Abstract
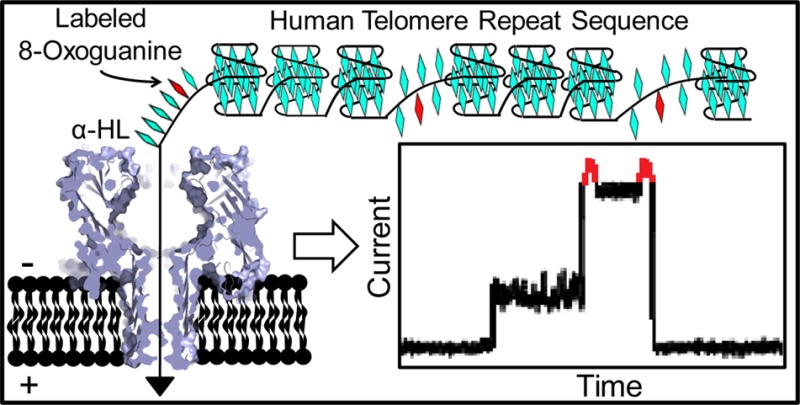
The human telomere repeat sequence 5′-TTAGGG-3′ is a hot spot for oxidation at guanine, yielding 8-oxo-7,8-dihydroguanine (OG), a biomarker of oxidative stress. Telomere shortening resulting from oxidation will ultimately induce cellular senescence. In this study, α-hemolysin (α-HL) nanopore technology was applied to detect and quantify OG in the human telomeric DNA sequence. This repeat sequence adopts a basket G-quadruplex in the NaCl electrolyte used for analysis that enters the α-HL channel, slowly unfolds, and translocates. The basket fold containing OG disrupts the structure, leading to >10× increase in the unfolding kinetics without yielding a detectable current pattern. Therefore, detection of OG with α-HL required labeling of OG with aminomethyl-[18-crown-6] using a mild oxidant. The labeled OG yielded a pulse-like signal in the current vs time trace when the DNA strand was electrophoretically passed through α-HL in NaCl electrolyte. However, the rate of translocation was too slow using NaCl salts, leading us to further refine the method. A mixture of NH4Cl and LiCl electrolytes induced the propeller fold that unravels quickly outside the α-HL channel. This electrolyte allowed observation of the labeled OG, while providing a faster recording of the currents. Lastly, OG distributions were probed with this method in a 120-mer stretch of the human telomere sequence exposed to the cellular oxidant 1O2. Single-molecule profiles determined the OG distributions to be random in this context. Application of the method in nanomedicine can potentially address many questions surrounding oxidative stress and telomere attrition observed in various disease phenotypes including prostate cancer and diabetes.
Keywords: human telomere, oxidative stress, G-quadruplex, α-hemolysin, 18-crown-6
Telomeres are located at the end of each chromosome for protection from cellular degradation and suppression of inappropriate activation of the double-strand break repair process.1 Human telomeric DNA has the repeat sequence 5′-TTAGGG-3′ copied for many kilobase pairs that reside in the duplex state. At the 3′ end, this repeat sequence persists as a single-stranded overhang of 100–200 nucleotides.2 Metaphase telomeres are bound by many proteins to generate the shelterin complex.1 The telomere is subject to shortening during the human lifespan and, therefore, has been referred to as a molecular clock for determination of cellular age. Increased telomere shortening has been correlated to chronic psychological stress, oxidative stress, and inflammation.3,4 The guanine (G)-rich nature of this sequence renders it prone to oxidative insults from reactive oxygen species (ROS) resulting from oxidative stress and inflammation.5 One of the chief G oxidation products found in the telomere is 8-oxo-7,8-dihydroguanine (OG).6 The telomeric concentration of OG increases with stress, because it evades the repair process.7,8 Persistent oxidative stress to the telomere leads to telomere attrition, which advances this molecular clock.9,10 However, the spatial distribution in which OG is formed in the telomere is not known; in other words, is OG preferentially formed in the single-stranded or duplex region of the telomere? Moreover, nuclease digestion of the genome followed by LC-MS can easily lead to inaccurate quantities of OG, because sample manipulation can cause G oxidation to OG.11 Therefore, a method for quantification of OG and determination of its spatial distribution in the telomere would provide a great benefit for molecular medicine, because these values provide a direct measure of the extent to which telomeres have been exposed to ROS. Further, understanding the distribution of OG will allow a better understanding of its role in telomere attrition. Knowledge of how OG concentrations increase with ROS is advantageous for providing preventative medicine to combat stressors that prematurely age the cell or lead to stress-induced diseases (e.g., cancer or diabetes).
Various methods have been developed to analyze human telomeric DNA;12 however, none of these methods can quantify OG in the telomere. Analysis of telomeres occurs via PCR-based methods or with fluorescently labeled probes, in which OG is silent.12 Single-molecule approaches to studying the human telomere repeat, such as optical tweezers13 and high-speed AFM,14 provide insight into these structures not available to averaged, bulk measurements. Another promising single-molecule platform for detection and quantification of OG in the telomere, as well as having the ability to potentially measure the telomere length, is nanopore technology. A commonly used biological nanopore is α-hemolysin (α-HL), which possesses a large nanocavity (vestibule) on the cis side, leading to a narrow β-barrel on the trans side with a central constriction separating these regions (Figure 1A).15 This nanopore senses single DNA or RNA strands while they are electrophoretically driven from the cis to the trans side of the channel, typically in KCl or NaCl electrolyte solution.16−19 The largest voltage drop occurs at the central constriction and β-barrel, providing the sensing capabilities. The similarity in diameter of single-stranded DNA (d = 1.0 nm)20 and the narrow β-barrel (d = 1.4 nm, Figure 1A)15 generates sequence-specific current–time signals as the DNA passes this narrow region.21,22 Active development in this field is applied to using these current–time patterns for single-molecule DNA sequencing.22−25 The single-molecule profiling capability of α-HL would be ideal for detection of OG in telomeres and to determine its distribution. DNA strands without secondary structure pass through the channel unabated, but the presence of hairpin and G-quadruplex (G4) structures impedes the movement of the strand.26−31 The electrophoretic force causes these secondary structures to unwind and eventually pass the channel, but this process can take >4 min.29 Most interestingly, the human telomere repeat sequence, in the absence of the complementary strand, adopts a G4 fold in the presence of KCl or NaCl salts.32 Therefore, an α-HL platform developed for analyzing human telomere sequences will need to address the ability of these G4-forming sequences to coordinate with the electrolyte cation that hinders movement of DNA as it is driven through the nanopore.
Figure 1.
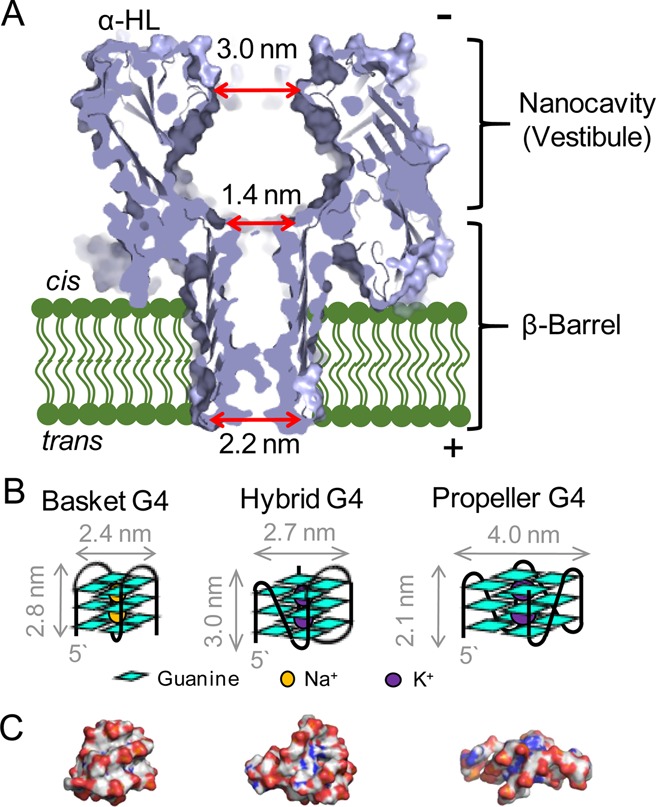
Structure of the α-HL nanopore and the observed hTelo G4 folds. (A) α-HL protein channel (pdb 7AHL)15 with critical regions and dimensions for this study labeled. (B) Cartoon drawings of three folds characterized from the hTelo sequence. (C) Space-filling models for the basket (pdb 143D),33 hybrid 1 (pdb 2JSQ),34 and propeller (pdb 1K8P)35 folds of the hTelo sequence.
Human telomeric DNA adopts hybrid, basket, or propeller G4 folds in the presence of K+, Na+, and K+ with high concentrations of Li+, respectively (Figure 1B and C).28 Previously, we demonstrated the ability of α-HL to analyze these three G4s and their drastically different unraveling kinetics.28 While the hybrid and basket folds with a 25-mer 5′-tail can enter tail first into the nanocavity of the protein and unravel slowly in this confined environment (∼0.1 to ∼240 s, respectively), the propeller fold is unable to enter because it is too big to fit through the opening of the vestibule (Figure 1).28 The size-selective properties of α-HL force the propeller fold to unravel outside of the protein nanocavity, where it can do so much faster (0.004 s) due to the greater degrees of freedom in this open space.28 Further, when the tail was removed, the hybrid and basket folds entered the nanocavity on the cis side, while the propeller fold did not.28,29,36 Interestingly, the hybrid folds without a tail, once trapped in the nanocavity, exited via the same side they entered (i.e., they could not unravel), while the basket folds are capable of unraveling and moving through the β-barrel to the trans side of the pore.28 Additional studies of the thrombin-binding aptamer G4 by Gu and co-workers demonstrated monovalent and divalent cation-dependent tuning of the unraveling kinetics for this G4 in the α-HL nanopore.30 Maglia and co-workers studied the thrombin-binding aptamer bound to thrombin in a large vestibule protein nanopore (ClyA) and demonstrated current modulations dependent on conformational heterogeneity of the complex.37 In this report, α-HL was used to detect and quantify a biomarker for oxidative stress, OG, in human telomeric G4s. Systems of increasing complexity were studied, starting from single G4 sequences bearing site-specifically synthesized OGs and expanding to a system that contains five G4s, in which OG was introduced nonspecifically via exposure to the ROS 1O2. In this final study, we demonstrate the ability to detect and quantify more than one OG lesion site. Achievement of this final step required repurposing of established chemistry for labeling OG with a detectable crown-ether marker developed in our laboratory.38 Furthermore, application of our knowledge of ion-dependent G4 unfolding kinetics allowed the development of an analytical method for detecting and quantifying these crown-ether markers covalently attached at OG in the human telomere sequence using α-HL.
Results and Discussion
Studies with Single G4s Bearing Site-Specifically Synthesized OG
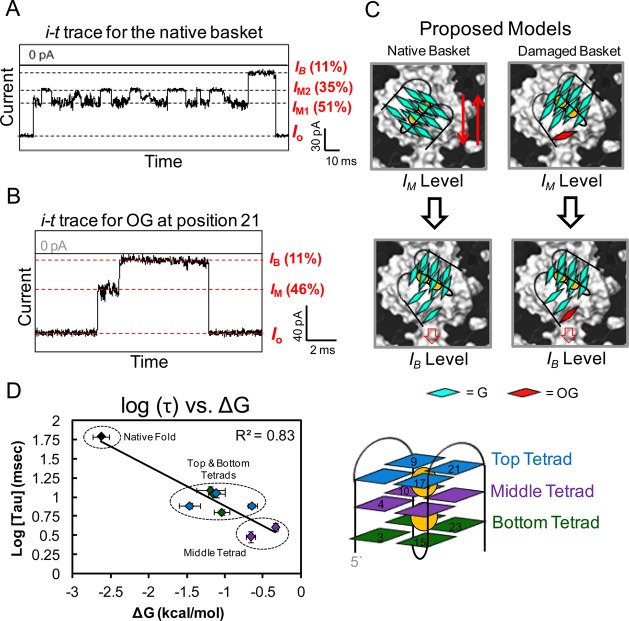
Preliminary studies were conducted with 24-mer synthetic DNA strands containing site-specifically synthesized OG in the human telomere sequence (hTelo). The natural form of the sequence was studied (5′-TAGGGTTAGGGTTAGGGTTAGGGTT-3′).34 The OGs were synthesized at sites demonstrated to be hot spots for G oxidation in this sequence (underlined Gs, Figure S1).39 The electrolyte NaCl (1 M) was selected, which induces the basket fold in the hTelo G4.28 Translocation of the OG-containing G4s at 120 mV (trans vs cis) yielded different ion current vs time (i–t) traces when they were driven through α-HL from the cis to trans side compared to the undamaged sequence (Figure 2A and B; Figures S2–9). The i–t traces recorded for the OG-containing G4s led to current patterns that initiated from the open channel current (Io) to an intermediate shoulder level (IM) followed by transitions to a clean deep blocking level ion current (I, Figure 2B) for >90% of the recorded events. Currents are reported as percent residual current vs the open channel current (i.e., %I/Io). As a result, the intermediate blocking level current showed a broad distribution of %IM/Io = 46 ± 5%, while the deep blockage current level was %I/Io = 11 ± 1%. The IM values showed stochastic fluctuations between many different current levels. In contrast to the observations for OG-containing G4s, the undamaged sequence presented i–t traces with unique features. First, distinct modulations between two intermediate current levels were observed (%IM1/Io = 51 ± 1%, and %IM2/Io = 35 ± 1%), and they modulated between these levels randomly. Eventually, the current transitioned to a deep blockage current level (%I/Io = 11 ± 1%) followed by a return to the open channel current (Figure 2A). These features were previously ascribed to the folded basket G4 moving up and down in the nanocavity to give two IM values, followed by an unfolding process that leads to translocation through the channel (Figure 2C).28,36 The native fold showed a second current pattern that we previously described as loop side entry, but these types of events were not observed in significant amounts (<10%) for the OG-containing strands.28,36 In contrast to the native fold, the OG-containing G4s gave a broad distribution of shoulder current levels, which is interpreted as damaged G4s entering the vestibule in a partially unfolded state that quickly progressed to the translocation state observed as a deep blockage to the current level (Figure 2B and C). On the basis of the current–time traces recorded for the OG-containing G4s, a triplex-like structure, which was previously characterized by the α-HL nanopore in our laboratory,29,36 might account for these changes from wild-type G4s. The triplex structure has also been characterized by optical tweezers40 and CD spectroscopy.41
Figure 2.
Nanopore analysis of hTelo basket folds with site-specifically incorporated OG damage. The hTelo DNA strand studied had OG synthesized at one of the underlined G’s in the sequence 5′-TAGGGTTAGGGTTAGGGTTAGGGTT-3′, in which OG placement in the top tetrad was at positions 9, 17, and 21 (blue), in the middle tetrad at positions 4 and 10 (purple), and in the bottom tetrad at positions 3, 15, and 23 (green). (A) Current vs time trace for the native fold. (B) Representative current vs time trace for OG incorporated at position 21 of the hTelo sequence. The Io, IM, and IB values are labeled as a percentage of the open channel current Io for panels A and B. (C) Proposed model to describe the data collected. (D) Plot of log τ vs ΔG for all damaged and undamaged hTelo sequences studied. The ΔG values were obtained by van’t Hoff analysis from the UV-monitored melting curves (Figure S10). All studies were conducted in 1 M NaCl and 25 mM Tris, pH 7.9, at 25 °C. The measurements were done under 120 mV (trans vs cis) bias. The data shown in panel A were previously described by our laboratory in An, N.; etal. Proc. Nat. Acad. Sci. U.S.A.2014, 111, 14325–14331.
The duration times for the events were measured (IM + IB) to provide a population that was plotted and fitted to a single-exponential decay model with a decay constant τ (Supporting Information Figures S2–9). The mean translocation time τ was compared between each OG position and to the undamaged fold. The undamaged basket fold, without a homopolymer tail, gave long τ values (∼110 ms), while those measured for the OG-containing G4s (τ = 1–13 ms) were much faster, indicating that the presence of oxidative damage decreased the stabilities of these G4s (Figure 2A, B, D and Figures S2–9). Because the hTelo G4s have three tetrads, the location of the oxidative damage should also be a factor in its destabilization. Incorporation of OG at various positions presented rather different translocation decay constants. Placement of OG in a top (positions 9, 17, and 21) or bottom (positions 2, 15, and 23) tetrad resulted in destabilization and unfolding of only that layer leading to the longest τ values, whereas formation of OG in the middle (positions 4 and 10) tetrad led to more substantial unfolding of the G4 and yielded the shortest τ values measured (Figure 2D). Additionally, the event durations for these G4s to traverse from the cis to trans side of the protein channel had τ values that exhibit a linear correlation with the unfolding free energy, ΔG. The ΔG values were determined from van’t Hoff analysis of thermal melting curves monitored by UV spectroscopy (Tm)42 that were measured for the native and each damaged G4 studied (Figure S10). Surprisingly, breaking a single hydrogen bond (G → OG) could dramatically destabilize the G4 structure [+1–2 kcal/mol (ΔΔG) or −20 °C (ΔTm)] and cause a >10× faster translocation time than the undamaged sequence (Figure 2D and Figure S11). The destabilization effects of OG were further confirmed by circular dichroism (CD) and Tm analysis (Supporting Information, Figures S2–9). In all cases, the introduction of OG in the hTelo sequence caused significant destabilization, leading to fast unraveling kinetics, an observation that complements previous NMR43 and Tm analyses,44,45 suggesting that the presence of OG disrupts the folded structure.
These observations in the singly oxidized G4 system demonstrate that the presence of OG could be detected from the significant reduction in the τ value. Further, the i–t trace patterns changed when OG was introduced, which leads to an observable pattern for determining the presence of OG in these sequence contexts. Next, these studies were extended to a system of greater complexity that contained a 5′-homopolymer tail to determine if the location of one or more OGs could be detected. The tail was composed of 25 nucleotides of 2′-deoxyadenosine (dA25) that aid in the capture and threading of the G4 into the nanopore vestibule.46 The 5′ tail was chosen based on how telomeres are analyzed. Telomere terminal fragments are made by clipping the chromosome end off with restriction endonucleases that yield the repeat sequence on the 3′ end with the heterosequence on the 5′ end.12 Consequently, as the first step, a 5′-poly(dA) tail was attached to a sequence that adopts a single G4 fold. The sequence was studied without damage (Q1-G, Figure 3A and B) and with OG positioned at the 5′-most G in an exterior tetrad (Q1-OG, Figure 3A and B), which is a reactive site for oxidation found in our earlier studies.39 One critical detail about the 5′-poly(dA) tail is that this sequence gives a broad distribution of intermediate current levels and durations times during entry into the pore;46 this drawback is outweighed by the fact that it will not interact with the G-rich sequence to which it is attached as would a poly(dC) tail or introduce two different populations of translocation times as observed with poly(dT) tails.46
Figure 3.
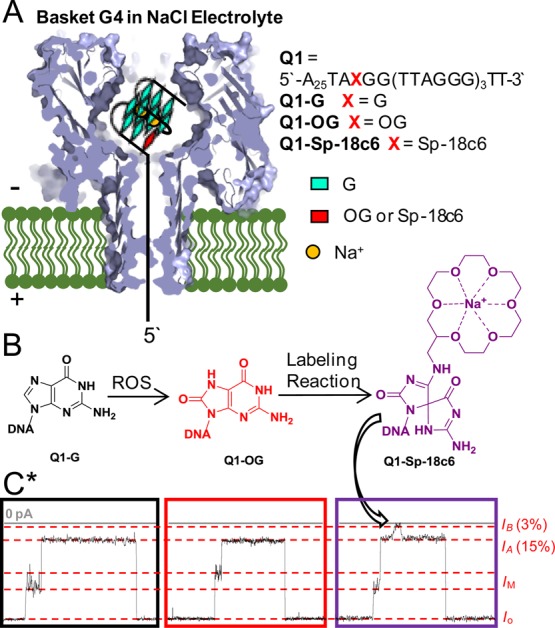
Translocation experiments for native, OG-containing, and 18c6-labeled G4s with a 5′-tail. (A) Scheme for experimental setup, including the G4 sequence studied and location of the OG and Sp-18c6-labeled OG. (B) Reaction scheme for G conversion to OG by ROS, followed by labeling of OG with aminomethyl-18c6 (18c6) in the presence of the mild one-electron oxidant K2IrBr6 to furnish the labeled OG product, Sp-18c6. (C) Current vs time traces for Q1-G (black box, left), Q1-OG (red box, middle), and Q1-Sp-18c6 (purple box, right). These traces demonstrate that OG does not yield a change in the current until it has been chemically labeled with 18c6. All studies were conducted in 1 M NaCl and 25 mM Tris, pH 7.9, at 25 °C. All measurements were done under 120 mV (trans vs cis). *The current vs time traces are on the same current axis, but they are on different time scales. These traces are intended to depict that labeling of OG with 18c6 in the hTelo G4 can result in a unique and characteristic modulation of the deep current level that was not observed for G or OG. See Figure S12 for the data shown on the correct scales.
Translocation of Q1-G and Q1-OG in NaCl electrolyte was studied. The 5′-polydA tail reinforced entry of these strands from the 5′ end (∼85%) while minimizing entry from the 3′ end with the G4 (∼15%) because the larger G4 was on the 3′ terminus. The current pattern for Q1-G and Q1-OG initiated with a broad distribution of shoulder current levels (%IM/Io = 48 ± 5%) that transitioned to a deep blockage current (%IA/Io = 15 ± 1%) and ended with a return to the open channel current (Figure 3C left and center, Figures S13–16). On the basis of the currents recorded for Q1-G and Q1-OG, a difference was not detected. Next, examination of translocation time revealed τ for Q1-G (τ ≈ 60 ms) to be 12 times longer than measured for Q1-OG (τ ≈ 5 ms, Figure S12). The decreased time observed for Q1-OG relative to Q1-G is consistent with the thermodynamic destabilization of OG identified above. The change in τ upon oxidation of G to OG suggests the damage could be determined based on translocation time; however, a telomere from a cellular source will possess ∼104 5′-TTAGGG-3′ repeats that can fold to series of ∼103 G4s. The predicted time for translocation of a relevant system would be >60 s, where the time distribution will become very broad due to diffusional and thermal factors.47 Further, biological samples will be heterogeneous and not allow a population of identical molecules to be studied, as we have done here. Based on our previous studies, a better approach for detection of damaged bases during single-molecule profiling is through the introduction of a marker that modulates the current.38 Under this proposed approach, changes in current levels can simply be observed and counted for the detection and quantification of telomeric OG. Therefore, to develop this method, a labeling strategy needed to be established.
Detection of OG through Chemical Labeling
Previously, we demonstrated the ability of α-HL to detect aminomethyl-[18-crown-6] (referred to as 18c6) attached to an abasic site (AP-18c6) in single-stranded DNA in NaCl electrolyte solution.38 The interaction between the 18c6-Na+ complex and the protein β-barrel allowed recording of the diagnostic pulse-like current signature. A mechanism was proposed for this signature. Due to its size and rigidity, 18c6-Na+ hesitates at the protein constriction, stalling the movement of the DNA molecule and permitting an accurate current recording of the nondamaged part of the molecule. After dissociation of the Na+ ion, the bulky 18c6 adduct passes through the constriction and generates a deeper current blockage that returns to a deep current level and then back to the open current value when the DNA has passed through the channel.38 These transitions in current levels result in a characteristic pulse-like pattern to the current that identifies the presence of the 18c6 adduct in NaCl electrolyte. When KCl was the electrolyte, the 18c6-K+ complex was too stable and prevented the DNA strand from passing through the channel and did not give an observable pulse-like signal.38 The ability to detect the 18c6-Na+ complex is particularly prominent when the DNA is threaded from its 5′ terminus. Labeling DNA with PEG,48 peptide,49 or benzo[a]pyrene50 adducts has also generated similar pulse-like signatures in the i–t traces. This knowledge was used as a starting point for labeling OG with an 18c6.38
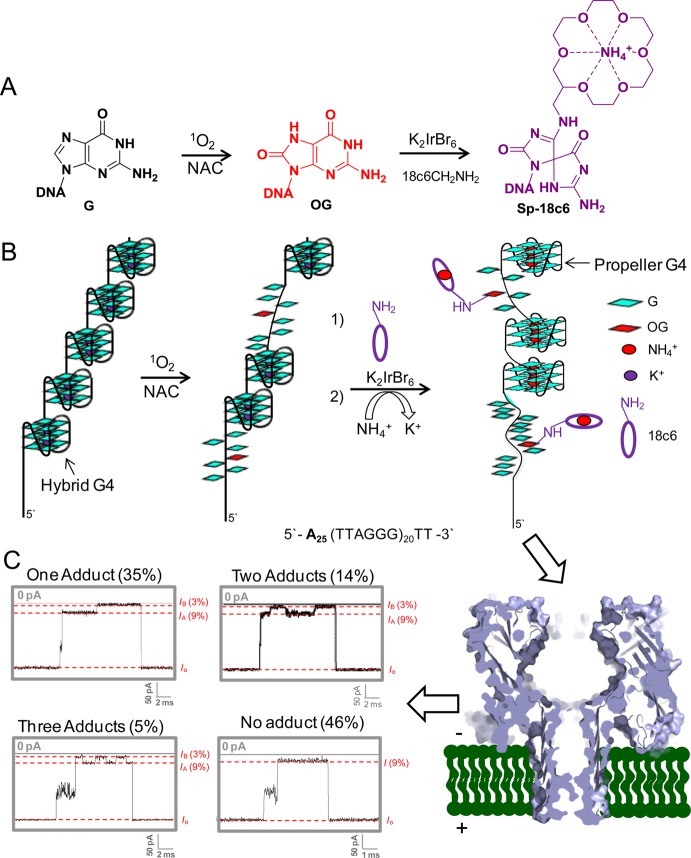
Before conducting the analysis, labeling of OG with 18c6 must be conducted. Installation of the 18c6 label was achieved using the mild one-electron oxidant K2IrBr6, which is selective for oxidizing OG. Oxidation of OG yields an electrophilic intermediate that is trapped by a nucleophilic primary amine in quantitative yield to furnish an adduct that rearranges to a stable spirocyclic structure.51,52 The 18c6 chosen has the desirable primary amine functionality, in which optimization reactions were conducted to furnish the labeled product, termed Sp-18c6, in >99% yield (Figure 3B and Figure S17). A test case was studied with an OG-containing G4 that was labeled with 18c6 using these conditions for which the yield was >99% (Figure S18); moreover, the structure of this adducted G4 sequence led to a CD spectrum that indicated a highly disrupted structure, similar to that observed for the triplex fold (Figure S18).29,40 A similar method for labeling of genomic OG was pursued by Greenberg and co-workers using a spermine-functionalized biotin.53,54
Next, nanopore analysis conditions were established to permit detection of the Sp-18c6 label in a control strand of the human telomere sequence. For this analysis, the Q1-OG strand was labeled with 18c6 using the best conditions identified above, to yield the product strand, termed Q1-Sp-18c6 (Figure 3C; Figures S17 and S18). Translocation of this strand through α-HL with 1 M NaCl provided a new i–t pattern that was not observed in the previous studies with Q1-G or Q1-OG. This new i–t trace changed from the open-channel current to a shoulder current level (%IM/I = 36 ± 5%) that progressed to an initial deep blockage current level (IA; %IA/I = 15 ± 1%). After a few milliseconds, the current flow decreased more to an even deeper blockage (IB; %IB/I = 3 ± 1%) that persisted for ∼1 ms and then returned to the IA current level that lasted many milliseconds before returning back to the open channel current (Figure 3C, Figure S19 and S20). The IA → IB → IA current pulse provides the readout for the labeled OG site (i.e., Sp-18c6); further, this observation is consistent with our previous results when 18c6 was used for detection of an AP site in DNA.38 The slow kinetics of ion release from the 18c6-Na+ complex provides ample time to obtain a good signal-to-noise ratio for this recording to ensure its accuracy. The overall translocation time for Q1-Sp-18c6 was faster (12× at 120 mV, Figure S12C) than that observed for the native Q1-G. This observation was expected because unraveling the G-quadruplex is a much slower process than ion release from 18c6, and based on CD analysis (Figure S18), the 18c6 label greatly destabilized the G-quadruplex, leading to the much faster time observed. This successful observation then pointed our attention to method development for handling a biological sample. A sample that originates from a cell will have ∼103 G4s, and the distribution and number of OG sites will not be known. Therefore, a method for increasing the rate of G4 unfolding while not interfering with cation release from 18c6 needed to be developed to handle a system of such large size.
Selection of a New Electrolyte System to Analyze Long hTelo Repeat Sequences
Human telomeric DNA can adopt various conformations upon coordination with different metal ions;55 additionally, 18c6 coordinates with many cations with variable binding constants.56 In the presence of 1 M NaCl or KCl solution, hTelo folds into a basket or hybrid G4, respectively, both of which are capable of entering the nanocavity of α-HL, where the kinetics of unfolding are slow, based on our previous results.28 Thus, these standard electrolytes at 1 M concentration will not aid in increasing the unfolding kinetics of the long runs of G4s. The hTelo can adopt the propeller fold in low concentrations of KCl (20 mM) under dehydrating conditions (5 M LiCl),28,57 which has the ideal G4 unraveling kinetics due to its large size, preventing this fold from entering the nanocavity of α-HL. However, when 18c6 is coordinated with K+, the binding constant is too large, preventing ion release that is required for yielding the pulse-like current signature.38 For these reasons, we searched the literature for a cation that has a moderate binding constant with 18c6, similar to Na+, while having a size similar to K+ that could potentially give propeller G4 folds. The NH4+ ion fits all of these requirements. NH4+ and Na+ have similar binding constants with 18c6 (Na+Keq = 6.3; NH4+Keq = 12.6; K+Keq = 115).56 The ionic radii for K+ and NH4+ are also similar (K+r = 1.33 Å; NH4+r =1.43 Å).58 Studies with NH4Cl solutions were conducted to determine if we could obtain the desired affects with regard to G4 folding and 18c6 ion release.
First, we tested the capability of 100 mM NH4+ with added LiCl to induce the propeller fold in the hTelo sequence that adopts one G4. The LiCl was added to increase the ionic strength that is preferable for recording the nanopore i–t traces, as well as the fact that LiCl in high concentration is a dehydrating agent that induces the propeller fold.28 CD spectroscopy was used to monitor the G4 folding topology when 100 mM NH4Cl was titrated with increasing amounts of LiCl. The addition of >2 M LiCl gave a CD peak of positive rotation at 260 nm that is diagnostic of the propeller fold.59 From these studies, the hTelo sequence adopts the preferred propeller fold with 100 mM NH4Cl and >2 M LiCl (Figure 4A).
Figure 4.
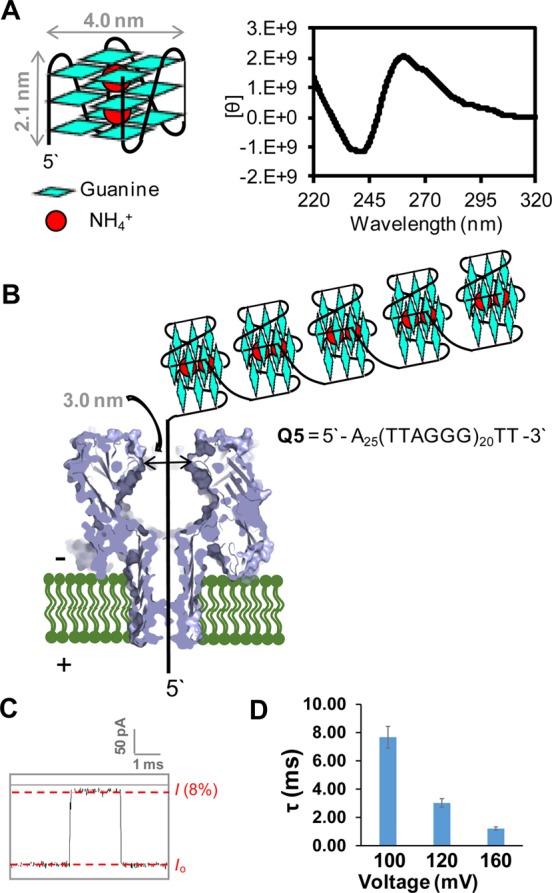
Propeller fold of the hTelo sequence observed in a mixture of NH4Cl and LiCl electrolyte and translocation studies of Q5 in this electrolyte system. (A) Propeller fold dimensions and characterization by CD spectroscopy. A peak with positive rotation at 260 nm in the CD spectrum was observed for hTelo in NH4Cl (100 mM) and 2 M (LiCl) characteristic of the propeller fold. (B) Scheme for translocation of Q5 that included up to five G4s in the propeller fold in a series with NH4Cl and LiCl electrolyte. (C) Typical current vs time trace for translocation of Q5 and the mean translocation time τ vs voltage for α-HL nanopore studies on Q5. (D) Voltage dependence in the mean translocation time τ studied at 100, 120, and 160 mV (trans vs cis). All studies were conducted in 25 mM Tris, pH 7.9, at 25 °C.
Second, the Q1-G sequence was studied with the NH4+/Li+ electrolyte system to ensure the translocation kinetic profile for the propeller fold was the same under these conditions as we previously measured using KCl (20 mM) and LiCl (5 M).28 This control study gave fast unzipping and translocation times, and the voltage-dependent study supports the translocation of this strand through the nanopore, because as the voltage was increased, the translocation time decreased (Figure 4D; Figures S21–S23). Further, in the NH4+/Li+ electrolyte, the Q1-G strand translocated ∼100× faster than observed in the NaCl electrolyte (Figures S12A and S23), as expected because the propeller fold (NH4+/Li+) unravels outside the nanocavity, while the basket fold (Na+) unravels inside the nanocavity.28 These results validated the conclusion that use of the NH4+/Li+ electrolyte composition led to the desired G4 unfolding rates. Next, a study was conducted with Q1-Sp-18c6 to determine if the crown ether binding constant was tuned to yield the characteristic pulse pattern that identifies the presence of the labeled OG (Figures S24 and S25). In this study, when Q1-Sp-18c6 was translocated through α-HL using 100 mM NH4Cl and 2 M LiCl as the electrolyte, the i–t traces had the IA → IB → IA current pulse signature characteristic of the OG adduct in the DNA strand. The pulse signature was a feature that was never observed in the Q1-G and Q1-OG samples; the pulse signature was observed only with 18c6 adducted DNA (i.e., Q1-Sp-18c6). The last control study was to address the 5′ vs 3′ entry bias for a sample with a 5′ propeller-folded G-quadruplex and for a system with a 5-tail and a 3′ triplex-like fold similar to the 18c6 adducted strand. First, with a propeller-folded G4 on the 3′ side, only 5′ tail entry was observed, consistent with our previous studies.28 Second, when a 3′ triplex-forming strand was studied with a 5′-poly(dA)25 tail, the 5′ entry was observed in ∼70% of the events (Figure S26). This observation is important because the pulse patterns were observed for Q1-Sp-18c6 in ∼50% of the total events. In our previous studies, the 18c6 label was observed only upon 5′ entry in ∼70% of the 5′ events,38 or put another way, at 120 mV bias, the 18c6 label was observed in ∼30% of the total events. In the present studies, the label was observed in a greater percentage (∼50%) of the total events due to the favorability of the 5′ entry. With these optimized conditions, experiments were conducted with hTelo sequences that contained a sufficient number of 5′-TTAGGG-3′ repeats to fold to more than one G4.
Analysis of a Long hTelo Sequence after Exposure to 1O2 to Detect and Quantify OG
There were two goals for the next set of studies. (1) Determine if the conditions developed to induce the propeller fold could be applied to the α-HL analysis of an hTelo sequence with enough repeats to yield two or more G4s. (2) Expose an hTelo sequence that can adopt up to five G4s to the ROS 1O2 and then determine if the 18c6 labeling and α-HL analysis conditions could detect and quantify the number of OGs in individual strands of the 120-mer hTelo sequence. To achieve the first goal, a series of strands were synthesized containing a 25-mer 5′-polydA tail with enough hTelo repeats to yield 1 (Q1-G), 2 (Q2-G), 4 (Q4-G), or 5 (Q5-G) G4s (see sequences in Figure S23 and data in Figures S27 and S28). Upon folding these strands in the NH4Cl/LiCl electrolyte solution, α-HL analysis determined that all systems of increased length could enter, unravel, and translocate through the nanopore. These claims are based on voltage-dependent studies that demonstrate for each system that the translocation time decreases as the voltage was increased (Figure 4B and C for Q5). Furthermore, as the strand became longer, so did the τ fitting value, as expected due to the longer length of the strands, and the increased number of G4s to be unraveled (Figure S23). These studies show that the increased length of the hTelo should not be an issue when analyzing long repeat systems capable of forming a series of G4s, as long as an electrolyte system is used to force the propeller topology, thus forcing the G4s to unravel outside the nanocavity where the rate of unfolding is much faster than inside.
To achieve the second goal, the Q5 strand was oxidized under physiologically relevant electrolyte concentrations, yielding OG that was labeled and then analyzed with the α-HL nanopore using NH4Cl/LiCl electrolyte. This Q5 strand represents the approximate length of the 3′-single-stranded region of the human telomere (∼100–200 nucleotides). The strand Q5 (10 μM) was allowed to fold under biologically relevant buffer and salt concentrations (20 mM phosphate buffer, pH 7.4, 12 mM NaCl, and 140 mM KCl at 37 °C).39 The presence of KCl under these conditions induces the hybrid folds that are proposed to exist in the cell.60 Oxidative damage to Q5 was incurred by the ROS singlet oxygen that was generated with the photooxidant Rose Bengal (0.1 mM); further, this reaction was supplemented with 3 mM N-acetylcysteine as a model for the cellular antioxidant glutathione (Figure 5A and B).39 After a 5 min exposure to 350 nm light, the reaction was stopped, and the reaction salts were removed with a desalting column. This oxidation is anticipated to mimic the oxidation reactions that occur inside of the cell that would produce a random distribution of the number of OGs and their positions in this G-rich strand. The damaged Q5 strand was then labeled with 18c6 using the optimized conditions found above, and the sample was submitted to α-HL nanopore analysis (Figure 5A and B).
Figure 5.
Oxidation of a 120-mer portion of the human telomere repeat sequence (Q5) with 1O2 to yield OG that was labeled with 18c6 followed by α-HL nanopore detection and quantification. (A) Reaction scheme for oxidation of hTelo by 1O2 to yield OG under physiological conditions (20 mM phosphate buffer, pH 7.4, 12 mM NaCl, 140 KCl, 37 °C). Scheme for the labeling reaction of OG by 18c6 in the presence of K2IrBr6. (B) Model of the Q5 strand in biologically relevant salts (hybrid G4) followed by oxidation labeling and refolding in NH4Cl (100 mM) and LiCl (2 M) electrolyte to yield the propeller fold. (C) Nanopore analysis and sample current vs time traces recorded by the α-HL nanopore that were conducted in 25 mM Tris, pH 7.9, 100 mM NH4Cl, and 2 M LiCl at 25 °C.
Examination of the oxidized and labeled Q5 strand in 2 M LiCl and 100 mM NH4Cl at 120 mV (trans vs cis) provided many different i–t trace types. Greater than half of the events contained pulse-like current modulations (IA → IB → IA). Overall, ∼35% of the events presented one modulation, ∼14% two modulations, and ∼5% three modulations per strand (Figure 5C and Figures S29–S32). A comparison of pulse patterns for a population of events with the same number of pulses provided considerable variability. For example, event types that had one pulse in the current showed an apparently random pattern of the pulse; examples of the pulse at the beginning, middle, or end of the event were observed. This stochastic nature was also observed with populations of events with two and three pulse patterns. The various locations of these modulations suggested different distributions of OG in the original sequence, but due to the stochastic nature of the analysis, this is only a qualitative observation. Analysis of the duration time vs the number of 18c6 pulses revealed that each pulse observed increased the mean translocation time τ by ∼1 ms (1 pulse, τ = 6.4 ms; 2 pulses, τ = 7.3 ms; 3 pulses, τ = 8.7 ms, Figure S33). Additionally, we noticed in our previous studies that strands with two 18c6 labels do not always produce multiple current modulations if the 18c6 labels are closely spaced.38 Thus, an exact count of how many OGs were present in a particular strand cannot be obtained. Nevertheless, the number of pulses does represent a lower limit of the number of OGs per strand. Furthermore, this method clearly identifies how random oxidative damage by 1O2, and likely any other oxidant, can be in the single-stranded portion of the telomere. By profiling these molecules one at a time, we can appreciate the great variability in which oxidation reactions occur to a single DNA strand by observing pulses to the current pattern. While not fully quantitative at this stage, this method is a vast improvement over other methods. Traditionally, gel electrophoresis would be used to monitor these reactions that yield a smear when random damage occurs, which we were able to recreate with this sequence. An alternative method to monitoring OG in long repeat sequences of the human telomere could be achieved with SMRT sequencing. This method relies on the kinetics of nucleotide insertion opposite OG to be different than that of G;61 however, when a single G4 sequence was in the template strand, the folded structure caused the polymerase insertion kinetics to change.62 Coupling a kinetic change for OG with a long series of G4s that alter the polymerase insertion kinetics of native DNA nucleotides will likely provide a challenge to the SMRT sequencing approach for identification of OG in the human telomere sequence. These observations further support the need for a method such as the one we have currently outlined.
Implications for Nanomedicine and Telomere Biology
A human cell has 46 chromosomes, each of which has two telomeres, leading to 96 telomeres per cell. Interestingly, no two telomeres have the same length, and this distribution can span many kilobases.63 Recent observations demonstrated that the greater the telomere length variability in prostate cancer cells, the more likely the cancer will metastasize or the patient will die from this cancer.64 Further, telomere length variability and oxidative stress show a casual correlation in prediabetic patients, who progress to Type II diabetes, in which increased oxidative stress decreases telomere length.65 More interestingly, the Greider laboratory demonstrated that cellular survival is determined by the shortest telomere and not the average length, which is typically measured.66 Further, the single-stranded region at the 3′ end of the telomere will likely be more reactive toward oxidation (>100×) when compared to the duplex region due to its greater solvent accessibility by diffusible free radicals.67 Telomere length distributions are determined by quantitative fluorescent in situ hybridization (Q-FISH),12 and OG levels are quantified by nuclease digestion followed by LC-MS analysis.11 These two methods do not overlap, and it requires both to answer these questions. Therefore, a method capable of addressing these questions simultaneously will be beneficial for studies on telomeres, oxidative stress, and their correlation with disease phenotypes. Single-molecule profiling experiments, such as the α-HL nanopore, could provide telomere length data while reading off the OG load.
The potential implications of the current work for nanomedicine applications are broad. Immediately, the current work can be put into production upon working out the conditions for harvesting telomere strands from the cell. One lingering question that this nanosensor could address surrounding telomeric OG is in regard to its distribution and how random or specific the location of damage occurs; more specifically, is the longer duplex region of the telomere exposed to more damaging oxidation reactions, or the shorter single-stranded region? The experiments and methods conducted in the present work provide a foundation using synthetic oligomers to study oxidative damage to the human telomere sequence. Once methods for extracting telomeres from a cell and preparing them for α-HL nanopore analysis have been developed, detailed studies to understand OG in the telomere can be commenced. We envision this method will struggle with determination of the absolute telomere length via translocation time, but these times will provide a first assessment of the length. The preference for 5′ entry over entry of a 3′ propeller G4 or triplex will enhance the pulse-like patterns observed. When kilobase-long DNA strands are eventually studied, the location of the pulse patterns will be more predictive than observed with the 120-mers in the current study. This straightforward analysis will determine if the single-stranded portion of the telomere sustains the greatest damage from oxidative stress. The utility of nanopore measurements for determining strand length via free translocation experiments is in its infancy;68,69 yet, this field is rapidly progressing, and this capability may soon be achieved. When these capabilities for determination of strand length via the α-HL nanopore are optimized, the labeling and analysis conditions outlined herein will benefit biologists and medical diagnosis. The oncological applications of this work are immense because telomeres are at the center of cancer’s immortality.70
Conclusions
Telomere attrition rates increase with oxidative and inflammatory stress.3,4 Oxidation of G to OG increases in telomeres exposed to oxidative insults.6 Herein, OG was detected by the α-HL nanopore in G4s that fold from the hTelo repeat sequence. Sequence lengths were increased to provide one to five G4s in a series. In the simplest single-G4 fold, OG caused significant distortion to the structure, leading to a >10× increase in the unfolding dynamics. Further, the tetrad where OG was placed (top/bottom vs middle) played a role in determining the degree of destabilization, with OG in the middle tetrad causing the greatest change (Figure 2D). When the G4 included a 5′ tail, the unfolding kinetics changed in the presence of OG, but this method of detection would not be suitable for hTelo sequences that can adopt many G4s. Therefore, a strategy for labeling OG with the methylamine derivative of 18c6 was established that, upon analysis with α-HL, demonstrated a pulse-like signature in the i–t trace for detection of OG (Figure 3C). This method was used to detect and provide a lower limit to the number of OGs found in a 120-mer stretch of the hTelo sequence exposed to 1O2 (Figure 5C). This study determined OG to be randomly formed under these conditions. Because the 5′ entry was reinforced with the presence of a 5′ homopolymer tail, the spatial distribution of the pulse signals allowed estimation of the relative position of OG, closer to the 5′ or the 3′ end. Analysis of telomeres for OG, a biomarker of oxidative stress, with the α-HL nanopore is an innovative approach that is envisioned to simultaneously allow quantification of OG and determination of telomere length in one experiment.
Methods
DNA Synthesis and Purification
All DNA strands studied were made by solid-phase synthesis by the DNA/Peptide core facility at the University of Utah following standard protocols. The OG phosphoramidites (Glen Research, Sterling, VA, USA) were incorporated via the manufacturer’s protocols at known sites of G oxidation in the hTelo sequence.39 After cleavage and deprotection of the strands they were purified on an ion-exchange HPLC column using the following mobile phases and method. Line A = 1.5 M LiOAc (pH 7) in 1:9 MeCN/ddH2O; B = 1:9 MeCN/ddH2O. The run was initiated at 35% B followed by a linear increase to 100% B over 20 min with a flow rate of 3 mL/min while monitoring the absorbance at 260 nm. Upon purification of the full-length product peak, the purification salts were removed by dialysis followed by lyophilization of the sample to dryness. The dry sample was resuspended in ddH2O to a working concentration of 500–1000 μM as determined by the 260 nm absorbance reading, using the primary sequences to determine the extinction coefficients.
Analysis of hTelo Strands with the α-HL Nanopore
A customized, low-noise amplifier and data acquisition system constructed by Electronic BioSciences Inc. (San Diego, CA, USA) was used for the ion channel recordings. All electrolyte solutions used in these studies were prepared from >18 MΩ/cm ultrapure H2O and filtered with a 0.22 μm Millipore vacuum filter. The wild-type α-HL protein nanopore was reconstructed in situ across a lipid bilayer supported by a glass nanopore membrane, which was made following a procedure previously described.71 The data were collected with a 100 kHz low-pass filter and an acquisition rate of 500 kHz. The sample i–t traces provided in the text were refiltered to 20 kHz for presentation purposes. QuB 1.5.0.31 and Igor Pro-6.1 were used to extract, analyze, and plot events that lasted longer than 10 μs. All nanopore measurements were conducted at 25 °C on samples with a concentration of 5 μM. Two independent trials (both nanopore and DNA sample) were conducted while collecting >500 events per trial.
Reaction for Labeling OG with 18c6 Methylamine
The labeling reaction for OG was conducted in a buffer and salt system that minimized folding of the hTelo sequences to any stable G4 structures (20 mM cacodylate pH 8.0, 100 mM LiCl). The DNA strand (10 μM) to be labeled was mixed with aminomethyl-[18-crown-6] (2 mM) in a 200 μL reaction vessel followed by incubation at 45 °C for 20 min prior to starting the reaction. The reaction was initiated by a bolus addition of K2IrBr6 (200 μM), and the reaction progressed for 30 min. The reaction salts and oxidant were removed by a NAP-5 column (GE Healthcare) following the manufacturer’s protocol. The column-purified sample was lyophilized and submitted to α-HL nanopore analysis.
Oxidation of Q5 with 1O2
Exposure of the 120-mer Q5 hTelo strand to the ROS 1O2 was conducted in a buffer salt mixture that mimicked cellular conditions. The Q5 strand (10 μM) was annealed in 20 mM phosphate buffer at pH 7.4 mixed with 12 mM NaCl and 140 mM KCl by heating to 90 °C for 5 min and then slowly cooling (∼4 h) to room temperature. The cooled sample was then exposed to oxidant. The Q5 strand (10 μM) was mixed with the photochemical oxidant Rose Bengal (0.1 μM) and N-acetylcysteine (3 mM) and then allowed to thermally equilibrate at 37 °C for 30 min prior to commencement of the reaction. The photosensitizer was initiated by exposing the sample to 350 nm light emitted by a sun lamp (300 W) that was held ∼7 cm above the reaction vessel with the lid open. After a 5 min reaction, the light was turned off. The Rose Bengal and reaction buffer were removed by a NAP-5 column (GE Healthcare) following the manufacturer’s protocols. Next, the oxidized Q5 strand containing OGs was labeled with methylamino-[18-crown-6] via the method outlined above. The conditions used to make OG in Q5 can lead to other products (e.g., spiroiminodihydantoin), and this reaction pathway was quenched with the N-acetylcysteine;39 however, it is anticipated that some amount of spiroiminodihydantoin does exist in the sample analyzed by the nanopore, which was not detectable by this method.
Acknowledgments
The authors gratefully acknowledge Electronic BioSciences (San Diego, CA, USA) for the donation of the instrument for recording the current–time traces, as well as Drs. Jan Riedl and Yun Ding for their critical reading of the manuscript. Funding for this work was provided by the National Institutes of Health (R01 GM093099).
Supporting Information Available
The hTelo sequences studied, example i–t traces, translocation time distributions, ion-current distributions, and HPLC analysis of the labeling reaction can be found in the Supporting Information file. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- de Lange T. How Telomeres Solve the End-Protection Problem. Science 2009, 326, 948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright W. E.; Tesmer V. M.; Huffman K. E.; Levene S. D.; Shay J. W. Normal Human Chromosomes have Long G-Rich Telomeric Overhangs at One End. Genes Dev. 1997, 11, 2801–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkowitz O. M.; Mellon S. H.; Epel E. S.; Lin J.; Dhabhar F. S.; Su Y.; Reus V. I.; Rosser R.; Burke H. M.; Kupferman E.; etal. Leukocyte Telomere Length in Major Depression: Correlations with Chronicity, Inflammation and Oxidative Stress - Preliminary Findings. PLoS One 2011, 6, e17837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan A.; Tomiyama A. J.; Lin J.; Puterman E.; Adler N. E.; Kemeny M.; Wolkowitz O. M.; Blackburn E. H.; Epel E. S. Stress Appraisals and Cellular Aging: A Key Role for Anticipatory Threat in the Relationship between Psychological Stress and Telomere Length. Brain Behav. Immun. 2012, 26, 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J.; Douki T.; Ravanat J.-L. Oxidatively Generated Damage to the Guanine Moiety of DNA: Mechanistic Aspects and Formation in Cells. Acc. Chem. Res. 2008, 41, 1075–1083. [DOI] [PubMed] [Google Scholar]
- O’Callaghan N.; Baack N.; Sharif R.; Fenech M. A qPCR-based Assay to Quantify Oxidized Guanine and Other FPG-sensitive Base Lesions Within Telomeric DNA. BioTechniques 2011, 51, 403–411. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Liu M.; Fleming A. M.; Burrows C. J.; Wallace S. S. Neil3 and NEIL1 DNA Glycosylases Remove Oxidative Damages from Quadruplex DNA and Exhibit Preferences for Lesions in the Telomeric Sequence Context. J. Biol. Chem. 2013, 288, 27263–27272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee D. B.; Ghosh A.; Lu J.; Bohr V. A.; Liu Y. Factors That Influence Telomeric Oxidative Base Damage and Repair By DNA Glycosylase OGG1. DNA Repair 2011, 10, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epel E. S.; Blackburn E. H.; Lin J.; Dhabhar F. S.; Adler N. E.; Morrow J. D.; Cawthon R. M. Accelerated Telomere Shortening in Response to Life Stress. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 17312–17315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zglinicki T. Oxidative Stress Shortens Telomeres. Trends Biochem. Sci. 2002, 27, 339–344. [DOI] [PubMed] [Google Scholar]
- Gedik C. M.; Collins A. Establishing the Background Level of Base Oxidation in Human Lymphocyte DNA: Results of an Interlaboratory Validation Study. FASEB J. 2005, 19, 82–84. [DOI] [PubMed] [Google Scholar]
- Aubert G.; Hills M.; Lansdorp P. M. Telomere Length Measurement-Caveats and a Critical Assessment of the Available Technologies and Tools. Mutat. Res. 2012, 730, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham Punnoose J.; Cui Y.; Koirala D.; Yangyuoru P. M.; Ghimire C.; Shrestha P.; Mao H. Interaction of G-Quadruplexes in the Full-Length 3′ Human Telomeric Overhang. J. Am. Chem. Soc. 2014, 136, 18062–18069. [DOI] [PubMed] [Google Scholar]
- Rajendran A.; Endo M.; Hidaka K.; Sugiyama H. Direct and Single-Molecule Visualization of the Solution-State Structures of G-Hairpin and G-Triplex Intermediates. Angew. Chem., Int. Ed. 2014, 53, 4107–4112. [DOI] [PubMed] [Google Scholar]
- Song L.; Hobaugh M.; Shustak C.; Cheley S.; Bayley H.; Gouaux J. Structure of Straphylococcal α-Hemolysin, a Heptameric Transmembrane Pore. Science 1996, 274, 1859–1866. [DOI] [PubMed] [Google Scholar]
- Branton D.; Deamer D. W.; Marziali A.; Bayley H.; Benner S. A.; Butler T.; Ventra M. D.; Garaj S.; Hibbs A.; Huang X.; etal. The Potential and Challenges of Nanopore Sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner J. E.; Balijepalli A.; Robertson J. W.; Campbell J.; Suehle J.; Kasianowicz J. J. Disease Detection and Management via Single Nanopore-Based Sensors. Chem. Rev. 2012, 112, 6431–6451. [DOI] [PubMed] [Google Scholar]
- Clamer M.; Hofler L.; Mikhailova E.; Viero G.; Bayley H. Detection of 3′-End RNA Uridylation with a Protein Nanopore. ACS Nano 2014, 8, 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell R.; Schmidt J. Discrimination of Single Base Substitutions in a DNA Strand Immobilized in a Biological Nanopore. ACS Nano 2009, 3, 2533–2538. [DOI] [PubMed] [Google Scholar]
- Drew H. R.; Wing R. M.; Takano T.; Broka C.; Tanaka S.; Itakura K.; Dickerson R. E. Structure of a B-DNA Dodecamer: Conformation and Dynamics. Proc. Natl. Acad. Sci. U.S.A. 1981, 78, 2179–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddart D.; Heron A.; Mikhailova E.; Maglia G.; Bayley H. Single-Nucleotide Discrimination in Immobilized DNA Oligonucleotides with a Biological Nanopore. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 7702–7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherf G. M.; Lieberman K. R.; Rashid H.; Lam C. E.; Karplus K.; Akeson M. Automated Forward and Reverse Ratcheting of DNA in a Nanopore at 5-Å Precesion. Nat. Biotechnol. 2012, 30, 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo A. H.; Derrington I. M.; Ross B. C.; Brinkerhoff H.; Adey A.; Nova I. C.; Craig J. M.; Langford K. W.; Samson J. M.; Daza R.; etal. Decoding Long Nanopore Sequencing Reads of Natural DNA. Nat. Biotechnol. 2014, 32, 829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wescoe Z. L.; Schreiber J.; Akeson M. Nanopores Discriminate among Five C5-Cytosine Variants in DNA. J. Am. Chem. Soc. 2014, 136, 16582–16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddart D.; Heron A. J.; Mikhailova E.; Maglia G.; Bayley H. Single-Nucleotide Discrimination in Immobilized DNA Oligonucleotides with a Biological Nanopore. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 7702–7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercoutere W.; Winters-Hilt S.; Olsen H.; Deamer D.; Haussler D.; Akeson M. Rapid Discrimination Among Individual DNA Hairpin Molecules at Single-Nucleotide Resolution Using an Ion Channel. Nat. Biotechnol. 2001, 19, 248–252. [DOI] [PubMed] [Google Scholar]
- Ding Y.; Fleming A. M.; White H. S.; Burrows C. J. Internal vs Fishhook Hairpin DNA: Unzipping Locations and Mechanisms in the Alpha-Hemolysin Nanopore. J. Phys. Chem. B 2014, 118, 12873–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An N.; Fleming A. M.; Middleton E. G.; Burrows C. J. Single-Molecule Investigation of G-Quadruplex Folds of the Human Telomere Sequence in a Protein Nanocavity. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 14325–14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An N.; Fleming A. M.; Burrows C. J. Interactions of the Human Telomere Sequence with the Nanocavity of the Alpha-Hemolysin Ion Channel Reveal Structure-Dependent Electrical Signatures for Hybrid Folds. J. Am. Chem. Soc. 2013, 135, 8562–8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J. W.; Tan Q.; Gu L.-Q. Single-Molecule Detection of Folding and Unfolding of the G-Quadruplex Aptamer in a Nanopore Nanocavity. Nucleic Acids Res. 2008, 37, 972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathe J.; Visram H.; Viasnoff V.; Rabin Y.; Meller A. Nanopore Unzipping of Individual DNA Hairpin Molecules. Biophys. J. 2004, 87, 3205–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J.; Phan A. T.; Kuryavyi V. Human Telomere, Oncogenic Promoter and 5′-UTR G-Quadruplexes: Diverse Higher Order DNA and RNA Targets for Cancer Therapeutics. Nucleic Acids Res. 2007, 35, 7429–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Patel D. J. Solution Structure of the Human Telomeric Repeat d[AG3(T2AG3)3] G-Tetraplex. Structure 1993, 1, 263–282. [DOI] [PubMed] [Google Scholar]
- Phan A. T.; Kuryavyi V.; Luu K. N.; Patel D. J. Structure of Two Intramolecular G-Quadruplexes Formed by Natural Human Telomere Sequences in K+ Solution. Nucleic Acids Res. 2007, 35, 6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson G. N.; Lee M. P. H.; Neidle S. Crystal Structure of Parallel Quadruplexes From Human Telomeric DNA. Nature 2002, 417, 876–880. [DOI] [PubMed] [Google Scholar]
- Wolna A. H.; Fleming A. M.; Burrows C. J. Single-Molecule Analysis of Thymine Dimer-Containing G-Quadruplexes Formed From the Human Telomere Sequence. Biochemistry 2014, 53, 7484–7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meervelt V.; Soskine M.; Maglia G. Detection of Two Isomeric Binding Configurations in a Protein-Aptamer Complex with a Biological Nanopore. ACS Nano 2014, 8, 12826–12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An N.; Fleming A. M.; White H. S.; Burrows C. J. Crown Ether-Electrolyte Interactions Permit Nanopore Detection of Individual DNA Abasic Sites in Single Molecules. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 11504–11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. M.; Burrows C. J. G-Quadruplex Folds of the Human Telomere Sequence Alter the Site Reactivity and Reaction Pathway of Guanine Oxidation Compared to Duplex DNA. Chem. Res. Toxicol. 2013, 26, 593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala D.; Mashimo T.; Sannohe Y.; Yu Z.; Mao H.; Sugiyama H. Intramolecular Folding in Three Tandem Guanine Repeats of Human Telomeric DNA. Chem. Commun. 2012, 48, 2006–2008. [DOI] [PubMed] [Google Scholar]
- Gray R. D.; Buscaglia R.; Chaires J. B. Populated Intermediates in the Thermal Unfolding of the Human Telomeric Quadruplex. J. Am. Chem. Soc. 2012, 134, 16834–16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen C. M.; Gmeiner W. H.; Marky L. A. Unfolding of G-quadruplexes: Energetic, and Ion and Water Contributions of G-Quartet Stacking. J. Phys. Chem. B 2006, 110, 6962–6969. [DOI] [PubMed] [Google Scholar]
- Lech C. J.; Cheow Lim J. K.; Wen Lim J. M.; Amrane S.; Heddi B.; Phan A. T. Effects of Site-Specific Guanine C8-Modifications on an Intramolecular DNA G-Quadruplex. Biophys. J. 2011, 101, 1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlícková M.; Tomasko M.; Sagi A. J.; Bednarova K.; Sagi J. 8-Oxoguanine in a Quadruplex of the Human Telomere DNA Sequence. FEBS J. 2011, 279, 29–39. [DOI] [PubMed] [Google Scholar]
- Chitranshi P.; Xue L. Utilizing G-Quadruplex Formation to Target 8-Oxoguanine in Telomeric Sequences. Bioorg. Med. Chem. Lett. 2011, 21, 6357–6361. [DOI] [PubMed] [Google Scholar]
- Butler T. Z.; Gundlach J. H.; Troll M. Ionic Current Blockades from DNA and RNA Molecules in the Alpha-Hemolysin Nanopore. Biophys. J. 2007, 93, 3229–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller A.; Nivon L.; Brandin E.; Golovchenko J.; Branton D. Rapid Nanopore Discrimination between Single Polynucleotide Molecules. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Wang Y.; Fricke B. L.; Gu L. Q. Programming Nanopore Ion Flow for Encoded Multiplex MicroRNA Detection. ACS Nano 2014, 8, 3444–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell N.; Howorka S. Chemical Tags Facilitate the Sensing of Individual DNA Strands with Nanopores. Angew. Chem., Int. Ed. 2008, 47, 5565–5568. [DOI] [PubMed] [Google Scholar]
- Perera R. T.; Fleming A. M.; Johnson R. P.; Burrows C. J.; White W. S. Detection of Benzo[a] Pyrene-Guanine Adducts in Single-Stranded DNA Using the a-Hemolysin Nanopore. Nanotechnology 2015, 26, 074002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.; Muller J. G.; Ye Y.; Burrows C. J. DNA-Protein Cross-Links between Guanine and Lysine Depend on the Mechanism of Oxidation for Formation of C5 vs C8 Guanosine Adducts. J. Am. Chem. Soc. 2008, 130, 703–709. [DOI] [PubMed] [Google Scholar]
- Hosford M. E.; Muller J. G.; Burrows C. J. Spermine Participates in Oxidative Damage of Guanosine and 8-Oxoguanosine Leading to Deoxyribosylurea Formation. J. Am. Chem. Soc. 2004, 126, 9540–9541. [DOI] [PubMed] [Google Scholar]
- Bajacan J. E. V.; Hong I. S.; Penning T. W.; Greenberg M. M. Quantitative Detection of 8-Oxo-7, 8-Dihydro-2′-Deoxyguanosine Using Chemical Tagging and qPCR. Chem. Res. Toxicol. 2014, 27, 1227–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.; Guo L.-H.; Greenberg M. M. Quantification of 8-OxodGuo Lesions in Double-Stranded DNA Using a Photoelectrochemical DNA Sensor. Anal. Chem. 2012, 84, 6048–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane A. N.; Chaires J. B.; Gray R. D.; Trent J. O. Stability and Kinetics of G-Quadruplex Structures. Nucleic Acids Res. 2008, 36, 5482–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesegang G. W.; Farrow M. M.; Arce Vazquez F.; Purdie N.; Eyring E. M. Ultrasonic Absorption Kinetic Studies of the Complexation of Aqueous Lithium(1+), Sodium(1+), Rubidium(1+), Thallium(1+), Silver(1+), Ammonium(1+), and Calcium(2+) Ions by 18-Crown-6. J. Am. Chem. Soc. 1977, 99, 3240–3243. [Google Scholar]
- Miller M. C.; Buscaglia R.; Chaires J. B.; Lane A. N.; Trent J. O. Hydration is a Major Determinant of the G-Quadruplex Stability and Conformation of the Human Telomere 3′ Sequence of d(AG3(TTAG3)3. J. Am. Chem. Soc. 2010, 132, 17105–17107. [DOI] [PubMed] [Google Scholar]
- Guschlbauer W.; Chantot J.-F.; Thiele D. Four-Stranded Nucleic Acid Structures 25 Years Later: From Guanosine Gels to Telomere DNA. J. Biomol. Struct. Dyn. 1990, 8, 491–511. [DOI] [PubMed] [Google Scholar]
- Heddi B.; Phan A. T. Structure of Human Telomeric DNA in Crowded Solution. J. Am. Chem. Soc. 2011, 133, 9824–9833. [DOI] [PubMed] [Google Scholar]
- Petraccone L.; Trent J. O.; Chaires J. B. The Tail of the Telomere. J. Am. Chem. Soc. 2008, 130, 16530–16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark T. A.; Spittle K. E.; Turner S. W.; J K. Direct Ddetection and Sequencing of Damaged DNA Bases. Genome Integr. 2011, 2, 2041–9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya S.; Boocock J.; Black M. A.; Gemmell N. J. Exploring Possible DNA Structures in Real-Time Polymerase Kinetics Using Pacific Biosciences Sequencer Data. BMC Bioinf. 2015, 16, 432–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdorp P. M.; Verwoerd N. P.; van de Rijke F. M.; Dragowska V.; Little M.-T.; Dirks R. W.; Raap A. K.; Tanke H. J. Heterogeneity in Telomere Length of Human Chromosomes. Hum. Mol. Genet. 1996, 5, 685–691. [DOI] [PubMed] [Google Scholar]
- Heaphy C. M.; Yoon G. S.; Peskoe S. B.; Joshu C. E.; Lee T. K.; Giovannucci E.; Mucci L. A.; Kenfield S. A.; Stampfer M. J.; Hicks J. L.; etal. Prostate Cancer Cell Telomere Length Variability and Stromal Cell Telomere Length as Prognostic Markers for Metastasis and Death. Cancer Discovery 2013, 3, 1130–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; Zhu Y.; Lin J.; Matsuguchi T.; Blackburn E.; Zhang Y.; Cole S. A.; Best L. G.; Lee E. T.; Howard B. V. Short Leukocyte Telomere Length Predicts Risk of Diabetes in American Indians: The Strong Heart Family Study. Diabetes 2014, 63, 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann M. T.; Strong M. A.; Hao L. Y.; Greider C. W. The Shortest Telomere, Not Average Telomere Length, Is Critical for Cell Viability and Chromosome Stability. Cell 2001, 107, 67–77. [DOI] [PubMed] [Google Scholar]
- Fleming A. M.; Muller J. G.; Dlouhy A. C.; Burrows C. J. Context Effects in the Oxidation of 8-Oxo-7,8-Dihydro-2′-Deoxyguanosine to Hydantoin Products: Electrostatics, Base Stacking, and Base Pairing. J. Am. Chem. Soc. 2012, 134, 15091–15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cracknell J. A.; Japrung D.; Bayley H. Translocating Kilobase RNA through the Staphylococcal Alpha-Hemolysin Nanopore. Nano Lett. 2013, 13, 2500–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon-Restrepo M.; Mikhailova E.; Bayley H.; Maglia G. Controlled Translocation of Individual DNA Molecules through Protein Nanopores with Engineered Molecular Brakes. Nano Lett. 2011, 11, 746–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J. W.; Zou Y.; Hiyama E.; Wright W. E. Telomerase and Cancer. Hum. Mol. Genet. 2001, 10, 677–685. [DOI] [PubMed] [Google Scholar]
- Zhang B.; Galusha J.; Shiozawa P.; Wang G.; Bergren A.; Jones R.; White R.; Ervin E.; Cauley C.; White H. Bench-Top Method for Fabricating Glass-Sealed Nanodisk Electrodes, Glass Nanpore Electrodes, and Glass Nanopore Membranes of Controlled Size. Anal. Chem. 2007, 79, 4778–4787. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.