons of experimental data with computational simulations suggested that for an unrepaired integration intermediate to induce cell death, infected scid lymphocytes must pass through the S phase (10). Therefore, it seemed possible that differences in transduction efficiencies might vary with culture density as cells become contact inhibited. We first established that the confluence density for these MEFs is 105 cells per well in a 96-well plate (not shown). As these cells are very sensitive to contact inhibition (25), the great majority of such confluent cells are expected to cease dividing under these conditions. In the experiment shown in Fig. 2, MEFs were plated at densities ranging from 103 to 5 × 104 cells per well and infected with the HIV-1 lacZ vector 24 h later. As these cells double within 24 h, the wells plated at the highest cell density reached confluence at the time of infection. Blue cells in each well were counted 2 days later as in Fig. 1. The results indicated that contact inhibition of cell growth was correlated with a substantial reduction in the number of transduced normal MEFs (grey columns). However, the numbers of transductants in the scid cultures (black columns) were significantly lower than in the control cultures at all cell densities (Fig. 2). Therefore, it appears that the relative difference between the normal and scid MEFs was maintained, regardless of cell culture status. From these results, we consider it unlikely that cell density differences can explain the discrepancy between our results and those of Baekelandt et al. (1) who reported using subconfluent MEF cultures in their assays.
FIG. 2.
Effect of cell density on the efficiency of transduction of MEFs by an HIV-1 vector. Normal MEFs (grey columns) and scid MEFs (black columns) (11th passage) were distributed in a 96-well plate at a density of 5 × 103 to 5 × 104 cells per well, two wells for each point. Cells were infected 24 h later with the HIV-1-lacZ vector at a MOI of ≈0.04 and scored 2 days later as described in the legend to Fig. 1. Mean numbers and standard deviations (error bars) are shown. ex, exponentially growing; sc, subconfluent; c, confluent.
The scid defect can be complemented by DNA-PKCS-coding DNA.
To confirm that the DNA-PKCS deficiency in scid cells was responsible for the reduced transduction, we took advantage of a murine scid cell line that contains a small fragment of human chromosome 8 that includes the DNA-PKCS gene and has been shown to restore V(D)J recombination in these scid cells (17). The parental scid line (ST.SCID) and the complemented derivative (100E) were infected with the HIV-1 lacZ vector. The results showed that the number of blue cells was about 10-fold higher in the 100E cultures than in the ST.SCID cultures (Fig. 3). From these results, we conclude that introduction of the chromosomal fragment carrying the DNA-PKCS into the scid background rescues the scid cell deficiency in stable transduction by the HIV-1 vector.
FIG. 3.
Effect of reintroduction of DNA-PKCS into the scid background on transduction efficiency by an HIV-1 vector. ST.SCID and 100E cells were distributed in a 96-well plate at a density of 104 cells per well. Cells were infected 24 h later with undiluted virus (MOI of ≈0.01) or the indicated dilutions of the HIV-1 lacZ vector preparations as described in the legend to Fig. 1. Two days postinfection, cells were stained using a β-galactosidase assay (Stratagene), blue cells were counted (two wells/datum point), and digital micrographs were taken of cells infected with the undiluted virus. ST.SCID cells (black columns) and 100E cells (shaded columns) are shown.
The scid defect can also be observed with different assay systems and quantitation methods.
All experiments described so far were performed with the same HIV-1 lacZ vector (22). To test whether the scid deficiency can be detected using another reporter and a different type of assay, we infected cells with an HIV-1 vector that encodes EGFP, allowing the transduction efficiency to be determined by flow cytometry (29). We first prepared integration-competent and integration-defective [IN(E152A)] stocks of this vector. In control experiments, we detected a significant number of weakly EGFP-positive MEFs by flow cytometry 48 h after infection with the integrase-defective vector. Previous studies have indicated that integration efficiencies are reduced by greater than 3 log units when cells are infected with HIV-1 vectors containing integrase with catalytic site substitutions (D64, D116, or E152 [18]). Therefore, our results are consistent with published data (22; noted above), which showed that low-level expression from unintegrated DNA can persist for several days postinfection (15). To reduce this apparent background, MEFs infected with the integration-competent vector were cultured for 12 days to dilute the unintegrated DNA. As illustrated in Fig. 4, fewer stable transductants were again observed with scid MEFs compared to normal MEFs using this vector and protocol. Because scid and wild-type MEFs grow at similar rates (Fig. 1), these results support the conclusion that expression of DNA-PKCS is required for efficient transduction of such cells.
As another test of this conclusion, we measured transduction by an ASV-derived vector (2) with a murine ecotropic envelope that mediates viral entry via direct membrane fusion. This ASV vector encodes a placental alkaline phosphatase reporter gene under control of the ubiquitous RNA polymerase II promoter. Our analyses (Fig. 5) showed that the number of scid MEF transductants was 10-fold lower than the control with this vector. These results are consistent with our previous observations that the scid DNA-PKCS mutation causes a significant reduction in the number of ASV and HIV-1 transductants (9) (Fig. 1, 3, and 4). Data obtained with this ASV vector therefore confirm our current and previous results with HIV-1, as well as other ASV vectors (9) that contain different envelope proteins, promoters, and reporter genes.
FIG. 5.
Transduction efficiency of scid and normal MEFs infected with an ASV vector. Normal (wild-type) and scid MEFs were distributed into the wells of 24-well plates at 2 × 104 cells per well. After overnight culturing, the cells were infected with undiluted (MOI of ≈0.2) or 1:5 diluted filtered stock of the ASV vector. Forty-eight hours postinfection, the cells were fixed and stained for alkaline phosphatase activity. (Top) Phase-contrast images of the alkaline phosphatase-stained fibroblasts from the cultures infected with the 1:5 dilution of virus. (Bottom) Stained cells were counted after overnight development, and the numbers per well were plotted. Each dilution was assayed in triplicate. The average number of transduced cells per well and standard deviation (error bar) are shown.
Xrcc4 and ligase IV are required for efficient stable transduction by an HIV-1 vector.
Xrcc4 is known to form a complex with ligase IV, thereby stimulating its activity; both Xrcc4 and ligase IV are components of the NHEJ repair pathway (see the review by Critchlow and Jackson [6]). We have shown previously that the transduction efficiency of an amphotropic ASV vector is greatly reduced in the CHO cell line XR-1, which lacks the Xrcc4 protein, compared to control CHO-K1 cells (9). One complication of these experiments is that CHO cells are relatively resistant to infection with vectors that enter via attachment to the amphotropic envelope receptor (12). Furthermore, it has been shown that Ty1 transposition in yeast cells, which requires Ku proteins, does not seem to require the yeast homolog of Xrcc4 (11). Therefore, we reexamined the transduction efficiency of these cells with our VSV G-protein-pseudotyped HIV-1 lacZ vector that enters cells via an endocytic pathway. As shown in the data summarized in Fig. 6, the number of Xrcc4-deficient cells that were stably transduced by this vector was ∼5- to 10-fold lower than the CHO-K1 control, confirming our previous results.
FIG. 6.
Transduction efficiency of Xrcc4−/− CHO cells infected with an HIV-1 vector. CHO-K1 and XR-1 cells were distributed in 60-mm-diameter dishes at a density of 105 cells per dish. Cells were infected 24 h later with the HIV-1 lacZ vector and scored 2 days postinfection as described in the legend to Fig. 1. The MOI at the 10−2 dilution was 0.0025. The average count of two dishes for each point and standard deviation (error bar) are shown.
Next we examined the transduction efficiency of the HIV-1 EGFP-expressing vector with the ligase IV-deficient Nalm-6 (LIG4−/−) human lymphoid cell line and parental, wild-type Nalm-6 (LIG4+/+) cells, monitoring transduction by flow cytometry. As with MEF infection (Fig. 4), we used EGFP readout as an indicator of stable integration. We first asked whether EGFP expression is a valid indicator for stable integration in these lymphoid cell lines. Our tests showed that the number of EGFP-positive parental Nalm-6 (LIG4+/+) cells was ∼10-fold lower (with weak intensity) after infection with a vector that carried an inactivating mutation in integrase than with the integration-competent vector (data not included). This result indicated that EGFP expression is strongly dependent upon integration in these cells.
For analysis of results of infection of the mutant cell line Nalm-6 LIG4−/−, two additional factors had to be considered. First, if these cells respond to integrase-competent virus infection like the NHEJ-deficient scid lymphoid cell lines, then a large percentage are likely to die by apoptosis (Fig. 7). Second, other ligases (or other repair pathways) may substitute in the absence of ligase IV. Both factors could affect the percentage of EGFP-positive mutant cells scored in our experiments. We have attempted to separate their effects by measuring net transduction efficiencies (Table 1) and cell killing (Fig. 7) independently.
The analyses summarized in Table 1 show that the transduction efficiency of Nalm-6 LIG4−/− cells is reduced approximately threefold compared to the wild-type control upon infection with the most diluted integration-competent vector samples (MOI of ≈0.01 or 0.1). The difference was less apparent with a higher MOI (∼1.0). We believe that this phenomenon can be explained as follows: at a higher MOI, a significant fraction of the infected ligase IV-deficient cells become apoptotic, most likely because the integration intermediate cannot be repaired. Such cells are lost from the population (Fig. 7B, left panel) and are not counted by flow cytometry. Surviving green cells in which “backup” pathways have repaired the integration site (9, 28) are expected to repopulate the culture over time (9, 28). The difference in apparent transduction efficiencies between LIG4+/+ and LIG4−/− cells would therefore be minimized, especially at the highest MOI.
We also observed another feature of the experimental system that may cause an underestimate of the apparent transduction efficiencies. This is a saturation effect or “saturation threshold” for the fraction of cells expressing EGFP. For example, when the parental Nalm-6 (LIG4+/+) cells were infected with the 10−2 dilution of virus, 10% of cells were EGFP positive, while addition of 10 times more virus (10−1 dilution) resulted in an increase in EGFP-positive cells of only fourfold (to 37.6%). In contrast, addition of the same amount of virus to Nalm-6 LIG4−/− cells resulted in an increase of EGFP-positive cells of approximately 10-fold (from 2.9 to 20.3%), indicating that a saturation threshold may not yet have been achieved. To obtain an accurate estimate of the relative differences among these cultures by flow cytometry, it is therefore important to measure transduction efficiency at a relatively low MOI, as results with a high MOI may be obscured by saturation artifacts and loss of a large proportion of the infected cells due to apoptosis.
Integrase-dependent killing of ligase IV-deficient cells.
As noted above, results of our flow cytometry analyses suggested that HIV-1 integration triggered cell death in a significant proportion of Nalm-6 LIG4−/− cells. To test this possibility directly, we infected Nalm-6 LIG4−/− and control cells with the HIV-1 EGFP vector or with a vector that contained integrase protein with an inactivating substitution (E152A) at a MOI of approximately 1 and 2 infectious units/cell under our standard assay conditions. We observed that the integration-competent vector induced cell death upon infection of the Nalm-6 LIG4−/− culture in a titer-dependent manner, whereas no effect of the integration-defective vector on the viability of these cells at either titer was detected (data not shown). Similar results were obtained with a vector that contained integrase with another inactivating substitution, D64V. In the experiment summarized in Fig. 7, we again infected Nalm-6 LIG4−/− cells with the integration-competent and IN(E152A)-containing vectors using our standard conditions at a MOI of 2. At the same time, portions of these cells were treated with the same viral inputs, but in this case, using spinoculation (23) conditions as described by Li et al. (20) in which the cell-virus mixtures are subjected to centrifugation.
Using standard conditions, we observed a sharp decrease in the viability of cells infected with the integration-competent vector by 16 h postinfection but not with the IN(E152A)-containing vector (Fig. 7A). As with infection of scid pre-B cells (9, 10), the percentage of viable cells in the ligase IV-deficient culture continued to decrease until about 24 h postinfection. However, uninfected cells and a fraction that survived the infection (owing to contribution of compensatory proteins or repair via an NHEJ-independent pathway) should continue to divide, and the number of viable cells in the culture starts to increase after this time, as expected. Therefore, these results are consistent with a role for ligase IV in repair of damage incurred during viral DNA integration and death of the fraction of cells in which repair cannot take place.
A different result was obtained with the cells that were subjected to spinoculation. In this case, the same viability decrease was seen with both integration-competent and integration-defective vectors (Fig. 7A). These method-dependent differences are also apparent in a comparison of cell numbers relative to the mock-infected and mock-spinoculated cultures (Fig. 7B). We conclude from these experiments that use of the spinoculation procedure can obscure the differences in the responses of these cells to integration-competent and integration-defective vectors.
DISCUSSION
In the work reported here, we have extended our initial findings, which indicated that the cellular NHEJ DNA repair pathway plays a role in completion of the retroviral DNA integration process (9). Subsequent to our initial report, we described several other studies that support this interpretation (7, 8, 10, 28). Furthermore, studies from several laboratories have indicated that there is critical interplay between retroviruses or retrotransposons and the NHEJ pathway (1, 11, 16, 20). In essentially all cases, deficiencies in NHEJ have resulted in deficiencies in either transposition, transduction, or cell survival. Furthermore, components of the NHEJ pathway have been shown to be physically associated with the retroviral preintegration complex or retrotransposon virus-like particles (11, 20). However, two apparent discrepancies have been reported in the literature related to the timing of retrovirus-mediated killing of NHEJ-deficient cells and the effect of an integration-deficient vector. These results have been interpreted to suggest that the NHEJ pathway is not required for efficient lentivirus DNA integration (1) or is acting solely on unintegrated viral DNA (20). In the present study, we have included analyses with HIV-1 and ASV vectors, using transduction of reporter genes as a readout for successful completion of the integration process and survival of the transduced cells. In addition to gaining further evidence for a role of NHEJ in the retroviral integration process, this work allowed us to address specific questions concerning these apparent discrepancies.
Effect of the scid mutation on the transduction efficiency of MEFs and fibroblast lines.
Our earlier experiments indicated that the efficiency of ASV vector-mediated transduction is decreased in cultured cells that lack DNA-PKCS. We hypothesized that a substantial percentage of such cells do not survive infection because they are unable to repair the damage incurred as a consequence of integration (9). In the present study, we have performed additional analyses using two VSV G-protein-pseudotyped HIV-1 vectors and DNA-PKCS-deficient scid MEFs. The results (Fig. 1, 2, and 4) indicate that, as we had observed earlier with an amphotropic ASV vector, transduction of the scid MEFs by the two HIV-1 vectors was 5- to 10-fold lower than with normal MEFs. We have also reexamined the effect of the scid mutation on transduction by an ASV vector with an ecotropic envelope that encodes a placental alkaline phosphatase reporter gene under control of the RNA polymerase II promoter. We have again observed that transduction of scid MEFs by this vector was reduced approximately 10-fold from that of normal MEFs (Fig. 5). Therefore, we conclude that DNA-PKCS is required for efficient transduction or survival of MEFs transduced by both ASV and HIV-1 vectors, that this requirement can be monitored by expression of a variety of reporter genes, and that it is independent of the cell surface receptor used for vector entry. Finally, we have shown that scid cells that have been complemented with a wild-type DNA-PKCS gene are transduced with increased efficiency (∼10-fold) (Fig. 3). These data therefore support our hypothesis that the reduced transduction efficiency of scid MEFs is due to the mutation in the DNA-PKCS gene.
To test additional aspects of our experimental design that might be a source of the noted discrepancies, we infected scid and normal MEFs at different cell densities and compared the transduction efficiencies with those of the HIV-1 vector. We reasoned that contact inhibition of cell growth might influence either viral infection, DNA repair, or the apoptotic response. We observed that infection of confluent, contact-inhibited normal MEFs by the HIV-1 lacZ vector is less efficient than infection of cycling MEFs, consistent with published data (19, 27). However, the same relative reduction in the number of stable transductants in scid MEFs compared to normal MEFs was observed at all cell densities tested (Fig. 5). These results show that contact inhibition of wild-type and scid cells had no effect on the relative differences in transduction efficiencies. We note that our conclusions regarding possible cell cycle effects are limited to this cell culture system. For example, it is possible that characteristics of differentiated cell types or their cycle status might be relevant to the requirement for NHEJ in retroviral transduction. With respect to culture systems in which cells are dividing, our computational modeling suggests that passage through S phase is required to trigger apoptosis in infected cycling scid pre-B cells (10). Baekelandt et al. (1) also reported that induction of scid cell death required cell passage. However, it is possible that some terminally differentiated, noncycling NHEJ-deficient cells do not die upon failure of an integration intermediate to be repaired via this pathway. In these circumstances, repair might eventually be completed via alternative pathways.
Results of our infections of MEFs with the lentiviral HIV-1 vector were all consistent with those from previous analyses with an alpharetroviral ASV vector. As we reproduced conditions described by Baekelandt et al. (1), it remains unclear why these researchers failed to observe a difference in transduction efficiency of scid and normal MEFs, as other results in their report supported our previous findings.
Xrcc4 and ligase IV are also required for survival and efficient transduction by HIV-1.
We found that transduction efficiency is also reduced in Xrcc4-deficient CHO cells infected with an HIV-1 vector (Fig. 6). Xrcc4 is known to form a complex with ligase IV and stimulates its activity in the NHEJ repair pathway (14). Our analysis of retroviral transduction of a ligase IV-deficient lymphoid cell line showed an approximately threefold reduction compared to the parental line (Table 1). This finding again supports our proposal that NHEJ proteins are required for efficient, stable retroviral transduction.
As reported by Li et al. (20), we have also observed that an HIV-1 vector carrying a wild-type integrase protein kills these ligase IV-deficient lymphoid cells. However, using the same cell lines and equivalent vectors, we find that an integration-defective vector does not kill such cells under standard infection conditions. One major difference in experimental procedures was the use of spinoculation (23) by these investigators. As they have noted, this method of infection, in which viral particles are forced onto the surfaces of cells by centrifugation, increases transduction efficiency 5- to 10-fold, presumably by increasing proportionately the number of particles that enter these cells. Our results (Fig. 7) show that spinoculation can obscure the differences between integration-competent and integration-deficient viruses. It is possible that in this case cell viability is compromised by some other mechanism; for example, exposure of cells to massive amounts of unintegrated viral DNA and/or vector-associated Vpr protein (24) may activate DNA damage responses under these conditions.
In summary, we present here additional evidence that cellular NHEJ DNA repair is required for efficient stable transduction and survival of lentivirus-infected cells. We note that in all of our experiments with NHEJ-deficient cells, including those deficient in ligase IV, some stably transduced cells are always obtained. Such cells can be transduced, although at lower efficiencies than NHEJ-proficient cells. These results indicate that there is an alternative, NHEJ-independent pathway(s) for repair of the retroviral DNA integration intermediate, as we have suggested previously (8, 9). A detailed understanding of the molecular mechanisms that govern the cellular response to damage incurred when retroviral DNA is joined to host chromatin will contribute to our knowledge of molecular aspects of both retroviral replication and cellular DNA damage response pathways and may suggest novel strategies for development of antiviral therapies.
Acknowledgments
This work was supported in part by National Institutes of Health grants AI40385, CA71515, CA06927, and CA98090, a Tobacco Formula Research Fund grant from the Pennsylvania Department of Health, and an appropriation from the Commonwealth of Pennsylvania.
We are grateful to M. Bosma (Fox Chase Cancer Center), M. Oettinger (Harvard Medical School), D. Roth (Baylor College of Medicine), E. Barsov (National Institutes of Health), M. Lieber (University of California at Los Angeles), and F. Bushman (Salk Institute) for providing cell lines and plasmids used in our studies. We also thank J. Taylor and W. Mason for critical reviews of our manuscript. The following Fox Chase Cancer Center Shared Facilities were used in the course of this work: Cell Culture Facility, Biochemistry and Biotechnology Facility (DNA Synthesis), and Research Secretarial Services.
The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute or any other sponsoring organization.
REFERENCES
- 1.Baekelandt, V., A. Clayes, P. Cherepanov, E. De Clerq, B. De Strooper, B. Nuttin, and Z. Debyser. 2000. DNA-dependent protein kinase is not required for efficient lentivirus integration. J. Virol. 74:11278-11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barsov, E. V., W. S. Payne, and S. H. Hughes. 2001. Adaptation of chimeric retroviruses in vitro and in vivo: isolation of avian retroviral vectors with extended host range. J. Virol. 75:4973-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, W. Y., X. Wu, D. N. Levasseur, H. Liu, L. Lai, J. C. Kappes, and T. M. Townes. 2000. Lentiviral vector transduction of hematopoietic stem cells that mediate long-term reconstitution of lethally irradiated mice. Stem Cells 18:352-359. [DOI] [PubMed] [Google Scholar]
- 4.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 5.Craig, N. L. 1996. V(D)J recombination and transposition: closer than expected. Science 271:1512. [DOI] [PubMed] [Google Scholar]
- 6.Critchlow, S. E., and S. P. Jackson. 1998. DNA end-joining from yeast to man. Trends Biochem. Sci. 23:394-398. [DOI] [PubMed] [Google Scholar]
- 7.Daniel, R., G. Kao, K. Taganov, J. G. Greger, O. Favorova, G. Merkel, T. J. Yen, R. A. Katz, and A. M. Skalka. 2003. Evidence that the retroviral DNA integration process triggers an ATR-dependent DNA damage response. Proc. Natl. Acad. Sci. USA 100:4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel, R., R. A. Katz, G. Merkel, J. C. Hittle, T. J. Yen, and A. M. Skalka. 2001. Wortmannin potentiates integrase-mediated killing of lymphocytes and reduces the efficiency of stable transduction by retroviruses. Mol. Cell. Biol. 21:1164-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniel, R., R. A. Katz, and A. M. Skalka. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644-647. [DOI] [PubMed] [Google Scholar]
- 10.Daniel, R., S. Litwin, R. A. Katz, and A. M. Skalka. 2001. Computational analysis of retrovirus-induced scid cell death. J. Virol. 75:3121-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Downs, J. A., and S. P. Jackson. 1999. Involvement of DNA end-binding protein Ku in Ty element retrotransposition. Mol. Cell. Biol. 19:6260-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eglitis, M. A., M. J. Kadan, E. Wonilowicz, and L. Gould. 1993. Introduction of human genomic sequences renders CHO-K1 cells susceptible to infection by amphotropic retroviruses. J. Virol. 67:1100-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2000. Principles of virology, molecular biology, pathogenesis, and control. ASM Press, Washington, D.C.
- 14.Grawunder, V., M. Wilm, X. Wu, P. Kulesza, T. E. Wilson, and M. Lieber. 1997. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388:492-495. [DOI] [PubMed] [Google Scholar]
- 15.Haas, D. L., S. S. Case, G. M. Crooks, and D. B. Kohn. 2000. Critical factors influencing stable transduction of human CD34+ cells with HIV-1-derived lentiviral vectors. Mol. Ther. 2:71-80. [DOI] [PubMed] [Google Scholar]
- 16.Jeanson, L., F. Subra, S. Vaganay, M. Hervy, E. Marangoni, J. Bourhis, and J. F. Mouscadet. 2002. Effect of Ku80 depletion on the preintegrative steps of HIV-1 replication in human cells. Virology 300:100-108. [DOI] [PubMed] [Google Scholar]
- 17.Kirchgessner, C. U., C. K. Patil, J. W. Evans, C. A. Cuomo, L. M. Fried, T. Carter, M. A. Oettinger, and J. M. Brown. 1995. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science 267:1178-1182. [DOI] [PubMed] [Google Scholar]
- 18.Leavitt, A. D., G. Robles, N. Alesandro, and H. E. Varmus. 1996. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J. Virol. 70:721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Z., T. Otevrel, Y. Gao, H.-L. Cheng, B. Seed, S. D. Stamato, G. E. Taccioli, and F. W. Alt. 1995. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell 83:1079-1089. [DOI] [PubMed] [Google Scholar]
- 22.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 23.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roshal, M., B. Kim, Y. Zhu, P. Nghiem, and V. Planelles. 2003. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J. Biol. Chem. 278:25879-25886. [DOI] [PubMed] [Google Scholar]
- 25.Sage, J., G. J. Mulligan, L. D. Attardi, A. Miller, S. Chen, B. Williams, E. Theodorou, and T. Jacks. 2000. Targeted disruption of the three Rb-related genes leads to loss of G1 control and immortalization. Genes Dev. 14:3037-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skalka, A. M. (ed.). 1999. Retroviral DNA integration, vol. 52. Academic Press, New York, N.Y.
- 27.Sutton, R. E., M. J. Reitsma, M. Uchida, and P. O. Brown. 1999. Transduction of human progenitor hematopoietic stem cells by human immunodeficiency virus type 1-based vector is cell cycle dependent. J. Virol. 73:3649-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taganov, K., R. Daniel, R. A. Katz, O. Favorova, and A. M. Skalka. 2001. Characterization of retroviral-host DNA junctions in NHEJ-deficient cells. J. Virol. 75:9549-9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, X., J. K. Wakefield, H. Liu, H. Xiao, R. Kralovics, J. T. Prchal, and J. C. Kappes. 2000. Development of a novel trans-lentiviral vector that affords predictable safety. Mol. Ther. 2:247-255. [DOI] [PubMed] [Google Scholar]